Abstract
Drug repositioning has been based largely on genomic signatures of drugs and diseases. One challenge in these efforts lies in connecting the molecular signatures of drugs into clinical responses, including therapeutic and side effects, to the repurpose of drugs. We addressed this challenge by evaluating drug‐drug relationships using a phenotypic and molecular‐based approach that integrates therapeutic indications, side effects, and gene expression profiles induced by each drug. Using cosine similarity, relationships between 445 drugs were evaluated based on high‐dimensional spaces consisting of phenotypic terms of drugs and genomic signatures, respectively. One hundred fifty‐one of 445 drugs comprising 450 drug pairs displayed significant similarities in both phenotypic and genomic signatures (P value < 0.05). We also found that similar gene expressions of drugs do indeed yield similar clinical phenotypes. We generated similarity matrixes of drugs using the expression profiles they induce in a cell line and phenotypic effects.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ A central premise in the computational approach to drug repositioning is that similar compounds induce similar clinical responses. Although, in clinical practice, drug administration is generally conducted based on phenotypic efficacy, whereas the computational prediction of novel drug indications has been based largely on genomic signatures of the drugs.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ Systematically, can we determine similar drugs by integrating drug‐associated gene expressions and known clinical phenotypes? Using cosine similarity approach, we have compared the drug‐drug similarity in terms of phenotypic terms and gene expression signatures. In overall, when a pair of drug showed significant similarity based on gene expression signatures, the pair also presented phenotypic similarity. In addition, we identified a promising drug repositioning candidate, thioridazine (anti‐schizophrenia drug) for metastasis of breast cancer, by integration of drug associated gene expressions and clinical phenotypes.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
☑ Using cosine‐similarity, relationships between drugs were evaluated based on high‐dimensional spaces, consisting of phenotypic terms of drugs and genomic signatures, respectively. Clinical phenotype or gene expression signatures of drugs are significantly nonindependent; similar gene expressions of drugs do indeed yield similar clinical phenotypes.
HOW MIGHT THIS CHANGE DRUG DISCOVERY, DEVELOPMENT, AND/OR THERAPEUTICS?
☑ This work is a generalized method that paves the way to leveraging drug‐induced gene expression profiles and term‐based phenotypic understandings for drug repositioning.
Drug repositioning is the process of identifying novel indications for approved drugs. This methodology in drug discovery has several advantages over novel drug discovery and development. For example, rising development costs, high attrition rates during clinical trials, and greater concerns about drug safety1, 2, 3 are among the many hurdles that hinder the success of novel disease therapies. However, drug repositioning has still not fully matured, and it generally relies on an unorganized process based largely on serendipity. For example, sildenafil (Viagra; Pfizer) was originally for cardiovascular indications and it was repositioned to erectile dysfunction because of side effects in human volunteers.4
Precise prediction of new indications could shorten development time and identify more potential uses for a single drug. Computational approaches to discover new indications or biological targets have been applied to generate novel repositioning opportunities.5, 6, 7, 8 Specifically, guilt‐by‐association is a well‐known approach that explores similar drug‐drug or disease‐disease pairs. These studies exploited relationships between drugs and target genes to infer novel drug indications.
Many studies have analyzed either molecular‐level states induced by disease or drugs, or phenotypic profiling from human individuals with the goal of drug repositioning.9, 10, 11 For example, the Connectivity Map elucidates relationships between small molecule drugs and diseases.12 In previous works, we proposed a method based on a guilt‐by‐association approach to predict new ones13 and integrated clinical phenotypes from electronic medical records.14 In this way, identification of similar drugs by considering diverse aspects, including molecular genomic profiles15 and phenotypes, such as clinical observations, indications, and side effects of a drug, can lead to repositioning. Meanwhile, exploring drug‐associated phenotypes (i.e., side effects and therapeutic indications) are promising for drug repositioning; however, large‐scale integration between phenotypic and genomic information‐derived drug repositioning remains as challenging issues and has been rarely attempted.16, 17
To date, drug‐associated phenotypic information has been used as either side effects or therapeutic indications without directionality.9, 10, 18 An example of directionality is that sildenafil reduced (downregulated) erectile dysfunction as an indication, and rarely induced (upregulated) headaches as an adverse effect. Typical application of drug‐associated phenotypes has mainly focused on side effects as Boolean values (i.e., has side effect “X” or not). Although the object of drug administration is to reverse disease phenotypes, using directional relationships between drug and phenotype signatures has not been reported thus far. Moreover, by connecting gene expression signatures to clinical phenotypes, such as efficacies (indications) and side effects, a systematic evaluation for drug‐drug relationships remains as a central promise for drug repurpose.
In this study, using cosine‐similarity measures, we compared drug‐drug relationships in terms of molecular and clinical levels, including gene expression signatures in a single cell line and known phenotypic terms in human individuals. Whereas in text mining field, the cosine‐similarity score is a well‐known method to quantify similarity between a pair of corpus, such as a set of clinical phenotypic terms (e.g., side effects) for drugs, also has been examined to detect gene‐based similarity of drugs.19, 20 We applied the cosine‐similarity score as a generalized measurement to determine relationships between drugs based on sparse and high dimensional feature vectors, such as corpus of phenotypic terms for drugs and identified gene signatures. Interestingly, 151 drugs had significant similarities in both phenotypic and gene expression profiles, revealing novel drug‐repositioning candidates.
METHODS AND DATA
Data
Drug‐associated phenotypic terms
We retrieved phenotype characteristics of drugs involving side effects and approved therapeutic indications using the side effect resource (SIDER2)21 and US Food and Drug Administration Adverse Event Reporting System (FAERS).22 We extracted side effects listed on labels and adverse effects that had been reported but were not listed on labels. In addition, we obtained approved indications of drugs from the DrugBank23 and the US Food and Drug Administration Orange Book.24 The Unified Medical Language System25 and MetaMap26 were used to systematically conduct a concept‐based integration9 of terms for side effects, drug indications, and names of drugs.
Gene expression profiles under drug treated conditions
The Connectivity Map (latest date of download: 19 August 2014)12 was used to prepare expression profiles under various drug‐treated conditions.
Drug‐associated phenotypic terms
Phenotypic terms (or concepts) are commonly represented by multidimensional feature vectors for each object (i.e., drug).27 We described j‐th drug using a vector of phenotypic terms Tdj as follows:
| (1) |
Here, tij grades the associations between i‐th phenotypic term and drug j (1 ≤ i ≤ n). The association value between phenotype and drug is very different by prevalence of phenotypic terms across drugs and the source of phenotypic information. For example, there are highly abundant side effects, such as nausea, and therapeutic indications from drug label information, such as a migraine headache. In addition, we also determined directionality of association between phenotypic terms and drugs, such as positive and negative relationships by every k‐th dataset. For example, the term “dizziness” for drug p in the side‐effect database has a positive relationship, whereas the same term for drug q in therapeutic indication resources has a negative relationship. To account for heterogeneity in different data sources (i.e., side effect and therapeutic indication databases), we normalized Dk(Tij) to measure the strength of an association between phenotype i and drug j by overall co‐occurrence of the phenotypic term i across all drugs in k‐th dataset. The relationship between phenotype i and drug j was determined by k‐th database as an index value Ik(Tij); −1 for negative relationship and +1 for positive relationship. By integrating phenotypic term values from various data sources, an association value of tij for drug j was defined using the following equation:
| (2) |
In Eq. 2, K denotes the number of integrated data sources and Dk(ni) denotes the number of drugs where the term i appears in the k‐th data source. Dk(Tij) is a Boolean value (0 or 1) due to our utilized data sources. For Ik(Tij), a database for the side effects of drugs (SIDER21 and FAERS22) has a positive index of +1, whereas a source for therapeutic indication (DrugBank23 and the US Food and Drug Administration Orange Book24) has a negative relation index of −1.
Drug‐associated gene expression signatures
In terms of the expression signature, we also described j‐th drug using a vector of expression signature Gdj:
| (3) |
Here, gpj denotes expression signatures of p‐th gene under j‐th drug‐treated conditions. Based on z‐score transformed expression signatures, we identified differentially expressed genes by comparing nontreated array expressions using the P value of the t test with adjusting the false discovery rate using the Benjamini‐Hochberg procedure for multiple comparisons (FDR <0.1). The detailed methods for our z‐score based methods have been described.28 In summary, we normalized gene expression signatures using z‐score transformation, and then filtered differentially expressed genes comparing nondrug treated conditions via t test (FDR <0.1). In this study, we utilized drug‐associated expression profiles with at least two arrays per cell line and drug in the Connectivity Map. With the genes selected from this work, we utilized mean values of z‐score transformed expression values for our cosine‐similarity analysis procedure. Because the Connectivity Map profiled expressions using five cell lines (MCF7, HL60, ssMCF7, PC4, and SKMEL5), we independently analyzed drug‐associated Gdj in each cell line. The Connectivity Map has a number of arrays and drug‐treated conditions for each cell line: 3,213 arrays for 1,294 drugs in MCF7 cells, 1,848 arrays for 1,182 drugs in PC3 cells, 1,281 arrays for 1,078 drugs in HL60 cells, 23 arrays for 16 drugs in ssMCF7 cells, and 22 arrays for 17 drugs in SKMEL5 cells.
Cosine similarity of drugs
Cosine‐similarity analysis is a widely used method for quantifying similarities between two concepts using high‐dimensional vectors. We analyzed the similarity of drugs using various features, including known phenotypic terms, and profiled expression signatures. The gene expression and phenotypic term‐based similarity between the vectors of two drugs x and y were calculated as follows:
| (4) |
| (5) |
Eq. 4 quantifies the similarity of drugs using associated phenotypic terms, and Eq. 5 quantifies the similarity of identical drug pairs based on gene expression signatures from the Connectivity Map. Cosine‐similarity scores range from −1 (opposite symptoms or gene expression patterns) to 0 (no common symptoms or gene expression patterns) to +1 (identical symptoms or expression patterns). Pairwise comparisons between phenotype and gene signature based cosine‐similarity scores were completed after quantile normalization process. We conducted random permutation analysis to evaluate statistical significance.
RESULTS
Overview of drug relationship analysis
We extracted drugs and phenotypic terms associated with them by integrating 1,513 therapeutic indications of 1,473 drugs (DrugBank), >13,000 side effects of >5,000 drugs (SIDER2 and FAERS) and therapeutic equivalence evaluations about 473 indications of 49,515 drugs (US Food and Drug Administration Orange Book).21, 22, 23, 24 The Unified Medical Language System25 and MetaMap26 were used to systematically conduct concept‐based integration of various terms for side effects and therapeutic indications from distinct resources,9 and were also used for the integration of drug names. Table 1 shows the number of drugs we analyzed and associated phenotypes for each resource. As described in Table 1, phenotypes of drugs comprise clinical phenotypes, such as side effects and disease indications.
Table 1.
Summary of data used
| Data level | Data resources | Features | Number |
|---|---|---|---|
| Drug‐associated clinical phenotypes | SIDER a |
No. of drugs
b
Type of associated side effects c |
996 4,492 |
| FAERS d |
No. of drugs
b
Type of reported side effects c No. of reported cases a |
5,689 9,268 223,178 |
|
| DrugBank |
No. of drugs
b
Type of approved therapeutic indications c |
1,473 1,513 |
|
| FDA Orange Book |
No. of drugs
b
Type of approved therapeutic use c |
49,515 473 |
|
| Integrated results |
No. of drugs
c
No. of associated phenotype terms |
1,631 (445
e
) 1,587 |
|
| Drug‐associated gene expression | The Connectivity Map |
Total no. of drugs Total no. of analyzed expression profiles Cell lines g |
1,309 (856
f
) 6,100 (3,204 f ) 5 |
FAERS, US Food and Drug Administration Adverse Event Reporting System; FDA, US Food and Drug Administration; SIDER, side effect resource.
aSIDER (sideeffects.embl.de/). bThis number was calculated by using the drug names or ingredients. cWe integrated the name of the drugs using concept identification of drugs in the Unified Medical Language System, and prepared drug lists by integrating drugs from the Connectivity Map. dAdverse events were selected if they had been reported in ≥30 cases. eNumber of drugs that have both the phenotypic terms and gene expression signatures from the Connectivity Map. fOwing to the z‐score based approach, drug‐associated expression profiles with single array data were excluded. Finally, of 1,309 drugs and 6,100 expression profiles, 856 drugs and 3,204 expression profiles were used for further analysis. gThe five cell lines were MCF7, HL60, PC3, ssMCF7, and SKMELS.
We analyzed >6,000 microarray profiles and used them to generate drug‐associated gene expression signatures from the Connectivity Map.12 Gene expression profiles under drug‐treated conditions were independently analyzed in five cell lines (MCF7, PC3, HL60, ssMCF7, and SKMEL5). Because we used a t test for selecting drug‐associated gene signatures after z‐score transformation of expression profiles, we excluded >3,000 arrays with single profiles for a drug‐treated condition, and only used arrays with at least two profiles per condition. Among them, we excluded over 1,000 drugs and associated expression profiles due to absence of phenotypic terms or gene expression profiles. The majority of treated drugs in the Connectivity map (759 of 1,309 drugs) were treated in MCF7. Thereby, we mainly utilized the drug associated gene signatures in MCF7 to compare the results of phenotype based drug similarity analyses. We selected the common drugs between 759 treated drugs in MCF7 and 1,631 drugs having phenotypic terms. This process left 445 drugs with 1,587 phenotypic terms and 1,099 expression profiles across 22,000 genes in MCF7 (Table 1).
Figure 1 shows our workflow for generating drug‐drug relationships. As described in the Methods section, we first computed term values of clinical phenotypes, such as side effects and disease indications, for each drug as a Boolean in each data resource (i.e., true or false relationships between drug A and phenotype X), and then determined the directionalities of these relationships between drugs and clinical phenotypes. To ensure the specificity of phenotypic terms, we normalized term values by overall frequencies of phenotype‐drug relationships, such as how many drugs shared clinical phenotype X. Each normalized and directional term value is identical within a data resource. We combined associated phenotypic terms for each drug and determined drug‐associated phenotypic signatures as a result. Next, we aggregated term values for each drug by calculating mean scores across various phenotype resources for the drugs (Figure 1 a).
Figure 1.
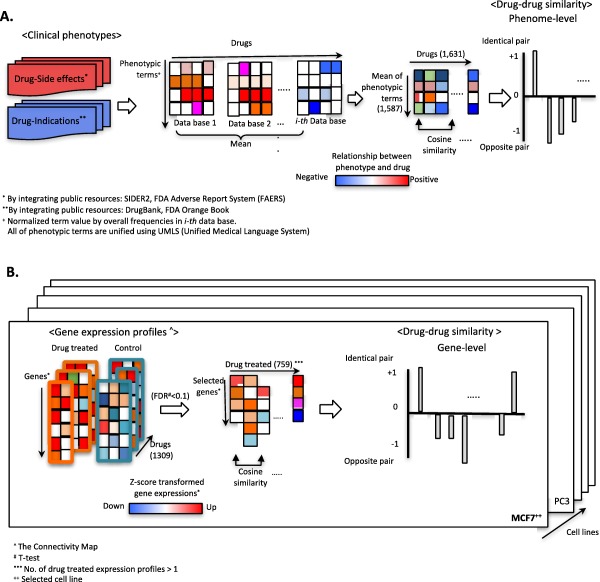
Pipeline for identifying drug‐drug relationships using phenotype and gene expression signatures. (a) Drug‐associated hybrid phenotypes, including side effects and therapeutic indications, were prepared via integration of multiple public resources, as noted. Directionality and normalized term values for each phenotype for each drug were determined as described in the Methods section. After aggregating term values, we computed cosine similarities between drug pairs (−1 or +1). (b) Data preparation and analysis procedure for gene‐signature based cosine similarity for two queried drugs. For direct comparisons of drug pairs, we transformed gene expression signatures as z‐scores. Drug‐associated gene signatures were prepared by t test analysis (false discovery rate [FDR] <0.1). Finally, transformed gene signatures for a drug consisted of high‐dimensional gene spaces to analyzed cosine similarity for drug pairs.
Figure 1 b starts with drug‐drug similarity analysis via z‐score transformation of gene expression profiles under drug‐treated and nontreated conditions in MCF7. Next, we examined P values of t tests and used them to select drug‐associated gene expression signatures in MCF7 (FDR <0.1), as described in the Methods section.28 Using z‐scores of selected genes in each drug‐treated MCF7, our cosine‐similarity analysis quantified relationships between drug‐drug pairs. The Supplementary Data has presented all of the normalized term values for all 445 drugs by utilized data resources, including DrugBank, Orange Book, FAERS, and SIDER2, and z‐score transformed gene expression profiles by treated drugs in MCF7 and PC3.
Using established phenotypic concepts and expression signatures for 445 drugs in MCF7, we compared cosine‐similarity analysis to determine similarity scores for queried drug pairs under distinct measures, such as clinical phenotypes and gene signatures in a cell line (Figure 1 a,b). Cosine‐similarity scores capture directionality of compared objects. A score of +1 denotes identical relationships, whereas −1 denotes opposite relationships for pairs of drugs based on phenotypic information and gene expression. For statistical significance of these scores, we used random permutation methods as described (P value < 0.05).29
Landscape of drug‐associated phenotypic effects and expression profiles
By integrating drug‐associated clinical phenotypic terms from public resources, as denoted in Table 1, we produced a bipartite network consisting of 3,218 nodes involving 1,631 drugs, 1,587 phenotypes, and 72,848 edges for significant positive and negative relationships. Negative relationships between drugs and phenotypic terms represent therapeutic effects of the drugs, and positive scores represent drug‐induced side effects. The number of clinical phenotypic terms for each drug followed a power‐law‐like distribution in terms of side effects and therapeutic indications (Supplementary Figure S1a–c).
We also established a network model of drugs and associated gene signatures in MCF7 using the Connectivity Map. As shown in Supplementary Figure S1d, 8,101 genes are differentially expressed across the effect of 759 drugs (FDR of t test <0.1), and these genes and drugs are represented as nodes linked with ∼17,000 edges in MCF7 cells. Using z‐score transformed gene expression signatures from drug‐treated and nontreated MCF7 cell lines,28 directionality of edges between drug and gene nodes was determined (negative association = z‐score <0; positive association = z‐score >0) and significance levels were computed using t tests with an FDR cutoff of 0.1. As with drug term relationships, expression and phenotypic signatures of drugs can be displayed as scale‐free networks (Supplementary Figure S1d,f).
Relationships between clinical phenotypic and genetic signature‐based drug similarity
We used drug‐associated gene signatures and phenotypic term values to determine and compare the drug‐drug relationships using cosine‐similarity analysis. By random permutation of drug‐gene expression and drug‐phenotypic term value matrixes, we generated background distributions of cosine‐similarity, respectively. Although both of the random models for gene expression and clinical phenotype‐based cosine‐similarity values converged upon zero, overall distributions were distinct (P value of t test 3.27E‐03; Figure 2 a). Using these random distributions, we independently evaluated statistical significances of cosine‐similarity based on phenotypic terms and gene signatures in MCF7 (P value of permutation < 0.05).
Figure 2.
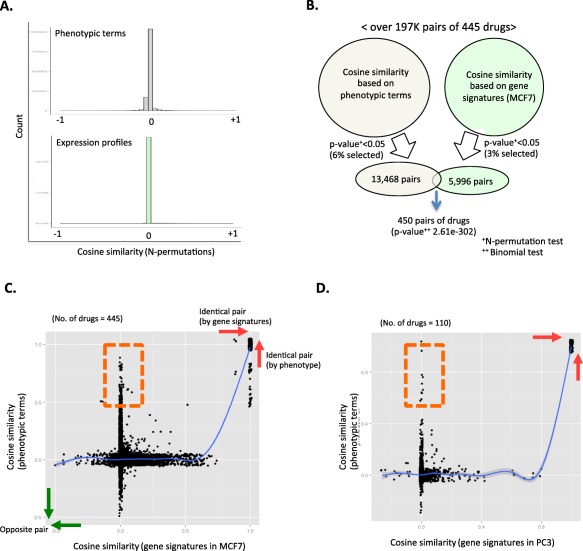
Comparison between phenotype and gene‐signature based cosine similarity. (a) Random distributions of cosine‐similarity measures using drug‐phenotypic term relations (upper chart) and gene expressions (lower part) by N‐permutation approach. (b) Vann diagram of selected similar drug pairs by using phenotypic terms (light gray) and gene signatures in MCF7 (light green). All lists of selected 450 drugs are presented in Supplementary Table S1. Gene‐signature‐based cosine similarity are in red. Prednisone (blue) is an example of disparity of phenotype and gene‐based cosine‐similarity analysis. Cosine similarity scores for phenotype and gene‐based results were quantile normalized for direct comparison. (c,d) Relationships between phenotype‐based and gene‐based cosine similarity in each cell line (c) comparison between cosine similarity based on phenotype and gene signatures in MCF7; (d) utilized gene signatures in PC3.
Among over 197,000 possible pairs of 445 drugs, we used cosine similarity to identify 5,996 pairs from among 409 drugs as having similar gene signatures (3% of over 197,000 possible pairs of 445 drugs; P value of permutation < 0.05). A total of 13,468 pairs from among 437 drugs were identified as phenotypically similar drugs (6% of over 197,000 possible pairs of 445 drugs, P value of permutation < 0.05; Figure 2 b). In order to maximize the number of comparisons between phenotypic terms and gene signature derived drug‐drug relationships, we introduced liberal P values to select sets of similar drug pairs without FDR corrections (P value < 0.05). A modest fraction (450 pairs of drugs consisting of 151 drugs) was found to be similar in both the phenotype‐based and gene‐based cosine‐similarity analysis (0.2% of 197,000 comparisons; P value of cosine similarity < 0.05). However, the P values of binomial tests show that selecting either phenotypic or expressional signatures based on similar drug pairs is significantly nonindependent (P value = 2.61E‐302; Figure 2 b). Likewise, in PC3 cell lines, we also identified that a selection of similar drugs based on gene signatures is associated with phenotypic similarity (P value = 5.3e‐65; Supplementary Figure S2). However, 25,752 pairs of drugs (13% of 197,580 pairs of drugs) were identified as similar drugs in terms of phenotype “without directionality” (Supplementary Table S3). The identified similar drug pairs in terms of phenotype “with directionality” (6% of over 197,000 possible pairs of 445 drugs) are a subset of these selected drugs without directionality (Supplementary Table S3). Interestingly, methotrexate and hydroflumethiazide are identified as a similar pair in term of phenotype “without directionality” (Supplementary Table S3). However, hydroflumethiazide is a known diuretic for liver cirrhosis and a known side effect of methotrexate is liver cirrhosis. In our cosine similarity of directional phenotype analysis, hydroflumethiazide and methotrexate are opposite pairs by the opposite relationship with liver cirrhosis (Figure 3 b). Thus, a pair of hydroflumethiazide and methotrexate is a false‐positive result. Thereby, identification of similar drug pairs by either nondirectional clinical phenotype or gene expression signatures in MCF7 is selected by chance (P value of binomial 0.19) due to the false‐positive results from the nondirectional phenotype signatures.
Figure 3.
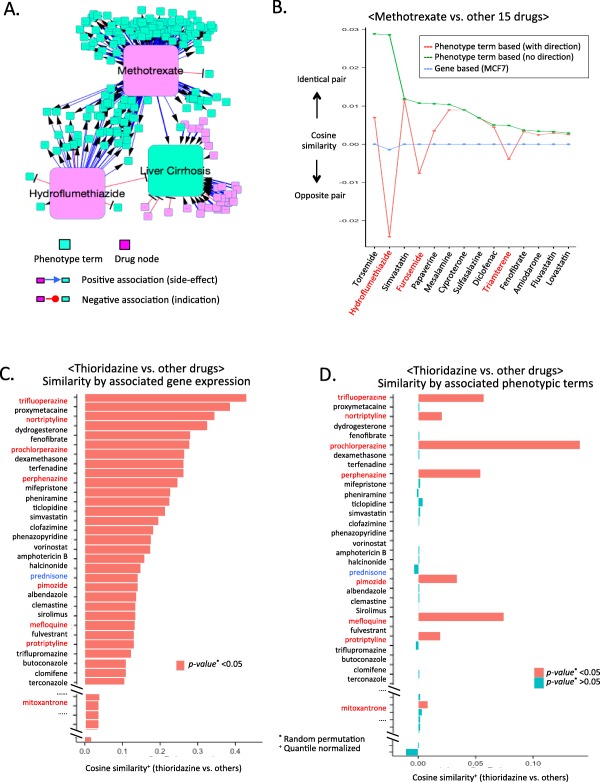
Evaluation of cosine‐similarity measures and selected repositioning candidate. (a) Example of shared clinical phenotypes between drugs in opposite directionality. Phenotype term “liver cirrhosis” has distinct relationships between methotrexate and hydroflumethiazide. (b) Comparison of cosine‐similarity scores with (red) or without phenotype directionality (green). (c,d) Computed cosine similarity between thioridazine and other drugs. Both charts were ordered in descending order by rank of gene‐signature based cosine similarity. The P values of cosine‐similarity values were determined by random permutations. Drugs similar to thioridazine in phenotype and gene‐signature‐based cosine similarity are red. Prednisone (blue) is an example of disparity of phenotype and gene‐based cosine similarity analysis. Cosine similarity scores for phenotype and gene‐based results were quantile normalized for direct comparison.
Figure 2 c,d present the relationship of drug similarity measures between gene‐based cosine similarities and phenotypic term‐based values in MCF7 and PC3. Interestingly, when pairs of drug have significantly shared gene expression signatures, phenotype‐based similarities have drastically increased (red arrow remarked). However, similar drug pairs, based on the phenotypic terms, have discrete trends for gene‐based similarity measures (orange boxed regions in Figure 2 c,d). Thus, similar gene expressions of drugs do indeed yield similar clinical phenotypes.
Based on the shared side effects between drugs, Campillos et al.30 also presented a prediction of similar drugs (1,017 pairs of drugs consisting of 424 drugs). Of these, 407 pairs of similar drugs consisting of 227 drugs are group 1, which is matched with known target shared drugs. Among our set of similar drugs in Supplementary Table S1 (Figure 2 b; 450 pairs of drugs consisting of 151 drugs), 48 pairs of drugs comprising 24 drugs involve the identical drugs in the group 1 predicted set of Campillos et al.30 In total, 25% of 48 pairs (12 pairs) are matched with the set of Campillos et al.30 Interestingly, other unmatched 36 pairs of similar drug in our prediction identified known similar drugs in terms of clinical uses (Supplementary Table S2). For example, our methods determined that raloxifene and tamoxifene are a similar pair of drug; these two drugs are well‐known drugs for treating breast cancer.31
By exploring established networks for drug and clinical phenotype in Supplementary Figure S1, we identified that 421 of clinical phenotypes can be regarded as either side effects or therapeutic effects of drugs. Meanwhile, 1,033 of clinical phenotypes can only be regarded as side effects of drugs. Otherwise, 133 terms have only been regarded as therapeutic effects. In Figure 3 a, we presented an example that shared clinical phenotype between a pair of drugs in opposite directionality. Methotrexate and hydroflumethiazide have shared a common clinical phenotype term, liver cirrhosis, in opposite directionality. Liver cirrhosis is a known therapeutic indication of hydroflumethiazide (i.e., negative association; red edge in Figure 3 a), whereas methotrexate has positive association with liver cirrhosis (side effects of methotrexate; blue edge in Figure 3 a). With directional associations between drugs and clinical phenotype terms, cosine‐similarity score determined hydroflumethiazide and methotrexate as an opposite pair of drugs, although it was identified as similar drugs without directionality information. In Figure 3 b, we compared similarities between methotrexate and other 15 drugs, including hydroflumethiazide, using clinical phenotypes with directionality (red line) and without directionality (green line). Cosine similarities in gene expression signatures in MCF7 also identified hydroflumethiazide and methotrexate as an opposite drug pair (blue line in Figure 3 b). Our method can identify the opposite relationships of drugs, whereas a method that compared cosine similarity with nondirectional phenotype vectors of drugs had overlooked the relationship. However, the contributions of utilized resources, such as side effects and therapeutic indications, remained unclear. Admittedly, comparing similarity results based on each phenotype resource might determine the benefit of the selected features. When we utilized only the therapeutic indication or side effect resources of 445 compared drugs, over 200 drugs had no associated phenotypic terms. Therefore, imbalance of data abundance hampered fair evaluations by the absence of identical sets.
Identification of drug repositioning candidates
The identified 450 similar drug pairs in Figure 2 b shared both clinical phenotype terms and gene expression signatures. These drugs are promising repositioning pairs and are presented in Supplementary Table S1. With stringent P values, such as P = 0.01 or 0.001, the majority of similar drugs are almost the same drugs, including nortriptyline (pain medication) and protriptyline (pain medication; Supplementary Table S1). In order to explore drug‐repositioning candidates, we applied a liberal P value of 0.05. As repurposing candidates among the 450 pairs of drug, we focused on pairs of similar drugs without known common indications.
Rank of phenotype and gene‐based cosine similarity of drugs between thioridazine and 445 other drugs showed disparity and partial matched trends, except in self‐comparison cases (Figure 3 c,d). Based on both of the phenotypic and gene signatures, thioridazine (antischizophrenia drug) displayed significant similarity with 16 drugs, including antimalarial and anticancer drugs (Supplementary Table S1). Among 16 drugs, 11 were antipsychotic drugs, such as trifluoperzine and pimozide (antipsychotic indications, including schizophrenia and depression; P value of hypergeometric test: 2.3E‐08). The therapeutic potential of thioridazine as an antimicrobial agent, including use for malaria, is supported by cell assay analysis.32 In addition, thioridazine has been cited as a promising candidate for cancer therapy based on cell‐based analysis.33 Owing to mitoxantrone's anticancer indication, we suggest thioridazine as a repurposing candidate for relapsed acute leukemia, metastasis of breast cancer, and non‐Hodgkin's lymphoma.
DISCUSSION
The pharmaceutical industry is under pressure to develop drugs at a time when costs related to validation are a major burden. This problem is compounded by soaring costs due to the high failure rate of de novo drug discovery; one resulting in a renewed interest in drug repositioning. An important challenge to repositioning efforts lies in selecting therapeutic areas to test drugs of interest. We have started to address this challenge by systematically compiling drug‐associated phenotypes, including side effects and therapeutic efficacies, as well as the transcriptomic effects of these compounds. We used public repositories and captured 1,587 approved drug indications and side effects, and 1,099 gene expression profiles of 445 drugs in the MCF7 cell line.
We applied a guilt‐by‐association data‐driven strategy to find drug‐repositioning candidates based on genomic and phenotypic features in our results. Although the disparity in relationship of drugs between known phenotypes (i.e., side effects) and molecular targets is well‐known,30 it depends on the expression profiles from a cancer cell line (MCF7) subset of compared drug pairs (∼0.2%), which showed that homogeneous trends between phenotype and gene signature had derived similarity. Interestingly, identifying similar drug pairs by either phenotype or gene signature‐based cosine‐similarity analysis was significantly nonindependent (P value of binomial test 2.61E‐302). In addition, identification of similar drug pairs by either of the nondirectional clinical phenotype or gene expression signatures in MCF7 is selected by chance (P value of binomial 0.19) due to the false‐positive results from the nondirectional phenotype signatures. However, imbalance of data abundance among utilized resources, such as side effects and therapeutic indications of drugs, hampered the contributions of utilized resources. We also compared our prediction results with previous work that utilized the side effects of drugs without directionality.30 Over 40% of a comparison set of drugs (12 pairs among 48 pairs consisting 24 drugs) has matched, and unmatched similar drugs of ours cover well‐known similar drugs, such as raloxifene and tamoxifen.31
As an example of the potential benefit of our approach in the drug‐repositioning task, we explored the suggested drug relationships for thioridazine for identifying drug‐repositioning candidates. Among 16 similar drugs, 11 identified several interesting repositioning candidates for thioridazine, including malaria and cancer, with previous studies.32, 33 Currently, a clinical trial for the use of thioridazine for refractory acute myeloid leukemia is recruiting (ClinicalTrials.gov: NCT02096289, phase I).
Our method demonstrates that analysis based on drug‐phenotype relations and molecular‐level pharmacological data can yield candidates for repositioning. In our previous work, we analyzed the similarity of diseases using only genomic signatures.28 The methods in our previous work (z‐score approach) utilized only continuous values, including gene expressions, and it is inappropriate for indexed (or categorical) values, such as term value of clinical phenotype. This study has focused on the examination of drug‐drug similarity regarding both gene expressions and clinical phenotypes using a simple and general method.
Some limitations of our approach should be recognized. Cosine‐similarity scores were based on expression profiles in a single cancer cell line, whereas drug‐associated phenotypes came from patients with diverse diagnoses. Thus, capturing drug‐associated gene signatures under the relevant disease conditions remains challenging. We acknowledge that phenotype‐based similarity analysis is biased toward older drugs that have diverse uses with many noted side effects. In addition, abundance of data among side effects and therapeutic indication‐associated resources are sparse and imbalanced. Thereby, zero values of cosine similarity of drugs using clinical phenotypes are substantial. Therefore, extraction of clinical phenotypes from medical records is the right direction of future studies. We acknowledge that a case study imperfectly displayed the benefit of directional relationships. However, a case study is a feasible way to deliver the potential benefit of our study. We note that there is no standard and feasible measure to evaluate the accuracy of drug‐drug relationships. In addition, except for clinical trials, there is no systematic guideline to evaluate drug‐repositioning candidates. Admittedly, large‐scale drug‐drug relationship analysis with/without directionality to prioritize drug‐repositioning candidates and experimental validation and then compare the success rate in clinical trials in population‐wide levels would be ideal. Nevertheless, practical constraints probably make such a systematic study unfeasible. However, we observed that many first‐line neuropsychiatric drugs, such as gabapentin for epilepsy and paroxetine for major depression, had the largest number of ranked side effects, indicating that they may be better‐targeted therapies to be considered as potential first‐line candidates for neuropsychiatric disease.
Nonetheless, we believe that our method, in conjunction with existing discovery approaches, such as clinical phenotype data and molecular drug signatures, will enable pharmaceutical companies and other medical associations to drive more testing of existing drugs and help expedite future drug repositioning efforts.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Source of Funding
This work was supported by Lucile Packard Foundation for Children's Health and a grant from the National Institute of General Medical Sciences (R01 GM079719).
Author Contributions
H.P., B.C., M.S., D.H., A.B., wrote the manuscript. H.P., A.B., designed the research. H.P., B.C., performed the research. H.P. analyzed the data.
Conflict of Interest
The authors declared no conflict of interest.
References
- 1. DiMasi, J.A. , Hansen, R.W. & Grabowski, H.G. The price of innovation: new estimates of drug development costs. J. Health Econ. 22, 151–185 (2003). [DOI] [PubMed] [Google Scholar]
- 2. Munos, B. Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov. 8, 959–968 (2009). [DOI] [PubMed] [Google Scholar]
- 3. Aronson, J.K. Old drugs–new uses. Br. J. Clin. Pharmacol. 64, 563–565 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashburn, T.T. & Thor, K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3, 673–683 (2004). [DOI] [PubMed] [Google Scholar]
- 5. Cheng, F. et al Prediction of drug‐target interactions and drug repositioning via network‐based inference. PLoS Comput. Biol. 8, e1002503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dudley, J.T. , Deshpande, T. & Butte, A.J. Exploiting drug‐disease relationships for computational drug repositioning. Brief. Bioinform. 12, 303–311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dudley, J.T. et al Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 3, 96ra76 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sirota, M. et al Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl. Med. 3, 96ra77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gottlieb, A. , Stein, G.Y. , Ruppin, E. & Sharan, R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 7, 496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iwata, H. , Sawada, R. , Mizutani, S. & Yamanishi, Y. Systematic drug repositioning for a wide range of diseases with integrative analyses of phenotypic and molecular data. J. Chem. Inf. Model. 55, 446–459 (2015). [DOI] [PubMed] [Google Scholar]
- 11. Bisgin, H. , Liu, Z. , Fang, H. , Kelly, R. , Xu, X. & Tong, W. A phenome‐guided drug repositioning through a latent variable model. BMC Bioinformatics 15, 267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lamb, J. et al The Connectivity Map: using gene‐expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006). [DOI] [PubMed] [Google Scholar]
- 13. Chiang, A.P. & Butte, A.J. Systematic evaluation of drug‐disease relationships to identify leads for novel drug uses. Clin. Pharmacol. Ther. 86, 507–510 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paik, H. et al Repurpose terbutaline sulfate for amyotrophic lateral sclerosis using electronic medical records. Sci. Rep. 5, 8580 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen, B. , Greenside, P. , Paik, H. , Sirota, M. , Hadley, D. & Butte, A.J. Relating chemical structure to cellular response: an integrative analysis of gene expression, bioactivity, and structural data across 11,000 compounds. CPT Pharmacometrics Syst. Pharmacol. 4, 576–584 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang, H. , Gu, Q. , Wei, J. , Cao, Z. & Liu, Q. Mining drug‐disease relationships as a complement to medical genetics‐based drug repositioning: where a recommendation system meets genome‐wide association studies. Clin. Pharmacol. Ther. 97, 451–454 (2015). [DOI] [PubMed] [Google Scholar]
- 17. Wang, F. , Zhang, P. , Cao, N. , Hu, J. & Sorrentino, R. Exploring the associations between drug side‐effects and therapeutic indications. J. Biomed. Inform. 51, 15–23 (2014). [DOI] [PubMed] [Google Scholar]
- 18. Yang, L. & Agarwal, P. Systematic drug repositioning based on clinical side‐effects. PLoS One 6, e28025 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou, X. , Menche, J. , Barabási, A.L. & Sharma, A. Human symptoms‐disease network. Nat. Commun. 5, 4212 (2014). [DOI] [PubMed] [Google Scholar]
- 20. Cheng, J. et al Evaluation of analytical methods for connectivity map data. Pac. Symp. Biocomput. 5–16 (2013). [PubMed] [Google Scholar]
- 21. Kuhn, M. , Campillos, M. , Letunic, I. , Jensen, L.J. & Bork, P. A side effect resource to capture phenotypic effects of drugs. Mol. Syst. Biol. 6, 343 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. US Food and Drug Administration . Adverse event reporting system. <http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/>.
- 23. Knox, C. et al DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 39(Database issue), D1035–D1041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. US Food and Drug Administration . Orange book: approved drug products with therapeutic equivalence evaluations. <http://www.accessdata.fda.gov/scripts/cder/ob/>.
- 25. Bodenreider, O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 32(Database issue), D267–D270 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aronson, A.R. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc. AMIA Symp. 17–21 (2001). [PMC free article] [PubMed] [Google Scholar]
- 27. Salton, G. , Wong, A. & Yang, C.S. A vector space model for automatic indexing. Commun. ACM 18, 613–620 (1975). [Google Scholar]
- 28. Suthram, S. , Dudley, J.T. , Chiang, A.P. , Chen, R. , Hastie, T.J. & Butte, A.J. Network‐based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput. Biol. 6, e1000662 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li, L. et al Disease risk factors identified through shared genetic architecture and electronic medical records. Sci. Transl. Med. 6, 234ra57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campillos, M. , Kuhn, M. , Gavin, A.C. , Jensen, L.J. & Bork, P. Drug target identification using side‐effect similarity. Science 321, 263–266 (2008). [DOI] [PubMed] [Google Scholar]
- 31. Vogel, V.G. et al Effects of tamoxifen vs raloxifene on the risk of developing invasive breast cancer and other disease outcomes: the NSABP Study of Tamoxifen and Raloxifene (STAR) P‐2 trial. JAMA 295, 2727–2741 (2006). [DOI] [PubMed] [Google Scholar]
- 32. Thanacoody, H.K. Thioridazine: resurrection as an antimicrobial agent? Br. J. Clin. Pharmacol. 64, 566–574 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Min, K.J. , Seo, B.R. , Bae, Y.C. , Yoo, Y.H. & Kwon, T.K. Antipsychotic agent thioridazine sensitizes renal carcinoma Caki cells to TRAIL‐induced apoptosis through reactive oxygen species‐mediated inhibition of Akt signaling and downregulation of Mcl‐1 and c‐FLIP(L). Cell Death Dis. 5, e1063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information


