Abstract
Background.
Astrocytomas are the most common primary human brain tumors. Receptor tyrosine kinases (RTKs), including tyrosine receptor kinase B (TrkB, also known as tropomyosin-related kinase B; encoded by neurotrophic tyrosine kinase receptor type 2 [NTRK2]), are frequently mutated by rearrangement/fusion in high-grade and low-grade astrocytomas. We found that activated TrkB can contribute to the development of astrocytoma and might serve as a therapeutic target in this tumor type.
Methods.
To identify RTKs capable of inducing astrocytoma formation, a library of human tyrosine kinases was screened for the ability to transform murine Ink4a−/−/Arf−/− astrocytes. Orthotopic allograft studies were conducted to evaluate the effects of RTKs on the development of astrocytoma. Since TrkB was identified as a driver of astrocytoma formation, the effect of the Trk inhibitors AZD1480 and RXDX-101 was assessed in astrocytoma cells expressing activated TrkB. RNA sequencing, real-time PCR, western blotting, and enzyme-linked immunosorbent assays were conducted to characterize NTRK2 in astrocytomas.
Results.
Activated TrkB cooperated with Ink4a/Arf loss to induce the formation of astrocytomas through a mechanism mediated by activation of signal transducer and activator of transcription 3 (STAT3). TrkB activation positively correlated with Ccl2 expression. TrkB-induced astrocytomas remained dependent on TrkB signaling for survival, highlighting a role of NTRK2 as an addictive oncogene. Furthermore, the QKI-NTRK2 fusion associated with human astrocytoma transformed Ink4a−/−/Arf−/− astrocytes, and this process was also mediated via STAT3 signaling.
Conclusions.
Our findings provide evidence that constitutively activated NTRK2 alleles, notably the human tumor-associated QKI-NTRK2 fusion, can cooperate with Ink4a/Arf loss to drive astrocytoma formation. Therefore, we propose NTRK2 as a potential therapeutic target in the subset of astrocytoma patients defined by QKI-NTRK2 fusion.
Key words: astrocytoma, CCL2, NTRK2, STAT3, QKI-NTRK2
Astrocytomas are the most common primary brain tumors and can occur in most parts of the brain. The World Health Organization (WHO) has established a grading system that assigns grades from I to IV for astrocytomas, with grades I and II being low-grade and grades III and IV being high-grade.1,2 Low-grade astrocytomas most commonly occur in children, whereas high-grade astrocytomas are most frequent in adults. Grade IV astrocytoma, also termed glioblastoma (GBM), is the most common and most malignant type of brain tumor. Although astrocytomas in children and adults share related histological characteristics, pediatric and adult astrocytoma have substantially different molecular features, tumor locations in the brain, and distributions of histological grades.3 As astrocytomas are a heterogeneous collection of distinct diseases with multiple oncogenic drivers, even within the same subtype,4–6 it is crucial to develop specific tailored therapies to improve drug response and patient outcomes for each molecular subtype.7
Receptor tyrosine kinases (RTKs) are key regulators of critical cellular processes, including proliferation and differentiation, survival, and metabolism of both normal and malignant cells of the CNS. RTKs function by transducing extracellular signals into cells to engage multiple cellular processes.8 Comprehensive genomic analysis of multiple tumor types, including low-grade and high-grade gliomas, has identified acquisition of somatic genetic alterations in RTKs as one of the most common sources of aberrant intracellular signaling.5,6 RTK signaling is normally tightly controlled; however, genetic and epigenetic alterations can change its activity, abundance, cellular distribution, or regulation. Abnormal RTK signaling can confer oncogenic potential by activating multiple proliferation and survival signaling cascades, including the phosphoinositide 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) signaling pathways, which play major roles in tumor development and progression.9 Furthermore, RTK alterations such as genomic amplification have proven to be remarkably heterogeneous in astrocytomas.10,11 The spectrum of RTK amplifications can vary from patient to patient and even among unique subpopulations within any one tumor. In fact, recent single-cell sequencing studies have shown that subclones within a single patient's GBM can harbor multiple unique mutations or alterations in the epidermal growth factor receptor gene (EGFR).12 Furthermore, some of the most common fusion/rearrangement events in astrocytomas and other gliomas involve RTKs, including EGFR and the genes for platelet-derived growth factor receptor alpha polypeptide (PDGFRA), fibroblast growth factor receptor 1/3 (FGFR1/3), and neurotrophic tyrosine kinase receptor type 2 (NTRK2).
Given the important role(s) of RTKs in astrocytomas, we developed an in vivo system to systematically identify RTKs whose activation can transform normal astrocytes. Activating RTK events frequently co-occur with loss-of-function mutations and/or genomic deletions involving tumor suppressor genes, such as phosphatase and tensin homolog (PTEN) and cyclin-dependent kinase inhibitor 2A (CDKN2A), which encodes both INK4a and ARF.13 Here, INK4a/ARF activity was selected as the “first hit” because loss-of-function events occur in 60% of malignant astrocytomas and are oncogenic when combined with mutant EGFR (Ink4a−/−/Arf−/−/EGFRvIII).14 By utilizing an Ink4a−/−/Arf−/− background but replacing EGFR variant III (EGFRvIII) with other activating RTK events, we screened a tyrosine kinase (TK)–TEL library that contains all RTKs to test their ability to drive astrocytoma formation in vivo. We identified multiple Ink4a−/−/Arf−/− cooperating events that were capable of inducing intracranial tumor formation by astrocytes in mice, including Ink4a−/−/Arf−/−/NTRK2-TEL. Importantly, NTRK2 fusions are present in 2%–3% of low-grade and high-grade astrocytomas.6,15–17 We further demonstrated that one QKI-NTRK2 fusion mutant with clinical relevance in humans is a driver in astrocytoma, suggesting that NTRK2 fusions might serve as potential therapeutic targets.
Materials and Methods
Primary Astrocyte Cell Culture
Primary astrocytes were isolated from the brain cortex of 5-day-old mouse pups as described.18 Briefly, brain cortices were dissected free of the surrounding brain, and single cells were obtained by mechanical dissociation followed by trypsin incubation. The cells were expanded in a conventional 2D culture flask until confluent and then further purified to remove oligodendrocytes. The cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (Invitrogen).
Plasmid Constructs, Retroviral and Lentiviral Vector Production
A cDNA library of TKs was fused to TEL (ETV6) with a flag tag that was generated in a retroviral pWZLneo vector. NTRK2-TEL-flag was amplified from pWZLneo-NTRK2-TEL-flag using primers P3 and P4 (Supplementary Table), digested with BamHI and EcoRI, and subcloned into pMSCV vector that had been digested with BglII and EcoRI.
To create a QKI-NTRK2 fusion, QKI 1-311 (encoded by QKI exons 1–6) and NTRK2 482-838 (encoded by NTRK2 exons 16–21) were amplified from a human mammary epithelial cell cDNA library using primers P5 with P6, and P7 with P8 (Supplementary Table), respectively. The 2 PCR products were mixed and served as the template for generation of the QKI-NTRK2 insert using primers P5 and P8. The resulting PCR products were digested with BamHI and EcoRI and subcloned into the BglII/EcoRI-digested pMSCV vector. The identity of all constructs was verified by sequencing. Retroviral production was performed as described previously.19
Lentiviral short hairpin (sh)RNA constructs in the pLKO.1 vector were obtained from the RNAi Core Facility at Dana-Farber Cancer Institute. The shRNA sequences targeting mouse Stat3 are listed in the Supplementary Table. Lentivirus preparation and transduction of cells were performed according to the online protocol of the RNAi Core Facility.20
Tyrosine Kinase Screen in Vivo
Primary Ink4a−/−/Arf−/− astrocytes were transduced with pools of retroviral vectors encoding 73 human TK-TEL fusion constructs (18 pools, each consisting of 3–5 unique TK-TELs). After tandem retroviral infection, the cells (1 × 106 cells in phosphate buffered saline with 30% Matrigel) were s.c. transplanted into nude mice. To determine which TKs contribute to gliomagenesis, the tumors that developed were harvested and genomic DNA was extracted. TK-TEL integrations were identified by PCR amplification using vector-specific primers P1 and P2 (Supplementary Table) followed by sequencing. In addition, each individual TK-TEL from the pool that formed tumors was transduced into primary Ink4a−/−/Arf−/− astrocytes, and the cells were s.c. transplanted into nude mice to further confirm the oncogenic effect.
Stereotactic Injection of Cells
Severe combined immunodeficient mice or nude mice (∼8 wk old; Taconic) were anesthetized with ketamine/xylazine and placed in a stereotactic frame using ear bars. Cells for injection were suspended in phosphate buffered saline (100 000 cells/μL), and 1 μL was intracranially injected into the right striatum (with bregma set as 0, 0, 0, the coordinates for cell injection were anterior 0, lateral 2 mm, ventral 2.5 mm). All animal experiments were performed in accordance with NIH animal use guidelines, and all protocols were approved by the Dana-Farber Cancer Institute Animal Care and Use Committee.
Compounds and Reagents
AZD1480 was obtained from Selleck Chemicals and MedChemexpress. Anti-Flag, anti–phosphorylated signal transducer and activator of transcription 3 (pSTAT3), anti-pMAPK, anti-pAkt, anti-STAT3, anti-MAPK, anti-Akt, and anti-TrkB antibodies were obtained from Cell Signaling Technology; anti–α-tubulin antibody was obtained from Sigma-Aldrich; anti–glial fibrillary acidic protein (GFAP) antibody was from Dako; anti-Ki67 antibodies were obtained from Vector Labs and Cell Signaling Technology. Epidermal growth factor (EGF), nerve growth factor (NGF), and brain-derived neurotrophic factor (BDNF) were purchased from Sigma-Aldrich. Fibroblast growth factor (hFGF2) was from Miltenyi Biotec.
In Vivo Tyrosine Kinase Inhibitor Treatment
AZD1480 was suspended in 0.5% hydroxypropylmethyl cellulose with 0.1% Tween 80 and given orally at a dose of 50 mg/kg daily. Animals were monitored daily for development of neurological defects.
Western Blot Analysis, Staining
Western blot analysis was performed as described previously.21 Hematoxylin and eosin staining was performed using standard techniques. Immunohistochemical (IHC) staining was performed as described previously.21
RNA Isolation and Real-Time PCR
Total RNA was isolated using Trizol (Invitrogen) and an RNeasy Mini Kit (Qiagen). Complementary DNA was generated using the Superscript III First-Strand Synthesis System (Invitrogen) according to the manufacturer's instructions. Real-time PCR was performed using SYBR Green mix (Applied Biosystems). Ccl2, Stat3, and Gapdh primers are listed in the Supplementary Table.
RNA Sequencing
Preparation of the whole transcriptome library in biological triplicate was performed using 5–10 ng of fragmented ribosomal RNA-depleted total RNA with the Ion Total RNA-Seq Kit V2 (Thermo Fisher) according to the manufacturer's protocol. Three libraries were multiplexed and clonally amplified to obtain template-positive ion sphere particles using the Ion OneTouch 2 System (Thermo Fisher), and then sequenced using an Ion Torrent Proton with one PI chip kit V2 (Thermo Fisher). Sequences were collected using Torrent Suite v4.4 software. Sequences were aligned to the genome using STAR (Spliced Transcripts Alignment to a Reference),22 and the remaining unmapped reads were aligned using Bowtie 2 in local mode,23 a 2-step method. Read counts were generated with htseq-count,24 and differential gene expression analysis was performed using the R, Bioconductor, and DESeq2 packages.25
Mouse Cytokine Array
Cytokine array experiments were performed according to the manufacturer's instructions (R&D). Spot intensities were quantified using Image Studio (LI-COR).
Chemokine C-C Ligand 2 Enzyme-Linked Immunosorbent Assay
Cells were cultured in a 6-well plate, and chemokine C-C ligand 2 (CCL2) concentration in the culture supernatant was measured with a sandwich enzyme-linked immunosorbent assay (ELISA; BioLegend) according to the manufacturer's protocol.
Statistical Analysis
An unpaired Student's t-test or ANOVA was performed using GraphPad Prism 6 software to determine statistical significance. Data were considered significant when P < .05.
Results
Identification of Astrocytoma-Initiating Receptor Tyrosine Kinases through in Vivo Screening of a TK-TEL Library
To assess the oncogenic potential of individual RTKs in astrocytoma formation, we performed a systematic in vivo pooled screen by infecting Ink4a−/−/Arf−/− astrocytes with a retroviral TK-TEL cDNA library encoding 73 individual human TKs fused to the TEL domain (Fig. 1 A). This domain induces kinase dimerization, resulting in constitutive ligand-independent activation and signaling of TK.26
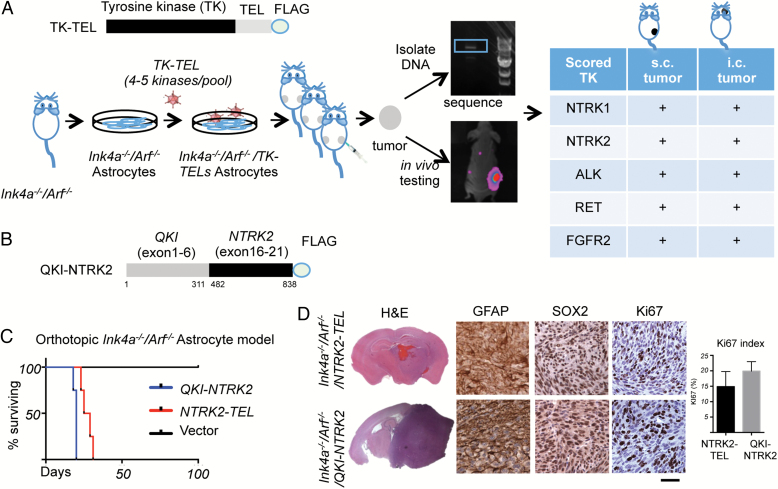
Fig. 1.
Identification of activated TrkB as a potential oncogenic tyrosine kinase by an in vivo screen. (A) Top panel, schematic structure of tyrosine kinase–TEL (TK-TEL) library; bottom panel, schematic flow chart of the in vivo tumorigenesis assay used to screen the TK-TEL library for transformation of Ink4a−/−/Arf−/− astrocytes in mice. (B) Schematic structure of the QKI-NTRK2 fusion protein. (C) Kaplan–Meier survival curves of mice bearing intracranially implanted Ink4a−/−/Arf−/−/QKI-NTRK2 (blue line, n = 4), Ink4a−/−/Arf−/−/NTRK2-TEL (red line, n = 4), or Ink4a−/−/Arf−/−/Vector (black line, n = 2) astrocytes. (D) Hematoxylin and eosin staining and IHC analysis of GFAP (glioma cell marker), Sox2 (neural stem cell marker), and Ki67 (proliferative marker) in the indicated astrocytomas. Scale bar, 50 μm. Bar graph represents quantification of Ki67 in tumors (mean ± SD).
Transduction of primary Ink4a−/−/Arf−/− astrocytes with 18 pools (3–5 unique TK-TELs/pool) resulted in the identification of 5 TK-TELs (NTRK1, NTRK2, RET, ALK, and FGFR2) that can cooperate with Ink4a/Arf loss to induce the formation of subcutaneous and intracranial astrocytomas in vivo (Fig. 1 A). Ink4a−/−/Arf−/− astrocytes alone did not form tumors in vivo. These findings highlight the oncogenic capacity of TK-TELs in Ink4a−/−/Arf−/− astrocytes.
Although several of these oncogenic TKs have been well studied (eg, the “rearranged during transfection” [RET] gene, anaplastic lymphoma kinase [ALK]),27–30 no functional data have been generated regarding the role of NTRK2 fusions in gliomagenesis. NTRK2 amplification and/or mutation events are uncommon in astrocytomas; however, recent studies have identified the presence of novel NTRK2 fusion proteins, including QKI-NTRK2 (Fig. 1 B).15 To determine whether NTRK2 fusion proteins can transform astrocytes, we cloned the QKI-NTRK2 fusion and transduced Ink4a−/−/Arf−/− astrocytes. Orthotopic transplants of Ink4a−/−/Arf−/−/QKI-NTRK2 astrocytes reproducibly formed gliomas in vivo, with ∼1-month survival and 100% penetrance, which is similar to the findings for Ink4a−/−/Arf−/−/NTRK2-TEL astrocytes (Fig. 1 C). Histological analysis of intracranial tumors revealed densely cellular gliomas with frequent mitoses. IHC characterization confirmed expression of glial lineage markers GFAP and sex determining region Y–box 2 (Sox2) and markedly elevated proliferation, highlighted by Ki67 expression, in both Ink4a−/−/Arf−/−/NTRK2-TEL and Ink4a−/−/Arf−/−/QKI-NTRK2 astrocytomas (Fig. 1 D).
Activated Tyrosine Receptor Kinase B Preferentially Signals via STAT3 but not PI3K or MAPK
RTKs are known to initiate multiple signal transduction pathways. To identify the major signaling pathways activated by oncogenic RTKs in Ink4a−/−/Arf−/− astrocytes, we assessed MAPK, PI3K, and STAT3 signaling under serum-negative growth conditions. Surprisingly, NTRK1-TEL and in particular NTRK2-TEL resulted in an increased level of pSTAT3, but not of pMAPK or pAkt (Fig. 2 A). Similarly, expression of QKI-NTRK2 also increased STAT3 signaling but reduced phosphorylation of Akt and MAPK (Fig. 2 A), suggesting that STAT3 signaling is a major event downstream of NTRK2 activation in this setting. Exposure of Ink4a−/−/Arf−/− astrocytes to BDNF, the ligand for TrkB, increased pSTAT3 signaling, whereas exposure to EGF, FGF, or NGF did not (Fig. 2 B), further supporting the specific activation of STAT3 signaling in response to TrkB activity. In addition, transcriptomic analyses revealed that the Janus kinase (JAK)/STAT3 gene set is selectively upregulated in Ink4a−/−/Arf−/−/NTRK2-TEL astrocytes compared with Ink4a−/−/Arf−/−/Vector astrocytes (Fig. 2 C).
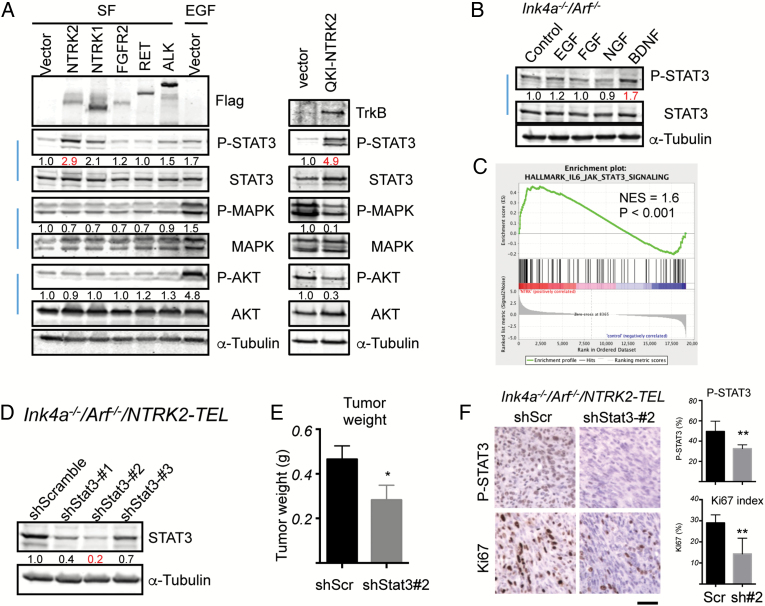
Fig. 2.
Upregulation of STAT3 signaling by constitutively activated TrkB. (A) Ink4a−/−/Arf−/− astrocytes expressing vector, RTK-TELs, or QKI-NTRK2 as indicated were serum starved (SF) overnight. SF starved Ink4a−/−/Arf−/− astrocytes expressing vector treated with 10 ng/mL EGF for 5 min as a control for signaling activation. Cell lysates were subjected to western blotting analysis with the indicated antibodies. Band intensities of phospho-proteins were quantified and normalized relative to the quantity of their respective total protein bands, and then expressed as fold difference compared with the vector-expressing samples. Images are representative of at least 2 independent experiments. (B) Ink4a−/−/Arf−/− astrocytes were serum starved overnight followed by 3-h stimulation with 10 ng/mL EGF, 10 ng/mL FGF2, 100 ng/mL NGF, or 100 ng/mL BDNF and western blot analyses with the indicated antibodies. Band intensities of phospho-STAT3 were quantified and normalized relative to the respective total STAT3 bands and expressed as the fold difference compared with the control sample. Images are representative of 2 independent experiments. (C) Gene Set Enrichment Analysis plot of interleukin-6–JAK-STAT3 gene set in Ink4a−/−/Arf−/−/NTRK2-TEL vs Ink4a−/−/Arf−/−/Vector astrocytes. Enrichment plot: bottom, plot of the ranked list of all genes. y-Axis, value of the ranking metric; x-axis, the rank for all genes. Normalized enrichment score (NES) was 1.6, nominal P < .001. (D) Knockdown of Stat3 in Ink4a−/−/Arf−/−/NTRK2-TEL astrocytes. Ink4a−/−/Arf−/−/NTRK2-TEL astrocytes stably expressed each indicated shRNA. Cell lysates were subjected to western blot analysis with the indicated antibodies. Band intensities of STAT3 were quantified and normalized relative to the quantity of α-tubulin bands, and then expressed as the fold difference compared with the shScramble sample, which was set as 1.0. (E) Tumor weight of Ink4a−/−/Arf−/−/NTRK2-TEL/shScr (shScr) and Ink4a−/−; Arf−/−/NTRK2-TEL/shStat3-#2 (shStat3-#2) s.c. allograft models in nude mice. Values represent means ± SEM, n = 6 per group. *P < .05. (F) IHC analysis of pSTAT3 and Ki67 in the indicated astrocytomas. Scale bar, 50 μm. Bar graphs represent mean ± SD of quantification of pSTAT3 and Ki67 in tumors (*P < .05, **P < .01).
To explore the role of STAT3 signaling in TrkB-induced glioma growth, we used lentiviral-delivered shRNAs to knock down Stat3 in Ink4a−/−/Arf−/−/NTRK2-TEL astrocytes, and confirmed a reduction in protein prior to s.c. implantation (Fig. 2 D). Ink4a−/−/Arf−/−/NTRK2-TEL/shStat3-#2 cells were chosen for in vivo studies because these cells had the greatest Stat3 knockdown. Although both shStat3 and shScramble (shScr) Ink4a−/−/Arf−/−/NTRK2-TEL astrocytes formed tumors in mice, we found a significant reduction in tumor mass among shStat3-suppressed tumors compared with shScr tumors (Fig. 2 E). In line with the decreased tumor mass, staining of pSTAT3 and Ki67 was diminished in Ink4a−/−/Arf−/−/NTRK2-TEL/shStat3-#2 tumors (Fig. 2 F). These findings suggest that in vivo growth of TrkB-induced astrocytoma is dependent on STAT3 signaling for optimal growth.
Tyrosine Receptor Kinase B Mediated STAT3 Signaling Upregulates CCL2 Production
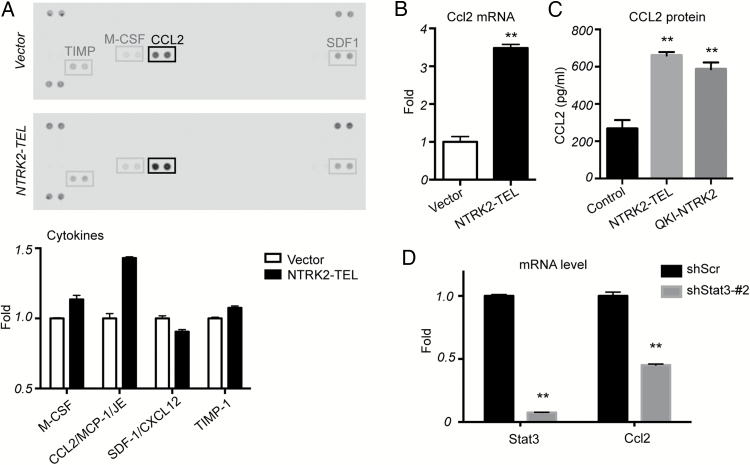
As JAK/STAT signaling represents a major downstream effector mechanism for cytokine signaling, we next assessed the role of TrkB in cytokine production. Using a murine cytokine array, we found that activated TrkB signaling in astrocytes resulted in significant upregulation of CCL2 compared with vector control (Fig. 3 A). We confirmed this finding by real-time PCR and ELISA, which showed increased Ccl2 mRNA and CCL2 protein levels, respectively, in NTRK2-TEL cells (Fig. 3 B and C). QKI-NTRK2 also significantly upregulated CCL2 protein levels, albeit to lower levels than NTRK2-TEL (Fig. 3 C). To determine whether CCL2 is induced by TrkB-mediated STAT3 signaling, we examined Ccl2 mRNA in shStat3 astrocytes. Indeed, suppression of Stat3 resulted in a nearly 50% reduction of Ccl2 mRNA (Fig. 3 D).
Fig. 3.
Upregulation of Ccl2 by activated TrkB is mediated by STAT3 signaling. (A) Supernatants from the indicated cells grown in serum-free medium for 1 day were collected and analyzed using a murine cytokine array. The bar graph represents spot intensity. Data from vector control cells were set as 1.0. (B) Quantitative PCR of mRNA expression levels of Ccl2 in NTRK2-TEL cells compared with those from control vector cells, which were set to 1.0. **P < .01. (C) CCL2 protein levels were measured by ELISA. Vector, NTRK2-TEL, and QKI-NTRK2 cells were serum starved for 2 days and supernatants were collected for analysis by ELISA. **P < .01. (D) Quantitative PCR of mRNA expression levels of Ccl2 in shStat3 cells compared with levels in shScr control cells, which were set to 1.0. **P < .01.
Inhibition of Tyrosine Receptor Kinase B Reduces Proliferation of TrkB-Induced Glioma
Having shown a clear role for TrkB-signaling in gliomagenesis, we next sought to determine whether glioma propagation and survival is dependent on continued TrkB-mediated pSTAT3 activity. To assess the dependency of continued TrkB signaling in TrkB-induced gliomas, we treated tumor cells with AZD1480 (an NTRK/JAK/STAT inhibitor that has previously been shown to have efficacy in glioma xenograft models)31–33 or RXDX-101 (an NTRK/ROS1/ALK inhibitor)34 and evaluated the cells for changes in growth and signaling profiles.
Both AZD1480 and RXDX-101 decreased pSTAT3 levels in a dose- and time-dependent manner (Fig. 4 A and B). Consistent with the role of CCL2 as a target of pSTAT3 signaling, a reduction in pSTAT3 was accompanied by decreased Ccl2 mRNA levels following AZD1480 or RXDX-101 treatment (Fig. 4 C). Mice with intracranial allografts of Ink4a−/−/Arf−/−/NTRK2-TEL gliomas treated with AZD1480 showed significantly prolonged survival with marked reductions in nuclear pSTAT3 staining, cell proliferation (Ki67), and Ccl2 mRNA levels compared with untreated control animals bearing the same tumors (Fig. 4 D–F).
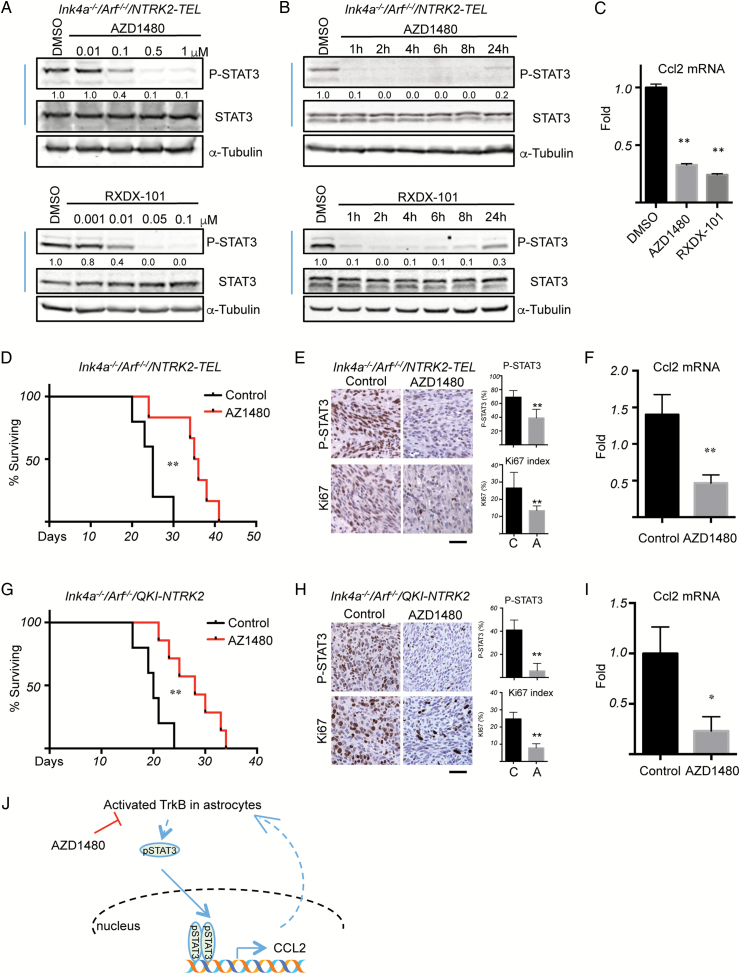
Fig. 4.
Inhibition of TrkB activity suppresses tumor growth initiated by activated TrkB. (A) Ink4a−/−/Arf−/−/NTRK2-TEL astrocytes were treated with different concentrations of AZD1480 (upper panel) or RXDX-101 (lower panel) for 1 h. (B) Cells were treated with 0.5 μM AZD1480 (upper panel) or 0.05 μM RXDX-101 (lower panel) for the indicated times. Western blot analyses were then performed with the indicated antibodies. Band intensities of pSTAT3 were quantified and normalized relative to the quantity of respective STAT3 bands, and then expressed as the fold difference compared with the dimethyl sulfoxide (DMSO) sample, which was set as 1.0. Images are representative of at least 2 independent experiments. (C) Quantitative PCR analysis of mRNA expression levels of Ccl2 in Ink4a−/−/Arf−/−/NTRK2-TEL astrocytes treated with 0.5 μM AZD1480 or 0.05 μM RXDX-101 for 4 h. Data from the DMSO treatment were set to 1.0; n = 3, data show mean ± SD. (D) Kaplan–Meier survival curves of mice bearing intracranially implanted Ink4a−/−/Arf−/−/NTRK2-TEL astrocytes. After 4 days of tumor engraftment, mice were treated with control (black line, n = 5) or AZD1480 (50 mg/kg, p.o., q.d.) (red line, n = 6). (E) IHC analyses of pSTAT3 and Ki67 expression in Ink4a−/−/Arf−/−/NTRK2-TEL allograft tumor samples of mice treated with control or AZD1480 (50 mg/kg, p.o., q.d.) for 4 days. Tumors were collected 2 h after the last dose, fixed, and subjected to IHC analyses. Scale bar, 50 μm. Bar graphs represent mean ± SD of quantification of pSTAT3 and Ki67 in tumors. (F) Quantitative PCR analysis of Ccl2 mRNA levels in glioma induced by activated TrkB after treatment with control or AZD1480. The value from one control sample was set to 1.0, n = 3 (control) or n = 5 (treatment), data show mean ± SEM. (G) Kaplan–Meier survival of mice bearing intracranially implanted Ink4a−/−/Arf−/−/QKI-NTRK2 astrocytes. After 4 days of tumor engraftment, mice were treated with control (black line, n = 5) or AZD1480 (50 mg/kg, p.o., q.d.) (red line, n = 7). (H) IHC analyses of pSTAT3 and Ki67 in Ink4a−/−/Arf−/−/QKI-NTRK2 allograft tumor samples of mice treated with vehicle or AZD1480 (50 mg/kg, p.o., q.d.). Tumors were collected 2 h after the last dose, fixed, and subjected to IHC analyses. Scale bar, 50 μm. Bar graphs represent mean ± SD of quantification of pSTAT3 and Ki67 in tumors. (I) Quantitative PCR of Ccl2 mRNA levels in gliomas induced by QKI-NTRK2 treated with control or AZD1480. The value from one control was set to 1.0; n = 5 (control) or n = 3 (treatment), data show mean ± SEM. *P < .05, **P < .01. (J) A working model of TrkB-STAT3 signaling in astrocytoma.
Similar to our observations with NTRK2-TEL, AZD1480 treatment significantly prolonged the survival of mice bearing orthotopic Ink4a−/−/Arf−/−/QKI-NTRK2 astrocytomas (Fig. 4 G). Consistent with these findings, nuclear pSTAT3 and Ki67 protein levels and Ccl2 mRNA levels were dramatically reduced by AZD1480 treatment (Fig. 4 H and I).
Together, our studies validate a role for NTRK2 as a potent oncogene in the setting of CNS cancers, implicating its potential clinical value as an anticancer target for the treatment of astrocytoma.
Discussion
NTRK2 and its preferred ligand, BDNF, are essential for neural development.35,36 NTRK2 is overexpressed in a number of aggressive human cancer types, including neuroblastoma and ovarian, pancreatic, and prostate cancers, where it has been shown to regulate cell migration, metastasis, anoikis, and tumor recurrence.37–40 Recent next-generation sequencing studies have identified gene rearrangements of NTRK1, 2, and 3 encoding novel oncogenic fusions in a variety of tumor types, including low-grade and high-grade astrocytoma.6,15–17 The NTRK2 fusion gene was first identified in pilocytic astrocytoma, with recent reports of additional fusions including QKI-NTRK2, NACC2-NTRK2, VCL-NTRK2, AGBL4-NTRK2, TRIM24-NTRK2, PAN3-NTRK2, AFAP1-NTRK2, SQSTM1-NTRK2, NAV1-NTRK2, and SLMAP-NTRK2.6,15–17,41,42 The mechanism of action of these fusion proteins in human gliomagenesis, progression, and drug resistance remains poorly understood. In this study we performed a systematic screen of TK-TELs, which suggested that activated TrkB could transform Ink4a−/−/Arf−/− astrocytes in vivo via upregulation of pSTAT3 signaling and Ccl2 production. We thus confirmed that the astrocytoma-associated QKI-NTRK2 fusion protein is capable of inducing astrocytoma and may serve as a potential therapeutic target in astrocytoma patients.
CCL2, a chemokine primarily produced by astrocytes in the brain, may also represent a unique target capable of disrupting the tumor microenvironment. Overexpression of CCL2 in astrocytoma resulted in microglial infiltration and promoted astrocytoma growth.43 CCL2 can recruit T cells into the tumor microenvironment by promoting lymphocyte extravasation.44 The use of CCL2-neutralizing antibodies holds promise for disrupting the TrkB-STAT-CCL2 axis.45,46
Precision medicine in cancer has gained prominence as a result of advances in sequencing technologies that readily identify “druggable” mutations. Oncogenic gene fusions, such as NTRK1 in sarcoma47 and ALK fusion in lung cancer, have now been targeted in clinical trials.48 Oncogenic fusions, deletions, and/or splice variants involving NTRK1, NTRK2, and NTRK3 represent novel and potentially targetable genetic events in astrocytoma.42 The JAK/NTRK inhibitor AZD1480 was advanced to a phase I clinical trial (NCT01112397), but the trial was terminated early due to unacceptable side effects on the CNS.49 Nonetheless, for cancers driven by novel NTRK2 fusion proteins, therapies engineered to attack these neoproteins may provide an effective anticancer strategy while limiting off-target toxicities. Attempts to identify these therapies are already under way, with multiple clinical trials ongoing. New NTRK inhibitors RXDX-101 and LOXO-101 are currently in phase II trials (NCT02568267 and NCT02576431, respectively) in subjects with solid tumors, including brain tumors harboring NTRK fusion.
Virtually all patients with RTK aberrations eventually acquire drug resistance to RTK-targeted inhibitors. A variety of mechanisms have been found to be responsible for the development of resistance to RTK-targeted therapies, including bypass mechanisms through genetic alteration and activation of other TK pathways. In this study, we relied on the established Ink4a−/−/Arf−/−/EGFRvIII tumor model, replacing EGFRvIII with other TKs to test their ability to support tumorigenesis. We identified multiple RTKs that can compensate for loss of EGFRvIII activity and are capable of bypassing the need for EGFRvIII in astrocytoma. Therefore, these RTKs might be promising therapeutic targets and warrant clinical trials using combined RTK inhibitors for patients with astrocytoma, especially those who have recurrence after EGFRvIII-inhibiting therapies. Taken together, our data warrant the development of mechanism-based combined targeted therapies against TrkB in tumors harboring oncogenic TrkB.
Supplementary Material
Supplementary material is available at Neuro-Oncology Journal online (http://neuro-oncology.oxfordjournals.org/).
Funding
This work was supported by NIH grants P01 CA142536 (C.D.S., R.A.S., K.L.L., W.C.H., and J.J.Z.), P50 CA165962 (C.D.S., T.M.R., K.L.L., and J.J.Z.), R01 CA188288 (K.L.L.), and K08 NS (S.H.R.); Pediatric Low-Grade Astrocytoma Foundation (C.D.S.); and DFCI Medical Oncology Grant (S.H.R. and K.L.L.).
Conflict of interest statement. Dr Roberts, Dr Hahn, and Dr Beroukhim are consultants of Novartis and have received research grants from Novartis. The remaining authors declare no competing financial interests.
Supplementary Material
Acknowledgments
We thank Dr Roderick Bronson and the Dana-Farber/Harvard Cancer Center Rodent Histopathology Core for histopathological analyses. We thank Dr Frank Pan (Life Technologies, Thermo Fisher) for assistance with RNA-seq using the Ion Torrent platform.
References
- 1. Louis DN, Ohgaki H, Wiestler OD et al. . The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114 (2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wen PY, Reardon DA. Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12 2:69–70. [DOI] [PubMed] [Google Scholar]
- 3. Jones C, Perryman L, Hargrave D. Paediatric and adult malignant glioma: close relatives or distant cousins? Nat Rev Clin Oncol. 2012;9 (7):400–413. [DOI] [PubMed] [Google Scholar]
- 4. Sturm D, Bender S, Jones DT et al. . Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14 (2):92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455 (7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu G, Diaz AK, Paugh BS et al. . The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46 (5):444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol. 2014;9:1–25. [DOI] [PubMed] [Google Scholar]
- 8. Carrasco-Garcia E, Saceda M, Martinez-Lacaci I. Role of receptor tyrosine kinases and their ligands in glioblastoma. Cells. 2014;3 (2):199–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141 (7):1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sottoriva A, Spiteri I, Piccirillo SG et al. . Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110 (10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Snuderl M, Fazlollahi L, Le LP et al. . Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20 (6):810–817. [DOI] [PubMed] [Google Scholar]
- 12. Patel AP, Tirosh I, Trombetta JJ et al. . Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344 (6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verhaak RG, Hoadley KA, Purdom E et al. . Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17 (1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bachoo RM, Maher EA, Ligon KL et al. . Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1 (3):269–277. [DOI] [PubMed] [Google Scholar]
- 15. Jones DT, Hutter B, Jager N et al. . Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45 (8):927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J, Wu G, Miller CP et al. . Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45 (6):602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bandopadhayay P, Ramkissoon LA, Jain P et al. . MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat Genet. 2016;48 3:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85 (3):890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Q, Boudot A, Ni J et al. . Cyclin D1 and C/EBPbeta LAP1 operate in a common pathway to promote mammary epithelial cell differentiation. Mol Cell Biol. 2014;34 (16):3168–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Broad Institute Genetic Perturbation Platform Protocols. http://www.broadinstitute.org/rnai/public/resources/protocols. [Google Scholar]
- 21. Ni J, Liu Q, Xie S et al. . Functional characterization of an isoform-selective inhibitor of PI3K-p110beta as a potential anticancer agent. Cancer Disc. 2012;2 (5):425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dobin A, Davis CA, Schlesinger F et al. . STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29 (1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9 (4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31 (2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 (12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jousset C, Carron C, Boureux A et al. . A domain of TEL conserved in a subset of ETS proteins defines a specific oligomerization interface essential to the mitogenic properties of the TEL-PDGFR beta oncoprotein. EMBO J. 1997;16 (1):69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morris SW, Kirstein MN, Valentine MB et al. . Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263 (5151):1281–1284. [DOI] [PubMed] [Google Scholar]
- 28. Chen Y, Takita J, Choi YL et al. . Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455 (7215):971–974. [DOI] [PubMed] [Google Scholar]
- 29. Santoro M, Carlomagno F, Romano A et al. . Activation of RET as a dominant transforming gene by germline mutations of MEN2A and MEN2B. Science. 1995;267 (5196):381–383. [DOI] [PubMed] [Google Scholar]
- 30. Vaishnavi A, Capelletti M, Le AT et al. . Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19 (11):1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ioannidis S, Lamb ML, Wang T et al. . Discovery of 5-chloro-N2-[(1S)-1-(5-fluoropyrimidin-2-yl)ethyl]-N4-(5-methyl-1H-pyrazol-3-yl)p yrimidine-2,4-diamine (AZD1480) as a novel inhibitor of the Jak/Stat pathway. J Med Chem. 2011;54 (1):262–276. [DOI] [PubMed] [Google Scholar]
- 32. McFarland BC, Ma JY, Langford CP et al. . Therapeutic potential of AZD1480 for the treatment of human glioblastoma. Mol Cancer Ther. 2011;10 (12):2384–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y, Holdbrooks AT, De Sarno P et al. . Therapeutic efficacy of suppressing the Jak/STAT pathway in multiple models of experimental autoimmune encephalomyelitis. J Immunol. 2014;192 (1):59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rolfo C, Ruiz R, Giovannetti E et al. . Entrectinib: a potent new TRK, ROS1, and ALK inhibitor. Expert Opin Investig Drugs. 2015;24 (11):1493–1500. [DOI] [PubMed] [Google Scholar]
- 35. Soppet D, Escandon E, Maragos J et al. . The neurotrophic factors brain-derived neurotrophic factor and neurotrophin-3 are ligands for the trkB tyrosine kinase receptor. Cell. 1991;65 (5):895–903. [DOI] [PubMed] [Google Scholar]
- 36. Squinto SP, Stitt TN, Aldrich TH et al. . TrkB encodes a functional receptor for brain-derived neurotrophic factor and neurotrophin-3 but not nerve growth factor. Cell. 1991;65 (5):885–893. [DOI] [PubMed] [Google Scholar]
- 37. Sinkevicius KW, Kriegel C, Bellaria KJ et al. . Neurotrophin receptor TrkB promotes lung adenocarcinoma metastasis. Proc Natl Acad Sci U S A. 2014;111 (28):10299–10304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kupferman ME, Jiffar T, El-Naggar A et al. . TrkB induces EMT and has a key role in invasion of head and neck squamous cell carcinoma. Oncogene. 2010;29 (14):2047–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Douma S, Van Laar T, Zevenhoven J et al. . Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430 (7003):1034–1039. [DOI] [PubMed] [Google Scholar]
- 40. Yin B, Ma ZY, Zhou ZW et al. . The TrkB+ cancer stem cells contribute to post-chemotherapy recurrence of triple-negative breast cancers in an orthotopic mouse model. Oncogene. 2015;34 (6):761–770. [DOI] [PubMed] [Google Scholar]
- 41. Qaddoumi I, Orisme W, Wen J et al. . Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016;131 6:833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5 (1):25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Platten M, Kretz A, Naumann U et al. . Monocyte chemoattractant protein-1 increases microglial infiltration and aggressiveness of gliomas. Ann Neurol. 2003;54 (3):388–392. [DOI] [PubMed] [Google Scholar]
- 44. Carrillo-de Sauvage MA, Gomez A, Ros CM et al. . CCL2-expressing astrocytes mediate the extravasation of T lymphocytes in the brain. Evidence from patients with glioma and experimental models in vivo. PloS One. 2012;7 (2):e30762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rafei M, Deng J, Boivin MN et al. . A MCP1 fusokine with CCR2-specific tumoricidal activity. Mol Cancer. 2011;10:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu X, Fujita M, Snyder LA et al. . Systemic delivery of neutralizing antibody targeting CCL2 for glioma therapy. J Neurooncol. 2011;104 (1):83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Doebele RC, Davis LE, Vaishnavi A et al. . An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov. 2015;5 (10):1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Solomon BJ, Mok T, Kim DW et al. . First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371 (23):2167–2177. [DOI] [PubMed] [Google Scholar]
- 49. Plimack ER, Lorusso PM, McCoon P et al. . AZD1480: a phase I study of a novel JAK2 inhibitor in solid tumors. Oncologist. 2013;18 (7):819–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.