We are currently living in the “genomic era.” In the last decade, high-throughput genomic analyses have generated massive quantities of data that can be used to explore gene function in specific biological settings. In the field of cancer research, studies performed in a variety of laboratories and by a number of large-scale projects, like The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/, accessed October 31, 2016) and the International Cancer Genome Consortium (ICGC),1 have given researchers an unparalleled amount of information on many different tumors through several platforms such as: mRNA expression arrays, comparative genomic hybridization arrays, DNA methylation arrays, exome sequencing, RNA sequencing, reverse protein phase arrays, and clinical parameters.2 Consolidating these data into an easily accessible and intuitive format is crucial for our ability to extrapolate any meaningful information.3
Although a few web portals, such as the cBioportal,4 UCSC,5 ICGC,1 Oasis,6 and others, have been developed to integrate and analyze multi-omics data from the various recent large-scale projects, these portals are missing most of the datasets and the thorough knowledge generated over the years by independent laboratories for specific tumor types. Here we introduce GlioVis (http://gliovis.bioinfo.cnio.es, accessed October 31, 2016), a powerful web-based tool to help researchers and clinicians to rapidly access data relevant to brain tumor research. The portal has been built with 2 extremely crucial issues in mind: easy use and reproducibility. GlioVis has a very intuitive interface, and no bioinformatics skills are required. For all the plots that are generated, the user can run different statistical tests, and the raw data to reproduce the plots can be downloaded for future analysis or publications.
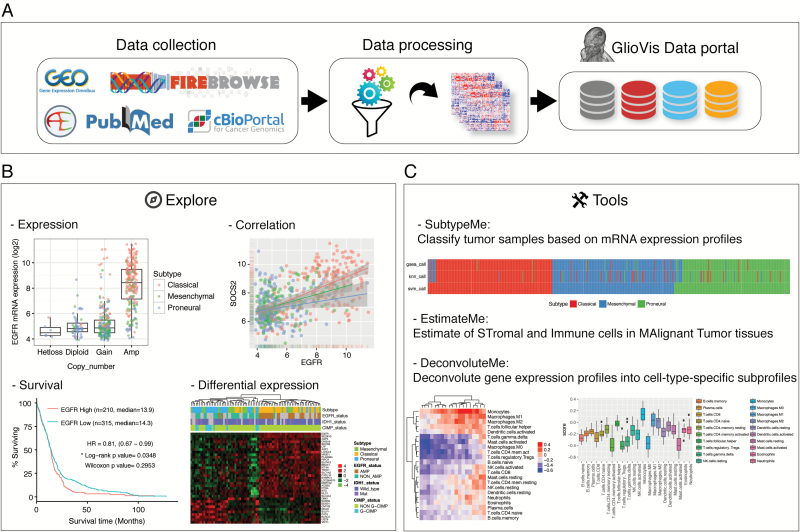
Currently GlioVis contains over 6500 tumor samples of approximately 50 expression datasets of a large collection of brain tumor entities (mostly gliomas), both adult and pediatric. Though at the moment only primary brain tumors are accessible through our portal, in the future we are also planning to include data for secondary/metastatic brain tumors, cell lines, and xenografts. A full list of the available datasets is presented on the GlioVis homepage. Raw expression data have been downloaded from various sources: Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/, accessed October 31, 2016), ArrayExpress (https://www.ebi.ac.uk/arrayexpress/, accessed October 31, 2016), and Firebrowse (http://firebrowse.org, accessed October 31, 2016). These data have then been processed using the R programming language7 and combined with manually curated phenotypic information to be visualized in a homogeneous and harmonized way (Fig. 1A). Details of the data processing can be found on the GlioVis help pages.
Fig. 1.
The GlioVis data portal. (A) Schematic representation of data collection, processing, and incorporation into the portal. (B) Examples of available data visualization. (C) A series of tools allow users to analyze their own datasets.
Through the GlioVis portal, users can explore the expression of a particular gene of interest taking advantage of all the information available for a specific dataset: histology, grade, subtype, copy number, etc (Fig. 1B). Data summaries allow users to obtain an overview of a given dataset or a specific gene in individual datasets. For glioblastoma multiforme (GBM) subtyping we have used the most recent 3-subtypes classification scheme.8 Briefly, after filtering out genes that are significantly highly expressed in tumor-associated microenvironment, unsupervised clustering grouped isocitrate dehydrogenase (IDH) wild-type GBM into 3 transcriptional subtypes. These subtypes, which are now based on gene expression in GBM cells only, recapitulated the proneural, classical, and mesenchymal subtypes but not the neural class.
In addition to gene expression exploratory studies, it’s possible to perform various other analyses: survival, correlation, mutation, etc (some examples are shown in Fig. 1B). For all the available analysis tools, users can restrict their studies to specific groups of interest, such as a particular tumor histology or subtype. For example, when performing survival analysis in GBM it’s possible to exclude the CpG island methylator phenotype (CIMP)+IDH wild-type samples and/or the recurrent tumors. CIMP status has been determined by supported vector machine (SVM), using GBM expression data from TCGA as the training dataset.
GlioVis also provides a series of tools that allow users to analyze their own datasets. SubtypeME, currently only available for gliomas, uses 3 different algorithms—SVM, K-Nearest Neighbor, and single sample gene set enrichment analysis—to classify the tumor samples by subtypes. These algorithms have been used for the classification of all the GBM samples available through the portal. EstimateME calculates tumor purity by estimating stromal and immune cells in malignant tumor tissues,9 and DeconvoluteME uses cell-type-specific signatures to deconvolute gene expression profiles into cell-type-specific subprofiles (Fig. 1C).
In conclusion, GlioVis offers the research community an unprecedented fast and intuitive portal to molecular profiles from a vast collection of brain tumor samples.
Funding
This work was supported by a grant from the Seve Ballesteros Foundation to M.S.
Acknowledgments
We are very grateful to all the laboratories that have generated and shared the data used in the portal.
References
- 1. International Cancer Genome Consortium , Hudson TJ, Anderson W, et al. International network of cancer genome projects. Nature. 2010;464(7291):993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cheng PF, Dummer R, Levesque MP. Data mining The Cancer Genome Atlas in the era of precision cancer medicine. Swiss Med Wkly. 2015;145:w14183. [DOI] [PubMed] [Google Scholar]
- 3. Schroeder MP, Gonzalez-Perez A, Lopez-Bigas N. Visualizing multidimensional cancer genomics data. Genome Med. 2013;5(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldman M, Craft B, Swatloski T, et al. The UCSC Cancer Genomics Browser: update 2013. Nucleic Acids Res. 2013;41(Database issue):D949–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fernandez-Banet J, Esposito A, Coffin S, et al. OASIS: web-based platform for exploring cancer multi-omics data. Nat Methods. 2015;13(1):9–10. [DOI] [PubMed] [Google Scholar]
- 7. R Core Team. R: A language and environment for statistical computing https://www.r-project.org/ Accessed October 31, 2016.
- 8. Wang Q, Hu X, Hu B, et al. Tumor evolution of glioma intrinsic gene expression subtype associates with immunological changes in the microenvironment. bioRxiv. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]