Abstract
Cytochrome P450 2A6 (CYP2A6) metabolizes several clinically relevant substrates, including nicotine, the primary psychoactive component in cigarette smoke. Smokers vary widely in their rate of inactivation and clearance of nicotine, altering numerous smoking phenotypes. We aimed to characterize independent and shared impact of genetic and nongenetic sources of variation in CYP2A6 mRNA, protein, and enzyme activity in a human liver bank (n = 360). For the assessment of genetic factors, we quantified levels of CYP2A6, cytochrome P450 oxidoreductase (POR), and aldo-keto reductase 1D1 (AKR1D1) mRNA, and CYP2A6 and POR proteins. CYP2A6 enzyme activity was determined through measurement of cotinine formation from nicotine and 7-hydroxycoumarin formation from coumarin. Donor DNA was genotyped for CYP2A6, POR, and AKR1D1 genetic variants. Nongenetic factors assessed included gender, age, and liver disease. CYP2A6 phenotype measures were positively correlated to each other (r values ranging from 0.47–0.88, P < 0.001). Female donors exhibited higher CYP2A6 mRNA expression relative to males (P < 0.05). Donor age was weakly positively correlated with CYP2A6 protein (r = 0.12, P < 0.05) and activity (r = 0.20, P < 0.001). CYP2A6 reduced-function genotypes, but not POR or AKR1D1 genotypes, were associated with lower CYP2A6 protein (P < 0.001) and activity (P < 0.01). AKR1D1 mRNA was correlated with CYP2A6 mRNA (r = 0.57, P < 0.001), protein (r = 0.30, P < 0.001), and activity (r = 0.34, P < 0.001). POR protein was correlated with CYP2A6 activity (r = 0.45, P < 0.001). Through regression analyses, we accounted for 17% (P < 0.001), 37% (P < 0.001), and 77% (P < 0.001) of the variation in CYP2A6 mRNA, protein, and activity, respectively. Overall, several independent and shared sources of variation in CYP2A6 activity in vitro have been identified, which could translate to variable hepatic clearance of nicotine.
Introduction
Cytochrome P450 2A6 (CYP2A6) metabolizes several clinically relevant substrates, including nicotine, tegafur, letrozole, efavirenz, valproic acid, and pilocarpine (Messina et al., 1997; Ikeda et al., 2000; Kiang et al., 2006; Endo et al., 2007; di Iulio et al., 2009; Murai et al., 2009). Nicotine metabolism by CYP2A6 is of interest as nicotine is the primary psychoactive component in cigarette smoke and the main source of tobacco dependence (Stolerman and Jarvis, 1995). Smokers can vary widely in their rate of inactivation and clearance of nicotine (Malaiyandi et al., 2005), with variations in the rate of nicotine metabolism associating with differences in smoking behaviors, cessation, and response to cessation pharmacotherapies (Schoedel et al., 2004; Patterson et al., 2008; Schnoll et al., 2009). The major pathway of nicotine inactivation is its conversion to cotinine, primarily catalyzed by CYP2A6 (Messina et al., 1997).
Genetic and nongenetic factors contribute to variation in CYP2A6 enzyme activity and rate of nicotine metabolism. The CYP2A6 gene, encoding the CYP2A6 enzyme, is highly polymorphic, and CYP2A6 genetic variation is associated with variable rates of nicotine metabolism in vitro and in vivo (Al Koudsi et al., 2010; Binnington et al., 2012), and accordingly with differences in smoking behavior (Wassenaar et al., 2011; Chen et al., 2014). A significant proportion of variation in CYP2A6 activity can be attributed to CYP2A6 genetic variation (heritability estimates of 60–80%) (Swan et al., 2009; Loukola et al., 2015); however, unaccounted for variation remains even after additional factors (age, sex, race/ethnicity, body mass index, cigarettes per day, and total nicotine equivalents) are controlled for (Chenoweth et al., 2014a; Park et al., 2016). Unidentified CYP2A6 genetic variation, or variation in other genes regulating cytochrome P450 (P450) expression or function, may account for the 35% missing variation.
Aldo-keto reductase 1D1 (AKR1D1), an enzyme involved in bile acid synthesis (Schuetz et al., 2001; Lee et al., 2009), has been identified as a potential regulator of P450 activity (Yang et al., 2010). AKR1D1 single-nucleotide polymorphism (SNP) rs1872930, associated with higher AKR1D1 mRNA expression, is associated with increased expression and activity of several P450s in vitro (Chaudhry et al., 2013). To our knowledge, the relationship between rs1872930 and CYP2A6 has not yet been investigated. P450 oxidoreductase (POR), an enzyme that donates electrons to P450s during their catalytic cycle (Hu et al., 2012), also contributes to variation in multiple P450 activities, including those of CYP2A6 (Gomes et al., 2009; Chenoweth et al., 2014b; Lv et al., 2016).
Combined effects of CYP2A6, AKR1D1, and POR genetic variation should also be considered. POR SNP rs1057868 interacts with CYP2A6 genotype, with rs1057868 only associating with faster CYP2A6 activity among individuals not possessing known CYP2A6 reduced-function genetic variants (Chenoweth et al., 2014b). Owing to potential shared effects of each genetic factor on CYP2A6 activity, we aimed to elucidate both the independent and combined influences of CYP2A6, POR, and AKR1D1 genetic variation on CYP2A6 mRNA expression, protein levels, and enzyme activity. We also aimed to investigate relationships between POR and AKR1D1 expression, independent of genotype, with variation in CYP2A6 expression and activity.
Several nongenetic factors, including gender, age, and liver disease, have been associated with altered CYP2A6 expression, function, and nicotine pharmacokinetics. Here we will examine independent and combined impacts of these variables in a large human liver bank (n = 360) with extensive CYP2A6 phenotyping. Relative to men, females had higher microsomal CYP2A6 protein levels in a smaller human liver bank (n = 67) (Al Koudsi et al., 2010), and women smokers exhibited greater CYP2A6-mediated nicotine metabolism (Benowitz et al., 2006), consistent with estrogen-mediated induction of CYP2A6 transcription (Benowitz et al., 2006; Higashi et al., 2007). Individuals 65 years and older have been shown to have lower nonrenal nicotine clearance than those aged 22–43 (Molander et al., 2001); however, there was no association between age and CYP2A6 protein or activity when investigated in a smaller human liver bank (n = 67) (Al Koudsi et al., 2010). Nonalcoholic fatty liver disease was associated with higher CYP2A6 mRNA expression and enzyme activity in vitro (Fisher et al., 2009).
Despite several known sources of variation in CYP2A6 activity, substantial variation remains to be characterized (Swan et al., 2009). Considering the clinical relevance of CYP2A6, it is important to further characterize causes of variability in CYP2A6 activity, and thus the rate of nicotine metabolism, for its utility in tailoring smoking cessation treatment (Lerman et al., 2015). We aimed to assess independent and shared impacts of CYP2A6, POR, and AKR1D1 genotypes and levels, as well as gender, age, and liver disease, on CYP2A6 mRNA expression, protein expression, and enzyme activity in a large human liver bank.
Materials and Methods
Chemicals and Reagents
Lodoacetamide, dithiothreitol, and sequencing-grade trypsin were purchased from ThermoFisher Scientific (Pierce Protein Biology, Rockford, IL). Ammonium bicarbonate was purchased from Acros Organics (Geel, Belgium). Sodium deoxycholate (98% purity) was obtained from MP Biomedicals (Santa Ana, CA). Synthetic light peptides for CYP2A6 and POR quantification were procured from New England Peptides (Boston, MA), with purity established by amino acid analysis. Heavy stable isotope–labeled amino acids [13C615N2]-lysine and [13C615N4]-arginine were purchased from ThermoFisher Scientific (Pierce Protein Biology). Liquid chromatography–mass spectrometry–grade acetonitrile (99.9% purity) and formic acid (≥99.5% purity) were purchased from Fischer Scientific (Fair Lawn, NJ). (–)-Nicotine hydrogen tartrate, (–)-cotinine, coumarin, 7-hydroxycoumarin, and 4-hydroxycoumarin were purchased from Sigma-Aldrich (St. Louis, MO); chemical structures are illustrated in Emami and Dadashpour (2015). Nicotine-D4 and cotinine-D3 were purchased from Toronto Research Chemicals (Toronto, ON, Canada), chemical structures illustrated in Hukkanen et al. (2005).
Human Liver Bank
Human liver tissue samples are from two liver banks: 1) the St. Jude Liver Resource at the St. Jude Children’s Research Hospital (Memphis, TN) (n = 295), and 2) the University of Washington Human Liver Bank (Seattle, WA) (n = 65). The St. Jude Liver Resource human liver tissues were obtained through the Liver Tissue Cell Distribution System, Minneapolis, MN, and Pittsburgh, PA, which was funded by National Institutes of Health contract no. HHSN276201200017C. Details on the selection of the livers and investigator blinding for sample analyses have been described previously (Shirasaka et al., 2016). Age, gender, and ethnicity were known for most (≥90%) of the liver donors. The donors ranged in age from 0–87 years (mean 40 years, S.D. ± 22 years). Of the donors with known gender, 58% were male. The liver bank consists of 92% Caucasian, 3% African American, <1% Asian, <1% Hispanic, and 5% unknown ethnicity donors. Cause of death, medications used, and liver pathology was known for less than 50% of donors. Smoking status was unknown for >88% of donors and therefore was not assessed as a predictor of CYP2A6 phenotypes in the present study.
CYP2A6, POR, and AKR1D1 mRNA Quantification
RNA Isolation.
Liver RNA was isolated and purified using a NucleoSpin miRNA kit (Macherey-Nagel, Duren, Germany; Clontech Laboratories, Mountain View, CA), according to manufacturer’s protocol. Briefly, ∼30 mg liver tissue was combined with 4°C lysis buffer, homogenized using a TissueLyser LT (Qiagen, Valencia, CA), and allowed to sit at room temperature for 5 minutes. The solution was then added to a column. After centrifugation to bind the large RNA to the column, the column was treated with an rDNAse solution at room temperature for at least 15 minutes. Meanwhile, the flow-through containing the small RNA was treated to precipitate out the protein. The small RNA was then bound to a new column. Following three wash steps of each column, the resulting large and small RNA were each eluted, quantitated, and bioanalyzed for quality control using a Bioanalyzer 2100 (Agilent, Santa Clara, CA). Only RNA with an RNA integrity score greater or equal to 7.0 was submitted for sequencing.
TruSeq Stranded mRNA Preparation.
Next-generation sequencing libraries were prepared from 1.25 μg of total RNA in an automated, high-throughput format using the TruSeq Stranded mRNA kit (Illumina, San Diego, CA). All the steps required for sequence library construction have been automated and performed on a Sciclone NGSx Workstation (PerkinElmer, Waltham, MA). During library construction, ribosomal RNA was depleted by means of a poly-A enrichment, and first- and second-strand cDNA syntheses were performed. Each library was then uniquely barcoded using the Illumina adapters and amplified using a total of 13 cycles of polymerase chain reaction (PCR). After amplification and cleanup, library concentrations were quantified using the Quant-it dsDNA Assay (Life Technologies/ThermoFisher Scientific, Carlsbad, CA). Libraries were subsequently normalized and pooled on the basis of Agilent 2100 Bioanalyzer results (Agilent Technologies, Santa Clara, CA). Pooled libraries were size-selected using a Pippin Prep (Sage Science, Beverly, MA) and then balanced by mass and pooled in batches of 96 with a final pool concentration of 2–3 nM for sequencing on the HiSEq 2500 (Illumina).
Read Processing and Analysis Pipeline.
The Northwest genome sequencing laboratory (University of Washington, Seattle, WA) processing pipeline included the following elements: 1) base calls generated in real time on the HiSeq or NextSeq instrument; 2) Illumina RTA-generated BCL files converted to FASTQ files; 3) custom scripts developed in-house and used to process the FASTQ files and to output de-multiplexed FASTQ files by lane and index sequence; 4) sequence read and base quality checked using the FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/); 5) sequences aligned to hg19 with reference transcriptome Ensembl v67 (http://www.ensembl.org) using TopHat (Johns Hopkins University Center for Computational Biology) (Kim et al., 2013) followed by mate-fixing; and 6) custom scripts for quality assessment generate metrics. All aligned read data were subject to the following steps: 1) lane level bam data files were merged using the Picard MergeSamFiles tool and suspected PCR duplicates were marked, not removed, in the alignment files using the Picard MarkDuplicates tool (http://broadinstitute.github.io/picard/); 2) local realignment performed around indels, and base quality score recalibration was run using GATK tools (Broad Institute) (McKenna et al., 2010); 3) variant detection performed with the GATK Unified Genotyper version 2.6.5 (DePristo et al., 2011); 4) aligned data were used for isoform assembly and quantitation with Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/) (Kim et al., 2013; Trapnell et al., 2013); genomic features were quantitated with featureCounts (Walter and Eliza Hall Institute, Parkville Victoria, Australia) (Liao et al., 2014); and 5) gene-specific quantitation data were used for further analysis.
CYP2A6 and POR Protein Quantification
Simultaneous quantification of CYP2A6 and POR was carried out using a liquid chromatography–tandem mass spectroscopy (LC-MS/MS) proteomics method (Prasad and Unadkat, 2014). The surrogate peptides were selected and light and heavy peptides (Table 1) containing labeled [13C615N2]-lysine or [13C615N4]-arginine residues were procured. Liver microsomal samples were diluted to 2 mg/ml, and 40 μg microsomal protein was digested as described before (Shuster et al., 2014). Briefly, microsomal protein was denatured and reduced with 4 μl of 100 mM dithiothreitol, 10 μl of sodium deoxycholate (2.6% w/v), and 10 μl of ammonium bicarbonate buffer (100 mM) at 95°C for 5 minutes. The denatured protein was then alkylated by 4 μl of 200 mM iodoacetamide at room temperature. The digestion was performed by addition 10 μl of trypsin (protein/trypsin ratio, 25:1) at 37°C for 22 hours. The reaction was quenched by the addition of 20 μl of peptide internal standard cocktail (prepared in 50% acetonitrile in water containing 0.1% formic acid) and 10 μl of the neat solvent, i.e., 50% acetonitrile in water containing 0.1% formic acid. The samples were vortexed and centrifuged at 3500g for 5 minutes. The calibration curves were generated using serial dilutions of light peptide standard in phosphate buffer (50 mM Kpi, 0.25 M sucrose, 10 mM EDTA, pH 7.4) to replace microsomal sample.
TABLE 1 .
| Predictor Variable | B | Beta | 95% CI | % Variation Accounted forc | P Value |
|---|---|---|---|---|---|
| CYP2A6*9 genotyped | −117.9 | −0.09 | −264.8 to 25.2 | 0.8% | >0.1 |
| Gender (M = 1, F = 0) | −12.0 | −0.01 | −118.5 to 98.5 | 0.01% | >0.1 |
| AKR1D1 mRNA (FPKM) | 6.0 | 0.4 | 4.3 to 7.6 | 16.0% | <0.001 |
Cases excluded pairwise.
R2 = 0.17, P < 0.001.
% Variation accounted for by each variable is determined by: (Part Correlation)2 × 100
CYP2A6*9 genotype coded as 0 for CYP2A6*1/*1 genotype, 1 for CYP2A6*1/*9 genotype, and 2 for CYP2A6*9/*9 genotype.
Triple-quadrupole LC-MS instrument (Agilent 6460A) coupled to an Agilent 1290 Infinity LC system (Agilent Technologies), in positive electrospray ionization mode was used for quantification. 2 μg of the trypsin digest was injected onto the column (Kinetex 1.7 μm, C18 100Å; 100 × 2.1 mm; Phenomenex, Torrance, CA). Mobile phase and gradient program were exactly the same as described before (Shuster et al., 2014). Surrogate light and heavy (internal standards) peptides were monitored using instrument parameters provided in Supplemental Table 1. The LC-MS/MS data were processed using MassHunter (Agilent Technologies) and Skyline (University of Washington) software.
CYP2A6 Enzyme Activity Assays
Human liver microsomes were prepared, and total protein concentrations were quantified, as described previously (Shirasaka et al., 2016). CYP2A6 enzyme activity was determined by quantifying the rate of metabolism of two known substrates of this enzyme, nicotine and coumarin. Linear conditions for the rate of nicotine metabolism (i.e., the rate of cotinine formation from nicotine) were established for the following assay conditions: 0.5 mg/ml microsomal protein, 50 μM Tris-HCl buffer (pH 7.4), 30 μM nicotine, 1 mM NADPH, 10 μl of cytosol (source of aldehyde dehydrogenase), and water to a final volume of 100 μl, for an incubation time of 20 minutes at 37°C. The reaction was terminated with 20 μl of 20% Na2CO3, and 20 ng of nicotine-D4 and cotinine-D3 internal standards were added. Samples were extracted and analyzed using LC-MS/MS, as described previously (Jacob et al., 2011; Craig et al., 2014).
To allow for adequate detection of 7-hydroxycoumarin and avoid substrate depletion, linear conditions for the rate of coumarin metabolism (i.e., the rate of 7-hydroxycoumarin formation from coumarin, a very rapidly metabolized CYP2A6 substrate) were established at multiple incubation times and protein concentrations using a number of substrate concentrations. For donors who exhibited a relatively slow rate of in vitro nicotine metabolism (cotinine formation velocity of <0.1 nmol/min per milligram), 0.05 mg/ml microsomal protein and 15-minute incubation was used. For donors exhibiting intermediate rates of nicotine metabolism (cotinine formation velocity of 0.1–0.3 nmol/min per milligram), the assay was adjusted to 0.02 mg/ml microsomal protein for a 10-minute incubation, and for donors with fast rates of nicotine metabolism (cotinine formation velocity of >0.3 nmol/min per milligram), 0.01 mg/ml microsomal protein for a 7-minute incubation was used. All other assay conditions were identical among slow, intermediate, and fast metabolizers, including 50 mM Tris-HCl buffer (pH 7.4), 2 μM coumarin, 1 mM NADPH, and water to a final volume of 200 μl with an incubation at 37°C. The reactions were terminated with 40 μl of trichloroacetic acid (20% w/v), and 25 ng of 4-hydroxycoumarin internal standard was added. Samples were extracted and analyzed using high-performance liquid chromatography, as described previously (Li et al., 1997), with minor modifications. Limits of quantification for nicotine, cotinine, coumarin, and 7-hydroxycoumarin were 1 ng/ml, 1 ng/ml, 50 ng/ml, and 10 ng/ml, respectively.
CYP2A6, POR, and AKR1D1 Genotyping
DNA was extracted using the DNeasy tissue kit (Qiagen). DNA from all donors was genotyped for the CYP2A6 alleles *2, *4, *9, and *12, whereas DNA from African-American and unknown ethnicity donors were also genotyped for CYP2A6 *17, *20, *23, *25, *28, and *35, and Asian and unknown ethnicity donors were additionally genotyped for CYP2A6 *7, *8, and *10, as these alleles have zero to extremely low frequencies among Caucasians (Mwenifumbo and Tyndale, 2007). Genotyping for the CYP2A6 alleles was conducted using a two-step allele-specific polymerase chain reaction approach, except for CYP2A6*2, which was genotyped using a TaqMan SNP genotyping assay (Applied Biosystems/ThermoFisher Scientific) and real-time polymerase chain reaction; CYP2A6 genotyping approaches have been described in detail previously (Wassenaar et al., 2016). All donors were genotyped for POR SNPs rs17148944, rs2868177, rs1057868 and AKR1D1 SNP rs1872930 using allele-specific TaqMan SNP genotyping assays (Applied Biosystems/ThermoFisher Scientific).
Statistical Analyses
The following data were non-normally distributed, and therefore nonparametric statistical tests were used: CYP2A6 mRNA, AKR1D1 mRNA, POR mRNA, CYP2A6 protein, POR protein, nicotine metabolism, and coumarin metabolism. Correlations were determined via Spearman rank correlations. We used the Mann Whitney test to analyze the association of gender, liver disease, and AKR1D1 genotype with CYP2A6 mRNA, CYP2A6 protein, and CYP2A6 activity. The Kruskal-Wallis test was used to determine associations between CYP2A6 genotype and CYP2A6 mRNA, CYP2A6 protein, and CYP2A6 activity, as well as between POR SNP genotypes and POR mRNA, POR protein, and CYP2A6 activity. We ran separate linear regression models for CYP2A6 mRNA, CYP2A6 protein, and CYP2A6 activity to calculate the proportion of variation in each that is accounted for by each variable in the model. The liver bank is composed of samples collected and processed at two different research sites; the phenotype means were different between sites. For the purposes of illustrating the combined data we have corrected for overall site differences using the following conversion factor: mean phenotype measure (CYP2A6 mRNA, AKR1D1 mRNA, POR mRNA, CYP2A6 protein, POR protein, rate of nicotine metabolism, and rate of coumarin metabolism) from University of Washington site divided by the mean phenotype measure from St. Jude site, which was multiplied by each sample from the University of Washington site (the smaller liver bank). Findings are illustrated using the site-corrected measures. Additional regression models were run in which the original, noncorrected phenotype data were included, along with the liver bank site (University of Washington or St. Jude) as a covariate. Both versions of each model (corrected versus uncorrected data with site as covariate) produced very similar results, indicating that the conversion factor had a minimal effect but allowed for combining the data sets for illustration purposes. Analyses were conducted with GraphPad Prism (v6.0; LaJolla, CA) and SPSS (v22.0; IBM), and statistical tests were considered significant for P < 0.05.
Results
Moderate to Strong Correlation between CYP2A6 mRNA, Protein, and Activity.
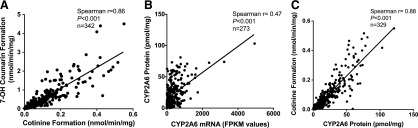
The two in vitro measures of CYP2A6 activity, velocity of cotinine formation from nicotine and velocity of 7-hydroxycoumarin formation from coumarin, were strongly positively correlated (Spearman r = 0.86, P < 0.001; Fig. 1A). The rate of nicotine metabolism serves as the primary measure of CYP2A6 activity in all analyses, and all coumarin metabolism data has been included in the supplemental materials. CYP2A6 mRNA and protein expression were moderately positively correlated (Spearman r = 0.47, P < 0.001; Fig. 1B). CYP2A6 protein was strongly positively correlated with both measures of CYP2A6 activity (nicotine metabolism: Spearman r = 0.88, P < 0.001; Fig. 1C; coumarin metabolism: Spearman r = 0.81, P < 0.001; Supplemental Fig. 1). All correlations were similar among males and females, and among wild-type (CYP2A6 *1/*1) donors and the whole sample (donors with wild-type and variant genotypes included); the rates of nicotine and coumarin metabolism were strongly correlated in females (r = 0.86, P < 0.001) and males (r = 0.86, P < 0.001); CYP2A6 mRNA and CYP2A6 protein were moderately correlated in females (r = 0.53, P < 0.001) and males (r = 0.42, P < 0.001); CYP2A6 protein and CYP2A6 enzyme activity (cotinine formation) were strongly correlated in females (r = 0.88, P < 0.001) and males (r = 0.88, P < 0.001); CYP2A6 mRNA and protein for the wild-type donors only were moderately correlated (r = 0.50, P < 0.001), CYP2A6 mRNA and enzyme activity (cotinine formation from nicotine) were moderately correlated (r = 0.49, P < 0.001), and CYP2A6 protein and enzyme activity were strongly correlated (r = 0.87, P < 0.001). The mean, standard deviation, and range for each CYP2A6 phenotype measure were as follows: CYP2A6 mRNA 373 ± 481 (0.2–4940) fragments per kilobase per million reads (FPKM) values, CYP2A6 protein 22.5 ± 19.4 (0.0–121.1) pmol/mg, CYP2A6 activity (nicotine metabolism) 0.10 ± 0.10 (0.0007–0.55) nmol/min per milligram, and CYP2A6 activity (coumarin metabolism) 0.52 ± 0.68 (0.01–4.51) nmol/min per milligram.
Fig. 1.
(A) Correlation of two measures of CYP2A6 enzyme activity (the velocity of cotinine formation from nicotine versus the velocity of 7-OH-coumarin formation from coumarin, nmol/min per milligram). (B) Correlation between CYP2A6 mRNA levels (FPKM values, fragments per kilobase per million reads) and CYP2A6 protein levels (pmol/mg). (C) Correlation between CYP2A6 protein levels and CYP2A6 enzyme activity (cotinine formation from nicotine). Values for r and P were determined on the basis of Spearman correlations.
Impact of Nongenetic Factors on CYP2A6 mRNA, Protein, and Activity.
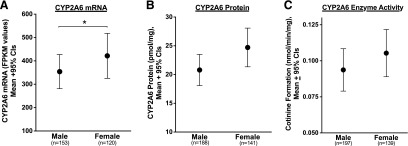
Gender was associated with differences in CYP2A6 mRNA expression, protein levels, and enzyme activity. Female liver donors exhibited higher CYP2A6 mRNA levels compared with males (P < 0.05; Fig. 2A). Although not statistically significant, there was a trend for higher CYP2A6 protein (P < 0.1; Fig. 2B) and enzyme activity (P < 0.1 for both nicotine and coumarin metabolism; Fig. 2C and Supplemental Fig. 2) among females relative to males.
Fig. 2.
Association of gender with (A) CYP2A6 mRNA levels (FPKM values), (B) CYP2A6 protein levels (pmol/mg), and (C) CYP2A6 enzyme activity (cotinine formation from nicotine, nmol/min per milligram). *P values of < 0.05 on the basis of Mann Whitney tests.
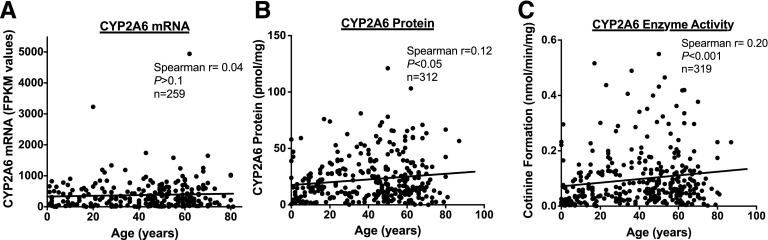
There was no association between donor age and CYP2A6 mRNA expression (Spearman r = 0.04, P>0.1; Fig. 3A), but age was weakly positively correlated with both CYP2A6 protein expression (Spearman r = 0.12, P < 0.05; Fig. 3B) and enzyme activity (nicotine metabolism: Spearman r = 0.20, P < 0.001; Fig. 3C; coumarin metabolism: Spearman r = 0.13, P < 0.05; Supplemental Fig. 3).
Fig. 3.
Correlation of age with (A) CYP2A6 mRNA levels (FPKM values), (B) CYP2A6 protein levels (pmol/mg), and (C) CYP2A6 enzyme activity (cotinine formation from nicotine, nmol/min per milligram). r and P values on the basis of Spearman correlations.
Liver disease was defined as being positive for at least one of the following conditions: hepatitis, liver injury, biliary atresia, cirrhosis, fat accumulation, fibrosis, or hepatoma. There was no association between liver disease state and CYP2A6 mRNA (normal versus disease, means = 383 and 299 FPKM values, respectively, P > 0.1), protein (normal versus disease, means = 21.6 and 20.0 pmol/mg, respectively, P > 0.1), or enzyme activity (normal versus disease, nicotine means = 0.10 and 0.09 nmol/min per milligram, respectively, coumarin means = 0.55 and 0.47 nmol/min per milligram, respectively, P > 0.1 for both nicotine and coumarin metabolism).
CYP2A6 Genotype Is Associated with CYP2A6 Protein and Enzyme Activity, but not CYP2A6 mRNA.
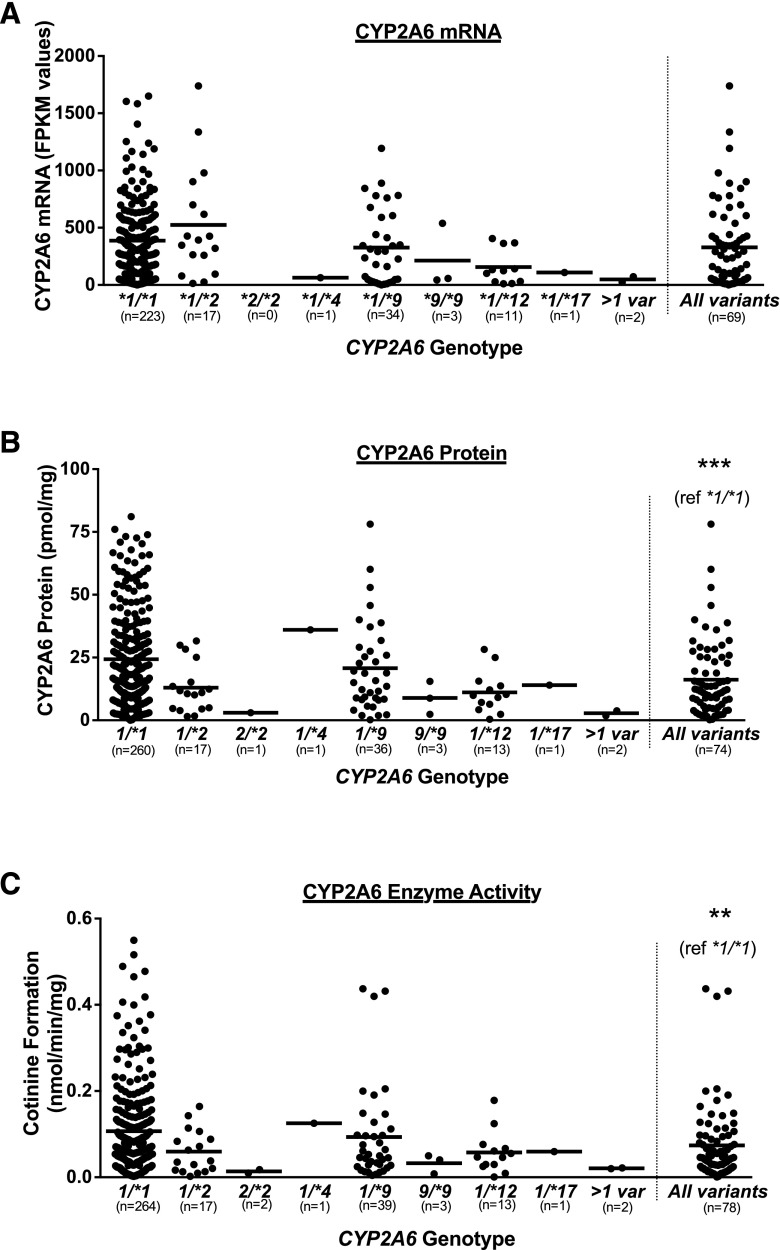
The CYP2A6 genetic variants *2, *4, *9, *10, *12, and *17 were identified among one or more liver donors, whereas *7, *8, *20, *23, *25, *28, or *35 were not. Liver donors were grouped according to their CYP2A6 genotype (e.g., *1/*9), where wild-type (*1/*1) donors were those who did not possess any tested CYP2A6 genetic variant, and the “all variants” group included all donors who possessed one or more of the CYP2A6 genetic variants. CYP2A6 mRNA expression did not differ across all genotypes (P > 0.1; Fig. 4A), and there was no difference between the wild-type and the all variants groups (P > 0.1; Fig. 4A). There was an apparent gene-dose effect on CYP2A6 mRNA expression with increasing copies of the CYP2A6*9 allele (mean FPKM values: *1/*1 386, *1/*9 326, *9/*9 213); this TATA box variant has previously been associated with decreased CYP2A6 transcription (Pitarque et al., 2001). Although no individual genotypes were significantly different with respect to CYP2A6 mRNA, protein, or activity, CYP2A6 genotype was associated with differences in CYP2A6 protein and activity such that liver donors possessing one or more of the CYP2A6 genetic variants exhibited lower CYP2A6 protein expression (P < 0.001; Fig. 4B) and enzyme activity (P < 0.01 for both nicotine and coumarin metabolism; Fig. 4C and Supplemental Fig. 4) relative to the wild-type CYP2A6*1/*1 donors. Additionally, there was a gene-dose effect on the CYP2A6 phenotypes for CYP2A6*9 (CYP2A6 protein: *1/*1 24.3, *1/*9 20.8, *9/*9 8.9 pmol/mg; CYP2A6 activity: *1/*1 0.11, *1/*9 0.09, *9/*9 0.03 nmol/min per milligram; Fig. 4, B–C), and CYP2A6*2 (CYP2A6 protein: *1/*1 24.3, *1/*2 13.0, *2/*2 3.0 pmol/mg; CYP2A6 activity: *1/*1 0.11, *1/*2 0.06, *2/*2 0.01 nmol/min per milligram; Fig. 4, B–C). A wide degree of variation in CYP2A6 mRNA (0.9–4940 FPKM values), protein (0.0–121.1 pmol/mg), and enzyme activity (nicotine metabolism: 0.002–0.55 nmol/min per milligram; coumarin metabolism: 0.02–4.51 nmol/min per milligram) was observed in the wild-type genotype group.
Fig. 4.
Association of CYP2A6 genotype with (A) CYP2A6 mRNA levels (FPKM values), (B) CYP2A6 protein levels (pmol/mg), and (C) CYP2A6 enzyme activity (cotinine formation from nicotine, nmol/min per milligram). Horizontal lines represent the mean for each genotype. Three data points exceed the y-axis limit for CYP2A6*1/*1 group in (A), two data points exceed the y-axis limit for CYP2A6*1/*1 group in (B), and two data points exceed the y-axis limit for CYP2A6*1/*1 group in (C); all points were included in the mean and statistical tests. **P < 0.01, ***P < 0.001, on the basis of Mann Whitney tests.
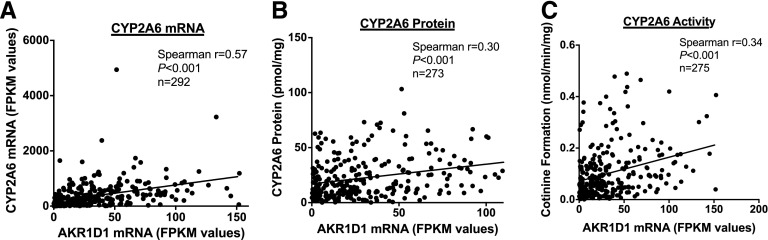
AKR1D1 mRNA Expression, but not Genotype, Is Associated with CYP2A6 mRNA, Protein, and Activity.
On the basis of work by Chaudhry and colleagues (Yang et al., 2010; Chaudhry et al., 2013), we investigated the correlation between AKR1D1 mRNA expression and CYP2A6 mRNA, protein, and enzyme activity, and further the association between the AKR1D1 SNP rs1872930 with AKR1D1 mRNA expression and CYP2A6 mRNA, protein, and enzyme activity. AKR1D1 mRNA expression was moderately correlated with CYP2A6 mRNA expression (Spearman r = 0.57, P < 0.001; Fig. 5A), protein levels (Spearman r = 0.30, P < 0.001; Fig. 5B), and enzyme activity (nicotine metabolism: Spearman r = 0.34, P < 0.001; Fig. 5C; coumarin metabolism: Spearman r = 0.30, P < 0.001; Supplemental Fig. 5). There was no difference in the association between AKR1D1 mRNA and CYP2A6 mRNA, protein, or enzyme activity in females versus males; AKR1D1 mRNA and CYP2A6 mRNA were moderately correlated in females (r = 0.49, P < 0.001) and males (r = 0.60, P < 0.001); AKR1D1 mRNA and CYP2A6 protein were weakly to moderately correlated in females (r = 0.26, P < 0.001) and males (r = 0.34, P < 0.001); AKR1D1 mRNA and CYP2A6 enzyme activity (nicotine metabolism) were moderately correlated in females (r = 0.30, P < 0.001) and males (r = 0.34, P < 0.001). There was no association between rs1872930 and AKR1D1 mRNA (TT versus TC genotype, means = 33.8 and 32.5 FPKM values, respectively, P > 0.1), CYP2A6 mRNA (TT versus TC genotype, means = 363 and 389 FPKM values, respectively, P > 0.1), CYP2A6 protein (TT versus TC genotype, means = 22.1 and 23.2 pmol/mg, respectively, P > 0.1), or CYP2A6 activity (TT versus TC genotype, nicotine means = 0.10 and 0.10 nmol/mg per minute, respectively, coumarin means = 0.51 and 0.52 nmol/mg per minute, respectively, P > 0.1 for both nicotine and coumarin metabolism). The mean, standard deviation, and range of AKR1D1 mRNA expression was as follows: 33.7 ± 32.3 (0.1–152) FPKM values.
Fig. 5.
(A) Correlation between AKR1D1 mRNA expression (FPKM values) and (A) CYP2A6 mRNA expression (FPKM values), (B) CYP2A6 protein levels (pmol/mg), and (C) CYP2A6 activity (cotinine formation from nicotine, nmol/min per milligram). Values for r and P determined on the basis of Spearman correlations.
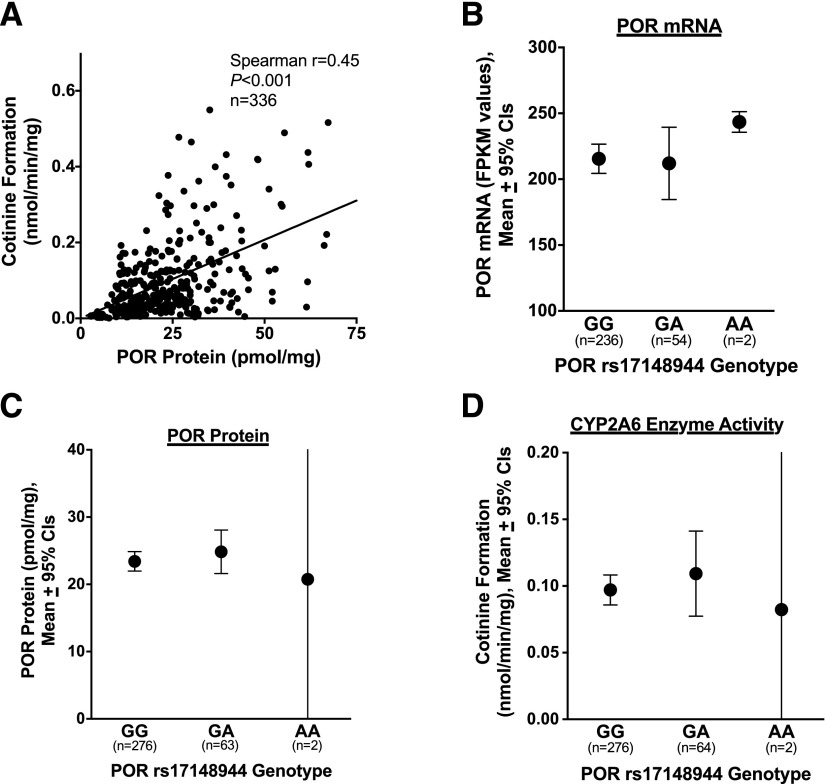
POR Protein Levels, but not Genotype, Are Positively Correlated with CYP2A6 Activity.
POR protein and CYP2A6 activity were moderately correlated (r = 0.45, P < 0.001 for both nicotine and coumarin metabolism; Fig. 6A and Supplemental Fig. 6). On the basis of the literature (Gomes et al., 2009; Chenoweth et al., 2014b; Lv et al., 2016), we investigated the association of three POR SNPs (rs17148944, rs2868177, rs1057868) with POR mRNA expression, POR protein levels, and, owing to the observed association between POR protein and CYP2A6 activity, with CYP2A6 enzyme activity. POR SNP rs2868177 was associated with higher POR mRNA expression (AA, AG, GG genotype means = 202, 221, and 240 FPKM values, respectively, P < 0.05); however, there were no other associations between POR genotypes and POR and CYP2A6 phenotypes (P values provided in Supplemental Table 2). Associations between POR SNP rs17148944 and POR mRNA, POR protein, and CYP2A6 activity are shown in Fig. 6, B–D as representative examples. There was a weak correlation between AKR1D1 mRNA and POR mRNA expression (Spearman r = 0.17, P < 0.01) and POR protein levels (Spearman r = 0.21, P < 0.001), suggesting minimal overlap in the influence of POR and AKR1D1 on CYP2A6 activity. The mean, standard deviation, and range for each POR phenotype measure were as follows: POR mRNA expression 215 ± 89 (95–795) FPKM values, and POR protein 23.7 ± 12.3 (1.2–67.3) pmol/mg.
Fig. 6.
(A) Correlation between POR protein levels (pmol/mg) and CYP2A6 activity (cotinine formation from nicotine, nmol/min per milligram). Values for r and P determined on the basis of Spearman correlations. (B–D) Association between POR SNP rs17148944 and (B) POR mRNA expression (FPKM values), (C) POR protein levels (pmol/mg), and (D) CYP2A6 activity (cotinine formation from nicotine, nmol/min per milligram). P values for B–D all >0.1, on the basis of Kruskal-Wallis tests.
A Significant Proportion of Variation in CYP2A6 mRNA, Protein, and Activity Is Accounted for by Genetic and Nongenetic Predictors.
Using linear regression analyses, we assessed the individual and combined contribution of genetic and nongenetic variables to variation in CYP2A6 mRNA, protein, and enzyme activity. In each regression model, we included only those variables that were significantly (P < 0.05), or trending (P < 0.1) toward being, associated with each CYP2A6 phenotype in univariate analyses, or variables that have been previously associated with CYP2A6 mRNA, protein, or enzyme activity in the literature. All models presented here are derived from phenotype data corrected for liver bank site differences, as illustrated above and described in the Materials and Methods. We have compared this to models in which uncorrected phenotype data were used with the addition of site as a covariate and found only minor insignificant differences between the two modeling approaches.
When modeling influences on CYP2A6 mRNA expression in the human livers, AKR1D1 mRNA expression was the only significant independent contributor to variation in CYP2A6 mRNA, accounting for 16.0% of this variation (P < 0.001; Table 1); neither gender or genotype were significant predictors despite both estrogen and CYP2A6*9 working at a transcriptional level (Pitarque et al., 2001; Higashi et al., 2007). Overall, this model accounted for 17% of the variation in CYP2A6 mRNA expression (R2 = 0.17, P < 0.001; Table 1).
To investigate factors that influence CYP2A6 protein levels, we ran two versions of the regression model, one without and one with CYP2A6 mRNA included as a predictor variable, to identify factors that influence CYP2A6 protein independently or as a byproduct of their influence on CYP2A6 mRNA. In model 1, genotype, AKR1D1 mRNA levels, and age were all significant predictors (Table 2). When CYP2A6 mRNA was added (model 2), the overall proportion of variation in CYP2A6 protein that we were able to account for increased from 17% (R2 = 0.17, P < 0.001) to 37% (R2 = 0.37, P < 0.001; Table 2). Additionally, as expected the CYP2A6*9 allele was no longer a significant independent predictor of CYP2A6 protein, with the proportion of variation accounted for by this genetic variant decreasing from 1.3% (P < 0.05) to 0.5% (P > 0.1; Table 2). There was a substantial decrease in the contribution of AKR1D1 mRNA to variation in CYP2A6 protein when CYP2A6 mRNA was included as a predictor in the model. The proportion of variation accounted for by AKR1D1 mRNA decreased from 9.1% (P < 0.001) to 1.0% (P < 0.05; Table 2). CYP2A6 mRNA independently accounted for 20.0% of the variation in CYP2A6 protein levels (P < 0.001; Table 2).
TABLE 2 .
Linear regression analysis of CYP2A6 protein levels (pmol/mg)a
| Predictor Variable | B | Beta | 95% CI | % Variation Accounted ford | P Value | |
|---|---|---|---|---|---|---|
| 1 Model 1b | CYP2A6*2 genotypee | −11.8 | −0.2 | −20.0 to –3.7 | 2.7% | <0.01 |
| CYP2A6*9 genotypee | −6.1 | −0.1 | −12.2 to –0.08 | 1.3% | <0.05 | |
| CYP2A6*12 genotypee | −11.4 | −0.1 | −23.2 to 0.5 | 1.2% | <0.1 | |
| Gender (M = 1, F = 0) | −2.2 | −0.06 | −6.8 to 2.3 | 0.3% | >0.1 | |
| AKR1D1 mRNA (FPKM) | 0.2 | 0.3 | 0.1 to 0.3 | 9.1% | <0.001 | |
| Age | 0.1 | 0.1 | 0.01 to 0.2 | 1.6% | <0.05 | |
| 2 Model 1 + CYP2A6 mRNAc | CYP2A6*2 genotypee | −13.6 | −0.2 | −20.8 to –6.4 | 3.5% | <0.001 |
| CYP2A6*9 genotypee | −3.8 | −0.07 | −9.1 to –1.5 | 0.5% | >0.1 | |
| CYP2A6*12 genotypee | −8.0 | −0.08 | −18.4 to 2.3 | 0.6% | >0.1 | |
| Gender (M = 1, F = 0) | −2.1 | −0.05 | −6.1 to 1.9 | 0.3% | >0.1 | |
| AKR1D1 mRNA (FPKM) | 0.07 | 0.1 | 0.001 to 0.1 | 1.0% | <0.05 | |
| Age | 0.1 | 0.1 | 0.01 to 0.2 | 1.2% | <0.05 | |
| CYP2A6 mRNA (FPKM) | 0.02 | 0.5 | 0.02 to 0.02 | 20.0% | <0.001 |
Cases excluded pairwise.
R2 = 0.17, P < 0.001.
R2 = 0.37, P < 0.001.
% Variation accounted for by each variable is determined by: (Part Correlation)2 × 100.
CYP2A6*2, CYP2A6*9, and CYP2A6*12 genotypes coded as 0 for CYP2A6*1/*1 genotype, 1 for CYP2A6*1/*2, CYP2A6*1/*9, or CYP2A6*1/*12 genotypes, and 2 for CYP2A6*2/*2 or CYP2A6*9/*9 genotypes.
Last, we quantified the contribution of predictor variables to CYP2A6 enzyme activity. We ran three versions of this model: 1) model 1, 2) model 1 with the addition of CYP2A6 mRNA as a predictor, and 3) model 1 with the further addition of CYP2A6 mRNA and CYP2A6 protein as predictors. Overall, each model accounted for 35%, 46%, and 77% of the variation in CYP2A6 enzyme activity, respectively (nicotine metabolism, R2 = 0.35, 0.46, 0.77, P < 0.001; Table 3; coumarin metabolism, R2 = 0.26, 0.32, 0.57, P < 0.001; Supplemental Table 3). In model 1 genotype, AKR1D1 mRNA, age, and POR protein were all significant predictors (Table 3). We again observed decreased contributions of both the CYP2A6*9 allele and AKR1D1 mRNA toward CYP2A6 enzyme activity when CYP2A6 mRNA was added (model 2). In the third model, with CYP2A6 protein included as a predictor variable, the independent contribution of all other variables decreased below 1%, except for CYP2A6 protein, which independently contributed to 31.9% of the variation in nicotine (nicotine metabolism P < 0.001; Table 3, coumarin metabolism 25.0% P < 0.001; Supplemental Table 3). Aside from CYP2A6 protein, POR protein was the only remaining significant independent predictor of CYP2A6 enzyme activity, accounting for 0.6% (nicotine metabolism, P < 0.05; Table 3; coumarin metabolism, 0.8% P < 0.05; Supplemental Table 3).
TABLE 3 .
Linear regression analysis of CYP2A6 enzyme activity (cotinine formation from nicotine, nmol/min per milligram)a
| Predictor Variable | B | Beta | 95% CI | % Variation Accounted fore | P Value | |
|---|---|---|---|---|---|---|
| 1) Model 1b | CYP2A6*2 Genotypef | −0.04 | −0.1 | −0.077 to –0.001 | 1.0% | <0.05 |
| CYP2A6*9 Genotypef | −0.03 | −0.1 | −0.059 to –0.003 | 1.2% | <0.05 | |
| CYP2A6*12 Genotypef | −0.05 | −0.09 | −0.105 to 0.004 | 0.8% | <0.1 | |
| Gender (M = 1, F = 0) | 0.004 | 0.02 | −0.02 to 0.03 | 0.03% | >0.1 | |
| AKR1D1 mRNA (FPKM) | 0.001 | 0.2 | <0.001 to 0.001 | 3.3% | <0.001 | |
| Age | 0.001 | 0.2 | <0.001 to 0.001 | 3.5% | <0.001 | |
| POR Protein | 0.004 | 0.5 | 0.003 to 0.005 | 21.5% | <0.001 | |
| 2) Model 1 + CYP2A6 mRNAc | CYP2A6*2 Genotypef | −0.05 | −0.1 | −0.08 to –0.01 | 1.5% | <0.01 |
| CYP2A6*9 Genotypef | −0.02 | −0.08 | −0.047 to 0.005 | 0.5% | >0.1 | |
| CYP2A6*12 Genotypef | −0.04 | −0.07 | −0.09 to 0.01 | 0.4% | >0.1 | |
| Gender (M=1, F=0) | 0.003 | 0.02 | −0.02 to 0.02 | 0.03% | >0.1 | |
| AKR1D1 mRNA (FPKM) | <0.001 | 0.07 | <0.001 to 0.001 | 0.4% | >0.1 | |
| Age | 0.001 | 0.2 | <0.001 to 0.001 | 2.6% | <0.01 | |
| POR Protein | 0.003 | 0.4 | 0.003 to 0.004 | 14.0% | <0.001 | |
| CYP2A6 mRNA (FPKM) | <0.001 | 0.4 | <0.001 to < 0.001 | 10.0% | <0.001 | |
| 3) Model 1 + CYP2A6 mRNA + CYP2A6 proteind | CYP2A6*2 Genotypef | 0.03 | 0.009 | −0.02 to –0.03 | 0.008% | >0.1 |
| CYP2A6*9 Genotypef | <0.001 | <0.001 | −0.02 to 0.02 | <0.001% | >0.1 | |
| CYP2A6*12 Genotypef | 0.007 | 0.01 | −0.03 to 0.04 | 0.01% | >0.1 | |
| Gender (M = 1, F = 0) | 0.009 | 0.05 | −0.003 to 0.022 | 0.2% | >0.1 | |
| AKR1D1 mRNA (FPKM) | <0.001 | 0.02 | <0.001 to < 0.001 | 0.04% | >0.1 | |
| Age | <0.001 | 0.03 | <0.001 to < 0.001 | 0.06% | >0.1 | |
| POR Protein | 0.001 | 0.09 | <0.001 to 0.001 | 0.6% | <0.05 | |
| CYP2A6 mRNA (FPKM) | <0.001 | 0.4 | <0.001 to < 0.001 | 0.1% | >0.1 | |
| CYP2A6 Protein | 0.004 | 0.8 | 0.004 to 0.005 | 31.9% | <0.001 |
Cases excluded pairwise.
R2 = 0.35, P < 0.001
R2 = 0.46, P < 0.001
R2 = 0.77, P < 0.001
% Variation accounted for by each variable is determined by: (Part Correlation)2 × 100.
CYP2A6*2, CYP2A6*9, and CYP2A6*12 genotypes coded as 0 for CYP2A6*1/*1 genotype, 1 for CYP2A6*1/*2, CYP2A6*1/*9, or CYP2A6*1/*12 genotypes, and 2 for CYP2A6*2/*2 or CYP2A6*9/*9 genotypes.
Discussion
In a large human liver bank, we have identified several genetic and nongenetic factors that contribute to variation in CYP2A6 mRNA, protein, and ultimately CYP2A6 enzyme activity, which is an important determinant of the rate of nicotine metabolism. We confirmed that our CYP2A6 phenotype measures were correlated with, and predictive of one another. Using regression models we were able to account for over 75% of the variation in CYP2A6 activity, as well as assessing the novel roles of AKR1D1 and POR in these CYP2A6 phenotypes.
Consistent with the literature, female gender was associated with higher CYP2A6 mRNA expression, relative to male donors in the liver bank, in univariate analyses with similar trends for protein levels and enzyme activity. However, gender was not a significant independent predictor of any CYP2A6 phenotype on the basis of regression modeling. It is possible that other factors are blunting the relationship between gender and CYP2A6 in our models, or that there is a relatively large impact of pre- and postmenopausal women in this analysis, where previous data suggests similar levels of activity (Benowitz et al., 2006). In this liver bank 20.1% and 42.5% of females are below 16-years- and above 50-years-old, respectively.
The age of liver donors was positively, but weakly, associated with CYP2A6 protein levels and enzyme activity. It is unclear if this relationship denotes a true age effect on CYP2A6, or if this increase results from unknown covariates, for example greater inducer exposure among older donors. For example, there is an overall increase in polypharmacy with increasing age (Hajjar et al., 2007), and several drugs are known CYP2A6 inducers, such as phenobarbital, dexamethasone, or rifampin (Maurice et al., 1991; Rae et al., 2001).
Additionally, there may be dietary differences between age groups, with elderly individuals consuming more CYP2A6-inducing foods, or a role for estrogen in increasing levels over puberty (Hakooz and Hamdan, 2007; Higashi et al., 2007). However, when modeling predictors of CYP2A6 phenotypes, age was a significant independent predictor of both CYP2A6 protein and activity, even when accounting for gender, suggesting that a female puberty effect is not responsible for the observed positive association between age and CYP2A6 protein levels and enzyme activity.
CYP2A6 genotype was associated with reduced CYP2A6 protein and enzyme activity in both univariate and regression analyses. We observed a step-wise decrease in CYP2A6 mRNA expression, protein levels, and enzyme activity with increasing copies of the CYP2A6*9 allele (i.e., from *1/*1 to *1/*9 to *9/*9). CYP2A6*9 is an SNP present in the TATA box of the CYP2A6 promoter and is associated with decreased CYP2A6 transcription (Pitarque et al., 2001), consistent with our results. There was also a step-wise decrease in CYP2A6 protein and enzyme activity associated with CYP2A6*2, which is a nonsynonymous SNP in exon 3, resulting in the failure of the enzyme to incorporate heme necessary for catalytic function, resulting in a less stable enzyme, vulnerable to degradation (Yamano et al., 1990). Both the CYP2A6*2 and CYP2A6*9 alleles were significant independent predictors of CYP2A6 protein levels and enzyme activity in our regression models. However, when CYP2A6 mRNA was included as a covariate in both the protein and activity models, the impact of the *9 allele on each phenotype decreased, and was no longer significant, suggesting that *9 exhibits a secondary effect on CYP2A6 protein and activity, via its direct influence on CYP2A6 mRNA. Conversely, the *2 allele remains a significant predictor of both CYP2A6 protein and activity when CYP2A6 mRNA was added to the model, suggesting a mechanism independent of CYP2A6 mRNA expression. Within the wild-type (*1/*1) group, which possesses no tested CYP2A6 genetic variants, we still observed a large range in CYP2A6 mRNA, protein, and enzyme activity. This implies that there are additional uncharacterized sources of variation, which may result from environmental exposures, or undetected genetic variation present at the CYP2A6 gene locus or potentially other regulatory loci.
AKR1D1 has been implicated as a potential regulator of the expression of P450s (Chaudhry et al., 2013), and our findings support a relationship between AKR1D1 and CYP2A6 mRNA expression. AKR1D1 mRNA expression was associated with increasing CYP2A6 mRNA, protein, and enzyme activity, whereas the specific genetic variation (AKR1D1 SNP rs1872930) was not. AKR1D1 mRNA levels accounted for a significant proportion (16%) of the variation in CYP2A6 mRNA expression (Table 1). AKR1D1 was also a significant predictor of CYP2A6 protein levels and enzyme activity; however, the proportion of variation in CYP2A6 protein and activity accounted for by AKR1D1 mRNA decreased more than 8-fold when CYP2A6 mRNA was added to each model. This suggests that the relationship between AKR1D1 mRNA and CYP2A6 phenotypes largely results from the influence of AKR1D1 mRNA on CYP2A6 mRNA expression. The association between AKR1D1 on CYP2A6 mRNA expression may result from the role of AKR1D1 in bile acid synthesis and/or the reduction of steroid hormones, which can act as ligands of nuclear hormone receptors. For example, AKR1D1 is responsible for 5β-reduction of progesterone, resulting in the activation of nuclear hormone receptor pregnane X receptor (Bertilsson et al., 1998). Bile acids can activate nuclear hormone receptors, such as pregnane X receptor and constitutive androstane receptor, which can regulate the transcription of several genes, including P450s (Schuetz et al., 2001). These data confirm a significant role for AKR1D1 on the regulation of multiple P450s, suggesting a broader role for it in altering drug metabolism. Whether the variation contributes to altered nicotine pharmacokinetics and subsequent smoking behaviors remains to be tested.
POR protein was positively correlated with CYP2A6 activity, remaining a significant independent predictor of CYP2A6 activity when other sources of variation were accounted for. There was no association between the POR genotypes tested and CYP2A6 enzyme activity. The role of POR as an electron donor to P450s during the process of substrate metabolism is consistent with the observed positive correlation of POR and CYP2A6 enzyme activity. However, the contribution of POR to CYP2A6 activity decreased from 21.5% to 14.0% to less than 1% when CYP2A6 mRNA and CYP2A6 protein were added to the models, respectively. This suggests a potential role for POR in regulating mRNA or protein levels, or that POR and CYP2A6 may have a common regulatory feature, as opposed to a direct influence on CYP2A6 enzyme activity.
Although this liver bank was extensively characterized, this study did not assess several genetic and environmental variables that have been associated with CYP2A6 and nicotine metabolism and may account for a portion of the 23% missing variation in CYP2A6 activity. For example, we have focused on the assessment of common (MAF > 1%) established CYP2A6 genetic variants; however, the majority of genetic variants found in pharmacogenes, including CYP2A6, are thought to be rare (MAF < 1%) (Kozyra et al., 2016), suggesting that a substantial portion of the 23% unidentified variation in CYP2A6 activity may be the result of unknown CYP2A6 rare variants. Variation in other genes regulating P450 expression or function, including AKRs or nuclear hormone receptors, may also, in part, account for the 23% missing variation. Several enzymes in the AKR1 family, including AKR1C1-1C4, may be associated with variation in CYP2A6 expression owing to their contribution to metabolism and biosynthesis of steroids (estrogen, testosterone, progesterone, unavailable in this study), steroid hormones, and bile acids, ultimately controlling concentrations of active ligands at nuclear receptors, ligand occupancy, and trans-activation of receptors (reviewed by Rižner and Penning, 2014). Cigarette smoking, an environmental factor, is associated with a reduction in nicotine clearance in vivo, such that nicotine clearance increases 14% after 4 days of abstinence in regular smokers (Benowitz and Jacob, 1993; Benowitz and Jacob, 2000). Similarly, grapefruit juice reduces nicotine’s metabolism to cotinine, mediated by CYP2A6, by 15% (Hukkanen et al., 2006).
In conclusion, we have identified several sources of variation in CYP2A6 activity in vitro that could translate to variable hepatic clearance of several clinically important drugs, namely nicotine. We were able to account for 77% of the variation in CYP2A6 activity in our models. Unaccounted for variation may result from unknown genetic variation at the highly polymorphic CYP2A6 gene locus and/or in additional regulatory genes, as well as from environmental factors. Characterizing sources of variation in CYP2A6 activity is important, as variation in the rate of nicotine metabolism and clearance can ultimately influence smoking behavior, and affect a smoker’s ability to quit smoking.
Acknowledgments
The authors thank Dr. Zhao Bin and Maria Novalen for developing and/or performing bioanalytical assays (LCMS and HPLC analyses), Drs. Tim Thornton and Michael Wu for advice on the multivariate statistical analysis, and Qian Zhou for conducting much of the genotyping.
Abbreviations
- AKR
aldo-keto reductase
- FPKM
fragments per kilobase per million reads
- LC-MS/MS
liquid chromatography–tandem mass spectroscopy
- P450
cytochrome P450
- POR
cytochrome P450 oxidoreductase
- SNP
single-nucleotide polymorphism
Authorship Contributions
Participated in research design: Tanner, Chaudhry, Thummel, Tyndale.
Conducted experiments: Tanner, Prasad, Claw, Stapleton.
Contributed new reagents or analytic tools: Prasad, Claw, Schuetz.
Performed data analysis: Tanner, Prasad, Claw.
Wrote or contributed to the writing of the manuscript: Tanner, Prasad, Claw, Stapleton, Chaudhry, Schuetz, Thummel, Tyndale.
Footnotes
The authors acknowledge the support of the Endowed Chair in Addictions for the Department of Psychiatry (R.F.T.); National Institutes of Health Pharmacogenomics Research Network grants U01 DA020830 (R.F.T.), and P30 ES007033 (P.S., K.E.T.); Canadian Institutes of Health Research grant THM 109787 (R.F.T.); Campbell Family Mental Health Research Institute of the Centre for Addiction and Mental Health, the CAMH Foundation; the Canada Foundation for Innovation (nos. 20289 and 16014) (R.F.T.); and the Ontario Ministry of Research and Innovation. The work was also supported by the National Institutes of Health National Cancer Institute (Cancer Center Support Grant P30 CA21765) and the American Lebanese Syrian Associated Charities (ALSAC). Source of liver tissue: Liver Tissue Procurement and Distribution System (National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases Contract N01-DK92310) and the Cooperative Human Tissue Network. Conflicts of interest: Dr. Tyndale has consulted for Apotex. The remaining authors declare no conflicts of interest.
A preliminary account of this work was previously presented at the following meeting: Tanner J-A, Chaudhry A, Bhagwat P, Schuetz EG, Thummel KE, and Tyndale RF (2016) Determination of predictors of CYP2A6 protein levels and nicotine metabolism in a human liver bank: influence of genetic and non-genetic factors, presented at the annual meeting of the Society for Research on Nicotine and Tobacco; 2016 March 2–5; Chicago, IL. Society For Research on Nicotine and Tobacco, Madison, WI.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Al Koudsi N, Hoffmann EB, Assadzadeh A, Tyndale RF. (2010) Hepatic CYP2A6 levels and nicotine metabolism: impact of genetic, physiological, environmental, and epigenetic factors. Eur J Clin Pharmacol 66:239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz NL, Jacob P., 3rd (1993) Nicotine and cotinine elimination pharmacokinetics in smokers and nonsmokers. Clin Pharmacol Ther 53:316–323. [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Jacob P., 3rd (2000) Effects of cigarette smoking and carbon monoxide on nicotine and cotinine metabolism. Clin Pharmacol Ther 67:653–659. [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Lessov-Schlaggar CN, Swan GE, Jacob P., 3rd (2006) Female sex and oral contraceptive use accelerate nicotine metabolism. Clin Pharmacol Ther 79:480–488. [DOI] [PubMed] [Google Scholar]
- Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Bäckman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. (1998) Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci USA 95:12208–12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnington MJ, Zhu AZ, Renner CC, Lanier AP, Hatsukami DK, Benowitz NL, Tyndale RF. (2012) CYP2A6 and CYP2B6 genetic variation and its association with nicotine metabolism in South Western Alaska Native people. Pharmacogenet Genomics 22:429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry AS, Thirumaran RK, Yasuda K, Yang X, Fan Y, Strom SC, Schuetz EG. (2013) Genetic variation in aldo-keto reductase 1D1 (AKR1D1) affects the expression and activity of multiple cytochrome P450s. Drug Metab Dispos 41:1538–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LS, Bloom AJ, Baker TB, Smith SS, Piper ME, Martinez M, Saccone N, Hatsukami D, Goate A, Bierut L. (2014) Pharmacotherapy effects on smoking cessation vary with nicotine metabolism gene (CYP2A6). Addiction 109:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth MJ, Novalen M, Hawk LW, Jr., Schnoll RA, George TP, Cinciripini PM, Lerman C and Tyndale RF (2014a) Known and novel sources of variability in the nicotine metabolite ratio in a large sample of treatment-seeking smokers. Cancer Epidemiol Biomarkers Prev 23:1773–1782. [DOI] [PMC free article] [PubMed]
- Chenoweth MJ, Zhu AZ, Sanderson Cox L, Ahluwalia JS, Benowitz NL, Tyndale RF. (2014b) Variation in P450 oxidoreductase (POR) A503V and flavin-containing monooxygenase (FMO)-3 E158K is associated with minor alterations in nicotine metabolism, but does not alter cigarette consumption. Pharmacogenet Genomics 24:172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EL, Zhao B, Cui JZ, Novalen M, Miksys S, Tyndale RF. (2014) Nicotine pharmacokinetics in rats is altered as a function of age, impacting the interpretation of animal model data. Drug Metab Dispos 42:1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Iulio J, Fayet A, Arab-Alameddine M, Rotger M, Lubomirov R, Cavassini M, Furrer H, Günthard HF, Colombo S, Csajka C, et al. Swiss HIV Cohort Study (2009) In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genomics 19:300–309. [DOI] [PubMed] [Google Scholar]
- Emami S, Dadashpour S. (2015) Current developments of coumarin-based anti-cancer agents in medicinal chemistry. Eur J Med Chem 102:611–630. [DOI] [PubMed] [Google Scholar]
- Endo T, Ban M, Hirata K, Yamamoto A, Hara Y, Momose Y. (2007) Involvement of CYP2A6 in the formation of a novel metabolite, 3-hydroxypilocarpine, from pilocarpine in human liver microsomes. Drug Metab Dispos 35:476–483. [DOI] [PubMed] [Google Scholar]
- Fisher CD, Lickteig AJ, Augustine LM, Ranger-Moore J, Jackson JP, Ferguson SS, Cherrington NJ. (2009) Hepatic cytochrome P450 enzyme alterations in humans with progressive stages of nonalcoholic fatty liver disease. Drug Metab Dispos 37:2087–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AM, Winter S, Klein K, Turpeinen M, Schaeffeler E, Schwab M, Zanger UM. (2009) Pharmacogenomics of human liver cytochrome P450 oxidoreductase: multifactorial analysis and impact on microsomal drug oxidation. Pharmacogenomics 10:579–599. [DOI] [PubMed] [Google Scholar]
- Hajjar ER, Cafiero AC, Hanlon JT. (2007) Polypharmacy in elderly patients. Am J Geriatr Pharmacother 5:345–351. [DOI] [PubMed] [Google Scholar]
- Hakooz N, Hamdan I. (2007) Effects of dietary broccoli on human in vivo caffeine metabolism: a pilot study on a group of Jordanian volunteers. Curr Drug Metab 8:9–15. [DOI] [PubMed] [Google Scholar]
- Higashi E, Fukami T, Itoh M, Kyo S, Inoue M, Yokoi T, Nakajima M. (2007) Human CYP2A6 is induced by estrogen via estrogen receptor. Drug Metab Dispos 35:1935–1941. [DOI] [PubMed] [Google Scholar]
- Hu L, Zhuo W, He YJ, Zhou HH, Fan L. (2012) Pharmacogenetics of P450 oxidoreductase: implications in drug metabolism and therapy. Pharmacogenet Genomics 22:812–819. [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Jacob P, 3rd, Benowitz NL. (2005) Metabolism and disposition kinetics of nicotine. Pharmacol Rev 57:79–115. [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Jacob P, 3rd, Benowitz NL. (2006) Effect of grapefruit juice on cytochrome P450 2A6 and nicotine renal clearance. Clin Pharmacol Ther 80:522–530. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Yoshisue K, Matsushima E, Nagayama S, Kobayashi K, Tyson CA, Chiba K, Kawaguchi Y. (2000) Bioactivation of tegafur to 5-fluorouracil is catalyzed by cytochrome P-450 2A6 in human liver microsomes in vitro. Clin Cancer Res 6:4409–4415. [PubMed] [Google Scholar]
- Jacob P, 3rd, Yu L, Duan M, Ramos L, Yturralde O, Benowitz NL. (2011) Determination of the nicotine metabolites cotinine and trans-3′-hydroxycotinine in biologic fluids of smokers and non-smokers using liquid chromatography-tandem mass spectrometry: biomarkers for tobacco smoke exposure and for phenotyping cytochrome P450 2A6 activity. J Chromatogr B Analyt Technol Biomed Life Sci 879:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiang TK, Ho PC, Anari MR, Tong V, Abbott FS, Chang TK. (2006) Contribution of CYP2C9, CYP2A6, and CYP2B6 to valproic acid metabolism in hepatic microsomes from individuals with the CYP2C9*1/*1 genotype. Toxicol Sci 94:261–271. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozyra M, Ingelman-Sundberg M, Lauschke VM. (2016) Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet Med DOI:10.1038/gim.2016.33 (published ahead of print). [DOI] [PubMed] [Google Scholar]
- Lee WH, Lukacik P, Guo K, Ugochukwu E, Kavanagh KL, Marsden B, Oppermann U. (2009) Structure-activity relationships of human AKR-type oxidoreductases involved in bile acid synthesis: AKR1D1 and AKR1C4. Mol Cell Endocrinol 301:199–204. [DOI] [PubMed] [Google Scholar]
- Lerman C, Schnoll RA, Hawk LW, Jr, Cinciripini P, George TP, Wileyto EP, Swan GE, Benowitz NL, Heitjan DF, Tyndale RF, PGRN-PNAT Research Group (2015) Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. Lancet Respir Med 3:131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li NY, Sellers EM. (1997) Comparison of CYP2A6 catalytic activity on coumarin 7-hydroxylation in human and monkey liver microsomes. Eur J Drug Metab Pharmacokinet 22:295–304. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. [DOI] [PubMed] [Google Scholar]
- Loukola A, Buchwald J, Gupta R, Palviainen T, Hällfors J, Tikkanen E, Korhonen T, Ollikainen M, Sarin AP, Ripatti S, et al. (2015) A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism. PLoS Genet 11:e1005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Hu L, Zhuo W, Zhang C, Zhou H, Fan L. (2016) Effects of the selected cytochrome P450 oxidoreductase genetic polymorphisms on cytochrome P450 2B6 activity as measured by bupropion hydroxylation. Pharmacogenet Genomics 26:80–87. [DOI] [PubMed] [Google Scholar]
- Malaiyandi V, Sellers EM, Tyndale RF. (2005) Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clin Pharmacol Ther 77:145–158. [DOI] [PubMed] [Google Scholar]
- Maurice M, Emiliani S, Dalet-Beluche I, Derancourt J, Lange R. (1991) Isolation and characterization of a cytochrome P450 of the IIA subfamily from human liver microsomes. Eur J Biochem 200:511–517. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina ES, Tyndale RF, Sellers EM. (1997) A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther 282:1608–1614. [PubMed] [Google Scholar]
- Molander L, Hansson A, Lunell E. (2001) Pharmacokinetics of nicotine in healthy elderly people. Clin Pharmacol Ther 69:57–65. [DOI] [PubMed] [Google Scholar]
- Murai K, Yamazaki H, Nakagawa K, Kawai R, Kamataki T. (2009) Deactivation of anti-cancer drug letrozole to a carbinol metabolite by polymorphic cytochrome P450 2A6 in human liver microsomes. Xenobiotica 39:795–802. [DOI] [PubMed] [Google Scholar]
- Mwenifumbo JC, Tyndale RF. (2007) Genetic variability in CYP2A6 and the pharmacokinetics of nicotine. Pharmacogenomics 8:1385–1402. [DOI] [PubMed] [Google Scholar]
- Park SL, Tiirikainen MI, Patel YM, Wilkens LR, Stram DO, Le Marchand L, Murphy SE. (2016) Genetic determinants of CYP2A6 activity across racial/ethnic groups with different risks of lung cancer and effect on their smoking intensity. Carcinogenesis 37:269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson F, Schnoll RA, Wileyto EP, Pinto A, Epstein LH, Shields PG, Hawk LW, Tyndale RF, Benowitz N, Lerman C. (2008) Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther 84:320–325. [DOI] [PubMed] [Google Scholar]
- Pitarque M, von Richter O, Oke B, Berkkan H, Oscarson M, Ingelman-Sundberg M. (2001) Identification of a single nucleotide polymorphism in the TATA box of the CYP2A6 gene: impairment of its promoter activity. Biochem Biophys Res Commun 284:455–460. [DOI] [PubMed] [Google Scholar]
- Prasad B, Unadkat JD. (2014) Optimized approaches for quantification of drug transporters in tissues and cells by MRM proteomics. AAPS J 16:634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Lippman ME, Flockhart DA. (2001) Rifampin is a selective, pleiotropic inducer of drug metabolism genes in human hepatocytes: studies with cDNA and oligonucleotide expression arrays. J Pharmacol Exp Ther 299:849–857. [PubMed] [Google Scholar]
- Rižner TL, Penning TM. (2014) Role of aldo-keto reductase family 1 (AKR1) enzymes in human steroid metabolism. Steroids 79:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N, Lerman C. (2009) Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: a validation study. Pharmacol Biochem Behav 92:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. (2004) Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics 14:615–626. [DOI] [PubMed] [Google Scholar]
- Schuetz EG, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim RB, Ramachandran V, Komoroski BJ, et al. (2001) Disrupted bile acid homeostasis reveals an unexpected interaction among nuclear hormone receptors, transporters, and cytochrome P450. J Biol Chem 276:39411–39418. [DOI] [PubMed] [Google Scholar]
- Shirasaka Y, Chaudhry AS, McDonald M, Prasad B, Wong T, Calamia JC, Fohner A, Thornton TA, Isoherranen N, Unadkat JD, et al. (2016) Interindividual variability of CYP2C19-catalyzed drug metabolism due to differences in gene diplotypes and cytochrome P450 oxidoreductase content. Pharmacogenomics J 16:375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster DL, Risler LJ, Prasad B, Calamia JC, Voellinger JL, Kelly EJ, Unadkat JD, Hebert MF, Shen DD, Thummel KE, et al. (2014) Identification of CYP3A7 for glyburide metabolism in human fetal livers. Biochem Pharmacol 92:690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolerman IP, Jarvis MJ. (1995) The scientific case that nicotine is addictive. Psychopharmacology (Berl) 117:2–10, discussion 14–20. [DOI] [PubMed] [Google Scholar]
- Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF, Benowitz NL. (2009) Genetic and environmental influences on the ratio of 3′hydroxycotinine to cotinine in plasma and urine. Pharmacogenet Genomics 19:388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. (2011) Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst 103:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar CA, Zhou Q, Tyndale RF. (2016) CYP2A6 genotyping methods and strategies using real-time and end point PCR platforms. Pharmacogenomics 17:147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano S, Tatsuno J, Gonzalez FJ. (1990) The CYP2A3 gene product catalyzes coumarin 7-hydroxylation in human liver microsomes. Biochemistry 29:1322–1329. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang B, Molony C, Chudin E, Hao K, Zhu J, Gaedigk A, Suver C, Zhong H, Leeder JS, et al. (2010) Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res 20:1020–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]