Abstract
Evidence suggests that the nonpsychotropic cannabis-derived compound, cannabidiol (CBD), has antineoplastic activity in multiple types of cancers, including glioblastoma multiforme (GBM). DNA-damaging agents remain the main standard of care treatment available for patients diagnosed with GBM. Here we studied the antiproliferative and cell-killing activity of CBD alone and in combination with DNA-damaging agents (temozolomide, carmustine, or cisplatin) in several human GBM cell lines and in mouse primary GBM cells in cultures. This activity was also studied in mouse neural progenitor cells (NPCs) in culture to assess for potential central nervous system toxicity. We found that CBD induced a dose-dependent reduction of both proliferation and viability of all cells with similar potencies, suggesting no preferential activity for cancer cells. Hill plot analysis indicates an allosteric mechanism of action triggered by CBD in all cells. Cotreatment regimens combining CBD and DNA-damaging agents produced synergistic antiproliferating and cell-killing responses over a limited range of concentrations in all human GBM cell lines and mouse GBM cells as well as in mouse NPCs. Remarkably, antagonistic responses occurred at low concentrations in select human GBM cell lines and in mouse GBM cells. Our study suggests limited synergistic activity when combining CBD and DNA-damaging agents in treating GBM cells, along with little to no therapeutic window when considering NPCs.
Introduction
Standard of care treatment of glioblastoma multiforme (GBM; the predominant and devastating subtype of gliomas that develops in human adults) extends the median survival of patients from approximately 12 months to only 15–17 months (Stupp et al., 2005; Adamson et al., 2009; Omuro and DeAngelis, 2013; Ostrom et al., 2014). Patients diagnosed with GBM undergo surgical resection followed by radiotherapy and chemotherapy. The most commonly prescribed chemotherapeutics are the DNA-damaging agents temozolomide (TMZ) (Temodar; Merck, Kenilworth, NJ) and carmustine (BCNU), both of which have limited value at curbing GBM pathogenesis because they exhibit poor efficacy at stopping cell proliferation and eliminating tumor mass (Adamson et al., 2009). Importantly, high doses of DNA-damaging agents are required to treat patients diagnosed with GBM and result in significant debilitating side effects because of their poor cancer selectivity and ensuing detrimental effects on dividing cells (Adamson et al., 2009). The poor prognoses associated with GBM, along with the lack of a safe standard of care available to treat and ultimately cure this disease, advocate for an urgent need to develop much improved medicines to treat this devastating type of cancer (Brem et al., 1995; Westphal et al., 2003; Stupp et al., 2005; Adamson et al., 2009; Omuro and DeAngelis, 2013).
The genetic profiling of human glioma tissues by The Cancer Genome Atlas revealed a remarkable heterogeneity in driver mutations and gene amplification that led to the classification of GBM into three subtypes: proneural, mesenchymal, and classic (Verhaak et al., 2010; Dunn et al., 2012; Ozawa et al., 2014). Several genetic mouse models of GBM have revealed how driver mutations participate in its pathogenesis (Zhu et al., 2009; Halliday et al., 2014; Leder et al., 2014; Ozawa et al., 2014), and a recent study indicated that amplification of platelet-derived growth factor (PDGF) signaling is associated with the proneural subtype of GBM and occurs early during GBM pathogenesis (Ozawa et al., 2014). Thus, investigators must consider the genetic makeup of GBM tumors when developing novel therapeutic strategies to treat and ultimately cure this cancer.
It has been shown that cannabidiol (CBD) exhibits antineoplastic activity in multiple GBM cell lines in culture and in xenograft mouse models (Massi et al., 2004, 2006, 2008; Vaccani et al., 2005; Marcu et al., 2010; Torres et al., 2011; Nabissi et al., 2013; Solinas et al., 2013; Soroceanu et al., 2013). This antineoplastic activity is mediated through plasma membrane–associated receptors, including G protein–coupled receptor (GPR) 55 and transient receptor potential cation channel subfamily V member (TRPV) 1/2, and involves the production of reactive oxygen as well as induction of autophagy and apoptosis (Bisogno et al., 2001; Ligresti et al., 2006; Massi et al., 2006; Ford et al., 2010; Ramer et al., 2010; Yamada et al., 2010; Piñeiro et al., 2011; Anavi-Goffer et al., 2012). Interestingly, a recent study reported that the antineoplastic activity of CBD synergizes with that of TMZ and BCNU, suggesting that combination treatment regimens (combined modality therapy) could provide greater benefit to patients diagnosed with GBM when considering CBD; however, this study reported activity at a single concentration of CBD and in one human GBM cell line (U87MG) (Nabissi et al., 2013). Thus, a more detailed and quantitative evaluation of the combined responses induced by CBD and DNA-damaging agents in multiple cell culture models is still required to better understand the therapeutic potential of CBD in treating patients diagnosed with GBM.
Here we studied the antiproliferative and cell-killing activities of CBD alone and combined with three DNA-damaging agents—TMZ, BCNU, and cisplatin (CDDP) (Platinol; Bristol-Myers Squibb, New York, NY)—in three human GBM cell lines (e.g., T98G, U251, and U87MG), as well as in primary cells derived from a genetically engineered mouse model of GBM that carries amplified PDGF signaling (PDGF-GBM cells). Mouse neural progenitor cells (NPCs) in culture were used to assess central nervous system (CNS) toxicity. The drug interactions between treatments were analyzed using the checkerboard assay and the fractional inhibitory concentration (FIC), allowing for quantitative and statistical evaluations of each combination.
Materials and Methods
Reagents
CBD (PubChem CID 644019) was obtained from the National Institute on Drug Abuse Drug Supply Service (Bethesda, MD). TMZ (PubChem CID 5394, catalog no. 2706) and CDDP (PubChem CID 441203, catalog no. 2251) were obtained from Tocris Bioscience (Ellisville, MO). BCNU (PubChem CID 2578, catalog no. C0400), dimethylsulfoxide, trypsin-EDTA solution (catalog no. T3924), and laminin (catalog no. 11243217001) were purchased from Sigma-Aldrich (St. Louis, MO). CBD and TMZ were dissolved in dimethylsulfoxide, BCNU was dissolved in ethanol, and CDDP was dissolved in phosphate-buffered saline (PBS). HyClone Dulbecco’s modified Eagle’s medium (DMEM) (with l-glutamine and sodium pyruvate, catalog no. SH30243.01), PhenoRed-free DMEM (with high glucose, modified, without PhenoRed, without l-glutamine, with sodium pyruvate, catalog no. SH30604.01), and fetal bovine serum (FBS) (catalog no. SH30088) were obtained from GE Healthcare Life Sciences (Logan, UT). Gibco Dulbecco’s PBS (catalog no. 14040141), penicillin-streptomycin (catalog no. 15140122), sodium bicarbonate 7.5% solution (catalog no. 25080094), HEPES (catalog no. 15630106), human recombinant fibroblast growth factor (catalog no. 13256029), and epidermal growth factor (catalog no. PHG0314) were purchased from Thermo Fisher Scientific (Grand Island, NY). NeuroCult neural stem cell basal medium (mouse, catalog no. 05700), NeuroCult neural stem cell proliferation supplements (mouse, catalog no. 05701), heparin solution (0.2%, catalog no. 07980), and Accutase (catalog no. 07920) were purchased from StemCell Technologies (Vancouver, BC, Canada).
Generation of the Mouse PDGF-GBM Cells and NPCs
All experimental procedures were approved by the Institutional Animal Care and Use Committee of Fred Hutchinson Cancer Research Center. Mouse GBM cells harboring amplified PDGF signaling (PDGF-GBM) were generated by injecting RCAS (replication competent ALV LTR with a splice acceptor)–PDGFA viruses into Nestin (N)/tv-a;Ink4a-arf−/−;Ptenfl/fl mice, as previously described (Ozawa et al., 2014). Animals were euthanized with CO2 at age 72 days when they presented brain tumor symptoms (i.e., cranial swelling and lethargy) and PDGFA-Ink4a-arf−/− GBM cells (PDGF-GBM cells) were harvested by mechanical dissociation of the tumors that had developed in the mouse brain. Mouse NPCs were generated by the mechanical dissociation of the newborn pup forebrains of N/tv-a;Ink4a-arf−/−;Ptenfl/fl mice. Mouse PDGF-GBM cells and NPCs were initially maintained as floating neurosphere cultures and starting at passages three and five (PDGF-GBM cells and NPCs, respectively) were cultured as adherent monolayers on laminin-coated dishes.
Cell Culture
T98G, U251, and U87MG cells were expanded in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 0.01 M HEPES, and 0.075% sodium bicarbonate. For cell proliferation and viability assays, cells were switched to DMEM (PhenoRed free) supplemented with 3% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin when seeded onto 96-well plates.
Mouse PDGF-GBM cells and NPCs were expended in mouse NeuroCult neural stem cell basal medium supplemented with NeuroCult neural stem cell proliferation supplements, 2 µg/ml heparin, 20 ng/ml epidermal growth factor, 10 ng/ml fibroblast growth factor, 100 U/ml penicillin, and 100 µg/ml streptomycin in a humidified atmosphere (37°C and 5% CO2). Cells at passages four to six were used in the following experiment.
Drug Treatments
Each cell type was treated with CBD, TMZ, BCNU, or CDDP at concentrations ranging from 10−7.5 to 10−3 M in half log10 steps and proliferation and viability were measured 72 hours after treatment. All dose-response experiments were conducted in triplicate and repeated at least three times. Data analyses were performed using GraphPad Prism software (GraphPad Software Inc., La Jolla, CA) and each experiment was considered as n = 1.
Cell Proliferation and Viability Assays
Cell proliferation was measured using the 5-bromo-2′-deoxy-uridine (BrdU) cell proliferation ELISA (colorimetric) kit (Roche, Indianapolis, IN) following the manufacturer’s protocol. The cell density per well for each cell type was selected in the logarithmic growth range (Supplemental Fig. 1). Thus, cells were seeded in 96-well plates overnight and then cultured with compounds and BrdU labeling agents for 72 hours. BrdU incorporated in newly synthesized cellular DNA was measured using anti-BrdU antibodies (conjugated with peroxidase) and detected by the subsequent substrate reaction. Sulfuric acid was used to stop the substrate reaction and the absorbance at 450 nm was quantified using a SpectraCount multiwell spectrometer (Packard Instrument Company, Le Meriden, CT).
Cell viability was measured using the water-soluble tetrazolium salt 1 (WST-1) reagent (Roche) following the manufacturer’s protocol. The cell density per well for each cell type was selected in the logarithmic growth range (Supplemental Fig. 1). Thus, cells were seeded into 96-well plates and treated with compounds 24 hours later, and cell viability was measured 72 hours later with WST-1 by adding 10 µl reagent and measuring absorbance at 450 nm after an hour using a SpectraCount multiwell spectrometer.
Checkerboard Assay and Data Analyses
The interaction between CBD and DNA-damaging agents was tested using the checkerboard assay (Supplemental Fig. 2). Thus, serial half log10 dilutions of CBD (0.3–100 µM) were combined with TMZ (1 µM to 1 mM), BCNU (3 µM to 1 mM), or CDDP (0.1–100 µM). Drugs for combination were prepared separately and added into the cell separately, but at the same time. All combinations were performed in duplicate and repeated three times. Data analyses was performed using GraphPad Prism software and each experiment was considered as n = 1.
Different analytical methods of the checkerboard assay may lead to differences in the interpretations (Bonapace et al., 2002). Thus, two methods were used to analyze the results from the checkerboard assay to provide a better understanding of these interactions. Method 1 focused on the percentage of occurrence of synergy, additivity, and antagonism observed in all of the tested concentrations of each individual combination, whereas method 2 focused on the examination of the interactions of compounds based on their efficacies (when producing half maximal inhibition effects).
Method 1: Full-Plate FIC.
The full-plate FIC was calculated over all of the combinations tested for drug A and drug B on a checkerboard assay. For each combination, the formula is as follows:
CA+B is the concentration of drug A in the presence of drug B, and CA is the concentration of drug A alone when producing the same effect as in the combination, and vice versa for CB+A and CB. The concentration of drug A (or B) alone was calculated by determining the concentration (x) for the effect of the combination (y) from the dose-response curve of drug A (or B) using GraphPad Prism software. The FIC index (ΣFIC) was then calculated to determine the synergic interactions for each combination of drug A and drug B on a checkerboard assay. Synergy was defined as ΣFIC < 0.5, antagonism was defined as ΣFIC > 4, and additivity was defined when ΣFIC was in the range of 0.5 to 4, as previously described (Orhan et al., 2005). The percentage of occurrences of synergy, additivity, or antagonism that occurred throughout all of the combinations of drug A and drug B tested on a checkerboard assay was then calculated (Bonapace et al., 2002).
Method 2: Efficacy FIC Index.
This index was calculated based on the combinations that produced half maximal inhibition. Thus, to identify the combinations that produced half maximal effects, we fixed the concentration of drug A and plotted the dose-response curve of the inhibitory effects of the combinations (containing a serial half log10 dilution of drug B in the presence of the fixed concentration of drug A) against the concentration of drug B used in the combinations. The half maximal inhibitory concentration (IC50) of drug B (in the presence of the fixed concentration of drug A) was then calculated using GraphPad Prism software. Therefore, the combination of the fixed concentration of drug A and the IC50 of drug B produce a half maximal inhibition. The efficacy FIC was calculated using the following formula:
IC50A+B is the half maximal inhibitory concentration of drug A in the presence of drug B, IC50A is the half maximal inhibitory concentration of drug A alone, CB_fixed is the fixed concentration of drug B, and IC50B is the half maximal inhibitory concentration of drug B alone. Synergy was defined as ΣFIC < 0.5, antagonism was defined as ΣFIC > 4, and additivity was defined when ΣFIC was in the range of 0.5 to 4, as previously described (Orhan et al., 2005). For detailed calculation examples of both Methods, see Supplementary Material.
Statistical Analyses
Dose responses for each compound were analyzed using GraphPad Prism software (version 5.01). Chi-square analysis was used to identify the source of significant differences (e.g., differences among cell type for each individual drug combination; differences among all of the drug combinations in each cell type) from checkerboard results obtained from method 1. One-way analysis of variance followed by a Bonferroni post hoc test t test was used to identify the source of significant differences in the FIC indices from checkerboard results obtained from method 2. Statistical analyses were performed using GraphPad Prism software (version 5.01). P < 0.05 was considered statistically significant.
Results
Antineoplastic Activity of CBD
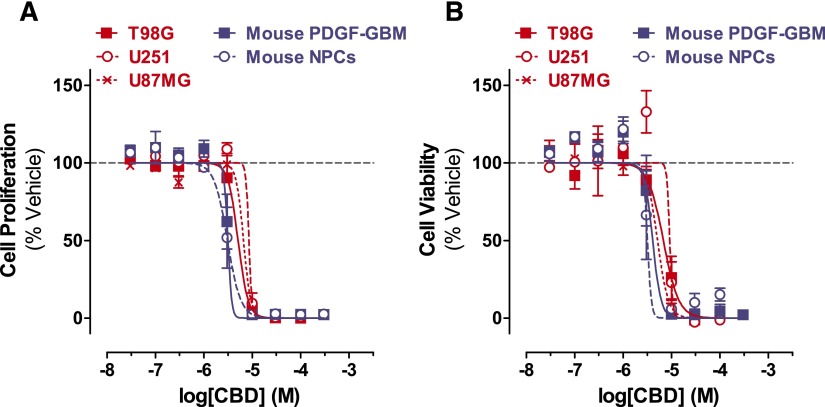
The antineoplastic activity of CBD was assessed by measuring both its antiproliferative and cell-killing activities in three human GBM cell lines (T98G, U251, and U87MG) and in mouse PDGF-GBM cells in primary culture. Its potential CNS toxicity was assessed by measuring the same readouts in mouse NPCs in culture. Thus, cells were treated with half log10 concentrations of CBD and changes in cell proliferation and cell viability were measured 3 days later by quantifying BrdU incorporation and mitochondrial activity with WST-1, respectively. We found that CBD inhibited cell proliferation and reduced cell viability of all cells with similar potencies (cell proliferation IC50s = 3.1–8.5 µM; cell viability IC50s = 3.2–9.2 µM) (Fig. 1; Supplemental Table 1). All dose responses were remarkably steep and Hill plot calculations indicate an allosteric mechanism of action for each response (Supplemental Table 2). Regarding the efficacy of this activity, we found that all cell types were greatly sensitive to both the antiproliferative and cell-killing activities of CBD (maximal efficacy = 94.19%–100%). Specifically, for both antiproliferative response and cell-killing activity, mouse PDGF-GBM cells and NPCs were the most sensitive (i.e., lower dose required for activity), whereas the three human GBM cell lines were the least sensitive (i.e., higher dose required for activity) (Supplemental Table 3). Our results confirm previous reports on the antineoplastic activity of CBD in GBM cells in culture and extend them by now emphasizing an allosteric mechanism of action that profoundly reduces both cell proliferation and viability with similar potency and efficacy in all human GBM cell lines (irrespective of their genetic makeup), as well as in mouse PDGF-GBM cells and NPCs.
Fig. 1.
CBD inhibits the cell proliferation and viability of GBM cells and NPCs. Dose response of CBD on inhibiting proliferation (A) and viability (B) of (red) human GBM cell lines (T98G, U251, and U87MG) and (blue) mouse PDGF-GBM cells and NPCs at the 72-hour time point. Data are expressed as means ± S.E.M. (n = 5–9 independent experiments, each in triplicate). Vehicle comprised 0.1% dimethylsulfoxide.
Antineoplastic Activity of DNA-Damaging Agents
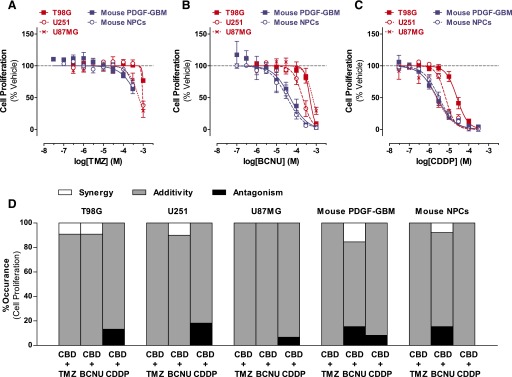
The antineoplastic activity of TMZ, BCNU, and CDDP was assessed using the same experimental design and readouts as with CBD. We found that these agents inhibited cell proliferation and reduced cell viability of all cell types but with remarkably different potencies and efficacies (Figs. 2, A–C, and 3, A–C; Supplemental Tables 1–3). Here, three sets of responses are worth noting. First, TMZ exhibited limited antiproliferative activity in all cells and limited cell-killing activity in the three human GBM cell lines. By contrast, the cell-killing activity of TMZ started at 30 µM in both mouse PDGF-GBM cells and NPCs (Figs. 2A and 3A). Second, mouse PDGF-GBM cells and NPCs were remarkably more sensitive to BCNU (IC50 = 34.1–48.1 µM) than the three human GBM cell lines (IC50s = 205–661 µM) (Figs. 2B and 3B). Third, mouse PDGF-GBM cells and NPCs, as well as U87MG cells, were more sensitive to CDDP (IC50s = 2.4–3.2 µM) than two of the human glioma cell lines (T98G and U251; IC50s = 6.8–25.2 µM, respectively) (Figs. 2C and 3C). Hill plot calculations of all dose responses indicated single-order nonallosteric responses triggered by DNA-damaging agents (Supplemental Table 2). Our results show that mouse PDGF-GBM cells and NPCs are in general more sensitive than human GBM cell lines to these DNA-damaging agents.
Fig. 2.
DNA-damaging agents on the proliferation of GBM cells and NPCs: single and CBD combination responses. (A–C) Dose response of TMZ (A), BCNU (B), and CDDP (C) on inhibiting proliferation of (red) human GBM cells (T98G, U251, and U87MG) and (blue) mouse PDGF-GBM cells and NPCs at the 72-hour time point. Data are expressed as means ± S.E.M. (n = 5–9 independent experiments, each in triplicate). Vehicle comprised 0.1% DMSO (A), 0.1% ethanol (B), or 0.1% PBS (C). (D) Interactions between CBD and DNA-damaging agents on inhibiting proliferation (method 1). Percentage of synergy (white), additivity (gray), and antagonism (black) that occurred in the checkerboard assay. Synergy was defined as ΣFIC < 0.5; antagonism was defined as ΣFIC > 4, and additivity was defined as 0.5 < ΣFIC < 4. Data are expressed as percentage of occurrence calculated using the mean ΣFIC from three independent experiments, each in duplicate. Vehicle comprised 0.05% DMSO (vehicle in lieu of CBD) plus 0.05% DMSO, ethanol, or PBS (vehicle in lieu of TMZ, BCNU, or CDDP, respectively). DMSO, dimethylsulfoxide.
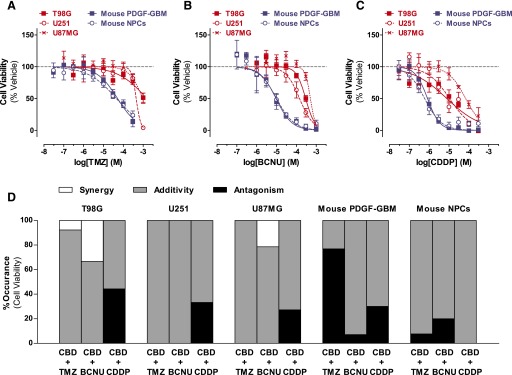
Fig. 3.
DNA-damaging agents on the cell viability of GBM cells and NPCs: single and CBD combination. (A–C) Dose response of TMZ (A), BCNU (B), and CDDP (C) on inhibiting viability of (red) human GBM cells (T98G, U251, and U87MG) and (blue) mouse PDGF-GBM cells and NPCs at the 72-hour time point. Data are expressed as means ± S.E.M. (n = 5 independent experiments, each in triplicate). Vehicle comprised 0.1% DMSO (A), 0.1% ethanol (B), or 0.1% PBS (C). (D) Interactions between CBD and DNA-damaging agents on inhibiting viability (method 1). Percentage of synergy (white), additivity (gray), and antagonism (black) occurred in the checkerboard assay. Synergy was defined as ΣFIC < 0.5, antagonism was defined as ΣFIC > 4, and additivity was defined as 0.5 < ΣFIC < 4. Data are expressed as percentage of occurrence calculated using the mean ΣFIC from three independent experiments, each in duplicate. Vehicle comprised 0.05% DMSO (vehicle in lieu of CBD) plus 0.05% DMSO, ethanol, or PBS (vehicle in lieu of TMZ, BCNU, or CDDP, respectively). DMSO, dimethylsulfoxide.
Interactions between CBD and DNA-Damaging Agents on Cell Proliferation
Combined modality therapy uses optimal low doses of two agents to achieve maximal antineoplastic effects while reducing potential side effects (Kummar et al., 2010). Here we performed quantitative and statistical analyses of the interactions between CBD and DNA-damaging agents on cell proliferation as measured by quantifying BrdU incorporation and performing checkerboard analyses.
We first studied the interactions between CBD and DNA-damaging agents over full dose responses [CBD (0.3–100 µM) combined with either TMZ (1 µM to 1 mM), BCNU (3 µM to 1 mM), or CDDP (0.1–100 µM)] and analyzed all possible combinations with the checkerboard method 1. On the basis of these results, we then calculated the percentage of occurrence of synergistic, additive, or antagonistic responses when evaluating all combinations. Figure 2D shows that the vast majority of CBD/DNA-damaging agent combinations resulted in additive antiproliferative responses in all cells. No significant difference in the frequency distribution of drug interactive effects was found among the three human GBM cell lines (P > 0.44) or between mouse PDGF-GBM cells and NPCs (P > 0.30) for each drug combination, suggesting similar responses in these groups of cells.
Four sets of interactions are worth emphasizing. First, low concentrations of CBD (1 µM) and TMZ (10 µM) exhibited a synergistic antiproliferative response in T98G cells (Fig. 2D). Second, synergistic responses occurred between low concentrations of CBD (0.3–10 µM) and BCNU (300 µM) in three GBM cells (T98G, U251, and mouse PDGF-GBM cells) but also in mouse NPCs (Fig. 2D). Third, cotreatment with CBD and BCNU led to all possible interactions (synergistic, additive, and antagonistic) depending on the concentrations in both mouse PDGF-GBM cells and NPCs (Fig. 2D). Forth, low concentrations of CBD (1–3 µM) and CDDP (100 µM) had antagonistic antiproliferative responses in all GBM cells, but not in mouse NPCs (Fig. 2D).
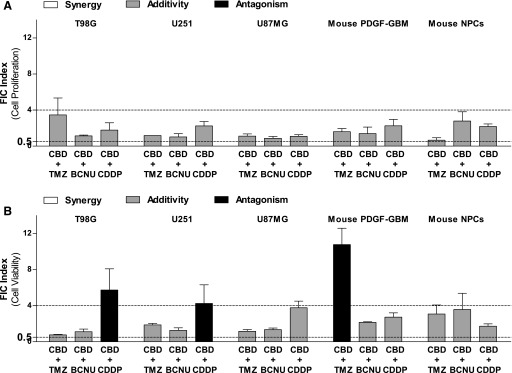
We then analyzed the interactions between CBD and DNA-damaging agents based on their efficacies by using checkerboard method 2 and calculated the FIC response of the combinations producing half maximal inhibition effects. Figure 4A and Table 1 show that all half maximal inhibitory combinations produced additive effects in all cells as indicated by their FIC indices, which were between 0.5 and 4. Thus, the combinations of CBD and DNA-damaging agents principally resulted in additive antiproliferative responses.
Fig. 4.
Efficacy FIC analyses on interactive responses between CBD and DNA-damaging agents on GBM cells and NPCs (method 2). (A and B) Efficacy FIC indices calculated from the combinations producing half maximal inhibitory effects on proliferation (A) and viability (B) when DNA-damaging agents were combined with CBD at a fixed concentration of 1 µM. Synergy (white) was defined as an FIC index (ΣFIC) < 0.5, additivity (gray) was defined as 0.5 < ΣFIC < 4, and antagonism (black) was defined as ΣFIC > 4. Data are expressed as means ± S.E.M. (n = 3 independent experiments, each in duplicate). Vehicle comprised 0.05% DMSO (vehicle in lieu of CBD) plus 0.05% DMSO, ethanol, or PBS (vehicle in lieu of TMZ, BCNU, or CDDP, respectively). DMSO, dimethylsulfoxide.
TABLE 1.
Quantitative efficacy FIC analyses of the interactive responses between CBD and DNA-damaging agents on cell proliferation of GBM cells and NPCs (method 2)
Data are expressed as means ± S.E.M. (n = 3 independent experiments, each in duplicate). CBD and IC50 values are given in molar concentrations (M).
| Cell | CBD | TMZ |
BCNU |
CDDP |
|||
|---|---|---|---|---|---|---|---|
| IC50 Value | FIC Index | IC50 Value | FIC Index | IC50 Value | FIC Index | ||
| Human cell lines | |||||||
| T98G | 1.00E-06 | 3.71E-03 ± 2.14E-03 | 3.46 ± 1.88 | 4.42E-04 ± 4.19E-05 | 1.13 ± 0.09 | 3.99E-05 ± 2.02E-05 | 1.78 ± 0.80 |
| 3.00E-06 | 1.25E-03 ± 2.25E-04 | 1.69 ± 0.20 | 3.61E-04 ± 1.08E-04 | 1.35 ± 0.23 | 3.74E-05 ± 1.10E-05 | 2.08 ± 0.44 | |
| U251 | 1.00E-06 | 1.01E-03 ± 4.08E-07 | 1.17 ± 0.00 | 1.84E-04 ± 7.38E-05 | 1.01 ± 0.36 | 1.43E-05 ± 3.35E-06 | 2.23 ± 0.49 |
| 3.00E-06 | 1.07E-03 ± 4.08E-06 | 1.47 ± 0.00 | 1.93E-04 ± 4.85E-05 | 1.29 ± 0.24 | 2.39E-05 ± 5.07E-06 | 3.89 ± 0.75 | |
| U87MG | 1.00E-06 | 5.73E-04 ± 1.53E-04 | 1.10 ± 0.25 | 4.65E-04 ± 1.38E-04 | 0.86 ± 0.21 | 2.19E-06 ± 4.26E-07 | 1.08 ± 0.18 |
| 3.00E-06 | 2.45E-04 ± 5.75E-05 | 0.86 ± 0.10 | 2.16E-04 ± 8.03E-05 | 0.78 ± 0.12 | 2.73E-06 ± 8.56E-07 | 1.62 ± 0.36 | |
| Mouse primary cells | |||||||
| PDGF-GBM | 1.00E-06 | 6.66E-04 ± 1.85E-04 | 1.59 ± 0.35 | 7.78E-05 ± 3.14E-05 | 1.40 ± 0.71 | 4.88E-06 ± 1.82E-06 | 2.24 ± 0.72 |
| 3.00E-06 | 1.44E-04 ± 1.39E-05 | 1.23 ± 0.03 | 6.22E-05 ± 1.65E-05 | 2.25 ± 0.34 | 2.85E-06 ± 1.73E-06 | 2.08 ± 0.68 | |
| NPCs | 1.00E-06 | 1.57E-03 ± 1.03E-03 | 0.66 ± 0.27 | 8.39E-05 ± 3.50E-05 | 2.78 ± 1.03 | 5.85E-06 ± 9.08E-07 | 2.17 ± 0.29 |
| 3.00E-06 | 1.09E-05 ± 5.39E-07 | 1.00 ± 0.00 | 5.43E-05 ± 5.33E-05 | 2.57 ± 1.56 | 1.98E-06 ± 1.01E-06 | 1.60 ± 0.32 | |
When DNA-damaging agents were combined with CBD at a fixed concentration of 1 or 3 µM, IC50 values of the DNA-damaging agents and the efficacy FIC indices were calculated from the combinations producing half maximal inhibitory effects on inhibiting cell proliferation of human GBM cell lines, primary mouse PDGF-GBM cells, and NPCs. Synergy was defined as ΣFIC < 0.5, additivity was defined as 0.5 < ΣFIC < 4, and antagonism was defined as ΣFIC > 4. The vehicle comprised 0.05% dimethylsulfoxide (vehicle in lieu of CBD) plus 0.05% dimethylsulfoxide, ethanol, or PBS (vehicle in lieu of TMZ, BCNU, or CDDP, respectively).
Interactions between CBD and DNA-Damaging Agents on Cell Viability
The interaction between CBD and DNA-damaging agents on cell viability was assessed using the same experimental design but measuring cell viability with WST-1. Figure 3D shows that the vast majority of combinations between CBD and DNA-damaging agents resulted in additive cell-killing activity in all cells. Similar to the effects on cell proliferation, no significant difference in the frequency distribution of drug interactive effects was observed among the three human GBM cell lines (P > 0.14) for each drug combination, suggesting an overall similar response in these cells. However, differences between mouse PDGF-GBM cells and NPCs were observed for the drug combinations of CBD/TMZ (P < 0.001) and CBD/CDDP (P < 0.05).
Five sets of interactions are worth noting. First, synergistic cell-killing responses occured when we combined low concentrations of CBD (1–10 µM) with either TMZ (30 µM) or BCNU (30 µM to 1 mM) in T98G cells, as well as with BCNU (10–100 µM) in U87MG cells (Fig. 3D). Second, combinations of CBD and BCNU at concentrations that produce synergistic antiproliferation responses of mouse PDGF-GBM cells and NPCs (Fig. 2D) resulted in merely additive reduction in cell viability (Fig. 3D). Third, combinations of CBD and BCNU produced antagonistic cell-killing responses in mouse PDGF-GBM cells and NPCs. Fourth, the majority of concentrations combining CBD and TMZ led to antagonistic cell-killing responses in mouse PDGF-GBM cells, but not in NPCs (P < 0.05). Fifth, combinations of CBD and CDDP antagonized their cell-killing activity in all GBM cell lines (Fig. 3D).
Analyses using method 2 indicated additive cell-killing responses for most combinations and clear antagonist killing responses between CBD/CDDP in T98G and U251 cells (Fig. 4B; Table 2). Analyses of the cell-killing activity of CBD and TMZ also showed profound antagonistic responses in mouse PDGF-GBM cells, consistent with the results obtained with method 1 (76.9%) (Fig. 4B; Table 2). These results extend the result obtained with method 1 by showing that a majority of cell-killing activity measured when combining CBD and DNA-damaging agents is additive, with several notable antagonist responses.
TABLE 2.
Quantitative efficacy FIC analyses of the interactive responses between CBD and DNA-damaging agents on cell viability of GBM cells and NPCs (method 2)
Data are expressed as means ± S.E.M. (n = 3 independent experiments, each in duplicate). CBD and IC50 values are given in molar concentrations (M).
| Cell | CBD | TMZ |
BCNU |
CDDP |
|||
|---|---|---|---|---|---|---|---|
| IC50 Value | FIC Index | IC50 Value | FIC Index | IC50 Value | FIC Index | ||
| Human cell lines | |||||||
| T98G | 1.00E-06 | 9.19E-04 ± 7.50E-05 | 0.76 ± 0.05 | 2.81E-04 ± 8.78E-05 | 1.10 ± 0.30 | 4.34E-05 ± 1.84E-05 | 5.73 ± 2.36 |
| 3.00E-06 | 6.83E-04 ± 1.55E-04 | 0.90 ± 0.10 | 1.94E-04 ± 1.08E-04 | 1.09 ± 0.37 | 3.92E-05 ± 2.12E-05 | 5.48 ± 2.73 | |
| U251 | 1.00E-06 | 7.78E-04 ± 8.00E-05 | 1.86 ± 0.18 | 1.66E-04 ± 4.25E-05 | 1.26 ± 0.29 | 4.42E-05 ± 2.22E-05 | 4.24 ± 2.08 |
| 3.00E-06 | 8.99E-04 ± 8.54E-07 | 2.34 ± 0.00 | 2.40E-04 ± 2.75E-05 | 1.99 ± 0.19 | 5.99E-05 ± 7.51E-06 | 5.93 ± 0.70 | |
| U87MG | 1.00E-06 | 9.17E-04 ± 1.79E-04 | 1.16 ± 0.19 | 5.86E-04 ± 8.58E-05 | 1.34 ± 0.17 | 2.48E-04 ± 5.15E-05 | 3.76 ± 0.74 |
| 3.00E-06 | 1.52E-03 ± 1.16E-03 | 2.19 ± 1.22 | 5.01E-04 ± 1.46E-04 | 1.57 ± 0.29 | 1.75E-04 ± 9.19E-06 | 3.11 ± 0.13 | |
| Mouse primary cells | |||||||
| PDGF-GBM | 1.00E-06 | 5.43E-04 ± 9.26E-05 | 10.78 ± 1.80* | 1.78E-05 ± 7.04E-07 | 2.14 ± 0.07 | 2.05E-06 ± 3.97E-07 | 2.72 ± 0.48 |
| 3.00E-06 | 1.20E-04 ± 6.86E-05 | 3.06 ± 1.33 | 1.47E-05 ± 4.44E-06 | 2.29 ± 0.47 | 3.14E-06 ± 5.68E-07 | 4.53 ± 0.69 | |
| NPCs | 1.00E-06 | 1.24E-04 ± 4.55E-05 | 3.07 ± 1.01 | 3.29E-05 ± 1.81E-05 | 3.58 ± 1.79 | 8.32E-07 ± 1.56E-07 | 1.72 ± 0.26 |
| 3.00E-06 | 4.99E-05 ± 4.84E-05 | 2.05 ± 1.08 | 8.71E-06 ± 4.49E-06 | 1.80 ± 0.45 | 6.00E-07 ± 2.74E-07 | 1.95 ± 0.46 | |
When DNA-damaging agents were combined with CBD at fixed concentration of 1 or 3 µM, IC50 values of the DNA-damaging agents and the efficacy FIC indices were calculated from the combinations producing half maximal inhibitory effects on inhibiting cell viability of human GBM cell lines, primary mouse PDGF-GBM cells, and NPCs. Synergy was defined as ΣFIC < 0.5, additivity was defined as 0.5 < ΣFIC < 4, and antagonism was defined as ΣFIC > 4. Vehicle comprised 0.05% dimethylsulfoxide (vehicle in lieu of CBD) plus 0.05% dimethylsulfoxide, ethanol, or PBS (vehicle in lieu of TMZ, BCNU, or CDDP, respectively).
P < 0.05, mouse PDGF-GBM cells versus mouse NPCs.
Discussion
We evaluated the antineoplastic activity of CBD alone and in combination with DNA-damaging agents in human GBM cell lines and mouse PDGF-GBM cells in culture, and we assessed the potential CNS toxicity of this treatment with mouse NPCs. We found that CBD both inhibits cell proliferation and kills all cells tested here with similar low micromolar potencies, suggesting that the molecular mechanism of action triggered by CBD that leads to the reduction of the proliferation and viability of all cells is not affected by their genetic makeup. Thus, it appears that CBD triggers a mechanism of action that does not preferentially target cancer cells and will likely be associated with NPC toxicity. Analyses of all dose responses triggered by CBD indicated a cooperative/allosteric mechanism of action. Together, these results suggest significant challenges when considering the poor therapeutic index and steep dose response of CBD used as a single agent to treat patients diagnosed with GBM.
Several studies have shown that the biologic activity of CBD is concentration dependent and likely mediated through multiple plasma membrane–associated receptors (Pertwee, 2008). At nanomolar concentrations, CBD binds to GPR55 as determined by in vitro radioligand binding assays (Ryberg et al., 2007). Accordingly, CBD modulates GPR55 activity as shown by 1–3 µM CBD that blocks the l-α-lysophosphatidylinositol–induced GPR55-mediated migration and polarization of breast cancer cells (Ford et al., 2010), as well as inhibits the proliferation of prostate cancer cells (Piñeiro et al., 2011). The pharmacological activity of CBD at GPR55 appears complex and likely cell dependent, as suggested by one study reporting that this compound promotes cell proliferation via the extracellular signal-regulated kinase pathway in glioma cells (Andradas et al., 2011). Cannabinoid receptors 1 and 2 (CB1 and CB2, respectively) are the key receptors responsible for the effects of cannabinoids (Mackie, 2006; Deng et al., 2015a,b). Previous studies show that low micromolar concentrations of CBD bind CB1 and CB2 receptors, as measured by radioligand binding assays (Petitet et al., 1998; Thomas et al., 2007; Pertwee, 2008). Accordingly, 1–10 µM CBD triggers apoptosis through CB2 receptors in leukemia cells (McKallip et al., 2006); 10 µM CBD suppresses glioma cell proliferation, and this response is partially blocked by a CB2 antagonist (Massi et al., 2004). Micromolar concentrations of CBD modulate the activity of several plasma membrane–associated ion channel receptors, including TRPV1 receptors with an EC50 of 3 µM (Bisogno et al., 2001; Iannotti et al., 2014), TRPV2 receptors when applied at 10–30 µM (Qin et al., 2008; Nabissi et al., 2013), and serotonin 1A (5-HT1A) receptors with an EC50 of 32 µM (Russo et al., 2005). CBD at 100 nM also modulates that activity of intracellular transcription factors such as peroxisome proliferator–activated receptor-γ (Esposito et al., 2011). It is important to emphasize that the lipophilic nature of CBD triggers biologic responses that are independent of protein-mediated mechanisms, including rapid changes in both membrane lipid raft and cholesterol metabolism when applied at 5–20 µM (Ligresti et al., 2006; Rimmerman et al., 2011, 2013). Additional examples of the protein-independent mechanism of CBD include increases in oxidative stress resulting in apoptosis, DNA damage, and autophagy in breast cancer cells (Shrivastava et al., 2011) and in glioma cells (Bisogno et al., 2001; Massi et al., 2004; Solinas et al., 2013; Soroceanu et al., 2013) when this compound is applied at 5–40 µM. Thus, there is a wide range of protein-dependent and protein-independent biologic activities induced by CBD applied in the micromolar range. Although the identification of the molecular target(s) and signaling step(s) that mediates the inhibition of cell proliferation and cell-killing activity of CBD reported here is beyond the scope of this work, the allosteric response of CBD that we measured in the 1- to 10-µM range in all cells suggests the involvement of a single class of proteins that mediate this activity.
Allosteric modulation plays an important role in the signal transduction mechanism of many proteins by favoring protein confirmations that enhance (i.e., positive allosteric modulator) or reduce (i.e., negative allosteric modulator) activities, and thus represents an active area of research for the development of new therapeutics (Changeux and Edelstein, 2005; Melancon et al., 2012). CBD at a concentration range of 0.1–5 µM is a negative allosteric modulator of CB1 receptors signaling when studied in human embryonic kidney 293 cells (Laprairie et al., 2015). CBD at 3– 30 µM affects TRPV1 and at 1–30 µM affects GPR55 activities (Bisogno et al., 2001; Ford et al., 2010; Piñeiro et al., 2011; Iannotti et al., 2014), both of which contain allosteric regulatory sites (Piñeiro et al., 2011; Anavi-Goffer et al., 2012; Cao et al., 2013; Maione et al., 2013). At 100 µM, CBD acts as a positive allosteric modulator of opioid μ receptors (Kathmann et al., 2006) but these high concentrations suggest that this target is unlikely to mediate the bioactivity of CBD. Thus, the 1–10 µM antineoplastic activity of CBD reported here could be mediated through at least three plasma membrane targets (GPR55, CB1, and TRPV1) that are known to both interact with CBD in this range of concentrations and have allosteric regulatory sites. Whether any of these targets mediate the response studied here still needs to be determined.
A common strategy to improve the therapeutic index of certain drugs is combined modality therapy, whereby treatment with reduced concentrations of two drugs known to be associated with side effects will result in enhancing overall therapeutic efficacy while reducing the incidence of side effects (Kummar et al., 2010). A recent study reported that 10 µM CBD exhibits synergistic GBM-killing activity when combined with either 400 µM TMZ or 200 µM BCNU and using U87MG cells as a model system (Nabissi et al., 2013). Although we reproduced this result, a more quantitative and unbiased analysis of the interaction between these modalities using two independent methods of calculations (method 1 analyzes all concentrations and method 2 analyzes the efficacy-related drug interactions) indicates that only a limited range of concentrations lead to synergistic responses. In fact, the predominant range of concentrations that we tested resulted in additive responses and a significant number of combinations resulted in antagonism. Thus, our analyses of the combined antineoplastic activity of CBD and DNA-damaging agents suggests little improvement in their respective therapeutic indices and, in some cases, loss in therapeutic efficacy by antagonism.
We identified several interactions between CBD and DNA-damaging agents occurring at select concentrations and in certain GBM cells that provide insights into the antineoplastic activity of these compounds. In mouse PDGF-GBM cells, we found that CBD/TMZ combinations antagonize their antiproliferative response while triggering an additive cell-killing response. This result suggests a clear dichotomy in the antiproliferative and cell-killing mechanism of action triggered when combining CBD and TMZ in mouse PDGF-GBM cells. In U87MG cells, combining CBD and DNA-damaging agents induced synergistic cell killing, but only additive inhibition of cell proliferation. This result suggests that the cell killing produced by these combinations does not rely on their ability to simply reduce cell proliferation and is likely independent. Thus, our results provide examples in which drug treatments exhibit dissociated and even opposite responses on cell proliferation and viability of GBM cells.
Several studies have shown that regimented treatment of glioma xenograft models with CBD significantly reduces tumor growth (Massi et al., 2004, 2008; Torres et al., 2011; Soroceanu et al., 2013). CBD is currently being tested in human clinical trials for the treatment of patients with GBM and the vast majority of this patient population will likely also be treated with standard-of-care DNA-damaging modalities (Saklani and Kutty, 2008). Our study provides a quantitative and unbiased evaluation of the antineoplastic activity of CBD alone and in combination with DNA-damaging agents in cell culture models of GBM, which suggests using caution when considering this phytocannabinoid for the treatment of patients diagnosed with GBM and treated with standard-of-care DNA-damaging agents.
Acknowledgments
The authors thank Dr. Eric C. Holland (University of Washington, Fred Hutchinson Cancer Research Center) for helpful discussions and expert input on the manuscript.
Abbreviations
- BCNU
carmustine
- BrdU
5-bromo-2′-deoxy-uridine
- CB1
cannabinoid receptor 1
- CB2
cannabinoid receptor 2
- CBD
cannabidiol
- CDDP
cisplatin
- CNS
central nervous system
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- FIC
fractional inhibitory concentration
- GBM
glioblastoma multiforme
- GPR
G protein–coupled receptor
- NPC
neural progenitor cell
- PBS
phosphate-buffered saline
- PDGF
platelet-derived growth factor
- TMZ
temozolomide
- TRPV
transient receptor potential cation channel subfamily V member
- WST-1
water-soluble tetrazolium salt 1
Authorship Contributions
Participated in research design: Deng, Stella.
Conducted experiments: Deng, Ng, Ozawa.
Performed data analysis: Deng.
Wrote or contributed to the writing of the manuscript: Deng, Ozawa, Stella.
Footnotes
This research was supported by the National Institutes of Health National Institute on Drug Abuse [Grant R01 DA026430].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, Bigner DD. (2009) Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs 18:1061–1083. [DOI] [PubMed] [Google Scholar]
- Anavi-Goffer S, Baillie G, Irving AJ, Gertsch J, Greig IR, Pertwee RG, Ross RA. (2012) Modulation of L-α-lysophosphatidylinositol/GPR55 mitogen-activated protein kinase (MAPK) signaling by cannabinoids. J Biol Chem 287:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andradas C, Caffarel MM, Pérez-Gómez E, Salazar M, Lorente M, Velasco G, Guzmán M, Sánchez C. (2011) The orphan G protein-coupled receptor GPR55 promotes cancer cell proliferation via ERK. Oncogene 30:245–252. [DOI] [PubMed] [Google Scholar]
- Bisogno T, Hanus L, De Petrocellis L, Tchilibon S, Ponde DE, Brandi I, Moriello AS, Davis JB, Mechoulam R, Di Marzo V. (2001) Molecular targets for cannabidiol and its synthetic analogues: effect on vanilloid VR1 receptors and on the cellular uptake and enzymatic hydrolysis of anandamide. Br J Pharmacol 134:845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonapace CR, Bosso JA, Friedrich LV, White RL. (2002) Comparison of methods of interpretation of checkerboard synergy testing. Diagn Microbiol Infect Dis 44:363–366. [DOI] [PubMed] [Google Scholar]
- Brem H, Piantadosi S, Burger PC, Walker M, Selker R, Vick NA, Black K, Sisti M, Brem S, Mohr G, et al. The Polymer-brain Tumor Treatment Group (1995) Placebo-controlled trial of safety and efficacy of intraoperative controlled delivery by biodegradable polymers of chemotherapy for recurrent gliomas. Lancet 345:1008–1012. [DOI] [PubMed] [Google Scholar]
- Cao E, Liao M, Cheng Y, Julius D. (2013) TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504:113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changeux JP, Edelstein SJ. (2005) Allosteric mechanisms of signal transduction. Science 308:1424–1428. [DOI] [PubMed] [Google Scholar]
- Deng L, Cornett BL, Mackie K, Hohmann AG. (2015a) CB1 knockout mice unveil sustained CB2-mediated antiallodynic effects of the mixed CB1/CB2 agonist CP55,940 in a mouse model of paclitaxel-induced neuropathic pain. Mol Pharmacol 88:64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Guindon J, Cornett BL, Makriyannis A, Mackie K, Hohmann AG. (2015b) Chronic cannabinoid receptor 2 activation reverses paclitaxel neuropathy without tolerance or cannabinoid receptor 1-dependent withdrawal. Biol Psychiatry 77:475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, et al. (2012) Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev 26:756–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Scuderi C, Valenza M, Togna GI, Latina V, De Filippis D, Cipriano M, Carratù MR, Iuvone T, Steardo L. (2011) Cannabidiol reduces Aβ-induced neuroinflammation and promotes hippocampal neurogenesis through PPARγ involvement. PLoS One 6:e28668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford LA, Roelofs AJ, Anavi-Goffer S, Mowat L, Simpson DG, Irving AJ, Rogers MJ, Rajnicek AM, Ross RA. (2010) A role for L-alpha-lysophosphatidylinositol and GPR55 in the modulation of migration, orientation and polarization of human breast cancer cells. Br J Pharmacol 160:762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday J, Helmy K, Pattwell SS, Pitter KL, LaPlant Q, Ozawa T, Holland EC. (2014) In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci USA 111:5248–5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannotti FA, Hill CL, Leo A, Alhusaini A, Soubrane C, Mazzarella E, Russo E, Whalley BJ, Di Marzo V, Stephens GJ. (2014) Nonpsychotropic plant cannabinoids, cannabidivarin (CBDV) and cannabidiol (CBD), activate and desensitize transient receptor potential vanilloid 1 (TRPV1) channels in vitro: potential for the treatment of neuronal hyperexcitability. ACS Chem Neurosci 5:1131–1141. [DOI] [PubMed] [Google Scholar]
- Kathmann M, Flau K, Redmer A, Tränkle C, Schlicker E. (2006) Cannabidiol is an allosteric modulator at mu- and delta-opioid receptors. Naunyn Schmiedebergs Arch Pharmacol 372:354–361. [DOI] [PubMed] [Google Scholar]
- Kummar S, Chen HX, Wright J, Holbeck S, Millin MD, Tomaszewski J, Zweibel J, Collins J, Doroshow JH. (2010) Utilizing targeted cancer therapeutic agents in combination: novel approaches and urgent requirements. Nat Rev Drug Discov 9:843–856. [DOI] [PubMed] [Google Scholar]
- Laprairie RB, Bagher AM, Kelly ME, Denovan-Wright EM. (2015) Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. Br J Pharmacol 172:4790–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder K, Pitter K, Laplant Q, Hambardzumyan D, Ross BD, Chan TA, Holland EC, Michor F. (2014) Mathematical modeling of PDGF-driven glioblastoma reveals optimized radiation dosing schedules. Cell 156:603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligresti A, Moriello AS, Starowicz K, Matias I, Pisanti S, De Petrocellis L, Laezza C, Portella G, Bifulco M, Di Marzo V. (2006) Antitumor activity of plant cannabinoids with emphasis on the effect of cannabidiol on human breast carcinoma. J Pharmacol Exp Ther 318:1375–1387. [DOI] [PubMed] [Google Scholar]
- Mackie K. (2006) Cannabinoid receptors as therapeutic targets. Annu Rev Pharmacol Toxicol 46:101–122. [DOI] [PubMed] [Google Scholar]
- Maione S, Costa B, Di Marzo V. (2013) Endocannabinoids: a unique opportunity to develop multitarget analgesics. Pain 154 (Suppl 1):S87–S93. [DOI] [PubMed] [Google Scholar]
- Marcu JP, Christian RT, Lau D, Zielinski AJ, Horowitz MP, Lee J, Pakdel A, Allison J, Limbad C, Moore DH, et al. (2010) Cannabidiol enhances the inhibitory effects of delta9-tetrahydrocannabinol on human glioblastoma cell proliferation and survival. Mol Cancer Ther 9:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massi P, Vaccani A, Bianchessi S, Costa B, Macchi P, Parolaro D. (2006) The non-psychoactive cannabidiol triggers caspase activation and oxidative stress in human glioma cells. Cell Mol Life Sci 63:2057–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massi P, Vaccani A, Ceruti S, Colombo A, Abbracchio MP, Parolaro D. (2004) Antitumor effects of cannabidiol, a nonpsychoactive cannabinoid, on human glioma cell lines. J Pharmacol Exp Ther 308:838–845. [DOI] [PubMed] [Google Scholar]
- Massi P, Valenti M, Vaccani A, Gasperi V, Perletti G, Marras E, Fezza F, Maccarrone M, Parolaro D. (2008) 5-Lipoxygenase and anandamide hydrolase (FAAH) mediate the antitumor activity of cannabidiol, a non-psychoactive cannabinoid. J Neurochem 104:1091–1100. [DOI] [PubMed] [Google Scholar]
- McKallip RJ, Jia W, Schlomer J, Warren JW, Nagarkatti PS, Nagarkatti M. (2006) Cannabidiol-induced apoptosis in human leukemia cells: a novel role of cannabidiol in the regulation of p22phox and Nox4 expression. Mol Pharmacol 70:897–908. [DOI] [PubMed] [Google Scholar]
- Melancon BJ, Hopkins CR, Wood MR, Emmitte KA, Niswender CM, Christopoulos A, Conn PJ, Lindsley CW. (2012) Allosteric modulation of seven transmembrane spanning receptors: theory, practice, and opportunities for central nervous system drug discovery. J Med Chem 55:1445–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabissi M, Morelli MB, Santoni M, Santoni G. (2013) Triggering of the TRPV2 channel by cannabidiol sensitizes glioblastoma cells to cytotoxic chemotherapeutic agents. Carcinogenesis 34:48–57. [DOI] [PubMed] [Google Scholar]
- Omuro A, DeAngelis LM. (2013) Glioblastoma and other malignant gliomas: a clinical review. JAMA 310:1842–1850. [DOI] [PubMed] [Google Scholar]
- Orhan G, Bayram A, Zer Y, Balci I. (2005) Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis. J Clin Microbiol 43:140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, Wolinsky Y, Kruchko C, Barnholtz-Sloan J. (2014) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro-oncol 16 (Suppl 4):iv1–iv63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, Charles N, Michor F, Holland EC. (2014) Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 26:288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee RG. (2008) The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: delta9-tetrahydrocannabinol, cannabidiol and delta9-tetrahydrocannabivarin. Br J Pharmacol 153:199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitet F, Jeantaud B, Reibaud M, Imperato A, Dubroeucq MC. (1998) Complex pharmacology of natural cannabinoids: evidence for partial agonist activity of delta9-tetrahydrocannabinol and antagonist activity of cannabidiol on rat brain cannabinoid receptors. Life Sci 63:PL1–PL6. [DOI] [PubMed] [Google Scholar]
- Piñeiro R, Maffucci T, Falasca M. (2011) The putative cannabinoid receptor GPR55 defines a novel autocrine loop in cancer cell proliferation. Oncogene 30:142–152. [DOI] [PubMed] [Google Scholar]
- Qin N, Neeper MP, Liu Y, Hutchinson TL, Lubin ML, Flores CM. (2008) TRPV2 is activated by cannabidiol and mediates CGRP release in cultured rat dorsal root ganglion neurons. J Neurosci 28:6231–6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramer R, Merkord J, Rohde H, Hinz B. (2010) Cannabidiol inhibits cancer cell invasion via upregulation of tissue inhibitor of matrix metalloproteinases-1. Biochem Pharmacol 79:955–966. [DOI] [PubMed] [Google Scholar]
- Rimmerman N, Ben-Hail D, Porat Z, Juknat A, Kozela E, Daniels MP, Connelly PS, Leishman E, Bradshaw HB, Shoshan-Barmatz V, et al. (2013) Direct modulation of the outer mitochondrial membrane channel, voltage-dependent anion channel 1 (VDAC1) by cannabidiol: a novel mechanism for cannabinoid-induced cell death. Cell Death Dis 4:e949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmerman N, Juknat A, Kozela E, Levy R, Bradshaw HB, Vogel Z. (2011) The non-psychoactive plant cannabinoid, cannabidiol affects cholesterol metabolism-related genes in microglial cells. Cell Mol Neurobiol 31:921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo EB, Burnett A, Hall B, Parker KK. (2005) Agonistic properties of cannabidiol at 5-HT1a receptors. Neurochem Res 30:1037–1043. [DOI] [PubMed] [Google Scholar]
- Ryberg E, Larsson N, Sjögren S, Hjorth S, Hermansson NO, Leonova J, Elebring T, Nilsson K, Drmota T, Greasley PJ. (2007) The orphan receptor GPR55 is a novel cannabinoid receptor. Br J Pharmacol 152:1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saklani A, Kutty SK. (2008) Plant-derived compounds in clinical trials. Drug Discov Today 13:161–171. [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Kuzontkoski PM, Groopman JE, Prasad A. (2011) Cannabidiol induces programmed cell death in breast cancer cells by coordinating the cross-talk between apoptosis and autophagy. Mol Cancer Ther 10:1161–1172. [DOI] [PubMed] [Google Scholar]
- Solinas M, Massi P, Cinquina V, Valenti M, Bolognini D, Gariboldi M, Monti E, Rubino T, Parolaro D. (2013) Cannabidiol, a non-psychoactive cannabinoid compound, inhibits proliferation and invasion in U87-MG and T98G glioma cells through a multitarget effect. PLoS One 8:e76918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroceanu L, Murase R, Limbad C, Singer E, Allison J, Adrados I, Kawamura R, Pakdel A, Fukuyo Y, Nguyen D, et al. (2013) Id-1 is a key transcriptional regulator of glioblastoma aggressiveness and a novel therapeutic target. Cancer Res 73:1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups. National Cancer Institute of Canada Clinical Trials Group (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996. [DOI] [PubMed] [Google Scholar]
- Thomas A, Baillie GL, Phillips AM, Razdan RK, Ross RA, Pertwee RG. (2007) Cannabidiol displays unexpectedly high potency as an antagonist of CB1 and CB2 receptor agonists in vitro. Br J Pharmacol 150:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres S, Lorente M, Rodríguez-Fornés F, Hernández-Tiedra S, Salazar M, García-Taboada E, Barcia J, Guzmán M, Velasco G. (2011) A combined preclinical therapy of cannabinoids and temozolomide against glioma. Mol Cancer Ther 10:90–103. [DOI] [PubMed] [Google Scholar]
- Vaccani A, Massi P, Colombo A, Rubino T, Parolaro D. (2005) Cannabidiol inhibits human glioma cell migration through a cannabinoid receptor-independent mechanism. Br J Pharmacol 144:1032–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Cancer Genome Atlas Research Network (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal M, Hilt DC, Bortey E, Delavault P, Olivares R, Warnke PC, Whittle IR, Jääskeläinen J, Ram Z. (2003) A phase 3 trial of local chemotherapy with biodegradable carmustine (BCNU) wafers (Gliadel wafers) in patients with primary malignant glioma. Neuro-oncol 5:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Ueda T, Shibata Y, Ikegami Y, Saito M, Ishida Y, Ugawa S, Kohri K, Shimada S. (2010) TRPV2 activation induces apoptotic cell death in human T24 bladder cancer cells: a potential therapeutic target for bladder cancer. Urology 76:509.e1–509.e7. [DOI] [PubMed] [Google Scholar]
- Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, Bronson RT, Chen JW, Weissleder R, Housman DE, et al. (2009) Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci USA 106:2712–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]