Abstract
We have shown that NADPH oxidase (NOX)5-S may mediate the acid-induced decrease in cell apoptosis. However, mechanisms of NOX5-S–dependent decrease in cell apoptosis are not fully understood. In this study, we found that silencer-of-death domain (SODD) was significantly increased in esophageal adenocarcinoma (EA) tissues, EA cell lines FLO and OE33, and a dysplastic cell line CP-B. Strong SODD immunostaining was significantly higher in low-grade dysplasia (66.7%), high-grade dysplasia (81.2%), and EA (71.2%) than in Barrett’s mucosa (10.5%). Acid treatment significantly increased SODD protein and mRNA expression and promoter activity in FLO cells, an increase that was significantly decreased by the knockdown of NOX5-S and nuclear factor κB (NF-κB)1 p50 with their small interfering RNAs. Similarly, acid-induced increase of SODD mRNA was blocked by knockdown of NOX5-S and p50 in a BE cell line CP-A. Overexpression of NOX5-S significantly increased SODD protein expression in FLO cells. Moreover, overexpression of NOX5-S or p50 significantly increased the SODD promoter activity and decreased the caspase 9 activity or apoptosis. NOX5-S overexpression-induced increase in SODD promoter activity was significantly decreased by knockdown of p50. In addition, acid treatment significantly decreased the caspase 9 activity, a decrease that was significantly inhibited by knockdown of SODD. Furthermore, chromatin immunoprecipitation assay showed that NF-κB1 p50 bound to SODD genomic DNA containing a NF-κB–binding element GGGGACACCCT. This binding element was further confirmed by a gel mobility shift assay. We conclude that acid-induced increase in SODD expression and decrease in cell apoptosis may depend on the activation of NOX5-S and NF-κB1 p50 in FLO cells.
Introduction
The major risk factor for esophageal adenocarcinoma (EA) is gastroesophageal reflux disease complicated by Barrett’s esophagus (BE) (Lagergren et al., 1999; Kahrilas, 2011; Pohl et al., 2013). The mechanisms of the progression from BE to EA are not fully understood. Acid reflux is thought to contribute to this progression (Fitzgerald et al., 1996; El-Serag et al., 2004; Zhang et al., 2009; Das et al., 2011; Kastelein et al., 2013).
NADPH oxidase (NOX)5-S has been shown by us to mediate acid-induced increase in H2O2 production and cell proliferation in EA cells (Fu et al., 2006; Si et al., 2007) because acid exposure–induced increase in H2O2 production and cell proliferation is blocked by knockdown of NOX5-S. The mechanisms of NOX5-S–dependent increase in cell proliferation and decrease in apoptosis are not fully understood. We have shown that cyclooxygenase 2 upregulation (Si et al., 2007) and p16 downregulation (Hong et al., 2010b) may partially mediate NOX5-S–dependent increase in cell proliferation.
Besides cyclooxygenase 2 and p16, other proteins may also be involved in NOX5-S–dependent increase in cell proliferation and decrease in apoptosis. Silencer-of-death domain (SODD) was identified in 1999 and belongs to a family of anti-apoptotic proteins of the BCL2 associated athanogene (BAG) family (Takayama et al., 1999). SODD contains a conserved region of approximately 45 amino acids near its C-terminal end, the so-called BAG domain. A potential role of SODD in carcinogenesis has been proposed based on increased mRNA levels in cancer tissues, for example, pancreatic cancers (Ozawa et al., 2000). The overexpression of SODD in various cancer cell lines is reported to suppress cell death (Jiang et al., 1999; Ozawa et al., 2000). We now show that acid increased SODD gene expression, which inhibited cell apoptosis in EA FLO cells. Acid-induced SODD expression may depend on activation of NOX5-S and nuclear factor κB (NF-κB)1 p50 in these cells.
Materials and Methods
Cell Culture and Acid Treatment.
Cell culture and acid treatment were similar to those we described previously (Hong et al., 2010a, 2013, 2016). Briefly, human esophageal squamous HET-1A cells were purchased from American Type Culture Collection (Manassas, VA) in 2011 and cultured in the bronchial epithelial cell medium (BEGM BulletKit; Cambrex, East Rutherford, NJ). Human Barrett’s cell line CP-A and Barrett’s dysplastic cell line CP-B were bought from American Type Culture Collection and cultured in wells precoated with collagen IV (1 μg/cm2; BD Biosciences, Bedford, MA) and in keratinocyte medium-2 (Ca2+-free solution; Cambrex, Rockland, ME) supplemented with 1.8 mM CaCl2, 5% fetal bovine serum, 400 ng/ml hydrocortisone, 20 ng/ml epidermal growth factor, 0.1 nM cholera toxin, 20 μg/ml adenine, 5 μg/ml insulin, 70 μg/ml bovine pituitary extract, and antibiotics.
Human Barrett’s adenocarcinoma cell line OE33 was bought from Sigma-Aldrich (St. Louis, MO) in 2012 and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum and antibiotics. Human Barrett’s adenocarcinoma cell line FLO was obtained in 2004 from D. Beer (University of Michigan Medical School, Ann Arbor, MI) (Hughes et al., 1997). FLO cells were cultured in DMEM containing 10% fetal bovine serum and antibiotics. All the cell lines were cultured at 37°C in a 5% CO2 humidified atmosphere.
For acid treatment, FLO cells were exposed to acidic DMEM (pH 4.0) or normal DMEM (pH 7.2, control) for 1 hour, washed, and cultured in fresh medium (pH 7.2, without phenol red) for an additional 24 hours. Finally, the culture medium and cells were collected for measurements. Acidic DMEM (pH 4.0, 300 μl) was added to each well in a 12-well plate, and the final pH was about 4.9 after a 1-hour incubation. CP-A cells were exposed to acidic culture medium (pH 6.5) or normal culture medium (pH 7.2, control) for 24 hours and then collected for measurements.
Human Esophageal Tissues.
Fresh normal esophageal mucosa and esophageal adenocarcinoma tissues were obtained from patients with esophageal adenocarcinoma undergoing esophagogastrectomy. Formalin-fixed and paraffin-embedded samples were collected between the years of 2011 and 2016 from the archives of the Department of Pathology at the Rhode Island Hospital. Twenty-one cases were included, as follows: 19 male patients and 2 female patients. The age ranged from 34 to 89 (average 63.8). The experimental protocols were approved by the Human Research Institutional Review Committee at Rhode Island Hospital. Informed consents were obtained from the participants.
Immunohistochemistry.
Immunohistochemistry for SODD was performed on 4-μm paraffin sections. Slides were stained with SODD antibody (1:200; Sigma-Aldrich) using the DAKO Envision + Dual Link System and the DAKO Liquid 3,3′-Diaminobenzidine Substrate Chromagen System (DAKO North America, Carpinteria, CA). Fallopian tubes were used as positive controls. Negative controls included replacement of the primary antibody with nonreacting antibodies of the same species.
Immunohistochemistry Assessment.
Cancers, dysplasia, and non-neoplastic mucosa that displayed a strong staining pattern for SODD were scored as 3+, moderately intense staining as 2+, and weak staining as 1+. The extent of staining (percentage of cell staining) was scored as follows: 1+, 1–30%; 2+, 31–60%; and 3+, 61–100%. A combined score of intensity and extent was calculated and categorized as follows: weak staining, 1–2; moderate staining, 3–4; and strong staining, 5–6.
Construction of pGL3-SODDP Reporter Plasmid.
The DNA fragment containing part of the promoter region (−988 to 19 from ATG) of SODD gene (GenBank accession number NM_004874.3) was amplified by polymerase chain reaction (PCR) from human genomic DNA. The primers used were as follows: SODDP sense, 5′-GGGGTACCTAGGTATTCCGATCCATCCAC-3′ (the introduced Kpn I is underlined) and SODDP antisense, 5′-GAAGATCTGAGCGCCTCAGGGCCGACATGG-3′ (the introduced BgLII is underlined). The obtained cDNA fragment was then cloned into pGL3-basic (Promega, Madison, WI ) between Kpn I and BgLII.
Small Interfering RNA and Plasmid Transfection.
The transfection protocol has been described previously by us (Hong et al., 2010a, 2013, 2016). Briefly, transfection of small interfering RNAs (siRNAs) was achieved by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). A total of 75 pmol NOX5-S siRNA (Ambion, Austin, TX), NF-κB p50 siRNA (Santa Cruz Biotechnologies, Santa Cruz, CA), or control siRNA formulated into liposomes was added to each well. Twenty-four hours after transfection, cells were treated with acidic culture medium, as described above.
The pCMV-tag5a-NOX5-S plasmid was obtained from D. Lambeth (Emory University School of Medicine, Atlanta, GA). Cells were transfected with 2 μg NOX5-S plasmid or control plasmid by using Amaxa-Nucleofector System (Lonza, Atlanta, GA).
Reverse-Transcription PCR.
Reverse-transcription PCR was performed, as we previously described (Hong et al., 2010a, 2013, 2016). Total RNA was extracted by TRIzol reagent, and 1.5 μg total RNA was reversely transcribed by using the kit SuperScript First-Strand Synthesis System for reverse-transcription PCR (Invitrogen).
Quantitative Real-Time PCR.
Real-time PCR was performed, as we previously described (Hong et al., 2010a, 2013, 2016), and carried out on a StepOne Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA). The primers used were as follows: SODD forward, 5′-GGGGTACCCAATGGTGCGATCTCGGCTCACTG-3′; SODD reverse, 5′-GAAGATCTCTCGAGGGGATCCGCTGCCCTGAAGCGCT-3′; 18S forward, 5′- CGGACAGGATTGACAGATTGATAGC-3′; and 18S reverse, 5′- TGCCAGAGTCTCGTTCGTTATCG -3′. PCRs were performed as follows: one cycle at 94°C for 5 minutes; 40 cycles at 94°C for 30 seconds; 59°C for 30 seconds; 72°C for 30 seconds; one cycle at 94°C for 1 minute; and one cycle at 55°C for 30 seconds. The transcript level of each specific gene was normalized to 18S amplification.
Western Blot Analysis.
Western blot analysis was performed, as described previously (Cao et al., 2003; Hong et al., 2010a, 2013, 2016). Primary antibodies used were as follows: SODD antibody (1:1000; Santa Cruz Biotechnologies) and glyceraldehyde-3-phosphate dehydrogenase antibody (1:2000; Santa Cruz Biotechnologies).
Luciferase Assay.
Luciferase assay was performed, as we previously described (Fu et al., 2006; Hong et al., 2013). Briefly, 0.1 μg renilla and 1.0 μg reporter plasmids pSODDP in combination with NOX5 siRNA, p50 siRNA, control siRNA, pcDNA3.1, or NOX5-S plasmid were transiently transfected in duplicate with Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were collected for the measurement of luciferase activity by using the Dual-Luciferase Reporter Assay System (Promega). For acid treatment, 12 hours after transfection FLO cells were treated with acidic DMEM (pH 4.0) for 1 hour, washed, and cultured in fresh medium (pH 7.2, without phenol red) for an additional 24 hours. Finally, the cells were collected for measurements. Luciferase activity was measured for 10 seconds after a 2-second delay using a BD Monolight 3010 luminometer (BD Biosciences, San Jose, CA). Variation in transfection efficiency was normalized by dividing the construct luciferase activity by the corresponding renilla luciferase activity.
Chromatin Immunoprecipitation Assay.
Chromatin immunoprecipitation (ChIP) assay was performed using the ChIP assay kit (Upstate, Charlottesville, VA) following manufacturer’s protocol, as we previously described (Fu et al., 2006; Hong et al., 2013). PCR was carried with the primer pairs that targeted the −1147 to −1019 region of the human SODD promoter (sense, 5′-CACTTCCTGTAACACGTGTGG-3′ and antisense, 5′-GAGCGCCTCAGGGCCGACATGG-3′) at 94°C for 5 minutes, 94°C for 30 seconds, 62°C for 30 seconds, and 72°C for 30 seconds for 35 cycles, followed by a 7-minute extension at 72°C.
Gel Mobility Shift Assay.
Gel shift assay was performed using the gel shift assay kit (LI-COR Biosciences, Lincoln, NE) following the manufacturer’s protocol, as we previously described (Fu et al., 2006; Hong et al., 2013). Briefly, human SODD promoter oligonucleotides (−310 to −300) were synthesized and labeled with IRDye 700 by Integrated DNA Technologies (Coralville, IA). A total of 1 μl 50 nM IRDye 700-labeled oligonucleotides were incubated with 5 μg HeLa nuclear extract or 0.25 μg recombinant NF-κB p50 protein (Cayman, Ann Arbor, MI). The wild-type competitor (−317 to −307) is 5′-CGTTGGGGGACACCCTTTCC-3′, and the mutant competitor (−317 to −307) is 5′-CGTTGGTTGACACTTTTTCC-3′. For a supershift assay, a NF-κB p50 antibody (1 μl; Upstate) was preincubated with the NF-κB p50 protein for 20 minutes at room temperature before addition of the IRDye700-labeled probes. The DNA–protein complexes were resolved by electrophoresis, and the gel was imaged immediately by LI-COR Odyssey imager system.
Enzyme-Linked Immunosorbent Apoptosis Detection Assay.
The levels of apoptosis are measured by using ApoStrand enzyme-linked immunosorbent assay apoptosis detection kit (Biomol, Plymouth Meeting, PA). This assay is based on the sensitivity of DNA in apoptotic cells to formamide denaturation, and the denatured DNA is detected with an antibody against single-stranded DNA. We have successfully used this assay in our published work (Fu et al., 2006).
Caspase-Glo 9 Assay.
pcDNA3.1, pCMV, p50, or NOX5-S plasmids were transiently transfected in duplicate with Lipofectamine 2000 (Invitrogen), according to manufacturer’s instruction. Twenty-four hours after transfection, caspase 9 activity was measured by using a Caspase-Glo 9 assay kit (Promega). For acid treatment, FLO cells were first transfected with SODD siRNA or control siRNA with Lipofectamine 2000. Twelve hours after transfection, FLO cells were treated with acidic DMEM (pH 4.0) for 1 hour, washed, and cultured in fresh medium (pH 7.2, without phenol red) for an additional 24 hours. Finally, the caspase 9 activity was measured.
The Caspase-Glo 9 assay is a homogeneous luminescent assay that measures caspase 9 activity. The assay provides a luminogenic substrate selective for caspase 9 in a buffer system optimized for caspase activity. The addition of a single Caspase-Glo 9 Reagent in an add-mix-measure format results in cell lysis, followed by caspase cleavage of the substrate, and generation of a glow-type luminescent signal. The signal generated is proportional to the amount of caspase 9 activity present.
Materials.
Other reagents were purchased from Sigma-Aldrich.
Statistical Analysis.
Data are expressed as mean ± S.E. Statistical differences between two groups were determined by Student t test. Differences among multiple groups were tested using analysis of variance and checked for significance using Fisher’s protected least significant difference test. For immunohistochemical data, statistical differences were determined by χ2 test.
Results
Expression of SODD in Different Esophageal Tissues and Cells.
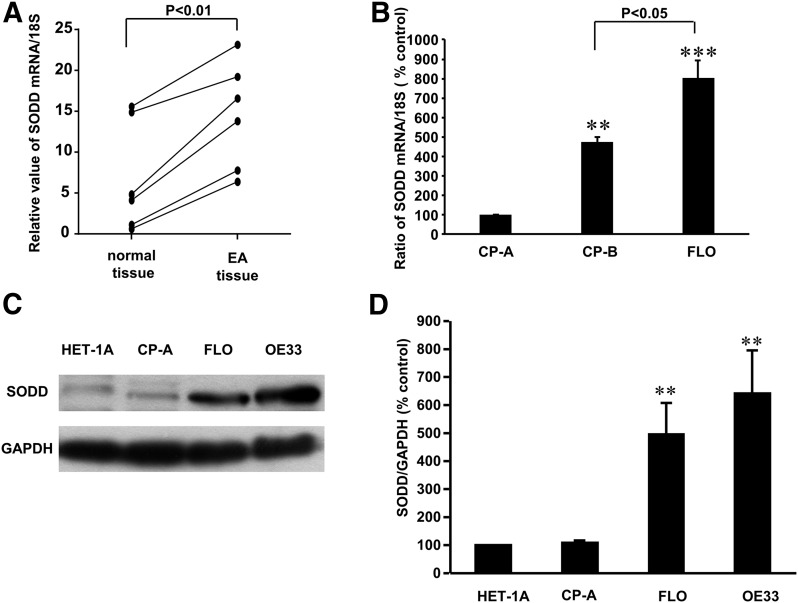
We found that SODD mRNA was significantly increased by 345.8% control in human EA tissue (Fig. 1A), when compared with normal mucosa. Real-time PCR showed that SODD mRNA was significantly higher in a dysplastic cell line CP-B and EA cells FLO than in CP-A cells (Fig. 1B). In addition, SODD mRNA was significantly increased in FLO cells, when compared with CP-B cells (Fig. 1B). Western blot analysis and summarized data showed that the protein levels of SODD were significantly higher in EA cell line FLO and OE33 than normal esophageal squamous epithelial cell line HET-1A or CP-A cells (Fig. 1, C and D). The data suggest that SODD may be important in the development of esophageal adenocarcinoma.
Fig. 1.
SODD expression in different cells and tissues. (A) Real-time reverse-transcription PCR showed that the levels of SODD mRNA were significantly increased in tumor tissues by 345.8% control, when compared with normal esophageal mucosa (N = 6, paired Student t test, P < 0.01). (B) Levels of SODD mRNA were significantly higher in CP-B and FLO cells than in CP-A cells and markedly higher in FLO cells than in CP-B cells. (C) A typical image of three Western blot analyses and (D) summarized data showed that the levels of SODD protein expression were significantly increased in FLO and OE33 EA cells (N = 3), when compared with normal squamous epithelial cells HET-1A and Barrett’s cells. The data suggest that SODD may be important in the development of EA. **P < 0.02, ***P < 0.001, compared with HET-1A cells or CP-A cells.
To further confirm these data, we performed immunohistochemical staining with SODD antibody. We found that strong SODD immunostaining was significantly higher in low-grade dysplasia (66.7%), high-grade dysplasia (81.2%), and esophageal adenocarcinoma (71.2%) than in Barrett’s mucosa (Fig. 2; Table 1). Although the strong immunostaining was slightly higher in high-grade dysplasia and EA than in low-grade dysplasia, the difference did not reach the statistical significance. The data suggest that SODD may be a potential marker for dysplasia and EA.
Fig. 2.
Representative images of BE mucosa, low-grade dysplasia, high-grade dysplasia, and EA with SODD immunostaining. Original magnification, 200×.
TABLE 1.
SODD expression in BE, low-grade dysplasia, high-grade dysplasia, and esophageal adenocarcinoma
| Negative | Mild | Moderate | Strong | |
|---|---|---|---|---|
| Barrett's cells (N = 19) | 1 (5.3%) | 2 (10.5%) | 14 (73.7%) | 2 (10.5%) |
| Low-grade dysplasia (N = 6) | 0 | 0 | 2 (33.3%) | 4 (66.7%)* |
| High-grade dysplasia (N = 16) | 0 | 0 | 3 (18.8%) | 13 (81.2%)* |
| EA (N = 7) | 0 | 0 | 2 (28.6%) | 5 (71.4%)* |
P < 0.01, compared with Barrett’s cells.
SODD Is Involved in Acid-Induced Decrease in Cell Apoptosis in EA Cells.
Because acid reflux may play an important role in the progression from BE to dysplasia and to adenocarcinoma (Fitzgerald et al., 1996; Ouatu-Lascar et al., 1999), we examined whether acid treatment affects levels of SODD protein. Acid treatment significantly increased SODD protein level in FLO cells (Fig. 3, A and B) and mRNA levels in CP-A cells (Fig. 3C). In addition, acid treatment significantly increased the luciferase activity in FLO cells transfected with SODD reporter plasmid pGL3-SODDP (Fig. 3D). The data suggest that acid treatment may increase SODD promoter activity and protein expression in FLO EA cells and mRNA expression in CP-A cells. Moreover, acid treatment significantly decreased caspase 9 activity in FLO EA cells (Fig. 3E), a decrease that was blocked by knockdown of SODD (Fig. 3F). At the basal condition, SODD siRNA significantly increased caspase 9 activity by 32.5% (N = 3, P < 0.02). Furthermore, acid-induced decrease in cell apoptosis was reversed by knockdown of SODD (Fig. 3G). These data indicate that SODD may contribute to acid-induced decrease in cell apoptosis in FLO cells.
Fig. 3.
The role of NOX5-S in acid-induced SODD expression. (A) A typical image of three Western blot analyses and (B) summarized data showed that acid treatment significantly increased SODD protein expression in FLO cells, an increase that was significantly decreased by knockdown of NOX5 with its siRNA (N = 3). (C) Acid treatment significantly increased SODD mRNA expression in a Barrett’s cell line CP-A cells, an increase that was significantly decreased by knockdown of NOX5 with its siRNA (N = 3). (D) Acid treatment remarkably increased the luciferase activity of SODD promoter in FLO cells transfected with SODD reporter plasmid pGL3-SODDP (N = 3), which was generated by ligating a SODD promoter fragment (−1008 to 20 from ATG) into the pGL3-basic vector, indicating that acid treatment may activate SODD promoter. Acid-induced increase in the luciferase activity was significantly decreased by knockdown of NOX5 (N = 3), suggesting that acid-induced activation of SODD promoter may depend on the activation of NOX5-S. (E) Acid treatment significantly decreased cell apoptosis in FLO EA cells (N = 3–5). (F) Acid-induced decrease in caspase 9 activity was significantly inhibited by knockdown of SODD, suggesting that SODD may mediate acid-induced decrease in cell apoptosis in FLO cells. (G) Acid-induced decrease in cell apoptosis was significantly inhibited by knockdown of SODD, suggesting that SODD may mediate acid-induced decrease in cell apoptosis in FLO cells. Percentage of decrease was calculated as follows: % decrease = (control-acid) × 100/control. Analysis of variance, *P < 0.02, ##P < 0.01, and ***P < 0.0001, compared with control siRNA group; **P < 0.01, compared with control siRNA + acid group; ▴▴P < 0.001, compared with control siRNA + acid group. t test, #P < 0.05; ▴P < 0.0001.
NOX5-S May Contribute to Acid-Induced SODD Expression in FLO Cells.
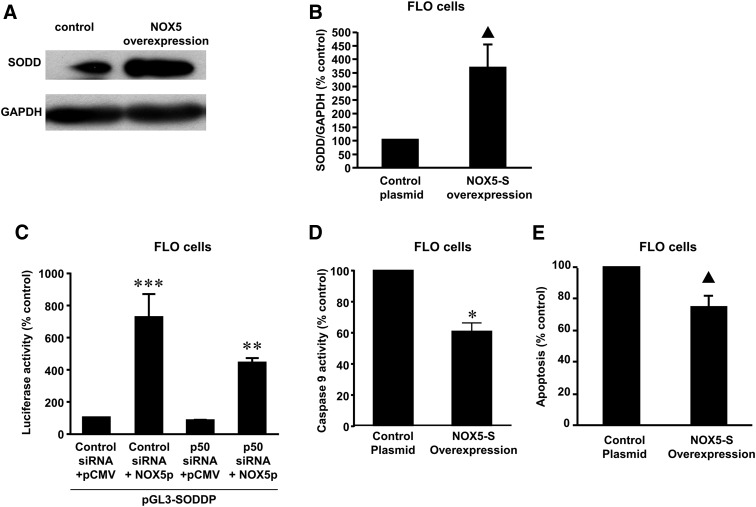
We have shown that NADPH oxidases may mediate acid-induced H2O2 production in Barrett’s mucosal biopsies (Fu et al., 2006) and that NADPH oxidase NOX5-S is the major isoform of NADPH oxidases in FLO EA cells (Hong et al., 2010a). Therefore, we examined whether NOX5-S participates in acid-induced increase in SODD expression. Western blot analysis and summarized data showed that knockdown of NOX5-S with its siRNA, which has been shown by us to effectively knock down NOX5-S expression (Fu et al., 2006), significantly decreased SODD expression in response to acid treatment in FLO cells (Fig. 3, A and B). Knockdown of NOX5-S also significantly decreased SODD mRNA expression in CP-A cells (Fig. 3C) and remarkably inhibited acid-induced increase in luciferase activity in FLO cells transfected with SODD reporter plasmid pGL3-SODDP (Fig. 3D). These data suggest that acid-induced SODD expression may be mediated by activation of NOX5-S. To further confirm this result, we transfected NOX5-S expression plasmid into FLO EA cells, which has been shown by us to overexpress NOX5-S protein (Si et al., 2007), and found that overexpression of NOX5-S significantly increased the protein expression of SODD (Fig. 4, A and B). Moreover, overexpression of NOX5-S dramatically increased the luciferase activity in FLO cells transfected with SODD reporter plasmid pGL3-SODDP (Fig. 4C) and significantly decreased caspase 9 activity (Fig. 4D) and cell apoptosis (Fig. 4E) at the basal condition in FLO EA cells. The data suggest that NOX5-S may be involved in SODD expression in FLO EA cells.
Fig. 4.
The role of NOX5-S in SODD expression. (A) A typical image of three Western blot analyses and (B) summarized data showed that overexpression of NOX5-S significantly increased SODD expression in FLO EA cells, indicating that NOX5-S may contribute to the SODD expression (N = 3). (C) Overexpression of NOX5-S by transfection of FLO cells with NOX5-S plasmid remarkably increased the luciferase activity of SODD promoter in FLO cells (N = 3), indicating that NOX5-S–derived ROS may activate SODD promoter. NOX5-S overexpression-induced increase in SODD promoter activity was significantly decreased by the knockdown of NF-κB1 p50 with its siRNA, suggesting that NOX5-S–dependent activation of SODD promoter may be mediated by NF-κB1 p50. (D) In FLO cells, overexpression of NOX5-S significantly decreased caspase 9 activity (N = 6), indicating that NOX5-S–derived ROS may inhibit cell apoptosis. (E) In FLO cells, overexpression of NOX5-S significantly decreased cell apoptosis (N = 3), indicating that NOX5-S–derived ROS may inhibit cell apoptosis. Analysis of variance, ***P < 0.0001, compared with control siRNA + pCMV group; **P < 0.01, compared with control siRNA + NOX5p group; t test, ▴P < 0.05; *P < 0.0001.
Role of NF-κB in Acid-Induced SODD Expression.
We have shown that NADPH oxidase NOX5-S mediates acid-induced cyclooxygenase-2 expression via activation of NF-κB in Barrett’s esophageal adenocarcinoma cells (Si et al., 2007). Next, we examined whether NF-κB participates in acid-induced SODD expression in FLO cells. We found that knockdown of p50 with its siRNA, which has been shown by us to effectively knock down p50 expression (Si et al., 2007), significantly decreased acid-induced increase in SODD protein expression in FLO cells (Fig. 5, A and B) and in SODD mRNA expression in CP-A cells (Fig. 5C). In addition, knockdown of p50 significantly decreased NOX5-S overexpression- or acid-induced increase in luciferase activity in FLO cells transfected with SODD reporter plasmid pGL3-SODDP (Fig. 4, C and D). Moreover, overexpression of NF-κB p50 significantly increased the luciferase activity in FLO cells transfected with SODD reporter plasmid pGL3-SODDP (Fig. 5E) and remarkably decreased the cell apoptosis in FLO cells (Fig. 5F). The data suggest that NF-κB p50 may be involved in acid-induced increase in SODD expression.
Fig. 5.
The role of NF-κB in acid-induced SODD expression. (A) A typical image of three Western blot analyses and (B) summarized data showed that knockdown of NF-κB1 p50 abolished acid-induced increase in SODD expression in FLO EA cells (N = 3), indicating that acid-induced expression of SODD may depend on activation of NF-κB in FLO EA cells. (C) Acid treatment significantly increased SODD mRNA expression in a Barrett’s cell line CP-A cells, an increase that was significantly decreased by knockdown of NF-κB1 p50 with its siRNA (N = 3). (D) Acid treatment remarkably increased the luciferase activity of SODD promoter in FLO cells transfected with SODD reporter plasmid pGL3-SODDP (N = 3), which was generated by ligating a SODD promoter fragment (−1008 to 20 from ATG) into the pGL3-basic vector, indicating that acid treatment may activate SODD promoter. Acid-induced increase in the luciferase activity was significantly decreased by knockdown of NF-κB1 p50 (N = 3), suggesting that acid-induced activation of SODD promoter may depend on the activation of NF-κB1 p50. (E) In FLO EA cells, overexpression of NF-κB1 p50 remarkably increased the luciferase activity in FLO cells transfected with SODD reporter plasmid pGL3-SODDP (N = 3), indicating that NF-κB1 p50 may activate SODD promoter. (F) In FLO cells, overexpression of NF-κB1 p50 significantly decreased cell apoptosis (N = 6), indicating that NF-κB1 p50 may inhibit cell apoptosis. Analysis of variance, *P < 0.05, ***P < 0.01, and ##P < 0.0001, compared with control siRNA group; **P < 0.01 and #P < 0.05, compared with control siRNA + acid group. t test, ▴P < 0.01, ▴▴P < 0.0001.
We identified one possible NF-κB–binding element, GGGACACCCT (positions −310 to −300), in the SODD promoter. To test whether NF-κB binds to SODD promoter, we performed a ChIP assay. Figure 6A showed that SODD genomic DNA was identifiable in the immunoprecipitate of FLO EA cell lysate with p50 antibody. The pair of primers used in the PCR targeted the −367 to +17 (position from ATG) region of the SODD promoter and covered the possible NF-κB binding site, as described above. The PCR products were sequenced and proved to be SODD genomic DNA. The data suggest that NF-κB may bind to the SODD promoter. A gel mobility shift assay was also done to confirm the above result. In the gel shift assay, we identified one prominent complex by using Hela nuclear extracts and IRdye 700-labeled SODD oligonucleotide, containing the NF-κB binding site GGGACACCCT (Fig. 6B). A high concentration of unlabeled SODD oligonucleotide significantly reduced the binding, whereas the mutant SODD oligonucleotide had less effect on the binding (Fig. 6B). In addition, one prominent complex was also identifiable with IRdye 700-labeled SODD oligonucleotide and recombinant p50 protein (Fig. 6C). The bands of these prominent complexes were supershifted with a p50 antibody (Fig. 6C). These data further suggest that NF-κB p50 may bind to the potential binding site GGGACACCCT.
Fig. 6.
(A) A typical example of three experiments showed that SODD DNA was detectable in the immunoprecipitated chromatin sample of FLO cells by using an antibody against NF-κB P50, suggesting that NF-κB binds to SODD promoter. Positive control, genomic DNA used as a positive control; rabbit IgG and c-Myc antibody were used as negative controls. (B) In gel mobility assay, a double-stranded IRdye 700-labeled oligonucleotide containing the sequence CGTTGGGGGACACCCTTTCC (SODD-pWT1) and HeLa nuclear extract provided by the kit were used. One prominent complex was detected (lane 2). Competition experiments with unlabeled DNMT1-pWT1 oligonucleotide significantly reduced binding (lane 3); however, the addition of the mutant oligonucleotide SODD-pMUT1 CGTTGGTTGACACTTTTTCC (lane 4) had no effect on binding. (C) In a supershift assay, one prominent complex was identifiable with IRdye 700-labeled SODD oligonucleotide and recombinant p50 protein. The bands of these prominent complexes were supershifted with a p50 antibody. These data suggest that NF-κB binds to the site GGGGACACCCT at the SODD promoter.
Discussion
Gastroesophageal reflux disease complicated by BE is a major risk factor for EA (Lagergren et al., 1999). However, mechanisms of the progression from BE (intestinal metaplasia) to EA are not fully understood. We have previously shown that acid, a major refluxate in patients with BE, increases reactive oxygen species (ROS) production in Barrett’s mucosal biopsies (Fu et al., 2006). This increase is blocked by NADPH oxidase inhibitor apocynin, suggesting that NADPH oxidases may mediate acid-induced increase in H2O2 production (Fu et al., 2006). NADPH oxidase has seven isoforms, NOX1–5, DUOX1, and DUOX2 (Suh et al., 1999; Banfi et al., 2000; Lambeth, 2004), whereas NOX5 has five isoforms, α, β, δ, and γ, and NOX5-S (Banfi et al., 2000; Vignais, 2002). NOX5 α, β, δ, and γ have helix-loop-helix motifs at its N-terminal (Banfi et al., 2000), whereas NOX5-S does not (Cheng et al., 2001). We have previously shown that acid-induced H2O2 production is mediated by the NADPH oxidase NOX5-S and that acid-induced NOX5-S expression depends on an increase in intracellular calcium and activation of cyclic AMP response element-binding protein (Fu et al., 2006). Overproduction of ROS derived from upregulation of NOX5-S increases cell proliferation and decreases apoptosis. However, the mechanism of acid-induced decrease in cell apoptosis is still not fully understood in EA cells.
In this study, we found that acid treatment remarkably increased SODD protein expression and promoter activity in FLO EA cells and SODD mRNA expression in CP-A cells, suggesting that acid treatment may upregulate SODD. Moreover, acid significantly decreased cell apoptosis, a decrease that was inhibited by knockdown of SODD, indicating that SODD may contribute to acid-induced decrease in cell apoptosis in FLO cells. It has been reported that pulsed acid treatment (pH 4) increased cell proliferation in mucosal tissues (Fitzgerald et al., 1996). We also found that pulsed acid treatment with pH 4.0 increased cell proliferation in FLO cells. Therefore, we used pH 4 in FLO cells. CP-A cells are less resistant to in vitro acid treatment. In our preliminary studies, we found that pH 4 and 5 caused cell apoptosis in CP-A cells, whereas pH 6.5 increased cell proliferation, as seen in FLO cells and in Barrett’s mucosal tissues. To mimic the findings obtained in mucosal tissues, which are much closer to the in vivo condition, we used pH 6.5 in CP-A cells.
SODD belongs to a family of anti-apoptotic proteins of the BAG family (Takayama et al., 1999) and contains a conserved region of approximately 45 amino acids near its C-terminal end, the so-called BAG domain. Overexpression of SODD has been reported to suppress cell death in various cancer cell lines (Jiang et al., 1999; Ozawa et al., 2000). SODD interacts with the ATPase domain of Hsc70/Hsp70 (Takayama et al., 1999) through its BAG domain and is associated with the cytoplasmic domain of the tumor necrosis factor receptor (TNFR)1 and death receptor-3, another member of the TNFR superfamily. Tumor necrosis factor treatment releases SODD from TNFR1, permitting the recruitment of proteins such as TNFR-associated death domain and TNFR-associated factor 2 to the active TNFR1 signaling complex (Jiang et al., 1999). Under unstimulated condition, SODD binds TNFR1 and prevents the ligand-independent oligomerization and spontaneous activation of TNFR1, thus inhibiting the cell apoptosis (Doong et al., 2002).
We also found that SODD expression was significantly increased in EA cells and tissues. By immunohistochemistry, strong SODD immunostaining was significantly higher in dysplasia and esophageal adenocarcinoma than in Barrett’s metaplastic cells, suggesting that SODD might be a potential marker for dysplasia and EA. Although the strong immunostaining was slightly higher in high-grade dysplasia and EA than in low-grade dysplasia, the difference did not reach the statistical significance, which might be due to our small sample size.
Acid-induced increase in SODD expression may depend on activation of NOX5-S because of the following: 1) knockdown of NOX5-S blocked acid-induced upregulation of SODD protein and mRNA; 2) knockdown of NOX5-S remarkably inhibited acid-induced increase in luciferase activity in FLO cells transfected with SODD reporter plasmid pGL3-SODDP; and 3) overexpression of NOX5-S significantly increased SODD promoter activity.
We have previously shown that NF-κB plays an important role in acid-induced cyclooxygenase-2 expression in Barrett’s esophageal adenocarcinoma cells (Si et al., 2007) and that acid treatment significantly decreased IκBα protein levels and increased luciferase activity in cells transfected with NF-κB reporter plasmid pNFκB-Luc, which contains five repeats of NF-κB–binding element GGGGACTTTCC in the enhancer element of the plasmid, suggesting that acid may activate NF-κB (Si et al., 2007). In addition, knockdown of NOX5-S significantly decreased acid-induced increase in luciferase activity in cells transfected with pNFκB-Luc, suggesting that activation of NF-κB may depend on activation of NOX5-S (Hong et al., 2013). Therefore, we examined the role of NF-κB in acid-induced SODD expression.
NF-κB has two isoforms, p50 and p52 (Karin et al., 2002). P50 is involved in lymphoid organogenesis and inflammation, whereas p52 mainly contributes to lymphoid organogenesis (Shih et al., 2011). Therefore, we focused on the role of p50 in acid-induced SODD expression.
We found that knockdown of p50 significantly decreased acid-induced increase in SODD protein expression in FLO cells and in SODD mRNA expression in CP-A cells. In addition, knockdown of p50 significantly decreased NOX5-S overexpression- or acid-induced increase in luciferase activity in FLO cells transfected with SODD reporter plasmid pGL3-SODDP. Moreover, overexpression of NF-κB p50 significantly increased SODD reporter activity and remarkably decreased the cell apoptosis in FLO cells. These data suggest that NF-κB1 p50 may be responsible for acid-induced SODD expression in EA cells.
One NF-κB–binding element GGGACACCCT (positions −310 to −300) was identified in the SODD promoter. Our data indicate that NF-κB may bind to this binding element because of the following: 1) SODD genomic DNA was identifiable in the immunoprecipitate of EA cell lysate with p50 antibody; 2) in the gel shift assay, IRdye 700-labeled SODD oligonucleotide containing the NF-κB binding site GGGACACCCT formed one prominent complex with Hela nuclear extracts or recombinant p50 protein; 3) a high concentration of unlabeled SODD oligonucleotide significantly decreased the binding, whereas the mutant SODD oligonucleotide had less effect on the binding; and 4) in the supershift assays, p50 antibody caused the supershift of the bands.
We conclude that acid-induced increase in SODD expression and decrease in cell apoptosis may depend on activation of NOX5-S and NF-κB1 p50. It is possible that acid reflux present in patients with BE may activate NOX5-S. High levels of ROS derived from NOX5-S may activate NF-κB1 p50 and upregulate SODD, which in turn decreases cell apoptosis, thereby contributing to the progression from BE to EA.
Abbreviations
- BAG
BCL2 associated athanogene
- BE
Barrett’s esophagus
- ChIP
chromatin immunoprecipitation
- DMEM
Dulbecco’s modified Eagle’s medium
- EA
esophageal adenocarcinoma
- NF-κB
nuclear factor κB
- NOX
NADPH oxidase
- PCR
polymerase chain reaction
- ROS
reactive oxygen species
- siRNA
small interfering RNA
- SODD
silencer of death domain
- TNFR
tumor necrosis factor receptor
Authorship Contributions
Participated in research design: Hong, Li, Cao.
Conducted experiments: Hong, Li.
Contributed new reagents or analytic tools: Cao.
Performed data analysis: Hong, Li, Cao.
Wrote or contributed to the writing of the manuscript: Hong, Cao.
Footnotes
This work was supported by National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01 DK080703].
References
- Bánfi B, Maturana A, Jaconi S, Arnaudeau S, Laforge T, Sinha B, Ligeti E, Demaurex N, Krause KH. (2000) A mammalian H+ channel generated through alternative splicing of the NADPH oxidase homolog NOH-1. Science 287:138–142. [DOI] [PubMed] [Google Scholar]
- Cao W, Sohn UD, Bitar KN, Behar J, Biancani P, Harnett KM. (2003) MAPK mediates PKC-dependent contraction of cat esophageal and lower esophageal sphincter circular smooth muscle. Am J Physiol Gastrointest Liver Physiol 285:G86–G95. [DOI] [PubMed] [Google Scholar]
- Cheng G, Cao Z, Xu X, van Meir EG, Lambeth JD. (2001) Homologs of gp91phox: cloning and tissue expression of Nox3, Nox4, and Nox5. Gene 269:131–140. [DOI] [PubMed] [Google Scholar]
- Das KM, Kong Y, Bajpai M, Kulkarni D, Geng X, Mishra P, Banerjee D, Hirshfield K. (2011) Transformation of benign Barrett’s epithelium by repeated acid and bile exposure over 65 weeks: a novel in vitro model. Int J Cancer 128:274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doong H, Vrailas A, Kohn EC. (2002) What’s in the ‘BAG’? A functional domain analysis of the BAG-family proteins. Cancer Lett 188:25–32. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Aguirre TV, Davis S, Kuebeler M, Bhattacharyya A, Sampliner RE. (2004) Proton pump inhibitors are associated with reduced incidence of dysplasia in Barrett’s esophagus. Am J Gastroenterol 99:1877–1883. [DOI] [PubMed] [Google Scholar]
- Fitzgerald RC, Omary MB, Triadafilopoulos G. (1996) Dynamic effects of acid on Barrett’s esophagus: an ex vivo proliferation and differentiation model. J Clin Invest 98:2120–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Beer DG, Behar J, Wands J, Lambeth D, Cao W. (2006) cAMP-response element-binding protein mediates acid-induced NADPH oxidase NOX5-S expression in Barrett esophageal adenocarcinoma cells. J Biol Chem 281:20368–20382. [DOI] [PubMed] [Google Scholar]
- Hong J, Behar J, Wands J, Resnick M, Wang LJ, DeLellis RA, Lambeth D, Souza RF, Spechler SJ, Cao W. (2010a) Role of a novel bile acid receptor TGR5 in the development of oesophageal adenocarcinoma. Gut 59:170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Li D, Cao W. (2016) Rho kinase ROCK2 mediates acid-induced NADPH oxidase NOX5-S expression in human esophageal adenocarcinoma cells. PLoS One 11:e0149735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Li D, Wands J, Souza R, Cao W. (2013) Role of NADPH oxidase NOX5-S, NF-κB, and DNMT1 in acid-induced p16 hypermethylation in Barrett’s cells. Am J Physiol Cell Physiol 305:C1069–C1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Resnick M, Behar J, Wang LJ, Wands J, DeLellis RA, Souza RF, Spechler SJ, Cao W. (2010b) Acid-induced p16 hypermethylation contributes to development of esophageal adenocarcinoma via activation of NADPH oxidase NOX5-S. Am J Physiol Gastrointest Liver Physiol 299:G697–G706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SJ, Nambu Y, Soldes OS, Hamstra D, Rehemtulla A, Iannettoni MD, Orringer MB, Beer DG. (1997) Fas/APO-1 (CD95) is not translocated to the cell membrane in esophageal adenocarcinoma. Cancer Res 57:5571–5578. [PubMed] [Google Scholar]
- Jiang Y, Woronicz JD, Liu W, Goeddel DV. (1999) Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science 283:543–546. [DOI] [PubMed] [Google Scholar]
- Kahrilas PJ. (2011) The problems with surveillance of Barrett’s esophagus. N Engl J Med 365:1437–1438. [DOI] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW. (2002) NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2:301–310. [DOI] [PubMed] [Google Scholar]
- Kastelein F, Spaander MC, Steyerberg EW, Biermann K, Valkhoff VE, Kuipers EJ, Bruno MJ, ProBar Study Group (2013) Proton pump inhibitors reduce the risk of neoplastic progression in patients with Barrett’s esophagus. Clin Gastroenterol Hepatol 11:382–388. [DOI] [PubMed] [Google Scholar]
- Lagergren J, Bergström R, Lindgren A, Nyrén O. (1999) Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N Engl J Med 340:825–831. [DOI] [PubMed] [Google Scholar]
- Lambeth JD. (2004) NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol 4:181–189. [DOI] [PubMed] [Google Scholar]
- Ouatu-Lascar R, Fitzgerald RC, Triadafilopoulos G. (1999) Differentiation and proliferation in Barrett’s esophagus and the effects of acid suppression. Gastroenterology 117:327–335. [DOI] [PubMed] [Google Scholar]
- Ozawa F, Friess H, Zimmermann A, Kleeff J, Büchler MW. (2000) Enhanced expression of silencer of death domains (SODD/BAG-4) in pancreatic cancer. Biochem Biophys Res Commun 271:409–413. [DOI] [PubMed] [Google Scholar]
- Pohl H, Wrobel K, Bojarski C, Voderholzer W, Sonnenberg A, Rösch T, Baumgart DC. (2013) Risk factors in the development of esophageal adenocarcinoma. Am J Gastroenterol 108:200–207. [DOI] [PubMed] [Google Scholar]
- Shih VF, Tsui R, Caldwell A, Hoffmann A. (2011) A single NFκB system for both canonical and non-canonical signaling. Cell Res 21:86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si J, Fu X, Behar J, Wands J, Beer DG, Souza RF, Spechler SJ, Lambeth D, Cao W. (2007) NADPH oxidase NOX5-S mediates acid-induced cyclooxygenase-2 expression via activation of NF-kappaB in Barrett’s esophageal adenocarcinoma cells. J Biol Chem 282:16244–16255. [DOI] [PubMed] [Google Scholar]
- Suh YA, Arnold RS, Lassegue B, Shi J, Xu X, Sorescu D, Chung AB, Griendling KK, Lambeth JD. (1999) Cell transformation by the superoxide-generating oxidase Mox1. Nature 401:79–82. [DOI] [PubMed] [Google Scholar]
- Takayama S, Xie Z, Reed JC. (1999) An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem 274:781–786. [DOI] [PubMed] [Google Scholar]
- Vignais PV. (2002) The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell Mol Life Sci 59:1428–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HY, Hormi-Carver K, Zhang X, Spechler SJ, Souza RF. (2009) In benign Barrett’s epithelial cells, acid exposure generates reactive oxygen species that cause DNA double-strand breaks. Cancer Res 69:9083–9089. [DOI] [PMC free article] [PubMed] [Google Scholar]