Abstract
Technologies such as genome sequencing, gene expression profiling, proteomic and metabolomic analyses, electronic medical records, and patient-reported health information have produced large amounts of data, from various populations, cell types, and disorders (big data). However, these data must be integrated and analyzed if they are to produce models or concepts about physiologic function or mechanisms of pathogenesis. Many of these data are available to the public, allowing researchers anywhere to search for markers of specific biologic processes or therapeutic targets for specific diseases or patient types.
We review recent advances in the fields of computational and systems biology, and highlight opportunities for researchers to use big data sets in the fields of gastroenterology and hepatology, to complement traditional means of diagnostic and therapeutic discovery.
Keywords: Big data, Translational bioinformatics, Drug repurposing, Precision medicine
Introduction
In 2003, the completion of the Human Genome Project—which culminated in the public release of the first sequenced and annotated genome derived from human DNA— was heralded as the dawning of the genomic era.1 Since that time, continued technological advances have enabled the rapid and cost-effective analysis of DNA, RNA, protein, and other biomolecules in large cohorts of patients. The integration of multiple types of omics* experiments (see Glossary for terms marked with *) across populations and conditions, made possible by the rapid accumulation of data generated using these technologies, has begun to yield clinically impactful discoveries by reanalysis of the deposited data.2 However, this surfeit of data has also made the analysis of omics studies an increasingly challenging task. Illustrating the scope of this problem, the European Bioinformatics Institute (EBI) reported in early 2016 housing 75 petabytes of publicly-accessible data3 (a quantity that would take more than nineteen years to download on an exceedingly fast 1 gigabit-per-second Internet connection), and between the two major public repositories of genomics data, ArrayExpress4 and the Gene Expression Omnibus5 (GEO), there are nearly two million samples currently available (for an overview of big data resources, see Table 1 and Supplementary Table 1). In addition to transcriptomic and genomic (i.e., DNA sequence/variants) datasets, additional types of omics data, assaying the proteome, metabolome, kinome, methylome, acetylome, lipidome, microbiome, phenome, exposome, meta-genome, and interactome, are increasingly being deposited for public use.
Table 1.
Selected publicly accessible big data analysis resources.
| Category | Name | Description | URL |
|---|---|---|---|
| Omics data repositories | Gene Expression Omnibus (GEO) |
Repository of raw and processed omics data. |
http://www.ncbi.nlm.nih.gov/geo/ |
| Sequence Read Archive (SRA) |
Repository of sequencing data. | http://www.ncbi.nlm.nih.gov/sra | |
| ArrayExpress | Repository of raw and processed omics data. |
https://www.ebi.ac.uk/arrayexpress/ | |
| The Cancer Genome Atlas |
Repository of genomic, proteomic, histologic, and clinical data for a wide variety of cancers. |
https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp | |
|
Functional perturbation data repository |
Connectivity Map (CMap) |
Database of drug perturbation gene-expression signatures.. |
https://www.broadinstitute.org/cmap/ |
| Library of Integrated Cellular Signatures (LINCS) |
Database of functional cellular responses to genetic and pharmacological perturbations measured in multiple types of biomolecules, e.g., transcriptome and kinome |
http://lincsportal.ccs.miami.edu/datasets/ | |
| ChemBank | Database/knowledgebase of high- throughput compound screens and other small molecule-related information. |
http://chembank.broadinstitute.org/ | |
|
Molecular pathway knowledgebase/analysis tool |
DAVID | Searchable/downloadable database of molecular pathway knowledgebase. |
https://david.ncifcrf.gov/ |
| NDEx | Biological network knowledgebase. |
http://www.home.ndexbio.org/ | |
| Molecular Signatures Database (MSigDb) |
Repository of molecular signatures from curated databases, publications, and research studies.. |
http://www.broadinstitute.org/msigdb | |
| Cheminformatics | DrugBank | Database of drug chemical, structural, pharmacologic, and target information. |
http://www.drugbank.ca |
| PubChem | Comprehensive database of structural, pharmacologic and biochemical activity data. |
https://pubchem.ncbi.nlm.nih.gov/ | |
| Protein Data Bank | Repository of protein structural data. |
http://www.wwpdb.org | |
| admetSAR | Web-tool predicting pharmacologic and toxicology parameters based on chemical structures. |
http://lmmd.ecust.edu.cn:8000/ | |
| The Drug Gene Interaction Database (DGIdb) |
Database of known drug-gene connections for selected genes. |
http://dgidb.genome.wustl.edu/ | |
| SIDER | Database of drug side effects. | http://sideeffects.embl.de/ | |
| Literature mining | PolySearch 2.0 | Web-based text mining tool. | http://polysearch.cs.ualberta.ca |
| Machine learning | Weka | Extensive library of machine learning algorithms with user- friendly interface. |
http://www.cs.waikato.ac.nz/ml/weka/ |
In parallel, the widespread adoption of electronic health records6 (EHR*) has also generated massive amounts of digitized personal health information, as has the increasing popularity of automatic serial data acquisition from wearable devices/technologies*7 and web applications that collect patient-reported health information* (e.g., the www.HepCure.org portal for hepatitis C patients and their physicians). Unprocessed clinical trial data will also soon become more widely accessible. Earlier this year, the International Committee of Medical Journal Editors issued a proposal that, if accepted, will require authors of clinical trials to make de-identified patient data publicly available after a 6 month embargo period, with the intention of increasing transparency and reproducibility of the trial results, and facilitating large-scale secondary analyses by external researchers.8 This and other open data initiatives—including the newly-launched Genomic Data Commons, which aims to serve as a hub for existing and future cancer research data9—will dramatically expand the public store of data available for both mining and integrated meta-analyses.
Developments like these have propelled biomedical research into the era of big data.* Given the hypothesis-free nature of data mining techniques, big data can be used to obtain a global perspective that complements the focused mechanistic studies typical of experimental biology, and enable the detection of high-level information patterns that would otherwise be impossible to perceive. Such approaches will help clarify the pathogenesis and proper classification of complex diseases, which typically involve a wide range of causal factors. For example, a recent integrated analysis of multiple datasets has defined three subtypes of type 2 diabetes that would not have been apparent based solely on clinical assessment.10
Additionally, the establishment of regional or national biobanks* (e.g., UK Biobank, www.ukbiobank.ac.uk) and large multicenter/national consortia (e.g., International Cancer Genome Consortium [ICGC], icgc.org) provide opportunities to more effectively integrate the breadth of human diversity (e.g., age group, sex, race/ethnicity, and environmental exposures) into biomarker and therapeutic discovery.
The potential value of big data in clinical medicine and basic science has been widely acknowledged.11 For instance, due to the practical limitations in designing and implementing randomized clinical trials (RCTs) to address many important clinical questions, the mining of retrospective EHR and other large-scale clinical outcomes data has been proposed as a supplement to RCTs in the generation of practice-guiding evidence.12 A restructured clinical taxonomy—one which moves away from the current organ/symptom-based classification system in favor of molecular descriptions of disease—has also been identified as a critical step toward precision medicine.*13 However, the current integration of molecular science into clinical medicine requires substantial progress before big data can make a meaningful impact on health care. The field of translational bioinformatics* has arisen over the past decade specifically to address this challenge, aiming to harness big data by developing statistical techniques and computational infrastructures capable of integrating and analyzing large, heterogeneous datasets, and ultimately deriving clinically-relevant insights that address unmet diagnostic and therapeutic needs across broad medical disciplines (see Figure 1 for an overview of the typical big data-driven workflow). Compared to a purely experimental approach, the incorporation of data mining and analytics tools into the biomedical pipeline is expected to shorten developmental timelines, reduce costs, and improve the success rate of candidate diagnostic and therapeutic tools (see Figure 2).
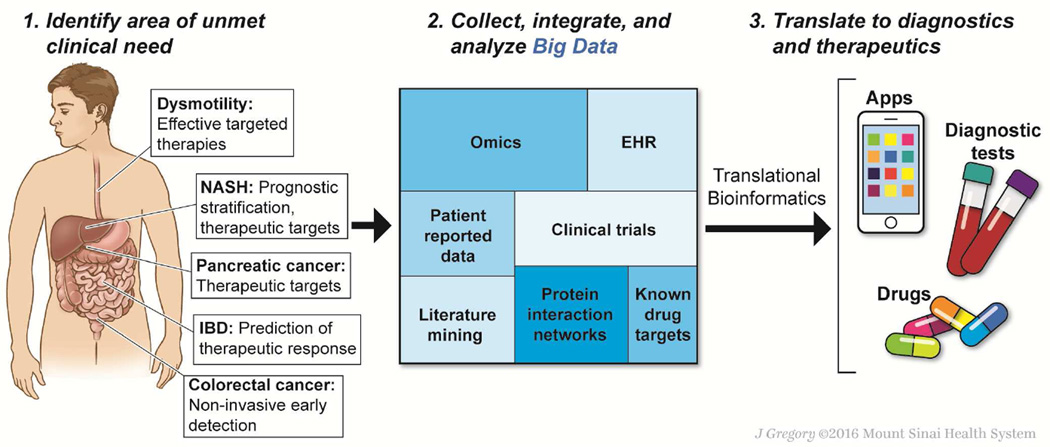
Figure 1. Big data-driven discovery in gastroenterology and hepatology.
Big data-driven discovery may provide new approaches to long-standing or emerging unmet needs in gastrointestinal and liver diseases (left panel). Multi-domain systematically and/or automatically collected data from patients and publicly or privately available databases are integrated into a highly rich and heterogeneous dataset (middle panel). Mining of the assembled big data by specialized methodologies (translational bioinformatics) more efficiently yields diagnostic devices, tools, and/or therapeutics (right panel).
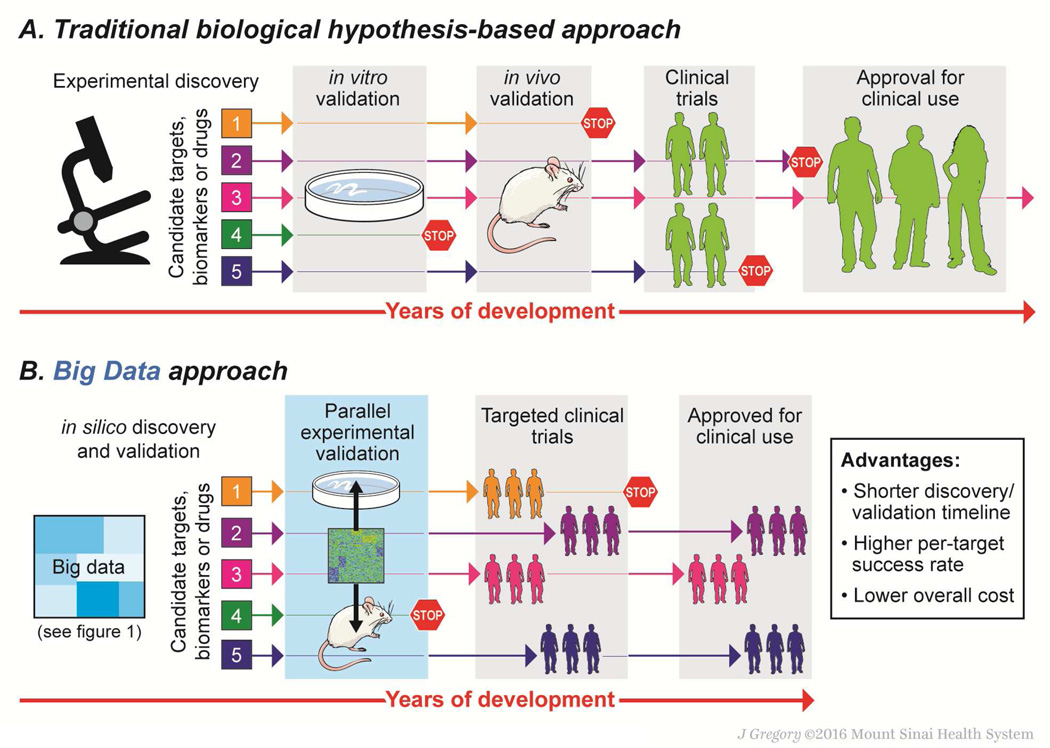
Figure 2. Advantages of the big data-driven approach.
In the traditional, biological hypothesis-driven approach for a specific disease (upper panel), candidate biomarkers and therapeutic targets go through lengthy and costly serial preclinical validations. Clinical evaluation is performed without incorporating genetic and environmental variations among enrolled patients, and a therapeutic benefit in a subset of patients can be missed. As a result, successful clinical translation suffers from lower efficiency and higher cost. In contrast, the big data-driven approach (lower panel) incorporates different data types, including both molecular and clinical information, and computationally derives candidate biomarkers and therapeutic targets/drugs without relying on any prior hypotheses. Subsequent preclinical and clinical validation can be simultaneously performed in parallel by incorporating computational cross-species analysis, thereby substantially reducing the required time and costs associated with biomarker/therapeutic development. Candidates may additionally be targeted to a specific niche patient subpopulation, further reducing the likelihood of translational failure.
Given these advantages, big data-based approaches are likely to have many productive applications within gastroenterology and hepatology, particularly for diseases in which diagnostic methods and/or treatments are imprecise. For example, despite several important therapies for inflammatory bowel diseases, due to our increased understanding of the associated immune dysregulation, its incidence is still increasing worldwide, and a significant proportion of patients do not achieve adequate remission of symptoms. Big data might also be analyzed in studies of gastrointestinal dysmotility, which is difficult to manage, or irritable bowel syndrome and other functional disorders, which are imperfectly understood and are difficult to diagnose and treat. There are also many sequence, gene expression, and proteomic and metabolomics data available on hepatic, colorectal, and gastric cancers, as well as pancreatic adenocarcinoma, that could be used to increase early detection or provide therapeutic targets
In liver, promising areas for data-driven discovery include viral hepatitis, where the complex interactions between viral heterogeneity, host genetic variations, and environmental factors in disease pathogenesis have not yet been satisfactorily integrated; liver cancer (hepatocellular carcinoma [HCC] and intrahepatic cholangiocarcinoma [ICC]), is increasing in incidence, yet therapies for HCC and ICC remain limited; progressive hepatic fibrosis, which likely shares core fibrosis pathways with other fibrotic diseases, yet for which there are no approved anti-fibrotic drugs; non-alcoholic fatty liver disease, in which recent epidemiological studies underscore the risk of developing liver cancer even in the absence of cirrhosis, emphasizing the need to identify biomarkers and targets for the “the next global liver disease epidemic”;15 and acute on chronic alcoholic hepatitis, where mortality is still extremely high (up to 50%) and treatment options are limited, and in which the precise identification of high-risk populations in need of therapy is still challenging.16
In this review, we will cover the currently available resources relevant to big data-driven research, and discuss future prospects for the integration of these resources within the fields of gastroenterology and hepatology.
Big data in diagnostic discovery
Biomarkers may be classified as diagnostic, prognostic, or therapeutic response-predictive depending on their intended use. Diagnostic biomarkers are used to determine the likelihood that a patient is suffering from a specific disease. Prognostic biomarkers inform physicians regarding the risk of clinical outcomes, such as cancer recurrence or disease development and progression, which may be used to assist patients and physicians in determining the appropriate aggressiveness of follow-up and/or care. Therapeutic response-predictive biomarkers are more specific because they are used to predict an individual’s response to specific treatments. Biomarker development follows the sequential processes of discovery, validation, and clinical implementation, with the eventual goal of establishing accessible tests that can be used to guide clinical decision making.17,18
Many of the candidate biomarkers reported recently have not been successfully translated into clinical practice, often because they did not pass the rigorous validation phase assessing technical/analytical validity (reproducibility and robustness of measurement) and clinical utility (replicated diagnostic, prognostic, or predictive capability in specific clinical contexts).17,19–21 Optimal study design is a key issue in maximizing the reliable discovery and successful validation of biomarkers. In many cases, sample availability may be limited, and both prospective enrollment and longitudinal follow-up studies to validate biomarkers over time can be costly and challenging to manage. In addition, cultural, environmental, and other variations across populations often necessitate large sample sizes to ensure generalizability, further complicating the design of appropriate studies.
With omics technologies, relatively rare genetic aberrations are increasingly identified as candidate predictive biomarkers of drug response, especially in the field of oncology.22 The therapeutic benefit of experimental therapies targeting such pathogenic aberrations often cannot be detected in the traditional “all comer” clinical trial design, which enrolls patients irrespective of the presence of these aberrations. Alternatively, new clinical trial designs, first stratifying the enrolled patients by molecular tests and then assigning a potentially effective therapy to each individual, have been evaluated (referred as “umbrella” or “basket” trial design).23 However, it is worth noting that performance of the biomarkers, e.g., positive/negative predictive value, should ideally be well defined prior to conducting biomarker-enriched clinical trials to ensure proper interpretation of a therapeutic benefit. The emergence of publicly available randomized controlled trial data (especially those that include -omic characterization of study participants) may allow post hoc assessment of predictive biomarkers before adopting them in prospective biomarker-enriched trials. Detailed molecular characterization of extreme responders has also been explored as an option of clinical biomarker-drug testing.24
Emerging public and private big data resources (including those listed in Table 1) will help overcome these challenges by enhancing the availability of data and/or samples. As the diversity and comprehensiveness of patient cohorts in these databases expands, ‘virtual’ patient enrollment will shorten the discovery process, improve reliability, and reduce costs of biomarker assessment in clinical trials through the incorporation of in silico validation* (Figure 3). The National Cancer Institute (NCI) has recommended improved sharing of existing specimens and data to create a NCI-wide inventory of specimens and cancer diagnosis data, and is funding pilot projects to support these efforts.25 The Institute of Medicine also encourages public sharing of clinical trial data while minimizing the risks and burdens of sharing.8,26 Although a number of ethical and legal challenges remain, these and related publicly–supported efforts to share data will accelerate biomarker development.
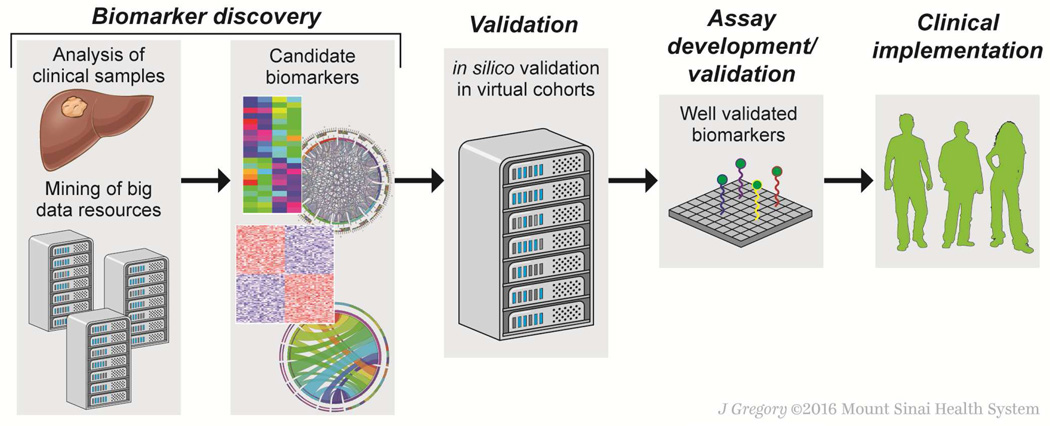
Figure 3. Big data-driven biomarker discovery.
Biomarker candidates may be identified from either analysis of newly-collected samples or in silico analysis of existing data from public and/or private big data repositories (left). Biomarker validation has traditionally been a costly process requiring assay development and prospective clinical evaluation with patients followed according to a strict protocol. By incorporating big data resources, in silico validation of a candidate biomarker can establish its clinical utility in multiple patient cohorts without conducting costly and lengthy prospective clinical trials. Only well-validated biomarkers are advanced to subsequent assay development and clinical evaluation with reduced risk of failure to demonstrate clinical utility (right).
Molecular diagnostics in gastroenterology
Within gastroenterology and hepatology, big data-driven approaches have identified several promising biomarkers (Table 2). Meta-analyses of publicly available genomic datasets have identified colorectal cancer diagnostic biomarkers,27 as well as molecular signatures that sub-classify pancreatic cancer,28 HCC,29 and colorectal cancer.30 An analysis of multiple transcriptomic profiles from cell types within liver has yielded a 122-gene signature that defines the presence hepatic stellate cells in fibrotic livers and correlates with clinical outcomes.31 Multi-cohort transcriptome analysis has also identified and validated a 186-gene hepatic signature predicting increased HCC risk and poorer prognosis in cirrhotic subjects.32–34 A reanalysis of 2,000 publicly available colorectal cancer transcriptomic profiles has led to identification of CDX2 as a prognostic biomarker predictive of disease-free survival, as well as predictive biomarker of response to adjuvant chemotherapy in stage II/III disease.35
Table 2.
Diagnostic strategies using publicly-available big data in liver disease - selected examples.
| Type of biomarker | Type of assay | Description | Clinical setting | Use of big data | Outcomes | Reference |
|---|---|---|---|---|---|---|
| Prognostic | Gene expression | 186-gene/32-gene prognostic liver signature |
HCV | Transcriptome datasets in public database |
Overall death, Progression to advanced cirrhosis, HCC |
33,34 |
| Hepatic injury and regeneration signature 65-gene signature |
HBV (89%) | Transcriptome datasets in public database |
HCC recurrence, | 130 | ||
| 27-gene activated hepatic stellate cell signature |
HBV (92%) | Transcriptome datasets in public database |
HCC recurrence and survival |
131 | ||
| 122-gene hepatic stellate cell signature |
HCV | Transcriptome datasets in public database |
Overall death, Decompensation, progression to advanced cirrhosis, HCC |
31 | ||
| SNP | 7-gene cirrhosis risk score |
HCV | Genome-wide SNP genotypes |
Progression to cirrhosis |
132 | |
| Disease classification | Gene expression | HCC subclasses | HCC from HBV, HCV, alcohol and cryptogenic |
Transcriptome datasets in public database |
3 HCC subclasses | 29 |
HCV, hepatitis C virus; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; SNP, single nucleotide polymorphism
Nevertheless, very few of the candidate biomarkers described in the literature have been implemented in clinical care because of several obstacles, including costly and lengthy assay development and prospective clinical utility validation, uncertain intellectual property regulations to protect omics/big data-driven biomarkers, and an unclear path toward regulatory approval and reimbursement.22 Instead, optimizing and validating biomarkers in silico using big data resources could reduce time and cost, and substantially lower the bar for clinical translation of molecular biomarkers.
Big data in therapeutic discovery
Integrating big data analytics and validating drugs in silico has the potential to improve the cost-effectiveness of the drug development pipeline. Here we review the two major drug discovery approaches—de novo development and drug repurposing—and the related computational techniques and resources that support them.
De novo drug discovery
Despite enormous investment in research and development (R&D) within the pharmaceutical industry, the rate at which new drugs are approved has not meaningfully increased over the past two decades.36 Further, the cost of developing a new drug remains high, ranging from $3 billion to more than $30 billion per approval between 2006 and 2014,37 reflecting the complex challenges involved in meeting current scientific, regulatory and commercial requirements. An over-reliance on in vitro high-throughput drug screening (HTS)* and the “one-drug-one-target-one-disease” concept is cited by some as a contributing factor in the abundance of late-stage R&D failures in recent years, many of which were the result of poor efficacy and unexpected toxicity of lead compounds developed using HTS technology.38,39 In contrast, certain experimental systems identify candidate drugs based on higher-level readouts of pharmacologic activity, in order to predict the effects of a compound in vivo.40,41 Phenotypic screens using animal or cell-based models of disease offer improved performance in this regard, but come with their own set of drawbacks, including relatively low throughput, high expense, mechanistic uncertainty, and limited coverage of the full spectrum of human disease.
Big data-driven strategies are being increasingly used to address these challenges. Computational prediction of drug toxicity and pharmacodynamic/pharmacokinetic properties, based on integration of multiple data types, helps prioritize compounds for in vivo and human testing, potentially reducing costs.42 In particular, computational exclusion of drugs that are likely to be toxic, prior to clinical assessment, will enhance patient safety while minimizing delays and expense, since drug toxicity is a major reason for failed clinical trials. For example, IL-17-targeting therapy, which has efficacy in rheumatic diseases, was found to be ineffective—and even harmful—in IBD, contrary to expectations based on the similar inflammatory features of these conditions. Global readouts of drug activity are expected to help clarify the causal relationships in such cases.
Similarly, chemical structure-based prediction of pharmacologic activity can identify more potent candidate compounds.43 Large-scale compound library screening datasets and cheminformatics* tools deposited in publicly available databases can enable in silico reanalysis for virtual drug exploration. The characterization of global transcriptional changes has been widely proposed as a universal readout to quantitatively assess disease states and drug responses. This approach allows drug-disease matching in a high-throughput, low-cost, and mechanistically revealing manner, while still providing the organism- or organ-system-level view of disease missing from target-driven studies.
Drug repurposing
A complementary approach to the discovery of new compounds is drug repurposing (also called drug repositioning), which entails the discovery of new indications for existing drugs. To date, successful drug repurposing has largely resulted from serendipity rather than systematic exploration.44 A classic example is sildenafil, which was repurposed from use in angina to erectile dysfunction based on an unexpected clinical effect. Similarly thalidomide, in spite of its well-known teratogenicity, was successfully repositioned as an effective treatment for multiple myeloma and leprosy.45 As public big data continues to accumulate, computational screening methods will foster a more systematic and comprehensive approach to drug repurposing (an example of the repurposing pipeline is outlined in Figure 4).
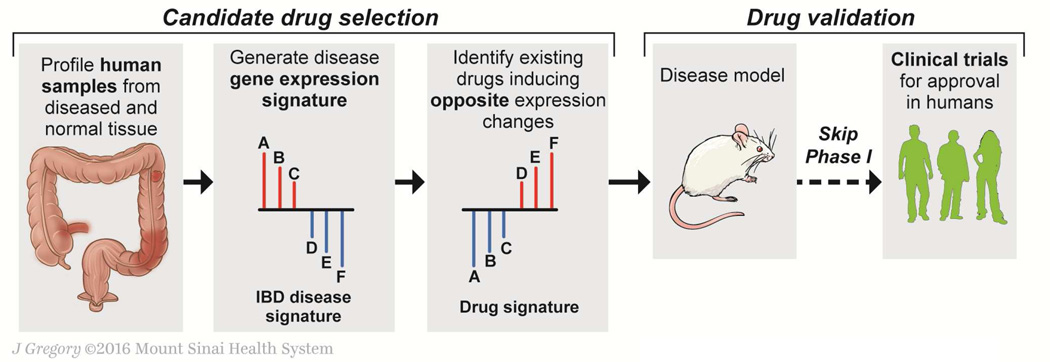
Figure 4. Big data-driven therapeutic discovery.
An example of the hypothesis-free, “signature inversion” therapeutic discovery approach for inflammatory bowel disease (IBD) is shown as an example of big data-driven drug discovery (e.g., Dudley et al55). A disease signature—a set of genes dysregulated in a coordinated manner in IBD patients—is first identified (left, genes A, B, and C are up-regulated, and genes D, E, and F are down-regulated). With the IBD disease signature, a database of drug perturbation gene signatures is queried to identify compounds that modulate the genes A-F in the opposite direction (i.e., suppress expression of genes A, B, and C, and induce expression of genes D, E, and F), and are thereby expected to antagonize the IBD disease signature. No mechanistic understanding of the associated gene dysregulation is needed for the computational compound identification. Subsequent experimental validation can confirm the predicted therapeutic effect and seek to uncover mechanism(s) of action before proceeding to further preclinical and clinical development (right). Because the screening is performed using data derived from approved drugs with known toxicity profiles, clinical testing can omit phase I and move immediately to phase II.
Conceptually, drug repurposing can be viewed as an optimization of the pharmacopoeia, aiming to maximize therapeutic efficiency within a fixed catalog of drugs and diseases.46 As such, repurposing has several attractive features as a complement to de novo drug development. First, the costs and time requirements associated with drug repurposing are greatly reduced,47 particularly for medications that have already been approved for clinical use in another indication or have cleared safety issues in phase I clinical trials.48 Additionally, in the proper clinical setting, off-label use prior to regulatory approval could further reveal the full clinical potential of repurposed compounds in a time- and cost-efficient manner. Second, reduced financial and regulatory barriers make drug repurposing an attractive option for rare and neglected diseases, which are generally less likely to be targeted by pharmaceutical companies due to lower profit potentials.49 The Orphan Drug Act, which incentivizes the development of drugs for rare diseases, has increased industry interest in this area, as has the recognition that so-called “niche busters” may mitigate the financial risk of pursuing large-market blockbusters,50 but there is still a significant unmet worldwide need. Drug repurposing may therefore serve a critical role in bring valuable treatments to underserved patients and populations. Third, the growing availability of publicly-accessible cheminformatics* data and advanced computational tools is allowing academic researchers to assist and even replace industry partners as the primary drivers of drug repurposing efforts.51 Finally, big data-based drug repurposing will be closely aligned with precision medicine, which has recently been established as a national priority.52 As stated earlier, increased characterization of the molecular mechanisms of disease has led to a rethinking of the traditional clinical taxonomy, moving from symptom-based descriptors to a molecular classification system.13 Omics-guided drug repurposing aims to discover molecular taxonomy-based therapeutic indications, which is an integral goal of precision medicine.
Big data-driven techniques for drug discovery
Integration of multi-layer omics information
The recent explosion of omics data has radically changed approaches to therapeutic discovery, particularly for drug repurposing. Cost-effective, high-throughput technologies can now characterize disease states at multiple levels to generate a multidimensional molecular “disease signature”.*44 Such a signature may include transcriptomic, proteomic or other changes as functional readouts of disease activity. In parallel, several search engines, most notably the Connectivity Map (CMap)53 and Library of Integrated Network-based Cellular Signatures (LINCS),54 have cataloged the effects of pharmacologic compounds on a variety of cell types. These databases may be queried in order to identify candidate compounds that are likely to either reverse a disease signature—a technique known as “signature inversion”44—or mimic desirable changes. Given that these databases contain data on many currently approved drugs, signature inversion studies can rapidly identify repurposing candidates while prioritizing widely available generic drugs. Several notable examples of this approach include the identification of topiramate as a potential treatment for inflammatory bowel disease,55 chlorpromazine, trifluoperazine56 and prenylamine57 to treat HCC, citalopram as a candidate therapy for colorectal cancer,58 and the HDAC inhibitor vorinostat for gastric cancer.59 Taking this concept further, Suthram et al derived a network of disease signatures mapped onto protein interaction data, and identified a subset of modules that were common to many diseases.60 Importantly, these shared modules were enriched with druggable proteins, confirming the potential of transcriptomic data to identify therapeutically relevant targets.
Genome Wide Association Studies (GWAS)—linking germline DNA polymorphisms with clinical phenotypes—as well as expression quantitative trait locus (eQTL) analyses that integrate gene expression data, have enabled identification of numerous disease susceptibility genes and more efficient discovery of therapeutic strategies, without relying on a priori biological hypotheses. These advances have been a driving force in the creation of large patient cohorts accompanied by archived biospecimens and omics databases.61 Similarly, the study of somatic DNA structural and/or chemical alterations has greatly accelerated cancer drug discovery and development.62
It should be noted that there are a number of challenges when analyzing omics data, including high dimensionality (i.e., many more variables than samples, which can lead to the emergence of spurious associations),63 marked heterogeneity in data attributes (e.g., diversity in assay platform, experimental conditions, analytical methodologies, etc.),64 and an imperfect concordance between different types of biomolecules (e.g., mRNA transcripts and the corresponding translated proteins).65 Other types of data, such as chemical modifications, enzymatic activities, and genotypes, can also be integrated as additional inputs for a multilayered omics characterization,66–68 adding valuable information but also increasing the complexity of analysis.
Similarity assessment across distinct data types/domains
Omics-based big data may also be used to link drugs and indications by making novel connections between distinct data types and domains (the “guilt-by-association” approach).44,69 These connections can be made in a number of different ways (e.g., common molecular dysregulations in disease states, shared indications between apparently unrelated compounds,70 and drug side-effect overlap71), based on the assumption that one type of similarity implies another. Unexpected drug side effects— which may or may not be undesirable, depending on the clinical context—provide a rich source of functional drug information, both as a means of discovering related groups of compounds through their shared effect profiles,72 or by directly leading to the identification of repurposing applications (for example, a drug with a side-effect of urinary retention might be rationally repurposed to the treatment of urge incontinence).73 To facilitate side-effects-driven analyses, Kuhn et al compiled a database of known drug-side-effect associations in an easily mineable format, including side-effect frequencies for many drugs.74 Additionally, several user-friendly tools have been developed for the quantification of disease and drug similarity (see Table 1, Supplemental Table 1, and 75). There have also been recent attempts to integrate multiple types of disease-disease and drug-drug similarity within a single analytic pipeline for drug discovery.76–80
Network biology
Human diseases are generally the result of complex interactions between a variety of biomolecules within a multi-scale biological system, ultimately conspiring to bring about the organism-level phenotypes that are observed clinically.81 Network biology/medicine*, which conceptually represents different aspects of the cellular environment as nodes, connected by edges and spatially interrelated into modules, attempts to capture the full complexity of biological interactions on the system level. Compared to a reductionist approach, network medicine is thought to better recapitulate the fundamental biological processes and machinery—rather than their components— which bring about complex disease states. A central implication of the network hypothesis is that many diseases with multi-level causal event interactions will be more effectively treated by promiscuous (i.e., multitarget) drugs than by highly-selective individual compounds, since multiple targets are engaged to ameliorate the disease phenotype.82
In addition to providing an intuitive means of representing the interactions that drive cellular biology, however, networks also enable the use of sophisticated computational comparisons, a reflection of the field’s mathematical roots as a derivative of graph theory. For instance, a curated network of drugs, proteins and side-effects may be used to visually explore the topological connections between compounds, which would otherwise be difficult to chemically relate.83 Similarly, networks have been used as a means of relating drugs and disease through common protein interactions.84
Omics data may also be integrated into networks. For example, Iorio et al generated a drug network by iteratively refining relationships between drug-induced transcriptional signatures, ultimately discovering “drug neighborhoods” which implied shared mechanisms and indications for member compounds.85 Networks additionally provide a convenient means of combining disparate data types into a unified analysis. In a recent study, Menche et al. attempted to relate diseases by topological network parameters using an integrated “interactome”, which they compiled from all known intracellular interactions. The authors considered diseases to be related if they were relatively adjacent on the network, and were able to demonstrate significant associations between this network proximity and externally-derived disease features, including gene expression and symptomatology.86
Literature mining
There is a vast amount of knowledge contained in the published literature, far more than could be assimilated by a single investigator through traditional means. Further, research often congregates in silos of specialized knowledge, limiting the dissemination of concepts between disparate areas of investigation. Literature mining,* which originally developed from Swanson’s ABC model (i.e., if A is connected to B, and B is connected to C, there is a further implied connection between A and C87), now aims to extract information from highly diverse semantic contexts through the use of natural language processing algorithms, and has recently been adapted specifically for drug discovery and repurposing. For example, an analogy-based literature mining approach was able to successfully predict the in vitro activity of nearly one-third of a small molecule library against prostate cancer cells, with the added advantage of uncovering plausible mechanisms of action.88 Similarly, an automated reasoning algorithm connected drug-target information obtained through database and literature mining with cancer target information, showing a significant ability to recover known drug-cancer connections.89 Further, literature mining can be used as a means of adding information content to separate analytic pipelines. For example, Gramatica et al used a combination of literature mining and graph theory to construct a network-based model capable of identifying non-obvious connections between drugs and diseases based on topological parameters.90 User-friendly literature mining tools have been developed; for instance, PolySearch is a web-based text mining application allowing users to quickly identify connections between a variety of biological entities, including drugs and diseases, using information drawn from a variety of literature sources and curated databases.91
Drug target analysis
The virtual screening of compounds based on molecular structure has advanced significantly over the past two decades. This approach seeks to predict likely interactions between drugs and target proteins, based on their respective structures. In silico structural screening can be valuable for both de novo drug design, by enabling promising compounds to undergo an initial selection process prior to experimental screening and validation, and for drug repurposing, being far more cost-effective than experimental HTS methods.92 Additionally, virtual screening can be conducted in either “forward docking” (screening a protein target against a library of compounds) or “reverse docking” (in which individual compounds are screened against a library of protein targets) formats, facilitating both disease- and drug-specific discovery.92,93 Several web-tools have been implemented to enable the prediction of connections between drugs and target proteins using forward/reverse docking (or the related concept of pharmacophore* mapping) for user-input source compounds.94–97 For instance, an online tool specifically designed for repurposing studies has used virtual docking to compare input compounds against a library of molecules with known indications and side-effects, helping to predict the potential uses and adverse effects for compounds of interest.98 An alternative approach, comparing compound structures against sets of ligands known to bind a variety of target proteins, has been used to predict several novel drug-protein connections.99 A related structural-similarity-based screening tool, TargetHunter, was implemented to enable the identification of multiple targets for a given compound of interest.100
Quantitative structure-activity relationship (QSAR) algorithms attempt to predict the therapeutic, toxic, and pharmacologic activities of compounds by inferring likely physicochemical properties from the compounds’ molecular structures.101 A user-friendly machine learning platform, AutoWeka, has been developed to aid in the implementation of QSAR studies.102 Other web-based tools are available for the prediction of pharmacodynamics/pharmacokinetic and toxicity profiles of compounds based on structural input data.103,104 Recently, several QSAR-based methods have been used in the discovery of novel agents for the treatment of IBD,105 uncovering compounds capable of inhibiting NF-κB106 and TNF-α converting enzyme,107 two molecules implicated in disease pathogenesis.
Drug combinations
With the enhanced ability to predict systems-level disease pathogenesis and drug effects, the rational design of combination therapies has become feasible. There are several potential advantages of drug combinations: better coverage of multiple disease mechanisms than with a single agent;108 dose reduction for a potentially toxic component of the combination while maintaining therapeutic efficacy;109 synergistic effects, including synthetic lethality in cancer110; and the prevention of innate and acquired drug resistance.111 Efficient, high-throughput identification of effective drug combinations is therefore an important component of a successful therapeutic discovery pipeline. To this end, a number of methods for computational prediction of synergistic compounds have been developed,112 based on side-effect profiles,108 chemical and pathway data,113 network analyses,114 and drug-induced gene expression patterns.109 Curated databases of reported drug combinations and other resources are also publicly available.115
Statistical methods, computational infrastructure
Machine learning*, an automated form of data modeling and inference, has become a dominant tool in computational drug discovery, particularly in the implementation of QSAR studies.116 Machine learning algorithms are generally categorized either as supervised (i.e., guided by external information) or unsupervised (i.e., exploring inherent data structure), with each individual algorithm within these categories having specific advantages and disadvantages.117 Traditional machine learning algorithms, such as support vector machine (SVM), have been successfully utilized in drug development analysis by integrating drug and protein structures, disease states, and drug toxicity for repurposing and sensitivity prediction.118 More recently, deep learning, which uses multi-layer artificial neural networks to extract meaning from data, has shown promise because of its robustness when working with complex, heterogeneous datasets.119 For example, a recent deep neural network analysis was able to categorize drugs into therapeutic categories using pathway-enriched transcriptional signatures, with improved predictive performance compared to SVM.120 Another study used deep learning to predict drug toxicity by correlating molecular structure to the risk of drug-induced liver injury.121 The simultaneous use of multiple machine learning algorithms (a technique known as ensemble learning) has been shown to combine the advantages of individual algorithms while minimizing their weaknesses, although the improved performance of ensembles often comes at the expense of computing time and poor interpretability.122 An ensemble learning approach recently proved effective in the prediction of drug sensitivity within a variety of human cancer cell lines.123
Although machine learning is showing great promise within drug discovery pipelines, the vast size and complexity of future big data could easily exceed the capability of currently available computational infrastructure. To address this challenge, several statistical measures that require less computational expense, such as the maximal information coefficient (MIC), have been developed to promote the efficient handling of big data.124 In parallel, development of computational infrastructure that can be rapidly expanded, such as the Hadoop file system,125 crowdsourcing,126 and massively parallel processing hardware (including the recruitment of graphical processing units127), are being actively explored and adopted.
Challenges
There are several challenges involved in the integration of a big data-driven pipeline for biomarker and drug discovery within gastroenterology and hepatology. As previously mentioned, the statistical complexity involved in the analysis of large, heterogeneous datasets is a major stumbling-block in the successful generation of data-driven discoveries.128 Adding layers of omics information may facilitate the identification of better molecular correlates, but potentially at the expense of larger required sample sizes to achieve proper statistical power. This in turn depends on the strength of association between molecular dysregulations and the phenotype of interest. In addition, significance assessment can be a challenging task when analyzing too many features. Thus, it is critical to: perform proper sample size calculation to ensure sufficient statistical power; correct for multiple hypotheses when estimating statistical significance; and reduce the dimensionality of molecular datasets by filtering out less informative features, extraction of representative information from multiple features (e.g., principal components), and/or prioritization of small subsets of molecular features for the analysis based on prior biological knowledge. Most importantly, external replication of the findings is the key because it is not practically feasible to eliminate the false positive associations that occur in a single high-dimensional dataset.
In addition, in order to fully realize the potential of big data in the clinical sphere, there is still a need for more and better data. Many diseases lack sufficient molecular characterization, and existing datasets are only infrequently linked to specific clinical features and outcomes. Currently available patient-derived omics data are heavily biased towards accessible organ systems such as blood and surgical tissues. In contrast, for example, there are very few genomic datasets for advanced HCC because tissue acquisition is not recommended as part of routine clinical practice. Additionally, the uneven quality of publicly available data can make valid interpretation difficult. A number of quality control measures have been devised for omics experiments, but integrating data from multiple experiments and/or different technological platforms and experimental conditions is an ongoing problem. Technical or clinical variation between individual experiments (so-called “batch effects”) can obscure or spuriously mimic the biological changes being sought through integrative analysis.64 It is therefore critical that publicly available data include detailed technical information regarding all factors that might contribute to experimental and clinical variation. On the other hand, given the growing body of big data automatically collected from sources like wearable devices and EHR, perfect data curation and quality control is an unrealistic expectation, and instead novel methods will need to be developed that are less sensitive to data heterogeneity. Finally, it should be emphasized that while big data-driven approaches promise to accelerate the discovery of new therapies and diagnostics, all computational predictions must still be thoroughly validated in experimental and clinical settings prior to general use.
Future Prospects
We are moving towards big data-based healthcare, including data-driven methodologies to accelerate the discovery of new diagnostics and drugs. To maximize the benefit of these big data-based approaches in gastroenterology and hepatology, it will be essential for clinical researchers to systematically collect specimens and clinical information in order to create centralized, comprehensive repositories of mineable data to address unmet needs. Routine collection of omics data such as whole genome or exome sequences may be an option once it is proven to be a cost-effective approach. This will require not only the incorporation of omics technologies into the clinical toolkit, but also the creation of medical information systems to regularly collect, curate, and analyze the data, and deliver results and interpretation to the clinic. Regulatory mechanisms for patient privacy protection that do not unrealistically hamper conduct of big data-based research will also be another critical requirement for the realization of precision medicine. Such plans are already evolving using secure, open formats, and investigators conducting clinical trials should become familiar with these resources.129 At the same time, a new breed of scientists and clinicians must emerge who are facile with big data approaches and can translate these data into novel biomarkers and drugs that prevent disease or improve the outcomes for patients with gastrointestinal and liver illnesses.
Supplementary Material
Acknowledgments
Grant support: This study was funded by National Institute of Health Grants 5T32DK007792, R01DK099558, RO1DK56621, the FLAGS foundation, the Nuovo-Soldati Cancer Research Foundation, an advanced training grant from Geneva University Hospital, the Irma T. Hirschl Trust, and the European Union grant ERC-AdG-2014 HEPCIR.
Abbreviations
- CMap
Connectivity Map
- DNA
deoxyribonucleic acid
- HCV
hepatitis C
- HDAC
histone deacetylase
- EHR
electronic health records
- EBI
European Bioinformatics Institute
- LINCS
Library of Integrated Network-based Cellular Signatures
- MIC
maximal information coefficient
- mRNA
messenger ribonucleic acid
- NCBI
National Center for Biotechnology Information
- RCT
randomized controlled trial
- GI
gastrointestinal
- HCC
hepatocellular carcinoma
- NCI
National Cancer Institute
- R&D
research and development
- RNA
ribonucleic acid
- HTS
high-throughput screening
- QSAR
quantitative structure-activity relationship
Glossary of terms
- Big data
Aggregate of various types of massive data, described by the following general attributes: volume (a large amount of data), velocity (rapid accumulation), variety (heterogeneity of relevant features), and veracity (the ability of data to faithfully represent its subject)
- Cheminformatics
Computational approaches to quantitative/predictive analysis of small molecules for their properties and performance
- Disease signature
A set of biomolecules (e.g., mRNA, protein) that changes in a coordinated manner in association with a specific disease condition such as presence or subtype of disease, poorer clinical outcome, and response to certain therapy
- Drug signature
A set of biomolecules (e.g., mRNA, protein) that changes in a coordinated manner in response to treatment by a drug in experimental systems
- Electronic health records
Clinical tool for gathering, storing, browsing, and analyzing patient health-related information. Widely implemented, though data format is not standardized, and access to data is controlled by regulatory policies and/or agencies such as Health Insurance Portability and Accountability Act (HIPAA) in the U.S
- High-throughput screening
Large-scale, often unbiased, assessment of biomolecule libraries to identify compounds eliciting biological activity against targets of interest
- In silico validation
The use of pre-collected biomedical data relevant to a specific research question, without generating new experimental and/or clinical data, to achieve a rapid and low-cost validation of new biological hypotheses, biomarkers, and therapies
- Machine learning
Computational approaches to identify, match, and predict patterns from highly complex data
- Omics data
Data comprehensively characterizing and/or quantifying a type of biomolecule, i.e., “-omes”, including the genome, transcriptome, proteome, methylome, epigenome, metabolome, microbiome, virome, interactome, and phenome
- Patient-reported health information
Health-related information reported or recorded by patients, beyond that normally recorded in routine clinical encounters, e.g., medication adherence, symptom tracking, voluntary genomic studies, etc
- Pharmacophore
Biologically relevant chemical substructures, often comprising the sites of activity between a compound and its substrates
- Precision medicine
An emerging paradigm in translational biomedicine aiming to deliver medical care that is tailored to the specific health needs of individuals, rather than populations
- Translational bioinformatics
Bioinformatics with special focus on clinical-basic translation
- Wearable technology
A range of devices wearable by study subjects, e.g., clothes with vital sign monitors, contact lens with glucose sensors, which regularly and automatically collect and store data, and/or transmit information to a data collection server
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: All authors participated in the composition and editing of this work.
Disclosures: The authors have no relevant conflicts.
References
Author names in bold designate shared co-first authorship.
- 1.Collins F, Green E, Guttmacher A, et al. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 2.Costa FF. Big data in biomedicine. Drug Discov. Today. 2014;19:433–440. doi: 10.1016/j.drudis.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Cook CE, Bergman MT, Finn RD, et al. The European Bioinformatics Institute in 2016: Data growth and integration. Nucleic Acids Res. 2016;44:D20–D26. doi: 10.1093/nar/gkv1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolesnikov N, Hastings E, Keays M, et al. ArrayExpress update-simplifying data submissions. Nucleic Acids Res. 2015;43:D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: Archive for functional genomics data sets - Update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pivovarov R, Elhadad N. Automated methods for the summarization of electronic health records. J. Am. Med. Informatics Assoc. 2015;22:938–947. doi: 10.1093/jamia/ocv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mezghani E, Exposito E, Drira K, et al. A semantic big data platform for integrating heterogeneous wearable data in healthcare. J. Med. Syst. 2015;39:185. doi: 10.1007/s10916-015-0344-x. [DOI] [PubMed] [Google Scholar]
- 8.Taichman DB, Backus J, Baethge C, et al. Sharing clinical trial data — a proposal from the International Committee of Medical Journal Editors. N. Engl. J. Med. 2016;374:384–386. doi: 10.1056/NEJMe1515172. [DOI] [PubMed] [Google Scholar]
- 9.National Cancer Institute Press Office. Newly launched Genomic Data Commons to facilitate data and clinical information sharing. 2016 Available at: http://www.cancer.gov/news-events/press-releases/2016/genomic-data-commons-launch.
- 10.Li LL, Cheng W-YW, Glicksberg BS, et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci. Transl. Med. 2015;7:1–16. doi: 10.1126/scitranslmed.aaa9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Issa NT, Byers SW, Dakshanamurthy S. Big data: the next frontier for innovation in therapeutics and healthcare. Expert Rev. Clin. Pharmacol. 2014;7:293–298. doi: 10.1586/17512433.2014.905201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badawi O, Brennan T, Celi LA, et al. Making big data useful for health care: A summary of the inaugural MIT critical data conference. J. Med. Internet Res. 2014;16:e22. doi: 10.2196/medinform.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: The National Academies Press; 2011. [PubMed] [Google Scholar]
- 14.Vanhove W, Nys K, Vermeire S. Therapeutic innovations in inflammatory bowel diseases. Clin. Pharmacol. Ther. 2016;99:49–58. doi: 10.1002/cpt.286. [DOI] [PubMed] [Google Scholar]
- 15.Sherif ZA, Saeed A, Ghavimi S, et al. Global epidemiology of nonalcoholic fatty liver disease and perspectives on us minority populations. Dig. Dis. Sci. 2016;61:1214. doi: 10.1007/s10620-016-4143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arsene D, Farooq O, Bataller R. New therapeutic targets in alcoholic hepatitis. Hepatol. Int. 2016;10:538–552. doi: 10.1007/s12072-015-9701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkinson DR, McCormack RT, Keating SM, et al. Evidence of clinical utility: an unmet need in molecular diagnostics for patients with cancer. Clin. Cancer Res. 2014;20:1428–1444. doi: 10.1158/1078-0432.CCR-13-2961. [DOI] [PubMed] [Google Scholar]
- 18.Sawyers CL, van’t Veer LJ. Reliable and effective diagnostics are keys to accelerating personalized cancer medicine and transforming cancer care: a policy statement from the american association for cancer research. Clin. Cancer Res. 2014;20:4978–4981. doi: 10.1158/1078-0432.CCR-14-2295. [DOI] [PubMed] [Google Scholar]
- 19.Poste G. Bring on the biomarkers. Nature. 2011;469:156–157. doi: 10.1038/469156a. [DOI] [PubMed] [Google Scholar]
- 20.Teutsch SM, Bradley LA, Palomaki GE, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: methods of the EGAPP Working Group. Genet. Med. 2009;11:3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Institute of Medicine. Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington, DC: The National Academies Press; 2012. [PubMed] [Google Scholar]
- 22.Goossens N, Nakagawa S, Sun X, et al. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015;4:256–269. doi: 10.3978/j.issn.2218-676X.2015.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redig AJ, Jänne PA. Basket trials and the evolution of clinical trial design in an era of genomic medicine. J. Clin. Oncol. 2015;33:975–977. doi: 10.1200/JCO.2014.59.8433. [DOI] [PubMed] [Google Scholar]
- 24.Gannon HS, Kaplan N, Tsherniak A, et al. Identification of an “exceptional responder” cell line to MEK1 inhibition: clinical implications for MEK-targeted therapy. Mol. Cancer Res. 2015:207–216. doi: 10.1158/1541-7786.MCR-15-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schully SD, Carrick DM, Mechanic LE, et al. Leveraging biospecimen resources for discovery or validation of markers for early cancer detection. J. Natl. Cancer Inst. 2015;107:djv012. doi: 10.1093/jnci/djv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mello MM, Francer JK, et al. Preparing for responsible sharing of clinical trial data. N. Engl. J. Med. 2013;369:1651–1658. doi: 10.1056/NEJMhle1309073. [DOI] [PubMed] [Google Scholar]
- 27.Jung Y, Lee S, Choi H-S, et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin. Cancer Res. 2011;17:700–709. doi: 10.1158/1078-0432.CCR-10-1300. [DOI] [PubMed] [Google Scholar]
- 28.Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 29.Hoshida Y, Nijman SMB, Kobayashi M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang DY, Goossens N, Guo J, et al. A hepatic stellate cell gene expression signature associated with outcomes in hepatitis C cirrhosis and hepatocellular carcinoma after curative resection. Gut. 2015;65:1754–1764. doi: 10.1136/gutjnl-2015-309655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoshida Y, Villanueva A, Sangiovanni A, et al. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology. 2013;144:1024–1030. doi: 10.1053/j.gastro.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King LY, Canasto-Chibuque C, Johnson KB, et al. A genomic and clinical prognostic index for hepatitis C-related early-stage cirrhosis that predicts clinical deterioration. Gut. 2015;64:1296–1302. doi: 10.1136/gutjnl-2014-307862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalerba P, Sahoo D, Paik S, et al. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N. Engl. J. Med. 2016;374:211–222. doi: 10.1056/NEJMoa1506597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munos B. Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov. 2009;8:959–968. doi: 10.1038/nrd2961. [DOI] [PubMed] [Google Scholar]
- 37.Schuhmacher A, Gassmann O, Hinder M, et al. Changing R&D models in research-based pharmaceutical companies. J. Transl. Med. 2016;14:105. doi: 10.1186/s12967-016-0838-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hart T, Xie L. Providing data science support for systems pharmacology and its implications to drug discovery. Expert Opin. Drug Discov. 2016;11:241–256. doi: 10.1517/17460441.2016.1135126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medina-Franco JL, Giulianotti MA, Welmaker GS, et al. Shifting from the single to the multitarget paradigm in drug discovery. Drug Discov. Today. 2013;18:495–501. doi: 10.1016/j.drudis.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scannell JW, Blanckley A, Boldon H, et al. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 2012;11:191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- 41.Swinney DC, Anthony J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Xing J, Xu Y, et al. In silico ADME/T modelling for rational drug design. Q. Rev. Biophys. 2015;1308:1–28. doi: 10.1017/S0033583515000190. [DOI] [PubMed] [Google Scholar]
- 43.Nantasenamat C, Prachayasittikul V. Maximizing computational tools for successful drug discovery. Expert Opin. Drug Discov. 2015;10:321–329. doi: 10.1517/17460441.2015.1016497. [DOI] [PubMed] [Google Scholar]
- 44.Li J, Zheng S, Chen B, et al. A survey of current trends in computational drug repositioning. Brief. Bioinform. 2016;17(1):2–12. doi: 10.1093/bib/bbv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 46.Power A, Berger AC, Ginsburg GS. Genomics-enabled drug repositioning and repurposing: insights from an IOM Roundtable activity. JAMA. 2014;311:2063–2064. doi: 10.1001/jama.2014.3002. [DOI] [PubMed] [Google Scholar]
- 47.Hurle MR, Yang L, Xie Q, et al. Computational drug repositioning: from data to therapeutics. Clin. Pharmacol. Ther. 2013;93:335–341. doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- 48.Chong CR, Sullivan DJ. New uses for old drugs. Nature. 2007;448:645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 49.Ekins S, Williams AJ, Krasowski MD, et al. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov. Today. 2011;16:298–310. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 50.Kakkar AK, Dahiya N. The evolving drug development landscape: From blockbusters to niche busters in the orphan drug space. Drug Dev. Res. 2014;75:231–234. doi: 10.1002/ddr.21176. [DOI] [PubMed] [Google Scholar]
- 51.Nair P. Second act. Proc. Natl. Acad. Sci. 2013;110:2430–2432. doi: 10.1073/pnas.201300188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collins FS, Varmus H. A new initiative on precision medicine. N. Engl. J. Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 54.Vidović D, Koleti A, Schürer SC. Large-scale integration of small molecule-induced genome-wide transcriptional responses, Kinome-wide binding affinities and cell-growth inhibition profiles reveal global trends characterizing systems-level drug action. Front. Genet. 2014;5:342. doi: 10.3389/fgene.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dudley JT, Sirota M, Shenoy M, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 2011;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen M-H, Yang W-LR, Lin K-T, Liu C-H, Liu Y-W, Huang K-W, et al. Gene expression-based chemical genomics identifies potential therapeutic drugs in hepatocellular carcinoma. PLoS One. 2011;6:e27186. doi: 10.1371/journal.pone.0027186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Li M, Wang Y, et al. Integrating subpathway analysis to identify candidate agents for hepatocellular carcinoma. Onco. Targets. Ther. 2016;9:1221–1230. doi: 10.2147/OTT.S97211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Noort V, Schölch S, Iskar M, et al. Novel drug candidates for the treatment of metastatic colorectal cancer through global inverse gene-expression profiling. Cancer Res. 2014;74:5690–5699. doi: 10.1158/0008-5472.CAN-13-3540. [DOI] [PubMed] [Google Scholar]
- 59.Claerhout S, Lim JY, Choi W, et al. Gene expression signature analysis identifies vorinostat as a candidate therapy for gastric cancer. PLoS One. 2011;6:e24662. doi: 10.1371/journal.pone.0024662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suthram S, Dudley JT, Chiang AP, et al. Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput. Biol. 2010;6:e1000662. doi: 10.1371/journal.pcbi.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 62.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zollanvari A. High-dimensional statistical learning: roots, justifications, and potential machineries. Cancer Inform. 2016;14:109. doi: 10.4137/CIN.S30804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mrna abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 66.Du J, Bernasconi P, Clauser KR, et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat. Biotechnol. 2009;27:77–83. doi: 10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bachovchin Da, Koblan LW, Wu W, et al. A high-throughput, multiplexed assay for superfamily-wide profiling of enzyme activity. Nat. Chem. Biol. 2014;10:656–663. doi: 10.1038/nchembio.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu C, Mannan AM, Yvone GM, et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat. Biotechnol. 2016;34:419–423. doi: 10.1038/nbt.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hodos RA, Kidd BA, Shameer K, et al. In silico methods for drug repurposing and pharmacology Wiley Interdiscip. Rev. Syst. Biol. Med. 2016;8:186–210. doi: 10.1002/wsbm.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiang AP, Butte AJ. Systematic evaluation of drug-disease relationships to identify leads for novel drug uses. Clin. Pharmacol. Ther. 2009;86:507–510. doi: 10.1038/clpt.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang L, Agarwal P. Systematic drug repositioning based on clinical side-effects. PLoS One. 2011;6:e28025. doi: 10.1371/journal.pone.0028025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye H, Liu Q, Wei J. Construction of drug network based on side effects and its application for drug repositioning. PLoS One. 2014;9:e87864. doi: 10.1371/journal.pone.0087864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang P, Wang F, Hu J, et al. Exploring the relationship between drug side-effects and therapeutic indications. AMIA Annu. Symp. Proc. 2013;2013:1568–1577. [PMC free article] [PubMed] [Google Scholar]
- 74.Kuhn M, Letunic I, Jensen LJ, et al. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016;44:D1075–D1079. doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu R, Li LL, Wang Q. Towards building a disease-phenotype knowledge base: extracting disease-manifestation relationship from literature. Bioinformatics. 2013;29:2186–2194. doi: 10.1093/bioinformatics/btt359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gottlieb A, Stein GY, Ruppin E, et al. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 2014;7:496–496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang P, Wang F, Hu J. Towards drug repositioning: a unified computational framework for integrating multiple aspects of drug similarity and disease similarity. AMIA Annu. Symp. Proc. 2014;2014:1258–1267. [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng L, Li J, Ju P, et al. SemFunSim: a new method for measuring disease similarity by integrating semantic and gene functional association. PLoS One. 2014;9:e99415. doi: 10.1371/journal.pone.0099415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang W, Yang S, Zhang X, et al. Drug repositioning by integrating target information through a heterogeneous network model. Bioinformatics. 2014;30:1–8. doi: 10.1093/bioinformatics/btu403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iwata H, Sawada R, Mizutani S, et al. Systematic drug repositioning for a wide range of diseases with integrative analyses of phenotypic and molecular data. J. Chem. Inf. Model. 2015;55:446–459. doi: 10.1021/ci500670q. [DOI] [PubMed] [Google Scholar]
- 81.Chan SY, Loscalzo J. The emerging paradigm of network medicine in the study of human disease. Circ. Res. 2012;111:359–374. doi: 10.1161/CIRCRESAHA.111.258541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 83.Von Eichborn J, Murgueitio MS, Dunkel M, et al. PROMISCUOUS: a database for network-based drug-repositioning. Nucleic Acids Res. 2011;39:D1060–D1066. doi: 10.1093/nar/gkq1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee H, Bae T, Lee J-H, et al. Rational drug repositioning guided by an integrated pharmacological network of protein, disease and drug. BMC Syst. Biol. 2012;6:80. doi: 10.1186/1752-0509-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iorio F, Shrestha RL, Levin N, et al. A semi-supervised approach for refining transcriptional signatures of drug response and repositioning predictions. PLoS One. 2015;10:e0139446. doi: 10.1371/journal.pone.0139446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Menche J, Sharma A, Kitsak M, et al. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347(6224):1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swanson DR. Medical literature as a potential source of new knowledge. Bull. Med. Libr. Assoc. 1990;78:29–37. [PMC free article] [PubMed] [Google Scholar]
- 88.Cohen T, Widdows D, Stephan C, et al. Predicting high-throughput screening results with scalable literature-based discovery methods. CPT pharmacometrics Syst. Pharmacol. 2014;3:e140. doi: 10.1038/psp.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tari L, Vo N, Liang S, et al. Identifying novel drug indications through automated reasoning. PLoS One. 2012;7:e40946. doi: 10.1371/journal.pone.0040946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gramatica R, Di Matteo T, Giorgetti S, et al. Graph theory enables drug repurposing – how a mathematical model can drive the discovery of hidden mechanisms of action. PLoS One. 2014;9:e84912. doi: 10.1371/journal.pone.0084912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Y, Liang Y, Wishart D. PolySearch2: a significantly improved text-mining system for discovering associations between human diseases, genes, drugs, metabolites, toxins and more. Nucleic Acids Res. 2015;43:W535–W542. doi: 10.1093/nar/gkv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ma DL, Chan DSH, Leung CH. Drug repositioning by structure-based virtual screening. Chem. Soc. Rev. 2013;42:2130–2141. doi: 10.1039/c2cs35357a. [DOI] [PubMed] [Google Scholar]
- 93.Lee A, Lee K, Kim D. Using reverse docking for target identification and its applications for drug discovery. Expert Opin. Drug Discov. 2016;11(7):707–715. doi: 10.1080/17460441.2016.1190706. [DOI] [PubMed] [Google Scholar]
- 94.Li H, Gao Z, Kang L, et al. TarFisDock: A web server for identifying drug targets with docking approach. Nucleic Acids Res. 2006;34:219–224. doi: 10.1093/nar/gkl114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Douguet D. e-LEA3D: A computational-aided drug design web server. Nucleic Acids Res. 2010;38:W615–W621. doi: 10.1093/nar/gkq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu X, Ouyang S, Yu B, et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010;38:W609–W614. doi: 10.1093/nar/gkq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X, Chen H, Yang F, et al. IDrug: A web-accessible and interactive drug discovery and design platform. J. Cheminform. 2014;6:1–8. doi: 10.1186/1758-2946-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Luo H, Chen J, Shi L, et al. DRAR-CPI: a server for identifying drug repositioning potential and adverse drug reactions via the chemical-protein interactome. Nucleic Acids Res. 2011;39:W492–W498. doi: 10.1093/nar/gkr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Keiser MJ, Setola V, Irwin JJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L, Ma C, Wipf P, et al. TargetHunter: an in silico target identification tool for predicting therapeutic potential of small organic molecules based on chemogenomic database. AAPS J. 2013;15:395–406. doi: 10.1208/s12248-012-9449-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nantasenamat C, Isarankura-Na-Ayudhya C, Naenna T, et al. A practical overview of quantitative structure-activity relationship. EXCLI J. 2009;8:74–88. [Google Scholar]
- 102.Nantasenamat C, Worachartcheewan A, Jamsak S, et al. AutoWeka: Toward an Automated Data Mining Software for QSAR and QSPR Studies. In: Cartwright H, editor. Artificial Neural Networks. 2nd. New York: Springer Science+Busines Media; 2015. pp. 119–147. [DOI] [PubMed] [Google Scholar]
- 103.Cheng F, Li W, Zhou Y, et al. AdmetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012;52:3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- 104.Pires DEV, Blundell TL, Ascher DB. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58:4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.García-Domenech R, Gálvez-Llompart M, Zanni R, et al. QSAR methods for the discovery of new inflammatory bowel disease drugs. Expert Opin. Drug Discov. 2013;8:933–949. doi: 10.1517/17460441.2013.800043. [DOI] [PubMed] [Google Scholar]
- 106.Gálvez-Llompart M, Recio MC, García-Domenech R. Topological virtual screening: A way to find new compounds active in ulcerative colitis by inhibiting NF-κB. Mol. Divers. 2011;15:917–924. doi: 10.1007/s11030-011-9323-4. [DOI] [PubMed] [Google Scholar]
- 107.Yang JS, Chun K, Park JE, et al. Structure based optimization of chromen-based TNF-α converting enzyme (TACE) inhibitors on S1’ pocket and their quantitative structure-activity relationship (QSAR) study. Bioorg. Med. Chem. 2010;18:8618–8629. doi: 10.1016/j.bmc.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 108.Huang H, Zhang P, Qu XA, et al. Systematic prediction of drug combinations based on clinical side-effects. Sci. Rep. 2014;4:7160. doi: 10.1038/srep07160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee J-H, Kim DG, Bae TJ, et al. CDA: combinatorial drug discovery using transcriptional response modules. PLoS One. 2012;7:e42573. doi: 10.1371/journal.pone.0042573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Prahallad a, Bernards R. Opportunities and challenges provided by crosstalk between signalling pathways in cancer. Oncogene. 2015;35:1–7. doi: 10.1038/onc.2015.151. [DOI] [PubMed] [Google Scholar]
- 111.Johannessen CM, Johnson LA, Piccioni F, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sun X, Vilar S, Tatonetti NP. High-throughput methods for combinatorial drug discovery. Sci. Transl. Med. 2013;5:205rv1. doi: 10.1126/scitranslmed.3006667. [DOI] [PubMed] [Google Scholar]
- 113.Chen L, Li B-Q, Zheng M-Y, et al. Prediction of effective drug combinations by chemical interaction, protein interaction and target enrichment of KEGG pathways. Biomed Res. Int. 2013;2013:723780. doi: 10.1155/2013/723780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang L, Li F, Sheng J, et al. DrugComboRanker: drug combination discovery based on target network analysis. Bioinformatics. 2014;30 doi: 10.1093/bioinformatics/btu278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu Y, Wei Q, Yu G, et al. DCDB 2.0: a major update of the drug combination database. Database (Oxford) 2014;2014:bau124. doi: 10.1093/database/bau124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lima AN, Philot EA, Trossini GHG, et al. Use of machine learning approaches for novel drug discovery. Expert Opin. Drug Discov. 2016;11:225–239. doi: 10.1517/17460441.2016.1146250. [DOI] [PubMed] [Google Scholar]
- 117.Smith TC, Frank E. Introducing machine learning concepts with WEKA. Methods Mol. Biol. 2016;1418:353–378. doi: 10.1007/978-1-4939-3578-9_17. [DOI] [PubMed] [Google Scholar]
- 118.Napolitano F, Zhao Y, Moreira VM, et al. Drug repositioning: a machine-learning approach through data integration. J. Cheminform. 2013;5:30. doi: 10.1186/1758-2946-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mamoshina P, Vieira A, Putin E, et al. Applications of deep learning in biomedicine. Mol. Pharm. 2016;13:1445–1454. doi: 10.1021/acs.molpharmaceut.5b00982. [DOI] [PubMed] [Google Scholar]
- 120.Aliper A, Plis S, Artemov A, et al. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol. Pharm. 2016;13:2524–2530. doi: 10.1021/acs.molpharmaceut.6b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu Y, Dai Z, Chen F, et al. Deep learning for drug-induced liver injury. J. Chem. Inf. Model. 2015;55:2085–2093. doi: 10.1021/acs.jcim.5b00238. [DOI] [PubMed] [Google Scholar]
- 122.Zhou Z-H. Ensemble Learning. In: S Li., editor. Encyclopedia of Biometrics. New York, NY: Springer Science+Busines Media; 2009. pp. 270–273. [Google Scholar]
- 123.Wan Q, Pal R. An ensemble based top performing approach for NCI-DREAM drug sensitivity prediction challenge. PLoS One. 2014;9:e101183. doi: 10.1371/journal.pone.0101183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Reshef DN, Reshef YA, Finucane HK, et al. Detecting novel associations in large data sets. Science. 2011;334:1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Agarwal P, Owzar K. Next generation distributed computing for cancer research. Cancer Inform. 2015;14:1–13. doi: 10.4137/CIN.S16344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Good BM, Su AI. Crowdsourcing for bioinformatics. Bioinformatics. 2013;29:1925–1933. doi: 10.1093/bioinformatics/btt333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jayaraj PB, Ajay MK, Nufail M, et al. GPURFSCREEN: a GPU based virtual screening tool using random forest classifier. J. Cheminform. 2016;8:12. doi: 10.1186/s13321-016-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Alyass A, Turcotte M, Meyre D. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med. Genomics. 2015;8:33. doi: 10.1186/s12920-015-0108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bierer BE, Li R, Barnes M, et al. A global, neutral platform for sharing trial data. N. Engl. J. Med. 2016;374:2411–2413. doi: 10.1056/NEJMp1605348. [DOI] [PubMed] [Google Scholar]
- 130.Kim JH, Sohn BH, Lee HS, et al. Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation. PLoS Med. 2014;11:e1001770. doi: 10.1371/journal.pmed.1001770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ji J, Eggert T, Budhu A, et al. Hepatic stellate cell and monocyte interaction contributes to poor prognosis in hepatocellular carcinoma. Hepatology. 2015;62:481–495. doi: 10.1002/hep.27822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang H, Shiffman ML, Friedman S, et al. A 7 gene signature identifies the risk of developing cirrhosis in patients with chronic hepatitis C. Hepatology. 2007;46:297–306. doi: 10.1002/hep.21695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.