Abstract
Most members of the kallikrein-related peptidase family have been demonstrated to be dysregulated in ovarian cancer and modulate tumor growth, migration, invasion, and resistance to chemotherapy. In the present study, we assessed the mRNA expression levels of KLK6 and KLK8 by quantitative PCR in 100 patients with advanced serous ovarian cancer FIGO stage III/IV. A pronounced correlation between KLK6 and KLK8 mRNA expression (rs = 0.636, P<0.001) was observed indicating coordinate expression of both peptidases. No significant associations of clinical parameters with KLK6, KLK8, and a combined score KLK6+KLK8 were found. In univariate Cox regression analysis, elevated mRNA levels of KLK6 were significantly linked with shortened overall survival (OS) (hazard ratio [HR] = 2.07, P=0.007). While KLK8 values were not associated with patients’ outcome, high KLK6+KLK8 values were significantly associated with shorter progression-free survival (HR = 1.82, P=0.047) and showed a trend towards significance in case of OS (HR = 1.82, P=0.053). Strikingly, in multivariable analysis, elevated KLK6 mRNA values, apart from residual tumor mass, remained an independent predictive marker for poor OS (HR = 2.33, P=0.005). Since KLK6 mRNA and protein levels correlate, KLK6 may represent an attractive therapeutic target for potent and specific inhibitors of its enzymatic activity.
Keywords: kallikrein-related peptidase, KLK6, KLK8, ovarian cancer, quantitative PCR
Introduction
Ovarian cancer represents the fifth most common cause of cancer death among women and the most lethal gynecological neoplasm (Prat, 2012). Annually, approximately 6.1 per 100,000 women are newly diagnosed with ovarian cancer (Ferlay et al., 2015). Owing to its intraperitoneal location, the biological behavior of most epithelial tumors, and lack of early symptoms, more than 70% of patients are diagnosed in late stages of the disease (International Federation of Gynecology and Obstetrics [FIGO] classifications: stage III-IV) in which the five-year survival rate is around 30% (Kipps et al; 2013; Prat, 2014). The most important clinical factor, and the only one to be influenced by the surgeon so far, is residual tumor mass after standard debulking operation (Dorn et al., 2011). Thus, valid biomarkers to predict prognosis and/or response to chemotherapy are urgently needed.
The family of the human kallikrein-related peptidases (KLK) serves as a promising pool of biomarkers for ovarian cancer (among other diseases). Its 15 members (KLK1-15) constitute a subgroup of the serine protease enzyme family and are encoded within a gene cluster localized on chromosome 19q13.3-4. Nearly all members of the KLK family are dysregulated in ovarian cancer and associated with prognosis. In fact, there is growing evidence that KLKs play an important role in cancer biogenesis including tumor growth, migration, invasion, and chemoresistance, thus, making them potential targets for anti-cancer therapeutic agents (Dorn et al., 2014; Kryza et al., 2016).
In human tissues, KLKs are often co-expressed and functionally linked to each other. For example, in the skin, KLKs 5, 6, 7, 8, and 14 are involved in corneocyte desquamation and/or regulation of inflammatory reactions or modulation of the lipid-permeability barrier. Prostatic KLKs, i.e. KLK3 in concert with KLK2 and KLK4, are responsible for semen liquefaction. In the central nervous system (CNS), 12 of the 15 members of the KLK family, including KLK6 (neurosin) and KLK8 (neuropsin), are expressed under physiological conditions (Prassas et al., 2015).
KLK6 seems to be the most abundant serine protease in the CNS and participates in numerous physiological and pathological processes. Together with KLK1 and KLK8, KLK6 seems to play an important role in brain injury (Scarisbrick, 2012). KLK6 levels are reduced in cerebrospinal fluid, serum, and plaques in Alzheimer’s disease, which can be employed to differentiate the disease from vascular dementia and pseudodementia patients (Bayani and Diamandis, 2012). KLK6 serum levels are significantly increased in multiple sclerosis (MS) and correlate with poor prognosis of MS patients (Bayani and Diamandis, 2012). In Parkinson’s disease, KLK6 was implicated in cleavage of α-synuclein preventing its polymerization in Lewy bodies (Tatebe et al., 2010). KLK6 expression is dysregulated in different cancers such as pancreatic (Ruckert et al., 2008), breast (Wang et al., 2008), colon (Vakrakou et al., 2014), and gastric cancer (Kolin et al., 2014). In ovarian cancer, KLK6 overexpression has been reported to be associated with poor prognosis, advanced clinical disease, shorter disease-free and overall survival, and chemotherapy resistance (Diamandis et al., 2003; Kountourakis et al., 2008; White et al., 2009; Loessner et al., 2012; Seiz et al., 2012).
KLK8 is moderately expressed in neurons of the hippocampus playing essential roles in synaptic plasticity as basis of learning and memory formation, while it displays elevated expression levels following cerebral injuries and in the hippocampus of Alzheimer patients (Shimizu-Okabe et al., 2001; Yoshida, 2003). KLK8 is overexpressed in the tumor tissue of cervix (Cane et al., 2004) and colon cancer (Yousef et al., 2004). Furthermore, its overexpression has been reported to have an - albeit controversial - impact on prognosis in lung cancer (Sher et al., 2006; Planque et al., 2010). In ovarian cancer, Kishi et al. (2003) reported high expression of KLK8 in serum, ascitic fluid, and tumor tissues obtained from primary ovarian cancer patients, being correlated with the protein expression of tumor-associated CA-125. High KLK8 protein expression was found to be associated with a favorable prognosis, lower nuclear grade, earlier clinical stage and extended overall and disease-free survival of ovarian cancer patients (Shigemasa et al., 2004; Borgono et al., 2006). On the contrary, Kountourakis et al. (2009) reported that low KLK8 protein expression was associated with better outcome in ovarian cancer. To date, there are no data describing the relationship between KLK8 mRNA expression and survival in patients with advanced serous ovarian cancer.
Since KLK6 has been described to be a valuable biomarker in ovarian cancer, and is often co-expressed with KLK8 in other organs such as the CNS, in the present study, we evaluated the predictive value of KLK6 and KLK8 mRNA levels as well as their combined expression score for progression-free (PFS) and overall (OS) survival in a cohort of patients with advanced high-grade serous ovarian cancer FIGO stage III/IV. Expression levels of KLK6 and KLK8 were determined by qPCR and the association of KLK mRNA levels with clinical parameters and with survival of the patients was analyzed.
Results
KLK6 and KLK8 mRNA expression in tumor tissue of advanced ovarian cancer patients and relation to patient and tumor characteristics
KLK6 and KLK8 mRNA levels were determined by qPCR in tumor tissues of a homogenous patient cohort (n = 100) encompassing patients with serous ovarian cancer FIGO stage III/IV, only. The relative KLK6 and KLK8 qPCR mRNA levels, normalized to the expression levels of the housekeeping gene HPRT, ranged from 0.38 to 1067.49 (median: 119.42) and from 0.02 to 86.42 (median: 10.75), respectively. As evident by Mann-Whitney test analysis (P < 0.001) and by nonparametric Spearman rank correlation analysis (rs = 0.636, P < 0.001), a highly significant, pronounced positive correlation between KLK6 and KLK8 mRNA expression is observed (Fig. 1A). Based on the observed co-expression of KLK6 and KLK8, we further categorized the expression levels of both factors in a KLK6+KLK8 low-expressing group (both KLK6 and KLK8 mRNA levels below the median) versus a high-expressing group (KLK6 and/or KLK8 mRNA values above the median) for statistical analysis (see below). In case of KLK6, protein expression values determined by ELISA in tumor tissue extracts were available for 51 patients (Dorn et al., 2015). As depicted in Figure 1B, a high positive correlation between KLK6 mRNA and KLK6 protein levels is found using both Mann-Whitney test (P < 0.001) and Spearman rank correlation (rs = 0.708, P < 0.001) analyses.
Figure 1. Correlations of KLK6 mRNA expression with KLK6 protein levels and KLK8 mRNA expression levels in tumor tissues of advanced ovarian cancer patients (FIGO III/IV).
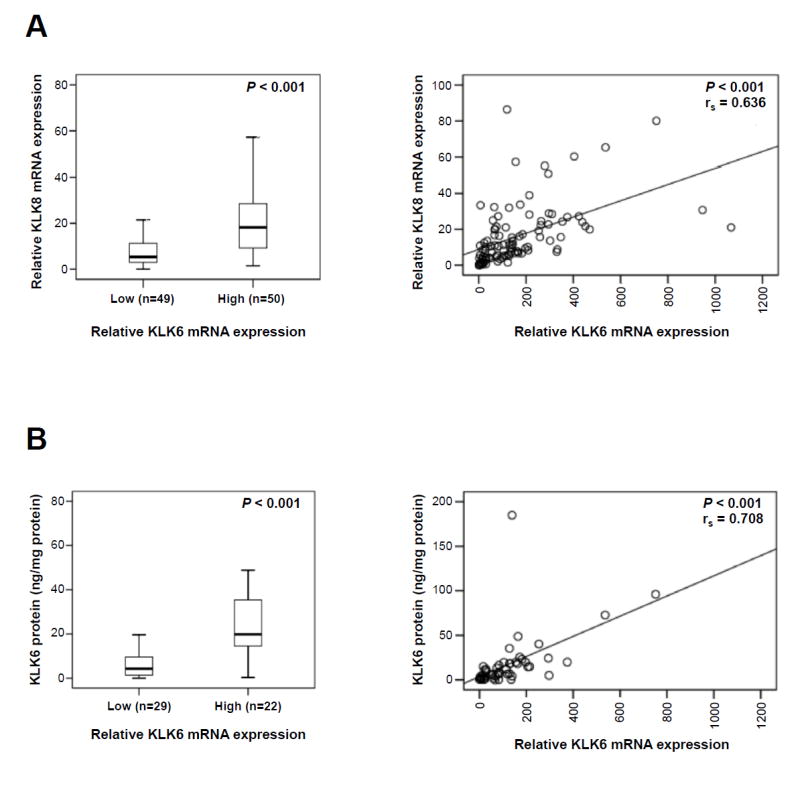
(A) KLK6 mRNA expression is significantly correlated with KLK8 mRNA levels in tumor tissue. KLK6 and KLK8 mRNA levels were determined by qPCR with normalization to HPRT mRNA levels. In the left panel, KLK6 mRNA levels were dichotomized into low and high by the median and analyzed for association with KLK8 mRNA values (Mann-Whitney test; P < 0.001). In the right panel, KLK6 mRNA and KLK8 mRNA levels were analyzed as linear factors (Spearman correlation; rs = 0.636, P < 0.001). (B) KLK6 mRNA expression is significantly correlated with its protein levels (determined by ELISA) in tumor tissue. In the left panel, KLK6 mRNA levels were dichotomized into low and high by the median and analyzed for association with KLK6 antigen levels (Mann-Whitney test; P < 0.001). In the right panel, KLK6 mRNA and antigen levels were analyzed as linear factors (Spearman correlation; rs = 0.708, P < 0.001).
Table 1 depicts the association between dichotomized KLK6 and KLK8 mRNA expression levels (low/high) and the combined KLK6 and KLK8 values (KLK6+KLK8) in relation to established clinical parameters in ovarian cancer such as age, pre-operative ascites fluid volume, and post-operative residual tumor mass. The mRNA levels of KLK6, KLK8, and KLK6+KLK8 do not differ significantly between tumors in relation to these clinical parameters, apart from a trend towards significance (P = 0.062) for low KLK6+KLK8 levels in optimally debulked patients versus patients displaying a residual tumor mass.
Table 1.
Association between clinical characteristics of advanced ovarian cancer patients (FIGO III/IV) and tumor biological factors.
| Clinical parameters | No. of patients | KLK6a Low/high | KLK8a Low/high | KLK6+KLK8b Low/high |
|---|---|---|---|---|
|
| ||||
| Total no. of patients | 100 | 49/50 | 50/50 | 34/65 |
|
| ||||
| Age | P = 0.919 | P = 0.841 | P = 0.942 | |
| ≤ 60 years | 49 | 24/25 | 25/24 | 17/32 |
| > 60 years | 51 | 25/25 | 25/26 | 17/33 |
|
| ||||
| Residual tumor mass | P = 0.230 | P = 0.359 | P = 0.062 | |
| 0 mm | 45 | 25/20 | 25/20 | 20/25 |
| > 0 mm | 54 | 23/30 | 25/29 | 14/39 |
|
| ||||
| Ascitic fluid volume | P = 0.875 | P = 0.551 | P = 0.264 | |
| ≤ 500 ml | 46 | 25/21 | 26/20 | 20/26 |
| > 500 ml | 38 | 20/18 | 19/19 | 12/26 |
Chi-square test (cutoff point = median)
High KLK6+KLK8: KLK6 and/or KLK8 above median; low KLK6+KLK8: KLK6 and KLK8 both below median
Association of KLK6, KLK8, and KLK6+KLK8 mRNA values with progression-free (PFS) and overall (OS) survival: univariate analysis
The strength of association between traditional clinical parameters, and KLK6, KLK8, and KLK6+KLK8 mRNA expression levels with patients’ 5-year PFS and OS by univariate analysis is summarized in Table 2. Patients with residual tumor mass after debulking surgery and those patients with high pre-operative amounts of ascitic fluid have a significantly shorter OS (HR = 4.63, 95 % CI = 2.53 – 8.48, P < 0.001 and HR = 2.17, 95 % CI = 1.22 – 3.85, P = 0.008, respectively) and PFS (HR = 3.31, 95 % CI = 1.89 – 5.78, P < 0.001 and HR = 2.27; 95 % CI = 1.27 – 4.05, P = 0.006, respectively). KLK6 mRNA levels dichotomized by the median represent a significant predictive factor for OS (HR = 1.78, 95 % CI = 1.04 – 3.03, P = 0.035) indicating an about two-fold increased probability of death in the KLK6 high-expressing group. Optimization of the KLK6 cut-off value (61st percentile) does slightly increase the predictive power for OS (HR = 2.07, 95% CI = 1.22 - 3.50, P = 0.007), whereas the association remains not significant concerning PFS. KLK8 mRNA expression is neither associated with OS nor PFS, yet, high KLK6+KLK8 values show a trend towards significance in case of OS (HR = 1.82, 95 % CI = 0.99 – 3.33, P = 0.053) and are significantly associated with shorter PFS (HR = 1.82, 95 % CI = 1.01 – 3.30 P = 0.047). These findings were also confirmed by Kaplan-Meier estimation: the association of KLK6, KLK8, and KLK6+KLK8 mRNA levels with OS and PFS is visualized by the respective survival curves (Fig. 2). Here, high KLK6+KLK8 mRNA levels are significantly associated with both shorter OS and PFS (P = 0.048 and P = 0.042, respectively).
Table 2.
Univariate Cox regression analysis of clinical outcome in advanced ovarian cancer patients (FIGO III/IV) with respect to clinical parameters and tumor biological factors
| Clinical parameters | OS | PFS | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| No.a | HR (95% CI)b | P | No.a | HR (95% CI)b | P | |
|
| ||||||
| Age | 0.061 | 0.228 | ||||
| ≤ 60 years | 43 | 1 | 41 | 1 | ||
| > 60 years | 48 | 1.66 (0.98-2.82) | 44 | 1.39 (0.82-2.35) | ||
|
| ||||||
| Residual tumor mass | < 0.001 | < 0.001 | ||||
| 0 mm | 41 | 1 | 40 | 1 | ||
| >0 mm | 49 | 4.63 (2.53-8.48) | 44 | 3.31 (1.89-5.78) | ||
|
| ||||||
| Ascitic fluid volume | 0.008 | 0.006 | ||||
| ≤500 ml | 41 | 1 | 39 | 1 | ||
| > 500 ml | 35 | 2.17 (1.22-3.85) | 31 | 2.27 (1.27-4.05) | ||
|
| ||||||
| KLK6 mRNA(med)c | 0.035 | 0.343 | ||||
| low | 44 | 1 | 43 | 1 | ||
| high | 46 | 1.78 (1.04-3.03) | 41 | 1.29 (0.76-2.18) | ||
|
| ||||||
| KLK6 mRNA(61st)d | 0.007 | 0.608 | ||||
| low | 54 | 1 | 53 | 1 | ||
| high | 36 | 2.07 (1.22-3.50) | 31 | 1.15 (0.67-1.97) | ||
|
| ||||||
| KLK8 mRNAc | 0.802 | 0.194 | ||||
| low | 44 | 1 | 41 | 1 | ||
| high | 47 | 1.07 (0.64-1.80) | 44 | 1.42 (0.84-2.40) | ||
|
| ||||||
| KLK6+KLK8 mRNAc | 0.053 | 0.047 | ||||
| low | 29 | 1 | 28 | 1 | ||
| high | 61 | 1.82 (0.99-3.33) | 56 | 1.82 (1.01-3.30) | ||
Number of patients
HR: hazard ratio (CI: confidence interval) of univariate Cox regression analysis
Dichotomized into high and low levels by the median
Dichotomized into high and low levels by the 61st percentile
Figure 2. Probability of overall survival and progression-free survival of patients with advanced ovarian cancer (FIGO III/IV) as stratified by KLK6, KLK8 and KLK6+KLK8 mRNA expression levels, respectively, in primary tumor tissues.
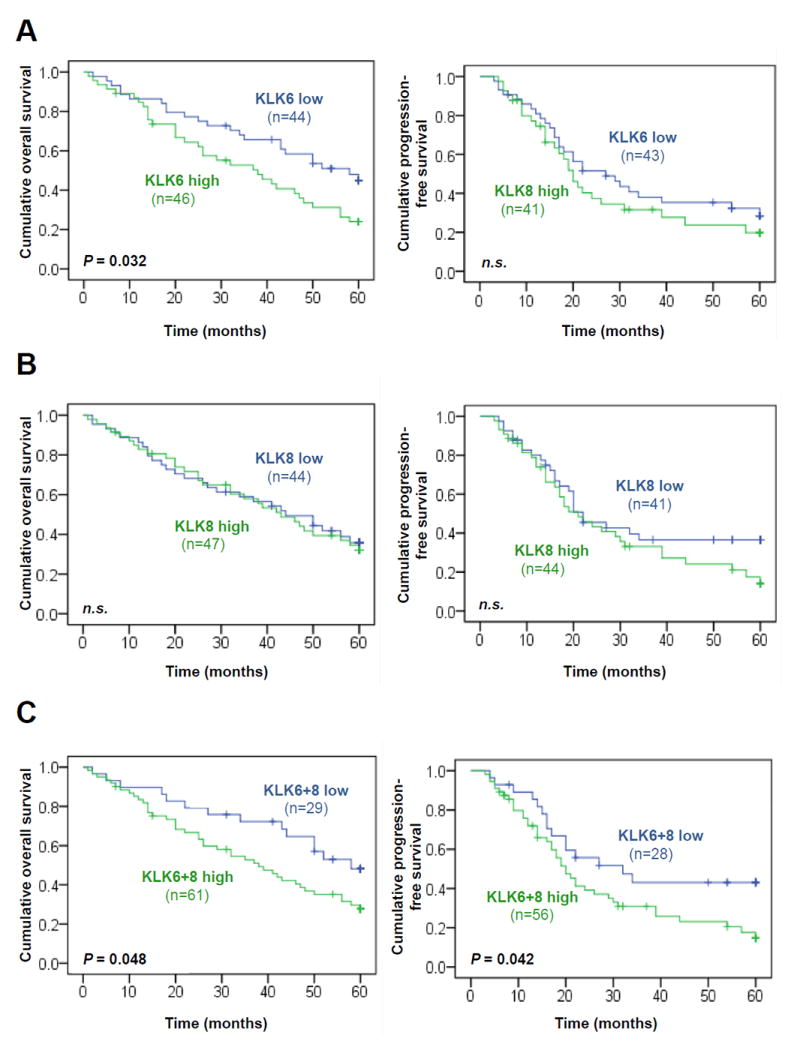
(A) Patients with low KLK6 mRNA expression show significantly better overall survival (Kaplan-Meier analysis, P = 0.032) (left panel) than the group of patients with high KLK6 mRNA expression, but not for progression-free survival (right panel). (B) KLK8 mRNA levels have no impact on both overall survival (left panel) and progression-free survival (right panel). (C) Patients with high KLK6 and/or high KLK8 expression (KLK6+8 high) display significantly shorter overall survival (Kaplan-Meier analysis, P = 0.048) (left panel) and progression-free survival (P = 0.042) (right panel), compared to patients with low mRNA expression levels of both KLK6 and KLK8 (KLK6+8 low). In all of the analyses, KLK6 and KLK8 mRNA levels were dichotomized into low and high by the median.
Association of KLK6, KLK8, and KLK6+KLK8 mRNA values with progression-free (PFS) and overall (OS) survival: multivariable analysis
The independent relationship of KLK6, KLK8, and KLK6+KLK8 with OS and PFS was studied by multivariable Cox hazard regression analysis, including the factors age, ascites fluid volume, and presence of residual tumor mass (base model). In this base model, residual tumor mass is the only clinical parameter representing a predictive marker for OS and PFS (HR = 3.56, 95 % CI = 1.63 – 7.78, P = 0.001 and HR = 2.30, 95 % CI = 1.06 – 4.96, P = 0.034, respectively), while the pre-operative ascitic fluid volume loses its predictive significance for both OS and PFS when subjected to multivariable analysis (Tab. 3). Among the tumor biological factors (added separately to the base model), only KLK6 mRNA levels (dichotomized by either the median or the 61st percentile) significantly contribute to the base model for OS (KLK6 [median]: HR = 1.85, 95 % CI = 1.03 – 3.31, P = 0.039 / KLK6 [61st percentile]: HR = 2.33, 95 % CI = 1.29 – 4.19, P = 0.005). KLK8 mRNA shows a trend towards significance for PFS (HR = 1.66, 95 % CI = 0.92 – 3.02, P = 0.094), while the combined KLK6+KLK8 mRNA values lose their predictive value in the multivariate model for both OS and PFS.
Table 3.
Multivariable Cox regression analysis of clinical outcome in advanced ovarian cancer patients (FIGO III/IV) with respect to clinical parameters and tumor biological factors.
| Clinical parameters | OS | PFS | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| No.a | HR (95% CI)b | P | No.a | HR (95% CI)b | P | |
|
| ||||||
| Age | 0.148 | 0.860 | ||||
| ≤ 60 years | 37 | 1 | 35 | 1 | ||
| > 60 years | 38 | 1.55 (0.86-2.82) | 34 | 1.06 (0.57-1.94) | ||
|
| ||||||
| Residual tumor mass | 0.001 | 0.034 | ||||
| 0 mm | 36 | 1 | 35 | 1 | ||
| > 0 mm | 39 | 3.56 (1.63-7.78) | 34 | 2.30 (1.06-4.96) | ||
|
| ||||||
| Ascitic fluid volume | 0.941 | 0.382 | ||||
| ≤ 500 ml | 41 | 1 | 39 | 1 | ||
| > 500 ml | 34 | 0.97 (0.48-1.98) | 30 | 1.38 (0.67-2.84) | ||
|
| ||||||
| KLK6 mRNA(med)c | 0.039 | 0.539 | ||||
| low | 40 | 1 | 39 | 1 | ||
| high | 35 | 1.85 (1.03-3.31) | 30 | 1.21 (0.66-2.21) | ||
|
| ||||||
| KLK6 mRNA (61st)d | 0.005 | 0.541 | ||||
| low | 49 | 1 | 48 | 1 | ||
| high | 26 | 2.33 (1.29-4.19) | 21 | 1.21 (0.65-2.27) | ||
|
| ||||||
| KLK8 mRNAc | 0.756 | 0.094 | ||||
| low | 40 | 1 | 37 | 1 | ||
| high | 35 | 0.91 (0.50-1.65) | 32 | 1.66 (0.92-3.02) | ||
|
| ||||||
| KLK6+KLK8 mRNAc | 0.317 | 0.113 | ||||
| low | 28 | 1 | 27 | 1 | ||
| high | 47 | 1.40 (0.73-2.68) | 42 | 1.68 (0.89-3.20) | ||
Number of patients
HR: hazard ratio (CI: confidence interval) of multivariable Cox regression analysis. Biological markers were separately added to the base model of clinical parameters: age, residual tumor mass, and ascitic fluid volume.
Dichotomized into high and low levels by the median.
Dichotomized into high and low levels by the 61st percentile.
Discussion
In ovarian cancer, co-expression of KLK6 and KLK8 has been observed (Yousef et al., 2003b; Zheng et al., 2007; Bayani et al., 2008; Avgeris et al., 2012) implicating that both proteases may contribute to tumor invasion and metastasis through cleavage of extracellular matrix components such as laminin, collagens, fibronectin, or vitronectin (Ghosh et al., 2004; Pampalakis and Sotiropoulou, 2007). In the current study, we did not only find co-expression of KLK6 and KLK8 in advanced ovarian cancer (FIGO III/IV), but there was clear evidence as well for coordinated regulation of expression of both factors on the mRNA level (rs = 0.636; P <0.001). Other KLKs such as KLK13 or KLK14 display only a weak correlation to either KLK6 or KLK8 or each other in the same ovarian cancer cohort (with rs-values around 0.4 or lower; N. Ahmed and L. Dettmar, personal communication). Also, on the protein level (Zheng et al., 2007), KLK6 expression was found to be strongly correlated with KLK8 expression in tumor tissue of ovarian cancer patients (rs = 0.829). The coordinated regulation of gene expression may, therefore, point towards an involvement of both proteases in shared cascades/pathways.
In fact, KLK6 and KLK8 are co-expressed in several human organs and tissues, a fact that militates in favor of common pathways of activation, upregulation and/or interrelated participation in numerous physiological and pathological activities. In CNS, KLK6 and KLK8 are expressed abundantly, regulating neural plasticity, development and demyelination. They contribute synergistically to degeneration of the spinal cord after injury, as highly reactive oligodendrocytes express both proteases (Yoshida, 2003; Yousef et al., 2003a; Terayama et al., 2004). Interestingly, Kishibe et al (2007) showed that both KLK6 and KLK8 are upregulated in a parallel manner in skin epidermis upon treatment with 12-O-tetradecanoyl-phorbol acetate (TPA), which induces epidermal hyperplasia similar to psoriatic lesions. Similarly, KLK6 and KLK8 are highly abundant in intercellular spaces of the stratum spinosum and stratum granulosum of the skin and it was proposed that they work together in a cascade-like pathway during wound healing, whereby KLK8 directly or indirectly upregulates and activates KLK6 contributing to keratinocytes migration and proliferation in early stages of wound healing (Kishibe et al., 2007; 2012). Subsequently, KLK6 will activate the protease activated receptor 2 (PAR2) which is believed to be responsible for further keratinocyte differentiation in the late stages of wound healing (Kishibe et al., 2007; 2012). Moreover, both KLKs have been implicated in pathological skin disorders such as psoriasis: here, high KLK6 and KLK8 protein levels were found in synovial fluid, skin lesions, and serum of psoriasis patients. In particular, elevated KLK8 levels were associated with disease severity (Eissa et al., 2013).
In the present study, KLK6 was found to be a strong predictor of OS (but not PFS), i.e. patients with high expression of KLK6 displayed an about two-fold increased risk of death. By multivariable analysis, KLK6 retained its statistical significance as an independent predictive biomarker. Likewise, several other independent studies previously reported that KLK6 overexpression is associated with unfavorable patients’ outcome in colon, pancreas, lung, and also ovarian cancer (Diamandis et al., 2003; Kountourakis et al., 2008; Ruckert et al., 2008; Nathalie et al., 2009; White et al., 2009; Seiz et al., 2012). Notably, KLK6 protein in serum and ascitic fluids of ovarian cancer patients - in contrast to KLK6 from cerebrospinal fluid of healthy individuals - was characteristically associated with a modification of the single N-glycosylation site by α2-6-linked sialic acid which may enable the development of tumor-specific antibodies to detect and/or target KLK6 (Kuzmanov et al., 2009).
Interestingly, in an immunohistochemical study, overexpression of KLK6 protein in ovarian cancer stromal cells was found to be significantly associated with unfavorable prognosis. This fact, together with the role of KLK6 in cleavage of ECM proteins, elucidate the contribution of KLK6 to tumor invasiveness and metastasis (Ghosh et al., 2004; Prezas et al., 2006; Seiz et al., 2012). Moreover, KLK6 in concert with KLKs 4, 5, and 7 provokes transforming growth factor (TGF-β) signaling which may be the cause for taxane drug resistance in ovarian cancer (Loessner et al., 2012). Thus, KLK6 expression may be linked to chemoresistence and this may be the reason for the shortened overall survival of the patients, which all had received adjuvant chemotherapy following surgery. Furthermore, in various in vitro cell culture model systems, KLK6 was shown to support cancer cell proliferation, to inhibit apoptosis, and to increase tumor cell invasiveness and migration through laminin and Matrigel matrices in gastric, colon, non-small cell lung, and pancreatic cancer cells by a wide variety of mechanisms, such as increase of ADAM10 expression, inhibition of E-cadherin expression, or PAR2-mediated activation of the epidermal growth factor receptor (EGFR) which in turn contributes to c-Myc expression regulation and interaction with angiogenic factors (Klucky et al., 2007; Henkhaus et al., 2008; Ruckert et al., 2008; Nathalie et al., 2009; Kim et al., 2011; Kryza et al., 2016).
KLK8 is upregulated in ovarian cancer compared to normal ovaries or ovaries with benign disease (Underwood et al., 1999; Kishi et al., 2003; Yousef et al., 2003b; Hibbs et al., 2004) and has previously been proposed as a favorable prognostic marker. Kishi et al. (2003), Shigemasa et al. (2004), and Borgono et al. (2006) demonstrated that KLK8 protein overexpression either detected by ELISA or immunohistochemistry in ovarian cancer was significantly associated with good prognosis. Maklara et al. (2001) determined KLK8 mRNA expression levels by qPCR and found a favorable prognostic impact of high mRNA expression as well. It is, however, important to note that all the studies mentioned above included also early stage patients (FIGO I and/or II) and other histologic subtypes in addition to high-grade serous carcinoma, and demonstrated that KLK8 expression is higher in early-stage and lower-grade tumors as compared to advanced-stage and high-grade tumors.
Among reports that analyzed the predictive impact of KLK8 expression in ovarian cancer, the study by Kountourakis et al. (2009) was the only one using a patient cohort similar to our study considering only advanced ovarian cancer patients FIGO stage III/IV and 69% high-grade serous subtype. Applying an automated quantitative analysis technique (AQUA), which evaluates tumor protein molecule expression per unit areas, high KLK8 expression was found to be associated with poor outcome (Kountourakis et al., 2009). Thus, elevated KLK8 may mediate some adverse effects in late stages of ovarian cancer. This would be in line with findings from our current study and others showing that expression levels of KLK6 and KLK8 are strongly and positively correlated (Zheng et al., 2007) and that KLK6 is associated with an unfavorable outcome (Diamandis et al., 2003; Kountourakis et al., 2008; White et al., 2009; Seiz et al., 2012). Although, in the present study, KLK8 alone showed no statistically significant impact on both OS and PFS, and, thus, may not represent a strong stand-alone biomarker in ovarian cancer, patients displaying both low KLK6 and low KLK8 intratumoral mRNA expression had a significantly longer OS and PFS compared to patients displaying high KLK6 and/or KLK8 levels (log-rank analysis). It should be stressed again that KLK6 levels alone were statistically significantly associated with OS, but not with PFS. Thus, KLK8, in combination with KLK6, may aid to identify patients with better outcome, especially concerning PFS. Interestingly, KLK6 and KLK8 are members of a multiparametric panel of KLKs (KLK’s 6, 8, 11, and 13), which has been demonstrated to predict patient progression at one year with good accuracy (Zheng et al., 2007).
In conclusion, our current study is the first to quantify KLK6 and KLK8 mRNA expression levels in a homogenous patient cohort of advanced serous ovarian cancer FIGO III/IV. Both mRNAs were found to be widely expressed and the mRNA expression levels were not associated with clinical factors. KLK6 was identified as a statistically independent unfavorable predictive factor for OS, while KLK8 mRNA levels were neither associated with OS nor with PFS. Nevertheless, the use of KLK8 in combination with KLK6 allowed for the identification of a patient group with better outcome (low KLK6 + low KLK8) for both OS and PFS. All in all, together with findings of other studies, KLK6 may be to be considered as validated predictive biomarker in ovarian cancer. Since elevated levels of this proteolytic enzyme is associated with poor patient outcome, KLK6 represents an attractive target for tumor therapy, e.g. via the development of potent and specific inhibitors of its enzymatic activity.
Material and methods
Patients
One hundred patients afflicted with advanced serous ovarian cancer FIGO stage III/IV, treated between 1990 and 2012 at the Department of Obstetrics and Gynecology, Klinikum rechts der Isar, Technische Universität München, Munich, Germany, were enrolled in this study. The study was approved by the local Ethics Committee and written informed consent was obtained from all patients. Median patients’ age at time of surgery was 61 years (range 23-88 years). All patients initially underwent standard stage-related primary radical debulking surgery. Forty-five patients (45%) were optimally debulked with complete removal of all macroscopically visible tumor manifestations. Following surgery, all of the patients received adjuvant treatment according to consensus recommendations at that time, including platinum-based chemotherapy. None of the patients received any neoadjuvant therapy before primary surgery. Median time of follow-up was 41 months for overall survival (OS; range 1 to 262 months after primary tumor resection) and 20 months for progression-free survival (PFS; range 3 to 262 months). Clinical and histomorphological parameters documented at the time of surgery included histologic subtype, absence or presence of residual tumor mass (0 mm [no visible or palpable tumor left after surgery] vs. > 0 mm [any residual tumor left after surgery]) and ascitic fluid volume (estimated preoperatively by vaginal ultrasound). During the analyzed follow-up time of 5 years, 57 of the 85 patients with available data for PFS had relapsed, and 57 of the 91 patients with available data for OS had died.
RNA isolation from cell lines
For preliminary establishment of the quantitative polymerase chain reaction (qPCR) assays for both KLK6 and KLK8, human ovarian cancer OV-MZ-6 cells stably transfected with pRcRSV-derived expression plasmids encoding the complete coding region (pre-pro-proteins) of either KLK6 or KLK8 (OV-KLK6 and OV-KLK8), were employed (Prezas et al., 2006). Total RNA was isolated from these cell lines using the RNeasy Mini Kit (Qiagen, Hilden, Germany).
RNA isolation from tumor tissue
Deep-frozen tumor tissue samples of ovarian cancer patients stored in liquid nitrogen were selected from the tissue storage facility of the Tumor Bank of the Medical Faculty of the Technical University of Munich, Germany. Tissue samples were sliced to obtain 10-20 μg of still-frozen tumor tissue material, which was immediately dissolved in 600 μl of RLT plus buffer (Qiagen) containing 6 μl 2-mercaptoethanol. Simultaneous purification of total DNA and RNA was performed on the automated QIAcube sample preparation machine (Qiagen) using the All Prep DNA/RNA Universal kit for RNA and DNA extraction from tissues (Qiagen) following the manufacturer’s instructions.
In brief, DNA was first purified by use of a DNA spin column, eluted, and stored at -20°C. The flow through of this purification step, containing the RNA, was supplemented with 150 μl chloroform to optimize the RNA purification for samples with higher content of fatty tissues. Samples were thoroughly vortexed and centrifuged (4°C for 3 min, 20,000 × g) to separate the phases. Then, the aqueous phase was carefully transferred to a 2 ml collection tube and an optimized RNA clean-up run on the QIAcube was performed together with a proteinase K digestion step and ethanol supplementation for optimized binding of total RNA to the RNeasy Mini spin column. A DNase I digestion step ensured high-yields of DNA-free RNA, which was eluted in 50 μl RNase-free H2O.
RNA concentration and purity was assessed using the Nano Drop 2000c spectrophotometer and the Nano Drop 2000/2000c software (Thermo Fisher Scientific, Wilmington, USA). Quality of RNA was assessed by determination of absorbance ratios at 260/280 nm and 260/230 nm according to the manufacturer’s instructions. RNA samples were stored at - 80°C until use.
Reverse transcription and cDNA synthesis
Reverse transcription of the isolated RNA (input: 1,000 and 500 ng per reaction for RNA of cell lines and tumor tissues, respectively) was conducted using the cloned AMV first strand cDNA Synthesis Kit (Invitrogen, Darmstadt, Germany) following the manufacturer‘s instructions and final elution of samples in RNAse-free H2O, resulting in a final cDNA concentration of 10 and 5 ng/μl for cell lines and clinical samples, respectively. cDNA samples were stored at - 20°C until further use.
Real-time polymerase chain reaction
To account for sample heterogeneity, different extraction/conversion efficiencies, and mastermix variations, the efficiency of KLK6 and KLK8 assays was validated by standard dilution series (Bustin and Nolan, 2013). A 2-fold dilution series of RNA from both cell lines, OV-KLK6 and OV-KLK8, respectively, with 5 dilution steps (DNA0-DNA4; range 30 - 1.875 ng), was used to establish a standard dilution curve for each KLK6, KLK8, and the housekeeping gene hypoxanthine phosphoribosyltransferase 1 (HPRT1). The logarithm of each known concentration in the dilution series (x-axis) was plotted against the cycle of threshold (Ct) value for the respective sample (y-axis). Linear regression analysis resulted in an R2 coefficient and efficiency (E) was calculated from the linear slope by using the following calculation formula: E = 10exp - (1/slope). 100% efficiency corresponds to an E-value of 2. A deltaE between KLK6 or KLK8 and HPRT1 was calculated for estimation of delta-efficiency-related error margins. Gene specific primers were designed with the Universal Probe Library Assay Design Center software (https://lifescience.roche.com/shop/products/universal-probelibrary-system-assay-design). The following primers (Metabion, Martinsried, Germany) and hydrolysis probes from the Universal Probe Library (Roche, Penzberg, Germany) were used:
KLK6 (numbers for the location of the primers are according to the NCBI entry NM_002774): KLK6-forward (261-280): TGGTGCTGAGTCTGATTGCT and KLK6-reverse (302-320): CGCCATGCACCAACTTATT (reaction concentration: 400 nM each); amplicon size: 60 bp. The assay detects KLK6 mRNA variants A and B, both encoding the identical, full length KLK6 protein. The other known variants C, D, and E, encoding a truncated version of KLK6 are not detected.
KLK8 (NM_007196): KLK8-forward (782-799): CAGCAAAGGGGCTGACAC, KLK8-reverse (869-886): GACCTCCCACAGGGGTCT (reaction concentration: 400 nM each), KLK8-probe: 5’-FAM-TGCCCTGG (reaction concentration: 200 nM); amplicon size: 105 bp. The assay detects the two major KLK8 mRNA transcript variants 1 and 2, encoding full length KLK8, plus variants 3, 5, and 6. mRNA variant 4 (potentially encoding a peptide of only 32 amino acids) is not detected.
HPRT1 (NM_000194): HPRT1-forward (218-241): TGACCTTGATTTATTTTGCATACC, HPRT1-reverse (300-319): CGAGCAAGACGTTCAGTCCT (reaction concentration: 400 nM each), HPRT1-probe: 5’-FAM-GCTGAGGA (reaction concentration: 200 nM); amplicon size: 102 bp.
For KLK6, the assay was optimized to use a SYBR-Green based real-time PCR assay using Brilliant III SYBR Green Mastermix (Agilent Technologies, Darmstadt), while for KLK8 and HPRT1, Taqman-based technology (FAM-labelled Universal Probe Library Taqman probes) with Brilliant III QPCR Master Mix with low ROX (Agilent Technologies, Darmstadt) was used. 10 μl of the respective 2x QPCR mastermix was supplemented with primer, probes and cDNA/controls and adjusted with water to an end volume of 20 μl.
All reactions were performed in triplicates (input: 15 ng/well and 30 ng/well for clinical samples and cell lines, respectively) in 96-well plates. The following cycling program was used: 95°C, 3 min (polymerase activation step), followed by 40 cycles at 95°C, 15 sec (denaturation) plus 60°C, 1 min (annealing/elongation) using the MxPro Software version 4.10. The instrument was set to detect and report fluorescence at each cycle during the 60°C annealing/extension step. Cycle threshold values (Ct) were determined automatically for each marker separately by the MXPro software (version 4.10; standard evaluation settings) by the course of the fluorescent signal readout during the PCR cycle program and including adaptive baseline correction and amplification-based threshold value (in the area of 5-60 % total fluorescence level, averaged over technical replicates). For specificity and sensitivity, two negative controls comprising a no-template control (RNA-free water) and total RNA (30 ng; no reverse transcriptase reaction) of ovarian cancer samples were included in duplicates in each run to test for false-positive results. Additional control runs were performed on a set of patient samples in independent repetitions over all sample batches to assess assay and sample robustness in the study. The different expression levels were normalized to human HPRT and a calibrator (OV-KLK6, OV-KLK8 for KLK6 and KLK8, respectively), which was included in each run to allow normalization of samples. Relative target gene expression was calculated using 2-∆∆Ct where ∆∆Ct = ∆Ct sample - ∆Ct calibrator and ∆Ct = Cttarget − CtHPRT (Pfaffl, 2012). Relative error propagation (EP) was calculated for each ∆Ct analysis step by the following formula: EP(∆Ct) = SQRRoot(((STDEVmarker)2 + (STDEVHPRT.)2)/2) and EP(∆∆Ct) = SQRRoot(((STDEV∆Ctsample)2 + (STDEV∆Ctcalibrator)2)/2). Absolute error was calculated by the following formula: ln 2*EP(∆∆Ct)* 2-∆∆Ct (sample). STDEV stands for standard deviation; SQRRoot for square root; ln for natural logarithm.
Due to possible detection limitation and variances of sample qualities and qPCR efficiencies, the following quality criteria were applied to exclude unassertive results. Samples were excluded from the study if (1) the Ct value for HPRT was >35, (2) the 2exp-∆∆Ct error progression% was >30% even after repetition, and (3) the % STDEV of the 2 exp-∆∆Ct for 2 valid runs was higher than 47.1%.
Quantification of KLK6 antigen concentrations in ovarian cancer tissue extracts
Preparation of extracts from ovarian cancer tissues and determination of KLK6 antigen concentrations in these tumor extracts by a non-commercial in-house ELISA test format were previously described (Dorn et al. 2015).
Statistical analyses
All statistical analyses were performed with the SPSS statistical analysis software (version 20.0; SPSS Inc., Chicago, IL, USA). The association of KLK6 and KLK8 mRNA expression levels with clinical characteristics of the patients was evaluated using the Chi-square test. Correlations between continuous variables of tumor biological markers (KLK6 mRNA, KLK8 mRNA, and KLK6 protein) were calculated using the Mann-Whitney U test and Spearman rank correlation (rs). Associations of tumor biological factors and clinical parameters with patient’s survival were analyzed by Cox univariate and multivariable proportional hazards regression models and expressed as hazard ratio (HR) and its 95% confidence interval (95% CI). The multivariable Cox regression model was adjusted for established clinical parameters in ovarian cancer such as age, presence of residual tumor mass, and ascitic fluid volume. For survival analyses, overall survival (OS) and progression-free survival (PFS) of ovarian cancer patients were used as follow-up end points. Survival curves were plotted according to Kaplan-Meier, using the log-rank model to test for differences. P-values ≤0.05 were considered statistically significant.
Acknowledgments
We thank Elisabeth Schüren for excellent technical assistance. Stefanie Avril is supported by the Clinical and Translational Science Collaborative of Cleveland, from the National Center for Advancing Translational Sciences (NCATS) component of the NIH (KL2TR000440). This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (DFG; DO 1772/1-1 and AV 109/4-1), the Austrian Science Fund (FWF; P25003-B21), and by a mobility grant (Personalized Medicine) from the German Academic Exchange Service (DAAD).
References
- Avgeris M, Mavridis K, Scorilas A. Kallikrein-related peptidases in prostate, breast, and ovarian cancers: from pathobiology to clinical relevance. Biol Chem. 2012;393:301–17. doi: 10.1515/hsz-2011-0260. [DOI] [PubMed] [Google Scholar]
- Bayani J, Paliouras M, Planque C, Shan SJ, Graham C, Squire JA, Diamandis EP. Impact of cytogenetic and genomic aberrations of the kallikrein locus in ovarian cancer. Mol Oncol. 2008;2:250–60. doi: 10.1016/j.molonc.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayani J, Diamandis EP. The physiology and pathobiology of human kallikrein-related peptidase 6 (KLK6) Clin Chem Lab Med. 2012;50:211–33. doi: 10.1515/CCLM.2011.750. [DOI] [PubMed] [Google Scholar]
- Borgono CA, Kishi T, Scorilas A, Harbeck N, Dorn J, Schmalfeldt B, Schmitt M, Diamandis EP. Human kallikrein 8 protein is a favorable prognostic marker in ovarian cancer. Clin Cancer Res. 2006;12:1487–93. doi: 10.1158/1078-0432.CCR-05-2106. [DOI] [PubMed] [Google Scholar]
- Bustin A, Nolan T. Analysis of mRNA expression by real-time PCR. In: Saunders A, Lee A, editors. Real-time PCR: advanced technologies and applications. Caister academic press; Norfolk, UK: 2013. pp. 51–88. [Google Scholar]
- Cané S, Bignotti E, Bellone S, Palmieri M, De las Casas L, Roman JJ, Pecorelli S, Cannon MJ, O’Brien T, Santin AD. The novel serine protease tumor-associated differentially expressed gene-14 (KLK8/Neuropsin/Ovasin) is highly overexpressed in cervical cancer. Am J Obstet Gynecol. 2004;190:60–6. doi: 10.1016/j.ajog.2003.07.020. [DOI] [PubMed] [Google Scholar]
- Diamandis EP, Scorilas A, Fracchioli S, Van Gramberen M, De Bruijn H, Henrik A, Soosaipillai A, Grass L, Yousef GM, Stenman UH, Massobrio M, Van Der Zee AG, Vergote I, Katsaros D. Human kallikrein 6 (hK6): a new potential serum biomarker for diagnosis and prognosis of ovarian carcinoma. J Clin Oncol. 2003;21:1035–43. doi: 10.1200/JCO.2003.02.022. [DOI] [PubMed] [Google Scholar]
- Dorn J, Harbeck N, Kates R, Gkazepis A, Scorilas A, Soosaipillai A, Diamandis E, Kiechle M, Schmalfeldt B, Schmitt M. Impact of expression differences of kallikrein-related peptidases and of uPA and PAI-1 between primary tumor and omentum metastasis in advanced ovarian cancer. Ann Oncol. 2011;22:877–83. doi: 10.1093/annonc/mdq462. [DOI] [PubMed] [Google Scholar]
- Dorn J, Beaufort N, Schmitt M, Diamandis EP, Goettig P, Magdolen V. Function and clinical relevance of kallikrein-related peptidases and other serine proteases in gynecological cancers. Crit Rev Clin Lab Sci. 2014;51:63–84. doi: 10.3109/10408363.2013.865701. [DOI] [PubMed] [Google Scholar]
- Dorn J, Bronger H, Kates R, Slotta-Huspenina J, Schmalfeldt B, Kiechle M, Diamandis EP, Soosaipillai A, Schmitt M, Harbeck N. OVSCORE - a validated score to identify ovarian cancer patients not suitable for primary surgery. Oncol Lett. 2015;9:418–424. doi: 10.3892/ol.2014.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa A, Cretu D, Soosaipillai A, Thavaneswaran A, Pellett F, Diamandis A, Cevikbas F, Steinhoff M, Diamandis EP, Gladman D, Chandran V. Serum kallikrein-8 correlates with skin activity, but not psoriatic arthritis, in patients with psoriatic disease. Clin Chem Lab Med. 2013;51:317–25. doi: 10.1515/cclm-2012-0251. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Ghosh MC, Grass L, Soosaipillai A, Sotiropoulou G, Diamandis EP. Human kallikrein 6 degrades extracellular matrix proteins and may enhance the metastatic potential of tumour cells. Tumour Biol. 2004;25:193–9. doi: 10.1159/000081102. [DOI] [PubMed] [Google Scholar]
- Henkhaus RS, Gerner EW, Ignatenko NA. Kallikrein 6 is a mediator of K-RAS-dependent migration of colon carcinoma cells. Biol Chem. 2008;389:757–64. doi: 10.1515/BC.2008.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs K, Skubitz KM, Pambuccian SE, Casey RC, Burleson KM, Oegema TR, Jr, Thiele JJ, Grindle SM, Bliss RL, Skubitz AP. Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol. 2004;165:397–414. doi: 10.1016/S0002-9440(10)63306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JT, Song EY, Chung KS, Kang MA, Kim JW, Kim SJ, Yeom YI, Kim JH, Kim KH, Lee HG. Up-regulation and clinical significance of serine protease kallikrein 6 in colon cancer. Cancer. 2011;117:2608–19. doi: 10.1002/cncr.25841. [DOI] [PubMed] [Google Scholar]
- Kipps E, Tan DS, Kaye SB. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat Rev Cancer. 2013;13:273–82. doi: 10.1038/nrc3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, Grass L, Soosaipillai A, Scorilas A, Harbeck N, Schmalfeldt B, Dorn J, Mysliwiec M, Schmitt M, Diamandis EP. Human kallikrein 8, a novel biomarker for ovarian carcinoma. Cancer Res. 2003;63:2771–4. [PubMed] [Google Scholar]
- Kishibe M, Bando Y, Terayama R, Namikawa K, Takahashi H, Hashimoto Y, Ishida-Yamamoto A, Jiang YP, Mitrovic B, Perez D, Iizuka H, Yoshida S. Kallikrein 8 is involved in skin desquamation in cooperation with other kallikreins. J Biol Chem. 2007;282:5834–41. doi: 10.1074/jbc.M607998200. [DOI] [PubMed] [Google Scholar]
- Kishibe M, Bando Y, Tanaka T, Ishida-Yamamoto A, Iizuka H, Yoshida S. Kallikrein-related peptidase 8-dependent skin wound healing is associated with upregulation of kallikrein-related peptidase 6 and PAR2. J Invest Dermatol. 2012;132:1717–24. doi: 10.1038/jid.2012.18. [DOI] [PubMed] [Google Scholar]
- Klucky B, Mueller R, Vogt I, Teurich S, Hartenstein B, Breuhahn K, Flechtenmacher C, Angel P, Hess J. Kallikrein 6 induces E-cadherin shedding and promotes cell proliferation, migration, and invasion. Cancer Res. 2007;67:8198–206. doi: 10.1158/0008-5472.CAN-07-0607. [DOI] [PubMed] [Google Scholar]
- Kolin DL, Sy K, Rotondo F, Bassily MN, Kovacs K, Brezden-Masley C, Streutker CJ, Yousef GM. Prognostic significance of human tissue kallikrein-related peptidases 6 and 10 in gastric cancer. Biol Chem. 2014;395:1087–93. doi: 10.1515/hsz-2014-0143. [DOI] [PubMed] [Google Scholar]
- Kountourakis P, Psyrri A, Scorilas A, Camp R, Markakis S, Kowalski D, Diamandis EP, Dimopoulos MA. Prognostic value of kallikrein-related peptidase 6 protein expression levels in advanced ovarian cancer evaluated by automated quantitative analysis (AQUA) Cancer Sci. 2008;99:2224–9. doi: 10.1111/j.1349-7006.2008.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kountourakis P, Psyrri A, Scorilas A, Markakis S, Kowalski D, Camp RL, Diamandis EP, Dimopoulos MA. Expression and prognostic significance of kallikrein-related peptidase 8 protein levels in advanced ovarian cancer by using automated quantitative analysis. Thromb Haemost. 2009;101:541–6. [PubMed] [Google Scholar]
- Kryza T, Silva ML, Loessner D, Heuze-Vourc’h N, Clements JA. The kallikrein-related peptidase family: Dysregulation and functions during cancer progression. Biochimie. 2016;122:283–99. doi: 10.1016/j.biochi.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Kuzmanov U, Jiang N, Smith CR, Soosaipillai A, Diamandis EP. Differential N-glycosylation of kallikrein 6 derived from ovarian cancer cells or the central nervous system. Mol Cell Proteomics. 2009;8:791–8. doi: 10.1074/mcp.M800516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner D, Quent VM, Kraemer J, Weber EC, Hutmacher DW, Magdolen V, Clements JA. Combined expression of KLK4, KLK5, KLK6, and KLK7 by ovarian cancer cells leads to decreased adhesion and paclitaxel-induced chemoresistance. Gynecol Oncol. 2012;127:569–78. doi: 10.1016/j.ygyno.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Magklara A, Scorilas A, Katsaros D, Massobrio M, Yousef GM, Fracchioli S, Danese S, Diamandis EP. The human KLK8 (neuropsin/ovasin) gene: identification of two novel splice variants and its prognostic value in ovarian cancer. Clin Cancer Res. 2001;7:806–11. [PubMed] [Google Scholar]
- Nathalie HV, Chris P, Serge G, Catherine C, Benjamin B, Claire B, Christelle P, Briollais L, Pascale R, Marie-Lise J, Yves C. High kallikrein-related peptidase 6 in non-small cell lung cancer cells: an indicator of tumour proliferation and poor prognosis. J Cell Mol Med. 2009;13:4014–22. doi: 10.1111/j.1582-4934.2009.00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampalakis G, Sotiropoulou G. Tissue kallikrein proteolytic cascade pathways in normal physiology and cancer. Biochim Biophys Acta. 2007;1776:22–31. doi: 10.1016/j.bbcan.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Prat J. Ovarian carcinomas: five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012;460:237–49. doi: 10.1007/s00428-012-1203-5. [DOI] [PubMed] [Google Scholar]
- Prat J. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int J Gynaecol Obstet. 2014;124:1–5. doi: 10.1016/j.ijgo.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Prezas P, Arlt MJ, Viktorov P, Soosaipillai A, Holzscheiter L, Schmitt M, Talieri M, Diamandis EP, Kruger A, Magdolen V. Overexpression of the human tissue kallikrein genes KLK4, 5, 6, and 7 increases the malignant phenotype of ovarian cancer cells. Biol Chem. 2006;387:807–11. doi: 10.1515/BC.2006.102. [DOI] [PubMed] [Google Scholar]
- Pfaffl W. Quantification strategies in real-time Polymerase Chain Reaction. In: Filion M, editor. Quantitative real-time PCR in applied microbiology. Caister Academic press; Norfolk, UK: 2012. pp. 53–61. [Google Scholar]
- Planque C, Choi YH, Guyetant S, Heuze-Vourc’h N, Briollais L, Courty Y. Alternative splicing variant of kallikrein-related peptidase 8 as an independent predictor of unfavorable prognosis in lung cancer. Clin Chem. 2010;56:987–97. doi: 10.1373/clinchem.2009.138917. [DOI] [PubMed] [Google Scholar]
- Prassas I, Eissa A, Poda G, Diamandis EP. Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat Rev Drug Discov. 2015;14:183–202. doi: 10.1038/nrd4534. [DOI] [PubMed] [Google Scholar]
- Ruckert F, Hennig M, Petraki CD, Wehrum D, Distler M, Denz A, Schroder M, Dawelbait G, Kalthoff H, Saeger HD, Diamandis EP, Pilarsky C, Grutzmann R. Co-expression of KLK6 and KLK10 as prognostic factors for survival in pancreatic ductal adenocarcinoma. Br J Cancer. 2008;99:1484–92. doi: 10.1038/sj.bjc.6604717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarisbrick IA. Kallikrein-related peptidases. Characterization, regulation, and interactions within the protease web. Vol. 1. DeGruyter; Berlin, Germany: 2012. Physiological and pathophysiological roles of kallikrein-related peptidases in the central nervous system; pp. 349–372. [Google Scholar]
- Seiz L, Dorn J, Kotzsch M, Walch A, Grebenchtchikov NI, Gkazepis A, Schmalfeldt B, Kiechle M, Bayani J, Diamandis EP, Langer R, Sweep FC, Schmitt M, Magdolen V. Stromal cell-associated expression of kallikrein-related peptidase 6 (KLK6) indicates poor prognosis of ovarian cancer patients. Biol Chem. 2012;393:391–401. doi: 10.1515/hsz-2011-0264. [DOI] [PubMed] [Google Scholar]
- Sher YP, Chou CC, Chou RH, Wu HM, Wayne Chang WS, Chen CH, Yang PC, Wu CW, Yu CL, Peck K. Human kallikrein 8 protease confers a favorable clinical outcome in non-small cell lung cancer by suppressing tumor cell invasiveness. Cancer Res. 2006;66:11763–70. doi: 10.1158/0008-5472.CAN-06-3165. [DOI] [PubMed] [Google Scholar]
- Shigemasa K, Tian X, Gu L, Tanimoto H, Underwood LJ, O’Brien TJ, Ohama K. Human kallikrein 8 (hK8/TADG-14) expression is associated with an early clinical stage and favorable prognosis in ovarian cancer. Oncol Rep. 2004;11:1153–9. [PubMed] [Google Scholar]
- Shimizu-Okabe C, Yousef GM, Diamandis EP, Yoshida S, Shiosaka S, Fahnestock M. Expression of the kallikrein gene family in normal and Alzheimer’s disease brain. Neuroreport. 2001;12:2747–2751. doi: 10.1097/00001756-200108280-00031. [DOI] [PubMed] [Google Scholar]
- Tatebe H, Watanabe Y, Kasai T, Mizuno T, Nakagawa M, Tanaka M, Tokuda T. Extracellular neurosin degrades alpha-synuclein in cultured cells. Neurosci Res. 2010;67:341–6. doi: 10.1016/j.neures.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Terayama R, Bando Y, Takahashi T, Yoshida S. Differential expression of neuropsin and protease M/neurosin in oligodendrocytes after injury to the spinal cord. Glia. 2004;48:91–101. doi: 10.1002/glia.20058. [DOI] [PubMed] [Google Scholar]
- Underwood LJ, Tanimoto H, Wang Y, Shigemasa K, Parmley TH, O’Brien TJ. Cloning of tumor-associated differentially expressed gene-14, a novel serine protease overexpressed by ovarian carcinoma. Cancer Res. 1999;59:4435–9. [PubMed] [Google Scholar]
- Vakrakou A, Devetzi M, Papachristopoulou G, Malachias A, Scorilas A, Xynopoulos D, Talieri M. Kallikrein-related peptidase 6 (KLK6) expression in the progression of colon adenoma to carcinoma. Biol Chem. 2014;395:1105–17. doi: 10.1515/hsz-2014-0166. [DOI] [PubMed] [Google Scholar]
- Wang SM, Mao J, Li B, Wu W, Tang LL. Expression of KLK6 protein and mRNA in primary breast cancer and its clinical significance. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2008;24:1087–9. [PubMed] [Google Scholar]
- White NM, Mathews M, Yousef GM, Prizada A, Popadiuk C, Dore JJ. KLK6 and KLK13 predict tumor recurrence in epithelial ovarian carcinoma. Br J Cancer. 2009;101:1107–13. doi: 10.1038/sj.bjc.6605280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S. Kallikrein-family serine protease in the central nervous system. Kaibogaku Zasshi. 2003;78:77–82. [PubMed] [Google Scholar]
- Yousef GM, Kishi T, Diamandis EP. Role of kallikrein enzymes in the central nervous system. Clin Chim Acta. 2003a;329:1–8. doi: 10.1016/s0009-8981(03)00004-4. [DOI] [PubMed] [Google Scholar]
- Yousef GM, Polymeris ME, Yacoub GM, Scorilas A, Soosaipillai A, Popalis C, Fracchioli S, Katsaros D, Diamandis EP. Parallel overexpression of seven kallikrein genes in ovarian cancer. Cancer Res. 2003b;63:2223–7. [PubMed] [Google Scholar]
- Yousef GM, Borgono CA, Popalis C, Yacoub GM, Polymeris ME, Soosaipillai A, Diamandis EP. In-silico analysis of kallikrein gene expression in pancreatic and colon cancers. Anticancer Res. 2004;24:43–51. [PubMed] [Google Scholar]
- Zheng Y, Katsaros D, Shan SJ, de la Longrais IR, Porpiglia M, Scorilas A, Kim NW, Wolfert RL, Simon I, Li L, Feng Z, Diamandis EP. A multiparametric panel for ovarian cancer diagnosis, prognosis, and response to chemotherapy. Clin Cancer Res. 2007;13:6984–92. doi: 10.1158/1078-0432.CCR-07-1409. [DOI] [PubMed] [Google Scholar]


