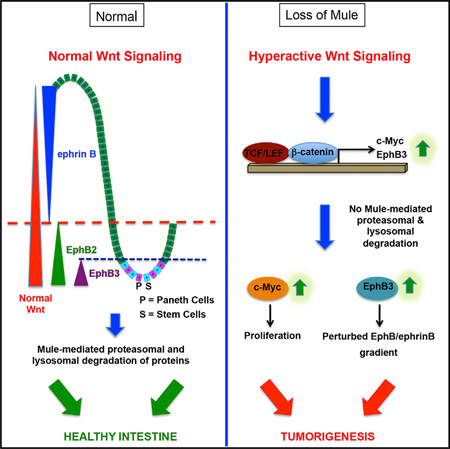
SUMMARY
The E3 ubiquitin ligase Mule is often overexpressed in human colorectal cancers, but its role in gut tumorigenesis is unknown. Here, we show in vivo that Mule controls murine intestinal stem and progenitor cell proliferation by modulating Wnt signaling via c-Myc. Mule also regulates protein levels of the receptor tyrosine kinase EphB3 by targeting it for proteasomal and lysosomal degradation. In the intestine, EphB/ephrinB interactions position cells along the crypt-villus axis and compartmentalize incipient colorectal tumors. Our study thus unveils an important new avenue by which Mule acts as an intestinal tumor suppressor by regulation of the intestinal stem cell niche.
Graphical Abstract
INTRODUCTION
Human colorectal carcinogenesis follows a defined sequence of histopathological changes associated with specific genetic alterations (Fearon and Vogelstein, 1990). These alterations most often affect Wnt signaling pathway components, notably the adenomatous polyposis coli (APC) gene (Groden et al., 1991; Nagase and Nakamura, 1993). Loss of APC function drives aberrant Wnt activation, resulting in β-catenin translocation from cytoplasm to nucleus and constitutive activation of β-catenin/ TCF target genes (Clevers, 2006). Two such targets relevant to our study are c-Myc and EphB3.
c-Myc is overexpressed in 70% of human colorectal cancers (Augenlicht et al., 1997). Transcription regulated by c-Myc is context dependent and drives responses ranging from increased proliferation to apoptosis (Dang et al., 2006). The abnormalities in intestinal cell proliferation, migration, differentiation, and apoptosis resulting from APC inactivation depend entirely on c-Myc (Sansom et al., 2007).
The EphB receptor tyrosine kinases are direct Wnt/β-catenin targets involved in patterning the intestinal crypt-villus axis (Batlle et al., 2002). Once engaged by membrane-bound ephrins, EphB receptors (EphB) mediate bi-directional signaling that dictates intestinal cell positioning (Himanen et al., 2001). In normal intestine, a gradient of EphB expression prevails, with the highest EphB levels at the crypt base. Conversely, an inverse gradient of ephrin expression exists, with the highest levels of these ligands at the villus tip (Batlle et al., 2002). EphB3-deficient (EphB3−/−) mice show mispositioned Paneth cells that are no longer concentrated in the crypts but instead scattered along the crypt-villus axis (Batlle et al., 2002).
The E3 ubiquitin ligase Mule, also known as Huwe1 and Arf-BP1, mainly targets substrates for degradation by attaching lysine 48 (K48)-linked polyubiquitin (pUb) chains. However, Mule-mediated K63-linked ubiquitination (Ubn) of c-Myc promotes its transcriptional activity (Adhikary et al., 2005), and that of the Wnt pathway component Disheveled (Dvl) inhibits the Dvl multimerization necessary for Wnt signaling (de Groot et al., 2014). The total-Mule-knockout mouse is embryonic lethal (Hao et al., 2012; Kon et al., 2012), and studies of conditional Mule mutants conflict on whether Mule is an oncogene or a tumor suppressor (Hao et al., 2012; Inoue et al., 2013; Kon et al., 2012; Zhao et al., 2008). In situ hybridization of colon tumor microarrays showed that Mule increased with tumor grade, suggesting that Mule is an oncogene (Adhikary et al., 2005). However, our previous study of Mule deficiency in a mouse model of 7,12-dimethylbenz[a]-anthracene (DMBA)/12-O-tetradecanoylphorbol-13-acetate (PMA)-induced skin cancer demonstrated that Mule is a tumor suppressor in skin (Inoue et al., 2013). Here, we show that Mule regulates intestinal cell sorting via EphB3 Ubn and also suppresses tumors by regulating c-Myc so as to fine-tune the Wnt pathway and dampen the proliferation of stem and progenitor cells.
RESULTS
Loss of Mule Exacerbates the APCmin Phenotype
To clarify Mule’s role in colon cancer, we crossed Mulefl/fl(y) (Mule cKO) mice (Hao et al., 2012) to Villin-Cre mice (Madison et al., 2002), in which Cre-mediated gene deletion occurs in intestinal epithelia starting at embryonic day 12.5 (Madison et al., 2002). We then crossed Mule cKO;Villin-Cre mice to APCmin mice, a mouse model of colon cancer (Su et al., 1992), to generate Mule cKO;Villin-Cre;APCmin mice (designated Mule cKO APCmin). We followed cohorts of control APCmin mice (n = 20) and Mule cKO APCmin mice (n = 33) and sacrificed them at predetermined humane endpoints. While APCmin mice lived for 4–9 months (mean 5 months), all Mule cKO APCmin mice had died by age 2–6 months (mean 4 months) (Figure 1A). Control Villin-Cre APCmin mice died with kinetics similar to APCmin mice, confirming that the Villin-Cre construct is not driving disease in Mule cKO APCmin mice. Mule cKO APCmin mice exhibited a ~9-fold increase in total adenoma numbers in small intestine and colon compared to age-matched APCmin littermates (Figures 1B–1E), with a 4-fold difference apparent when mice of all ages were compared (Figures S1A and S1B). Villin-Cre APCmin and APCmin mice had comparable numbers of adenomas (Figures S1A and S1B). Most adenomas were low-grade dysplasias, although some were high-grade dysplasias or intramucosal carcinomas (Figures S1C and S1D). Our finding that Mule deficiency accelerates adenoma development in APCmin mice indicates that Mule is a tumor suppressor in the gut.
Figure 1. Loss of Mule Exacerbates the APCmin Phenotype.
(A) Kaplan-Meier analysis of tumor-free survival of APCmin, Villin-Cre APCmin, and Mule cKO APCmin mice.
(B and C) Total adenoma numbers in small intestine (B) and colon (C) of the indicated strains. Data are mean ± SEM (n = 5).
(D and E) Representative H&E-stained sections of the small intestines in (B); scale bar, 1 mm.
See also Figures S1A–S1D.
Mule-Deficient APCmin Mice Exhibit Hyperactivated Wnt/β-Catenin Signaling and Develop Adenomas Populated by Intestinal Stem Cells and Paneth Cells
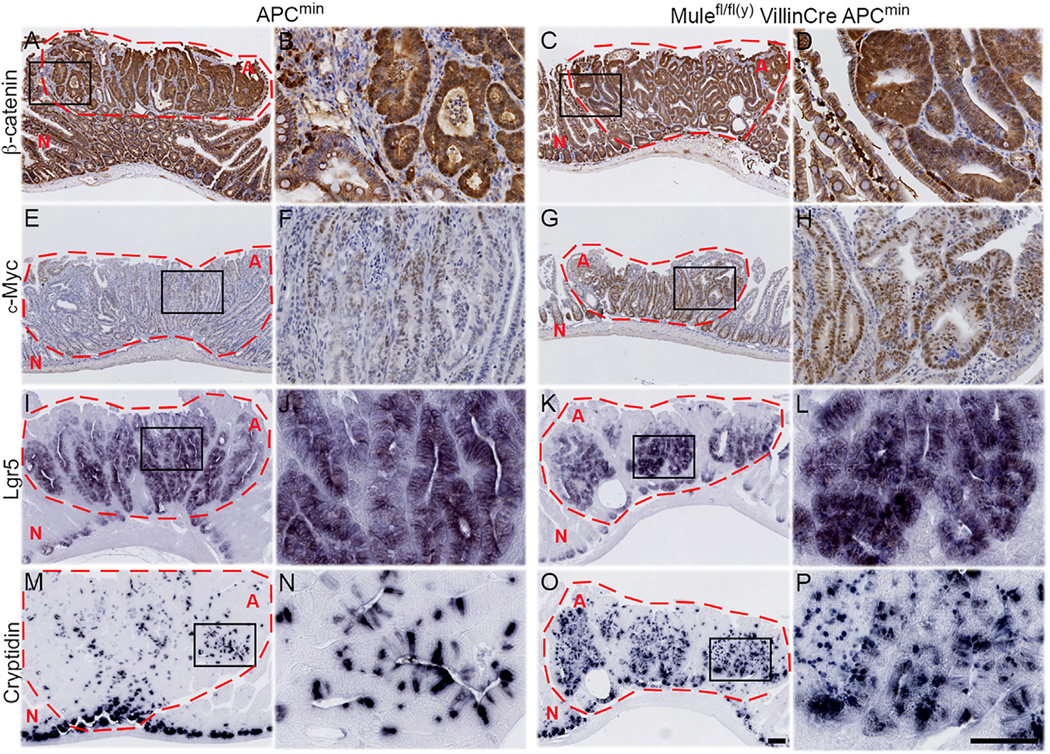
Because Wnt is the main driver of intestinal cell proliferation, we compared nuclear β-catenin levels in adenomas of APCmin and Mule cKO APCmin mice. Nuclear β-catenin was observed in adenomas in both strains (Figures 2A–2D). Mule cKO APCmin adenomas contained more Ki67+ proliferating cells than APCmin adenomas (Figures S1E–S1H) but showed no difference in apoptosis (Figures S1I–S1L). Most Mule cKO APCmin adenomas also contained more c-Myc+ cells than APCmin adenomas (Figures 2E–2H). Immunohistochemistry (IHC) to detect L-fatty acid binding protein (LFABP; a marker for enterocytes), periodic acid Schiff (PAS; goblet cells), chromogranin A (neuroendocrine cells), lysozyme or cryptidin (Paneth cells), and Olmf4 or Lgr5 (intestinal stem cells [ISCs]) revealed that Mule cKO APCmin adenomas were more heavily populated with ISCs (Figures 2I–2L, S1W, and S1X) and Paneth cells (Figures 2M–2P, S1S, and S1T) than APCmin adenomas. Thus, loss of Mule in the APCmin background promotes an increase in c-Myc that enhances cellular proliferation and increases numbers of adenomas populated by greater numbers of ISC and Paneth cells.
Figure 2. Mule Deficiency in the APCmin Background Hyperactivates the Wnt/β-Catenin Pathwayand Generates Adenomas Populated by ISC and Paneth Cells.
Adenomas from APCmin (A, B, E, F, I, J, M, and N) and Mule cKO APCmin (C, D, G, H, K, L, O, and P) littermates were immunostained to detect β-catenin (A–D,), c-Myc (E–H), Lgr5 (I–L; ISC marker), and cryptidin (M,N,O,P; Paneth cell marker). (A, C, E, G, I, K, M, and O)Low-magnification images; scale bar, 100µm. (B, D, F, H, J, L, N, and P) High-magnification images of the boxed areas within the adenomas in the low-magnification images; scale bar, 100 µm. Data are representative of five mice per group. Adenomas are circled in red and marked with red “A.” Normal tissue is marked with red “N.” See also Figures S1E–S1X.
Loss of Mule Alone Is Sufficient to Promote Tumorigenesis
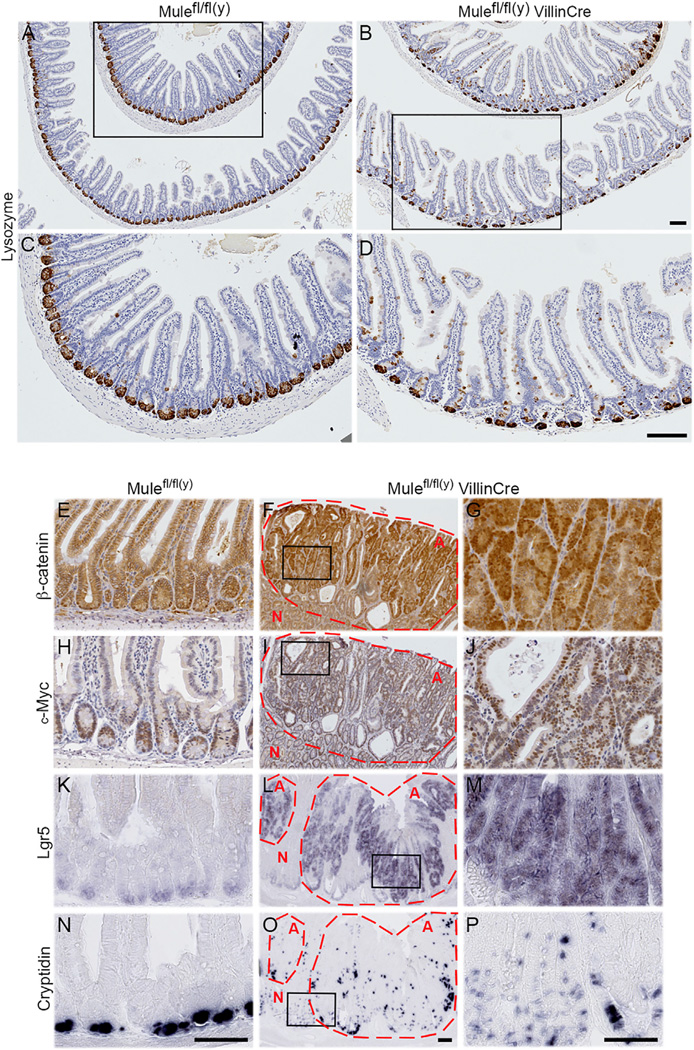
To isolate the contribution of Mule deficiency to the tumorigenesis observed in Mule cKO APCmin mice, we studied our Mulefl/fl(y) Villin-Cre (Mule cKO) mice in more detail. To determine Mule’s normal localization in the gut, we performed in situ hybridization of intestinal tissue from Lgr5-EGFP-CreERT2+ (Barker et al., 2007) Apcfl/fl mice and demonstrated that Mule is found within the ISC niche (Figure S2A). This result was confirmed by IHC for GFP, to mark Lgr5+ stem cells (Figure S2B), and lysozyme staining to mark Paneth cells (Figure S2C). Additionally, RNAscope in situ hybridization which employs specific RNA probes to detect mRNA expression of a gene of interest at the single cell level, showed that in WT tissue ~5-fold more crypt cells coexpress both Mule and Lgr5 mRNA puncta (Figures S2D and S2E) than Mule mRNA alone. Immunoblotting of isolated villus and crypt fractions from three pairs of Mule cKO (normal APC) and Mulefl/fl(y) wild-type (WT) littermates confirmed the presence of Mule in intestinal crypts (Figure S2F). However, although Mule cKO mutants appeared to develop normally, they showed an increase in the number of BrdU+ cells per crypt in small intestine and colon at 4 months of age compared to WT controls (Figures S2G and S2H). Consequently, Mule cKO intestine exhibited enlarged crypts and elongated villi (Figures 3A and 3B). No defects were detected in cell migration along the crypt-villus axis (Figure S2I), in cell death (Figures S2J–S2L), or in the differentiation of five major intestinal cell types, but Paneth cells were strikingly mislocalized in Mule cKO intestine (Figure 3A–D.
Figure 3. Loss of Mule Alone Promotes Tumorigenesis.
(A–D) Sections of small intestines of the indicated mouse strains (age 4 months, n = 3/group) stained to detect lysozyme. (A and B) Low-magnification images; scale bars, 100 µm. (C and D) High-magnification images of the boxed areas in (A) and (B); scale bars, 100 µm.
(E–P) Sections of normal WT intestine (E, H, K, and N) and spontaneous Mule cKO adenomas (F, G, I, J, L, M, O, and P) from 1- to 2-year-old mice (n = 30/group) stained to detect β-catenin (E–G), c-Myc (H–J), LGR5 (K–M), or cryptidin-4 (N–P). (E, H, K, and N) WT intestine; scale bar, 100 µm. (F, I, L, and O). Low-magnification images of Mule cKO adenomas; scale bar, 100 µm. (G, J, M, and P) High-magnification images of the boxed areas within the Mule cKO adenomas shown in (F, I, L, and O); scale bars, 100 µm. Adenomas are circled in red and marked with red “A.” Normal tissue is marked with red “N.” See also Figures S2–S4.
To determine if Mule cKO mice spontaneously developed adenomas, a cohort was aged for 2 years, by which time 50% had indeed developed abnormal growths (Figure S3A–S3D). Intriguingly, unlike Mule cKO APCmin adenomas, ~20% of the lesions in Mule cKO mice progressed to cancers. When we stained Mule cKO adenomas to detect β-catenin and histological markers, we observed abundant β-catenin in adenoma cell nuclei (Figures 3E–3G and S3E–S3P), suggesting that Mule’s absence hyperactivates Wnt signaling. Indeed, in Mule cKO crypt cells, the Wnt targets Axin-2, MMP7, Tiam1, Sox9, and CD44 were all upregulated compared to WT controls (Figure S3Q). RNAscope in situ hybridization revealed that Mule co-localizes with Lgr5+ stem cells in APCmin adenomas and accounts for ~45% of the cells within an adenoma (Figures S3R–S3U). Thus, Mule is expressed in cancer stem cells (CSCs) and may itself be amplified by Wnt signaling.
c-Myc is both a Mule substrate and a downstream Wnt target, so that APC inactivation that hyperactivates Wnt signaling increases c-Myc expression (Sansom et al., 2007). To assess the contribution of Mule deficiency to the increased c-Myc+ cells in Mule cKO APCmin intestine, we examined the guts of adult Mule cKO mice. The enlarged crypts of Mule-deficient intestines housed ~2-fold more c-Myc+ cells than WT crypts (Figures S4A–S4C). β-Catenin remained cytoplasmic in Mule cKO cells (Figures S4D and S4E). Adenomas in aged Mule cKO mice contained ~65% c-Myc+ cells (Figures 3H–3J and S4F) and were largely populated by Lgr5+Olmf4+ ISCs (Figures 3K–3M, S3O, and S3P) and cryptidin-4+lysozyme+ Paneth cells (Figures 3N–3P, S3K, and S3L).
Next, we generated intestinal organoids (Sato et al., 2011) as a model system to study stem cell-driven proliferation and gene expression in our Mule cKO versus WT mice in vitro. We found that Mule cKO organoids initially grew with faster kinetics than WT organoids, which was of interest since proliferation in organoids is driven by c-Myc expression in Lgr5+ stem cells. Mule cKO organoids, however, showed a growth deficit by day 44 (Figures S4G–S4O). The mutant organoids were highly disorganized and Paneth cells were mislocalized. By day 66, the Mule cKO organoids had become undifferentiated spherical cysts (Figures S4M–S4O), a morphology that correlates with elevated Wnt signaling and is observed in APC-deficient organoids (Fatehullah et al., 2013). RT-PCR examination of RNA from Mule cKO and WT organoids confirmed that the Wnt targets c-Myc, Axin-2, Sox9, CD44, and Ascl2 were all upregulated in mutant organoids, and that the stem cell markers Lgr5 and Olmf4 were increased 3-and 5-fold, respectively (Figure S4P). Thus, Mule regulates c-Myc in intestinal cells to dampen proliferation and promote differentiation, and thereby opposes ISC expansion.
Loss of c-Myc in Mule cKO Mice Restores Normal Cell Proliferation but Does Not Rescue Paneth Cell Mispositioning
c-Myc deficiency rescues phenotypes of APC ablation (Sansom et al., 2004, 2007); we therefore tested if loss of c-Myc could also mitigate the abnormalities of Mule cKO mice. Mule cKO mice were crossed with c-Mycfl/flVillin-Cre (c-Myc cKO) mice to generate Mule c-Myc cDKO mutants. Since Villin-Cre-mediated deletion of c-Myc is never complete and some c-Myc+ cells escape to repopulate the crypts (Muncan et al., 2006), we immunostained Mule cKO and Mule c-Myc cDKO intestines to compare c-Myc levels. At 4 months, crypts in double mutant intestines actually contained more c-Myc+ cells than in single mutant crypts (Figures S5A and S5B), possibly due to the increased Wnt pathway activity that occurs in the absence of Mule. The aberrant morphology of c-Myc-deficient crypts was consistent with c-Myc’s role in crypt formation (Bettess et al., 2005). Mule cKO intestine displayed ~15 more BrdU+ cells/crypt than WT controls (Figures 4A, 4B, and 4E), while c-Myc KO and Mule c-Myc cDKO crypts, despite their repopulation by c-Myc+ cells, both contained ~10 BrdU+ cells/crypt (Figures 4C–4E, S5C, and S5D). c-Myc cKO and Mule c-Myc cDKO crypt cells were also smaller than WT and Mule cKO crypt cells. These results are in line with published data showing that c-Myc-deficient crypt cells proliferate slowly and are much smaller than WT crypt cells (Muncan et al., 2006).
Figure 4. Loss of c-Myc in Mule cKO Mice Restores Intestinal Cell Proliferation but Does Not Rescue Paneth Cell Mispositioning.
(A–D) BrdU staining of intestines from the indicated mouse strains (age 4 months, n = 3/group). Scale bar, 100 µm.
(E) Quantitation of BrdU+ cells/crypt in the mice in (A)-(D). Data are the mean ± SEM (n = 3).
(F–M) Staining to detect Olmf4 (F–I) and cryptidin-4 (J–M) in intestines of the indicated strains (age ~2 years, n = 10/group). Scale bar, 100 µm.
See also Figures S5 and S6A–S6H.
We previously showed that Mule-mediated regulation of the c-Myc/Miz1 complex prevents transcriptional repression of the cell-cycle inhibitors p21 and p15 (Inoue et al., 2013). However, Peter et al. proposed that Mule continually targets Miz1 for degradation, derepressing c-Myc target genes that drive the growth of colon cancer cells in vitro and in vivo (Peter et al., 2014). When we compared Miz1 and c-Myc protein levels in Mule cKO and WT organoids by immunoblotting, there was more c-Myc protein after the loss of Mule but there was no difference in Miz1 (Figure S5E). Levels of both c-Myc and Miz1 were comparable in cancer stem cells (CSCs) derived from adenomas isolated from APCmin or aged Mule cKO mice and cultured as organoids (Figure S5F). Thus, an absence of Mule does not stabilize Miz1 in CSCs. P21 was not detectable by immunoblot in either WT or Mule cKO organoids, but IHC revealed p21 expression in WT and Mule cKO cells differentiating at the crypt-villus interface (Figures S5G and S5H). In addition, IHC revealed upregulation of the cell-cycle markers cyclin D, E, and A and the Myc target B23 in Mule cKO adenomas (Figures S5I–S5T). Finally, RNAscope in situ hybridization demonstrated that ~2-fold more cells coexpress c-Myc and Lgr5 mRNA puncta in Mule cKO APCmin adenomas than in APCmin adenomas (Figures S6A, S6B, S6D, S6E, and S6G). Even in normal intestine, Mule cKO crypts have ~2-fold more cells that coexpress Lgr5+ and c-Myc+ puncta than WT crypts (Figures S6C, S6F, and S6H). Thus, in our system, the loss of Mule leads to increased c-Myc at the transcriptional and protein level, which in turn activates c-Myc target genes and cell-cycle progression in both normal cells and CSCs. The fact that CSCs isolated from APCmin adenomas express c-Myc comparable to that seen in CSCs isolated from Mule cKO adenomas despite robust Mule expression is a bit puzzling. Thus, we acknowledge the possibility that in addition to our findings, other distinct mechanisms of c-Myc regulation may be involved.
We next investigated if the ISC expansion seen in aged Mule cKO mice was rescued by c-Myc deletion. No ISC expansion was observed in 2-year-old Mule c-Myc cDKO mice (Figures 4F–4I), but their Paneth cells remained mislocalized (Figures 4J–4M). In APCmin mice, Paneth cells are mislocalized because Wnt is hyperactivated, which increases EphB and c-Myc expression and disturbs the EphB/ephrinB gradient (Batlle et al., 2002; Sansom et al., 2004). Thus, we were surprised that ablation of c-Myc in Mule-deficient mice did not restore normal Paneth cell localization. We then surmised that Mule must have additional effects on the EphB/ephrinB gradient.
EphB3 Is Overexpressed in Mule cKO Intestinal Crypts
EphB2, EphB3, and their shared ligand ephrinB1 are differentially expressed along the crypt-villus axis in a pattern that is tightly regulated by Wnt signaling and dictates cell sorting (Batlle et al., 2002; Gregorieff et al., 2005). In Mule cKO intestine, ephrinB1 was restricted to differentiated cells while EphB2 was detected in a large proliferative zone, as expected (data not shown). However, EphB3, which is normally expressed only at the very base of the crypts, was detected higher up in Mule cKO crypts (Figures 5A and 5B). Moreover, Mule cKO adenomas showed EphB3 positivity throughout (Figure 5C). Large numbers of EphB3-expressing cells were also observed in Mule cKO colon (Figures 5D–5G). Indeed, the EphB3 expression domain was expanded 2-fold in Mule cKO small intestine and colon (Figure 5H). Immunoblotting of Mule cKO organoids to detect EphB/ephrinB gradient components revealed increased EphB2 and EphB3 expression, and ephrinB1 ran at a higher molecular weight suggestive of post-translational modification (Figure 5I). Thus, Mule has direct effects on components of the EphB/ephrinB gradient.
Figure 5. Mule Regulates EphB3 Protein Levels.
(A–C) Staining to detect EphB3 protein in small intestines of WT (A) and Mule cKO (B) mice, and in a Mule cKO adenoma (C). Black arrows, EphB3 positivity. Scale bars represent 25 µm (A and B) and 100 µm (C).
(D–G) Staining to detect EphB3 in colons of WT (D and E) and Mule cKO (F and G) mice (n = 5/group). (D and F) Low-magnification images; scale bar, 100 µm.
(G and H) High-magnification images of the boxed areas in the low-magnification images; scale bar, 25 µm. (H) Quantitation of EphB3 domains in small intestine and colon of the indicated mice (n = 3/ group). Data are mean ± SEM.
(I) Immunoblot to detect the indicated proteins in lysates of organoids generated from the indicated mouse strains (n = 3 mice/group). β-Actin, loading control.
Mule Promotes Ubiquitin-Mediated Sorting of EphB3 Pools
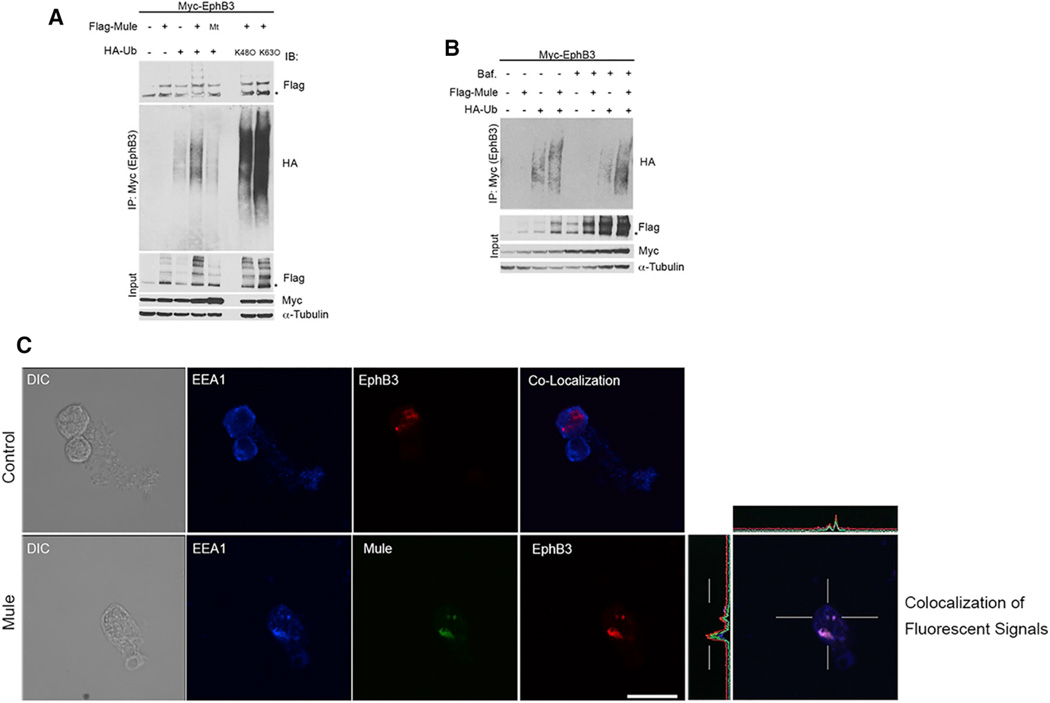
To investigate if Mule influences EphB3 turnover through Ubn leading to degradation, we transiently transfected HEK293T cells with vectors expressing Myc-tagged EphB3 and Flag-tagged Mule. Immunoprecipitation (IP) analysis revealed a physical interaction between Mule and EphB3 (Figure 6A). In the presence of hemagglutinin (HA)-tagged ubiquitin (Ub), EphB3 degradation was increased when Mule was overexpressed (Figures 6A and S6I). Next, we co-expressed a catalytically inactive Mule mutant (C4341A) with Myc-EphB3 and observed only background Ubn levels (presumably due to endogenous Mule or another E3 ligase) (Figure 6A). When WT Mule was overexpressed, a potential EphB3 cleavage product appeared (Figures S6I and S6J), suggesting that some EphB3 is cleaved prior to proteasomal degradation. In some cases, the overexpression of Mule increased the EphB3 protein suggesting it may stabilize, rather than degrade, a fraction of the EphB3 pool (Figure S6I).
Figure 6. Mule Promotes Proteasomal and Lysosomal Degradation of EphB3 as well as Its Ubiquitin-Mediated Internalization.
(A) IP analysis of HEK293T cells transfected with constructs expressing Flag-Mule or catalytically inactive Flag-Mule (C4341A; mutant, Mt), plus Myc-EphB3, and/ or WT HA-Ub, HA-Ub in which all lysine residues were mutated except K48 (K48O), or HA-Ub in which all lysines were mutated except K63 (K63O), as indicated. Cells were treated with MG132. EphB3 was immunoprecipitated with anti-Myc Ab and the blot probed with anti-Flag and anti-HA Abs. α-Tubulin, loading control.
(B) IP analysis as in(A) ofHEK293T cells that were transfected with constructs expressing Flag-Mule, Myc-EphB3 and/or WTHA-Ub, and treated with Bafilomycin A1, as indicated.
(C) Sequential scanning immunofluorescence co-localization analysis of control HEK293T cells and HEK293T cells overexpressing His-Mule that were transfected with SNAP-EphB3 plus ephrinB1, treated with Bafilomycin A1, and stained to detect all three proteins. Scale bar, 10 µm.
See also Figures S6I–S6L and S7.
We then co-expressed EphB3, Mule, and WT Ub or Ub in which all lysine residues were mutated to arginine except K48 (K480) or Ub in which all lysines were mutated to arginine except K63 (K630) in HEK293T cells treated with the proteasome inhibitor MG132. EphB3 was increased in cells expressing K480, which represented a pool of EphB3 targeted for proteasomal degradation (Figure 6A). There was also a pool of K63-Ub EphB3 that was presumably recycled. To determine if lysosomal inhibition had the same effect, we transfected HEK293T cells with Myc-EphB3, Flag-Mule and HA-Ub, and cultured these cells with/without the lysosome inhibitor Bafilomycin A1. EphB3-Ub accumulated in the presence of Bafilomycin A1, indicating lysosomal degradation, and Mule overexpression further increased this pool (Figure 6B). Similar results were obtained using HT29 human colon cancer cells, which lack endogenous EphB3 and express low levels of endogenous Mule (Figures S6K and S6L). Thus, Mule mediates both proteasomal and lysosomal EphB3 degradation.
To investigate how Mule influences EphB3 subcellular localization, we transfected HEK293T cells stably expressing His-Mule or empty vector with Myc-EphB3 and immunostained them at ~20 hr post-transfection. Although only minimal co-localization of Mule and EphB3 occurred at this time point, Mule overexpression induced a redistribution of EphB3 from the membrane to various intracellular locations (Figure S7A). To monitor EphB3 movement, we created a EphB3 construct with a SNAP-tag that binds membrane-impermeable SNAP Alexa Fluor 546. We transfected HEK293T cells stably expressing His-Mule with SNAP-EphB3, with/without ephrinB1. These cells were treated with Bafilomycin A1 and stained with anti-EEA1 to mark early endosomes and anti-His to visualize Mule. In control cells expressing Myc-EphB3 alone, EphB3 was concentrated normally at the plasma membrane (Figure S7A, control panel). In control cells co-expressing SNAP-EphB3 and ephrinB1 (but not Mule), SNAP-EphB3 accumulated mainly in the nucleus, with minor amounts in early endosomes (Figure 6C, top). However, in cells overexpressing both Mule and ephrinB1, SNAP-EphB3 co-localized with Mule in early endosomes (Figure 6C, bottom).
To determine if Mule influences EphB3 recycling, HEK293T cells stably expressing His-Mule were transfected with SNAP-EphB3 and Rab11-dsRed, a marker of sorting endosomes. Several such cells showed Mule, EphB3 and Rab11 to be co-localized or in very close proximity (Figure S7B). To confirm that Mule targets EphB3 for lysosomal degradation, HEK293T cells expressing His-Mule were transfected with SNAP-EphB3 and lamp1-RFP, a lysosomal marker. Co-localization of Mule, EphB3 and Lamp-1 was readily detected (Figure S7C). Thus, Mule promotes not only the proteasomal and lysosomal degradation of EphB3 but also its cell surface recycling.
Loss of One Allele of EphB3 Rescues the Paneth Cell Mislocalization in Mule cKO Mice
Paneth cell mislocalization can be caused either by Wnt-mediated EphB3 upregulation following APC loss (Sansom et al., 2004) or by EphB3 deficiency (Batlle et al., 2002), implying that it is perturbation of the EphB/ephrinB gradient that alters Paneth cell migration. We hypothesized that the elevated EphB3 in Mule cKO intestine disrupted the EphB/ephrinB gradient and thus impaired intestinal cell positioning. Since EphB3 is only one component sustaining the EphB/ephrinB gradient, we thought it unlikely that complete EphB3 ablation would prevent Paneth cell mislocalization in Mule cKO mice. Instead, because EphB3 is haplosufficient (Batlle et al., 2002), we crossed Mule cKO mice with EphB3+/− mice (mixed background) and looked for rescue of the Paneth cell defect. Intestines from 25 Mule cKO EphB3 mice (females: Mulefl/fl VillinCre EphB3−/− [n = 6] and Mulefl/+ VillinCre EphB3+/− [n = 7]; males: Mulefl/y VillinCre EphB3−/− (n = 6) and Mulefl/y VillinCre EphB3+/− [n = 6]) were examined. Paneth cells became localized normally in the absence of Mule only if one allele of EphB3 was ablated (Figures 7A–7E). Thus, the EphB/ephrinB gradient is highly sensitive to alterations in its components and regulators, including Mule.
Figure 7. EphB3 Restores Normal Localization of Mule cKO Paneth Cells, and Loss of Mule Favors Colon Cancer-Associated Mutations.
(A–D) Staining to detect lysozyme (Paneth cell marker) in small intestine of the indicated strains (n = 5–6 mice per group). Scale bar, 100 µm.
(E and F) Somatic mutations in Mule cKO adenomas. (E) Number of total somatic mutations, including synonymous and intronic variants (blue) and nonsynonymous coding variants (red) in five adenomas from two Mule cKO mice (116 and 784).
(F) Allele frequency distributions of mutant alleles observed for all somatic mutations (blue) and nonsynonymous coding mutations (red) for the five adenomas in (E).
(G) Model of proposed mechanism of Mule-mediated regulation of Wnt and EphB3. In the absence of Wnt signaling (left), β-catenin is recruited into the APC/Axin/GSK3b/CK1 destruction complex, which degrades β-catenin and prevents its translocation into the nucleus. The transcription of β-catenin target genes such as c-Myc and EphB3 is therefore blocked. When the Wnt pathway is active (right), Wnt binds to its receptor Frizzled and co-receptor LRP5/6 to activate Disheveled (Dvl), blocking APC/Axin/ GSK3b/CK1-mediated β-catenin degradation. Stabilized β-catenin translocates into the nucleus and together with TCF/LEF activates target gene transcription. Mule fine-tunes this pathway by inhibiting Dvl multimerization and thus the activation of the Wnt pathway and by controlling the production of the EphB3 and c-Myc proteins needed to regulate intestinal cell proliferation and positioning; these activities collectively contribute to tumor suppression. When the APC/Axin/ GSK3b/CK1 destruction complex cannot function, as occurs following APC mutation, Mule regulates c-Myc and targets EphB3 for proteasomal and/or lysosomal degradation in an attempt to restrain proliferation and maintain a proper EphB/ephrinB gradient.
See also Table S1.
Loss of Mule Favors Colon Cancer-Associated Mutations
Because our Mule cKO organoids became undifferentiated cysts, we investigated if loss of Mule alone resulted in activating mutations in the Wnt pathway or inactivating mutations of tumor suppressors linked to colon cancer. Whole-exome sequencing was performed on five matched adenomas and adjacent normal tissues from two Mule cKO mice (two adenomas in mouse 116 and three adenomas in mouse 784), and somatic mutations were identified using the MuTect algorithm. The distribution of variant allele frequencies for the numerous somatic mutations identified in all samples showed that the most were present at low frequencies (Figure 7E), consistent with the heterozygous mutations present in subclonal tumor cell populations and also with the presence of normal tissue in tumor samples. There were no significant differences in variant allele frequencies in these adenomas (Figure 7F).
To investigate mutations potentially contributing to adenoma development, we identified nonsynonymous coding mutations and found that the distribution of variant allele frequencies among these mutations was similar to that among total somatic mutations (Figure 7F). Nonsynonymous mutations were identified in Trp53, PIK3CA, and EphA2 (Table S1), which are linked to colon cancer progression. Thus, loss of Mule exerts a selective pressure favoring mutations driving gut tumorigenesis. The fact that Mule is expressed in the intestinal stem cell niche suggests that these mutations could facilitate the transition from stem cells to CSCs. CSCs are believed to drive tumor progression and has been linked to disease relapse in colorectal cancer (Zeuner et al., 2014).
DISCUSSION
The role of Mule in colon cancer has been controversial. In vitro, Mule polyubiquitinates the tumor suppressor p53 at K48, targeting it for proteasomal degradation (Chen et al., 2005), but Mule also transcriptionally activates the oncogene c-Myc via K63pUb attachment (Adhikary et al., 2005). In our in vivo study of intestine-specific Mule cKO mice, we observed enhanced intestinal cell proliferation due to elevated c-Myc as well as Paneth cell mislocalization due to perturbation of the EphB/ephrinB gradient. Loss of Mule amplifies the APCmin phenotype, establishing Mule as a true intestinal tumor suppressor with independent effects on Paneth cell regulators.
Constitutive Wnt signaling causes Paneth cells to disperse throughout the intestinal epithelium rather than home to the crypt base (Sansom et al., 2004; van Es et al., 2005). Our results imply that Mule has another target within the Wnt pathway (besides c-Myc) that can amplify β-catenin, resulting in elevated EphB2 and EphB3 in the crypts and a disproportionate amount of ephrinB1 in the villi. Notably, several Wnt target genes were upregulated in Mule cKO mice. During our studies, it was reported that Mule mediates the K63pUbn of Dvl to inhibit Wnt pathway activation (de Groot et al., 2014). We identified nuclear β-catenin in Mule cKO adenomas, implying that Mule acts on a substrate upstream of β-catenin, but found no difference in Dvl levels in WT and Mule cKO crypts (data not shown). Mule’s Ubn of Dvl therefore does not appear to target it for degradation but rather modifies its activity. We propose that Mule acts on at least two Wnt pathway components to fine-tune Wnt signaling and suppress gut tumorigenesis: (1) Dvl, the cytoplasmic Wnt pathway component; and (2) c-Myc, a downstream effector of β-catenin/TCF (Figure 7G).
Disruption of the EphB/ephrinB gradient results in Paneth cell mislocalization (Batlle et al., 2002). Mule regulates EphB3 pools, and so loss of Mule leads to an EphB3/ephrinB1 imbalance. Accordingly, Mule cKO mice expressed EphB3 not only in the base of the crypts but also higher up, leading to Paneth cell mislocalization. It should be noted that there was significant variability in EphB3 expression patterns among our mutants, perhaps reflecting the penetrance of Mule deletion. The fact that one EphB3 allele rescues the defect caused by the loss of Mule underlines the sensitivity of the EphB/ephrinB1 gradient to alterations in its components and regulators.
Our in vitro work has shown that Mule physically interacts with EphB3 and regulates its protein levels, as reflected by the altered Paneth cell positioning in Mule cKO intestine and organoids. Deregulated Paneth cell localization generates an environment encouraging ISC growth (Sato et al., 2011), which might expand nearby clones of mutated ISCs. ISCs and early progenitors express EphB3, suggesting that Mule-mediated EphB3 regulation may distinguish progenitor cells that differentiate and move up the crypt (i.e., contain K48pUb-EphB3) from Paneth cells and ISC that inhabit the base of the crypt (ie., contain K63pUb-EphB3). In vivo, ISCs can still differentiate in the absence of Mule, but we speculate that Mule-mediated regulation is needed to safeguard ISCs against unwanted expansion. The same may apply to the Paneth cells that sustain ISCs, especially in a highly proliferative setting. Significantly, none of our aged Mule c-Myc DKO mice developed adenomas despite partial crypt re-population by c-Myc+ cells, suggesting that Paneth cell mislocalization contributes to tumor growth only in a hyperproliferative backdrop.
In human cancers, Mule is subject to somatic mutation, homozygous deletion or amplification (http://www.cbioportal.org). In patients with colon and rectal adenocarcinomas, some mutations within Mule’s HECT domain have been identified (http://www.sanger.ac.uk). Our exome-sequencing data revealed that an absence of Mule exerts selective pressure on genes linked to colon cancer progression or involved in cell adhesion, prote-olysis, or the extracellular matrix. Because EphB3 compartmentalizes incipient tumors and inhibits their metastasis (Cortina et al., 2007), loss of Mule function leading to increased EphB3 protein may favor the transcriptional silencing of EphB3 that often occurs in colorectal cancer. This altered EphB3 expression might permit tumor cells to grow without constraints.
In conclusion, our in vitro and in vivo evidence indicate that Mule functions as a tumor suppressor in the gut rather than an oncogene. Furthermore, we have shown that Mule controls ISC numbers and proliferation via effects on c-Myc and maintains proper levels of EphB3 protein by targeting it for proteasomal and lysosomal degradation. Through this modulation of EphB3, Mule influences the intestinal cell sorting crucial for avoiding colorectal carcinogenesis.
EXPERIMENTAL PROCEDURES
Mice
Mulefl/fl(y) mice were as described previously (Hao et al., 2012) and crossed to Villin-Cre (Madison et al., 2002), APCmin(Su et al., 1992), c-Mycfl/fl (de Alboran et al., 2001), or EphB3−/− (Orioli et al., 1996) mice. All animal experiments were approved by the University Health Network Animal Care Committee (UH-NACC; ID: AUP985).
Histology and IHC
Small intestine and colon from sacrificed mice were placed immediately into 10% neutral buffered formalin (NBF). Intestines were opened longitudinally, cleaned, and swiss-rolled. Swiss rolls were fixed for ~24 hr in 10% NBF at room temperature (RT). Fixed tissues were embedded in paraffin and stained with H&E. Adenomas were graded by inspection of H&E-stained sections.
For immunohistochemistry (IHC), primary antibodies (Abs) recognized were β-catenin (Cell Signaling Technology, #2677), c-Myc (Santa Cruz, #SC764, N-262), lysozyme (#A0099), ephrinB1 (R&D Systems, clone AF473), EphB2 (R&D Systems, clone AF467), EphB3 (R&D Systems, clone AF432), cleaved caspase-3 (Cell Signaling Technology, #9661), Ki67 (Dako, Tec3 clone), chromogranin A (Immunostar, #20085), p21 (Santa Cruz Biotechnology, clone M19), GFP (Novus, 100–1678), cyclin D1 (Cell Signaling Technology, #2978), cyclin E1 (Abcam, ab7959), cyclin A1 (Santa Cruz Biotechnology, sc-751), and B23 (Sigma, B0556). Anti-LFABP antibody was the kind gift of Dr. Jeffrey Gordon (Washington University, St. Louis).
In Vivo Cell Proliferation
The BrdU In Situ Detection Kit (BD Pharmingen, #550803) was used according to the manufacturer’s instructions. Briefly, 4-month-old mice were intraperitoneally injected with 2.5 mg bromodeoxyuridine (BrdU) and intestines harvested 2 or 24 hr later, followed by processing for histology as described above. BrdU+ cells were quantified in 60 crypts per mouse (n = 3 mice per group per condition). Migration was determined by measuring the distance between the crypt base and the highest labeled cell along the crypt-villus axis in five different fields per genotype.
In Situ Hybridization
In situ probes targeting Lgr5, OLMF4, and cryptidin-4 were as described previously (Tian et al., 2011; van der Flier et al., 2009; Yilmaz et al., 2012). The Mule probe corresponded to expressed sequence tags of a partially sequenced mouse cDNA from Open Biosystems (MGI:1926884). To ensure probe specificity, both sense and antisense probes were generated by in vitro transcription using DIG RNA labeling mix (Roche) as described previously (Gregorieff et al., 2005).
RNAscope In Situ Hybridization
The RNAscope 2-plex Reagent kit (ACD, catalog number [Cat No.] 320700) was used according to the manufacturer’s instructions. Huwe1 (Mule)-C1 (Cat No. 434801), c-Myc-C1 (Cat No. 413451), and Lgr5-C2 (Cat No. 312171) were purchased from ACD. In the normal intestine, the number of cells containing the indicated probes was determined in ten crypts per mouse (n = 3). In adenomas, the % of cells containing the indicated probes was determined in two fields per adenoma (n = 6).
Overexpression Vectors
Human EPHB3 (Myc-DDK-tagged in the pCMV6-Entry vector) was purchased from Origene. The human Mule expression vector (pCI.GNeo-3xFlag-12xHis-mouse Mule FL) was the kind gift of Dr. Qing Zhong (University of Texas Southwestern Medical Center; Zhong et al., 2005). Plasmids pRK5-HA-Ub-WT, pRK5-HA-Ub-K48O, and pRK5-HAUb-K63O were from Addgene (#17608, 17605, and 17606, respectively). N-terminal SNAP-tagged EphB3 was generated by PCR-subcloning EphB3 into the SNAP-tag cloning vector (New England Biolabs). The human ephrinB1 expression vector was the generous gift of the late Dr. Tony Pawson (Mount Sinai Hospital, Toronto).
Cell Culture
HEK293T cells were cultured in DMEM H21 (Invitrogen), and HT29 cells (ATCC) were cultured in McCoy 5A Media Modified. Media were supplemented with 10% FBS (Sigma), 100 U/ml penicillin, and 100 mg/ml streptomycin.
Immunofluorescence
For lysosomal inhibition studies, HEK293T cells (control or stably expressing Mule) were grown on poly-D-lysine-coated chamber slides (BD Biosciences) for 16 hr prior to transfection with vectors expressing SNAP-EphB3 or ephrinB1. At 6 hr post-transfection, cells were treated with Bafilomycin A1 (10 nM; Sigma) overnight. To visualize EphB3 localization, transfected cells were labeled with 1 mM SNAP Surface Alexa Fluor 546 (New England Biolabs) for 15 min at RT, chased for 15 min at 37°C, and fixed in 4% paraformaldehyde. Fixed cells were incubated overnight with rabbit anti-His Ab (Cell Signaling Technology, #2365) to identify Mule, and mouse anti-EEA1 Ab (BD Bioscience, 610457) to identify early endosomes, followed by incubation with secondary Ab rabbit Alexa 488 or mouse Alexa Fluor 405, respectively (Invitrogen). Stained cells were mounted with Vectashield (Vector Laboratories) and examined using an Olympus FV-1000 confocal microscope. Sequential scanning minimized cross-talk between channels and generated a profile for each fluorescent signal. Co-localization was identified by direct overlay of fluorescence signals from individual channels.
For receptor internalization studies, control or Mule-overexpressing HEK293T cells were transfected with Myc-EphB3 vector. At 20 hr post-transfection, cells were fixed in ice cold 100% methanol for 15 min. Cells were incubated overnight with anti-His antibody (Mule), then for 1 hr at RT with anti-Myc-tag (Cell Signaling Technology, 9B11), and 1 hr with rabbit anti-Cy2 and mouse anti-Cy3 Abs (Vector Laboratories). Endosomal and lysosomal sorting experiments were conducted as above using Rab11-dsRed and Lamp1-RFP (Addgene).
Ubiquitination
HEK293T or HT29 cells were transfected with Myc-EphB3 and Flag-Mule, plus either HA-Ub, HA-Ub-K48O, or HA-Ub-K63O, using Lipofectamine 2000 (Invitrogen) as described previously (Inoue et al., 2013). Proteasomal degradation was determined by adding MG132 (20 mM; Sigma) at 24 hr post-transfection and culturing cells for 3 hr. Lysosomal degradation was determined by adding Bafilomycin A1 (10 nM; Sigma) at 6 hr post-transfection and culturing cells overnight. Lysates were prepared as described previously (Inoue et al., 2013). Supernatants were immunoprecipitated with anti-Myc-tag (Cell Signaling Technology, 9B11) for 2 hr at 4°C (overnight at 4°C for HT29 cells). Protein G-Sepharose beads were added with rocking for 1–2 hr at 4°C. Immunocomplexes were washed four times with lysis buffer and subjected to immunoblotting.
Immunoblotting
Standard immunoblotting was performed using Abs recognizing HA (Santa Cruz Biotechnology, clone Y-11), Flag (Sigma clone M2), Myc-tag (Cell Signaling Technology, 9B11), vinculin (Abcam, SPM227), Mule (Bethyl Laboratories, A300-486A), ephrinB1 (R&D Systems, clone AF473), EphB2 (R&D clone AF467), EphB3 (R&D Systems, clone AF432), Miz1 (Santa Cruz Biotechnology, sc-22837), c-Myc (Santa Cruz Biotechnology, N262), α-tubulin (Sigma), and β-actin (Sigma). Anti-mouse and anti-rabbit HRP-conjugated secondary Abs were from Thermo Fisher Scientific. Quantification of immunoblots was performed using Image J software (NIH).
RT-PCR
RNA was extracted using the NucleoSpin RNA II kit (Macherey-Nagel) and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was performed using these cDNAs as templates and Power SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7700 instrument (Applied Biosystems). All procedures were performed according to the manufacturer’s instructions. Each sample was assayed in triplicate and data were analyzed using SDS software (Applied Biosystems). Primer sequences are available upon request.
Organoids
Standard organoids were generated from intestines of Mule cKO and WT litter-mate mice (2–4 months old) as described previously (Sato et al., 2009). WT and Mule cKO organoids were passaged once a week. Crypt culture medium (Sato et al., 2009) was changed every other day.
CSC organoids were generated by incubating resected adenomas in DMEM plus 10% FBS containing Dispase (12.5 mg/50 ml medium; Life Technologies) and Collagenase from Clostridium histolyticum (12.5 mg/50 ml medium; Sigma) for 30 min to 1 hr (depending on adenoma number) at 37°C on a nutator with vortexing every 15 min. Digested tissues were passed through a 40-µm strainer and mashed with a plunger. Cells were collected by centrifugation at 1,000 rpm for 5 min and pellets were resuspended in 1–2 ml DMEM plus Y-27632 dihydrochloride monohydrate (10 µM, Sigma). A total of 5,000 cells/25 µl of Matrigel (BD Biosciences) in crypt culture medium were cultured in a 48-well dish, with Y-27632 dihydrochloride monohydrate added for the first week.
Quantitation of EphB3 Domains and c-Myc+ Cells
The EphB3 domain in 100 crypts per genotype (six fields per mouse; n = 3 per group) was determined using the Photoshop ruler tool (Adobe) to measure the distance between the lowest and highest EphB3+ cells in the crypt. A 250-mm scale bar was measured as 278 pixels and used as the reference. In normal tissue, c-Myc+ cells were counted in 60 crypts per mouse (n = 3 mice/group/condition). In adenomas, c-Myc+ cells were counted in four fields per adenoma (n = 6 mice). The mean number of c-Myc+cells/mouse was used to determine the total mean for the group.
Whole-Exome Sequencing
Whole-exome sequencing and data analysis were performed at the Princess Margaret Genomics Centre. DNA was extracted from freshly excised adenomas and adjacent normal gut using NucleoSpin Tissue XS (Macherey-Nagel). Read quality was checked using FASTQC (v. 0.11.2), and raw sequence data were aligned to the mouse genome (mm9) using BWA-MEM (v. 0.7.10). Alignment quality was assessed using Qualimap (v. 2.0). Bam files were pre-processed using Picard (v. 1.124) to mark duplicates and GATK (v. 3.2-2) to perform local realignment around indels and to perform base quality score recalibration. MuTect (v. 1.1.5) was used to identify somatic point mutations in tumor samples relative to paired normal tissue. Mutations were annotated using ANNOVAR (v. 2014-11–12).
Statistics
p values for intergroup comparisons and mouse survival were determined using the unpaired Student’s t test and the Mantel-Cox test, respectively. p < 0.05 is considered significant.
Supplementary Material
Highlights.
Mule regulates murine intestinal stem and progenitor cell proliferation
Mule modulates the Wnt pathway via c-Myc
Mule targets EphB3 to the proteasome and lysosome
Proper protein levels of EphB3 are required to maintain the EphB/ephrinB gradient
In Brief.
Dominguez-Brauer et al. show that the ubiquitin ligase Mule fine-tunes the Wnt pathway via c-Myc to control unwarranted proliferation and stem cell expansion. Mule also regulates the levels of EphB3 to maintain the EphB/ephrinB gradient, which is responsible for cell positioning along the crypt-villus axis.
Acknowledgments
This study is dedicated to the memory of C.D.-B.’s late mother, Maria Gloria Dominguez, who fought a valiant battle against colon cancer. C.D.-B. would like to thank all members of the Mak laboratory for their unstinting support. All authors are grateful to Kyle Gill of the genotyping facility and the animal resource center at the Princess Margaret Hospital (Toronto). Lastly, we thank Dr. Mary Saunders for scientific editing of the manuscript. This work was supported by a grant to T.W.M. from the Canadian Institutes of Health Research (CIHR), a postdoctoral fellowship to C.D.-B. from the Takeda Canada-CIHR-Canadian Association of Gastroenterology, and a postdoctoral fellowship to R.N. from EMBO and Marie Curie (ALTF 725-2015).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2016.04.002.
AUTHOR CONTRIBUTIONS
C.D.-B. designed and performed most experiments. Z.H., A.E., J.F., R.N, P.M.B., Y.S., M.D.M., I.I.C.C., J.H., A.P., R.C., L.T., S.I., C.R., A.M., J.S., M.C., A.W., O.H.Y., and J.V.E. performed or assisted with some experiments. D.M.S, O.H.Y., H.C., and T.S. provided technical advice. C.D.-B. and T.W.M. analyzed and interpreted the data. C.D.-B. wrote the manuscript.
REFERENCES
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Augenlicht LH, Wadler S, Corner G, Richards C, Ryan L, Multani AS, Pathak S, Benson A, Haller D, Heerdt BG. Low-level c-myc amplification in human colonic carcinoma cell lines and tumors: a frequent, p53-independent mutation associated with improved outcome in a randomized multi-institutional trial. Cancer Res. 1997;57:1769–1775. [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, Clevers H. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- Bettess MD, Dubois N, Murphy MJ, Dubey C, Roger C, Robine S, Trumpp A. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol. Cell. Biol. 2005;25:7868–7878. doi: 10.1128/MCB.25.17.7868-7878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Kon N, Li M, Zhang W, Qin J, andGu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Cortina C, Palomo-Ponce S, Iglesias M, Fernández-Masip JL, Vivancos A, Whissell G, Humà M, Peiro N, Gallego L, Jonkheer S, et al. EphB-ephrin-B interactions suppress colorectal cancer progression by compartmentalizing tumor cells. Nat. Genet. 2007;39:1376–1383. doi: 10.1038/ng.2007.11. [DOI] [PubMed] [Google Scholar]
- Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin. Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gärtner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- de Groot RE, Ganji RS, Bernatik O, Lloyd-Lewis B, Seipel K, Šedová K, Zdráhal Z, Dhople VM, Dale TC, Korswagen HC, Bryja V. Huwe1-mediated ubiquitylation of dishevelled defines a negative feedback loop in the Wnt signaling pathway. Sci. Signal. 2014;7:ra26. doi: 10.1126/scisignal.2004985. [DOI] [PubMed] [Google Scholar]
- Fatehullah A, Appleton PL, Näthke IS. Cell and tissue polarity n the intestinal tract during tumourigenesis: cells still know the right way up, but tissue organization is lost. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20130014. doi: 10.1098/rstb.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumor-igenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Gregorieff A, Pinto D, Begthel H, Destrée O, Kielman M, Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- Hao Z, Duncan GS, Su YW, Li WY, Silvester J, Hong C, You H, Brenner D, Gorrini C, Haight J, et al. The E3 ubiquitin ligase Mule acts through the ATM-p53 axis to maintain B lymphocyte homeostasis. J. Exp. Med. 2012;209:173–186. doi: 10.1084/jem.20111363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen JP, Rajashankar KR, Lackmann M, Cowan CA, Henkemeyer M, Nikolov DB. Crystal structure of an Eph receptor-ephrin complex. Nature. 2001;414:933–938. doi: 10.1038/414933a. [DOI] [PubMed] [Google Scholar]
- Inoue S, Hao Z, Elia AJ, Cescon D, Zhou L, Silvester J, Snow B, Harris IS, Sasaki M, Li WY, et al. Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev. 2013;27:1101–1114. doi: 10.1101/gad.214577.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N, Zhong J, Qiang L, Accili D, Gu W. Inactivation of arf-bp1 induces p53 activation and diabetic phenotypes in mice. J. Biol. Chem. 2012;287:5102–5111. doi: 10.1074/jbc.M111.322867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, Cole AM, Gregorieff A, de Alboran IM, Clevers H, Clarke AR. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/ Tcf-4 target gene c-Myc. Mol. Cell. Biol. 2006;26:8418–8426. doi: 10.1128/MCB.00821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H, Nakamura Y. Mutations of the APC (adenomatous polyposis coli) gene. Hum. Mutat. 1993;2:425–434. doi: 10.1002/humu.1380020602. [DOI] [PubMed] [Google Scholar]
- Orioli D, Henkemeyer M, Lemke G, Klein R, Pawson T. Sek4 and Nuk receptors cooperate in guidance of commissural axons and in palate formation. EMBO J. 1996;15:6035–6049. [PMC free article] [PubMed] [Google Scholar]
- Peter S, Bultinck J, Myant K, Jaenicke LA, Walz S, Müller J, Gmachl M, Treu M, Boehmelt G, Ade CP, et al. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol. Med. 2014;6:1525–1541. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon-Assmann P, Clevers H, Nathke IS, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesen-chymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, Thiele A, van den Born M, Begthel H, Brabletz T, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Katajisto P, Lamming DW, Gültekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuner A, Todaro M, Stassi G, De Maria R. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692–705. doi: 10.1016/j.stem.2014.11.012. [DOI] [PubMed] [Google Scholar]
- Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, Iavarone A, Lasorella A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.