Abstract
Glial cells are the most abundant cell type in our brains, yet we understand very little about their development and function. An accumulating body of work over the last decade has revealed that glia are critical regulators of nervous system development, function, and health. Based on morphological and molecular criteria, glia in Drosophila melanogaster are very similar to their mammalian counterparts, suggesting that a detailed investigation of fly glia has the potential to add greatly to our understanding of fundamental aspects of glial cell biology. In this article, we provide an overview of the subtypes of glial cells found in Drosophila and discuss our current understanding of their functions, the development of a subset of well-defined glial lineages, and the molecular-genetic tools available for manipulating glial subtypes in vivo.
BACKGROUND
Neurons are not the only cell type in the nervous system; ~90% of the cells in our brain are glia. Although they have long been thought of as simple support cells, work over the last two decades has revealed a number of critical roles for glia in the development, function, and maintenance of the nervous system. Glia perform diverse roles in the developing nervous system, such as modulating neural stem cell proliferation (Ebens et al. 1993), regulating the differentiation of neural precursors guiding axon pathfinding (Hidalgo and Booth 2000; Sepp et al. 2001; Gilmour et al. 2002), ensheathing nerves and individual axons (Barres 2008; Nave and Trapp 2008), delineating and isolating distinct lobes of the brain and their subcompartments (Oland and Tolbert 2003; Awasaki et al. 2008), supplying trophic support for neurons (Xiong and Montell 1995; Booth et al. 2000), engulfing neurons and debris that are eliminated during development (Sonnenfeld and Jacobs 1995; Freeman et al. 2003; Awasaki and Ito 2004; Watts et al. 2004), and promoting synapse formation and maturation (Barres 2008). In the mature nervous system, glia maintain a proper ionic balance in the central nervous system (CNS), take up neurotransmitters after synaptic signaling (Danbolt 2001), isolate and protect neurons by forming or regulating the blood-brain barrier (BBB; Abbott 2005), associate closely with synapses and likely modulate their activity (Barres 2008), and act as the major immune cell type in the brain. These lists that attempt to catalog the breadth of glial functions undoubtedly only partially represent the true range of glial roles in the developing and mature nervous system. Whenever glial biology is carefully explored in any new developmental or functional context, it seems glia reveal additional unexpected depths to their governance of nervous system morphogenesis and function.
Despite the widespread importance of glia in the nervous system, we know surprisingly little about the underlying molecular pathways that mediate glial biology in any organism. For example, excellent studies have described the morphology of glia as they ensheath individual synapses, but we understand almost nothing about how glia and synapses acquire this intimate relationship or the functional significance of such associations. Why are the majority of synapses covered by glial membranes? What drives glia to ensheath synapses? What is the in vivo function for glia at a glutamatergic, cholinergic, or GABAergic synapse? Do they aid in information processing through direct modulation of synaptic signaling?
There are a number of reasons to explain our ignorance of the molecular pathways involved in most aspects of glial biology. First, glia have not been studied as intensively as their excitable neighbors, neurons. The focus on neurons is due partly to their ability to fire action potentials, in contrast to most glia, which remain electrophysiologically silent. Second, mammalian glia are difficult to analyze in vivo. This is attributable mainly to a dearth of markers and reagents for manipulating glia. Instead, much of our understanding of neuron–glia interactions has been gleaned from studies of primary cultures. This approach can certainly provide an excellent first step toward identifying genes that mediate neuron–glia signaling. For example, recent work in primary cultures has identified glial-secreted molecules that are required for synapse formation and maturation, some of which have been verified by functional analysis in vivo (Christopherson et al. 2005; Stevens et al. 2007). Nevertheless, the vast majority of in vitro observations have not been repeated in experiments with the living organism. Confirming observations from in vitro studies in vivo is important because glia and neurons are in close association with each other soon after differentiation, both cell types become interdependent for survival, and they mature morphologically, and likely molecularly, in response to cues from each other.
It is abundantly clear that when glia are dissociated and plated in culture, they rapidly acquire properties that are different from those of glia in the intact brain. For example, astrocytes in vivo are tightly coupled to one another through gap junctions and essentially form a syncytium within the brain that acts as a sink for K+ ions (Kofuji and Newman 2009). Dissociation of astrocytes destroys this coupling and results in dramatic changes in the electrophysiological properties of these cells (Kimelberg 2009). Likewise, astrocytes display a tufted morphology and are profusely branched in three-dimensional space in vivo, but when dissociated and plated for culture in vitro, astrocytes lose this complex morphology and take on a fibroblast-like, flattened appearance. As such, it is reasonable to assume that the physiology of astrocytes in vitro may also be dramatically different from astrocytes in vivo. Until in vitro observations are also assayed for functional significance in the intact animal, the relevance of in vitro neuron–glia signaling events will remain in question.
Although the Drosophila nervous system is relatively simple in structure, it shares a number of quite sophisticated glial characteristics with its mammalian counterparts, including (1) mechanisms mediating reciprocal trophic support between neurons and glia; (2) glial pathways for recycling of synaptic neurotransmitters (e.g., glutamate and GABA [γ-aminobutyric acid]); (3) subtypes of glia that are capable of parsing axons into distinct fascicles and individually ensheathing axons (similar to Remak bundles in mammals); (4) a subtype of glia that bears striking morphological and molecular similarity to mammalian astrocytes; and (5) glia that act as the primary immune cell in the brain and can respond immunologically to neural trauma. As such, Drosophila glia seem well positioned to provide exciting insights into glial biology that will be relevant to glial functions in mammals.
This article is meant to provide an introduction to the study of glial cell biology in Drosophila. Flies offer the opportunity to study a diverse array of neuron–glia interactions in the intact organism while also exploiting the powerful molecular-genetic tools available in Drosophila to address fundamental questions in glial cell biology. We have divided this article into two broad sections: (1) glia in the embryonic nervous system and (2) glia in the larva and adult. We provide a list of methods and tools we find particularly useful for studying these cell types. Much of the detailed discussion of lineages, morphogenesis, and function focuses on embryonic glia because they have been studied much more extensively than glia in the larva, pupa, or adult. However, postembryonic stages offer a number of advantages with respect to molecular-genetic approaches to assay glial gene function or glial cell biology, and these are discussed.
GLIAL CELL DEVELOPMENT AND FUNCTION IN THE EMBRYONIC CNS
The embryonic ventral nerve cord (VNC) is an excellent model for studying early glial developmental events such as glial cell-fate specification and migration. The locations of and lineages generated by each of the 30 neuroblasts that form in each hemisegment of the VNC have been extremely well characterized at the cellular level (Doe 1992; Bossing et al. 1996; Schmidt et al. 1997; Schmid 1999). Of the 30 neuroectoderm-derived neuroblasts per hemisegment, eight (the so-called neuroglioblasts [NGBs]) produce mixed lineages of neurons and glia (NGB1-1A, NGB1-3, NGB2-2T, NGB2-5, NGB3-5, NGB5-6, NGB6-4A, and NGB7-4 [A refers to abdominal and T to thoracic; 6-4A is purely glial]) and one precursor produces only glia (Bossing et al. 1996; Schmidt et al. 1997; Schmid 1999). The Drosophila VNC contains on average 25–30 glia per hemisegment, and embryonic glia can be assigned to three broad categories based on their position and morphology: (1) surface glia, which form a layer around the CNS or peripheral nerves; (2) cortex glia, which ensheath neuronal cell bodies; and (3) neuropil glia, which associate directly with the neuropil (Ito et al. 1995). As discussed below, these broad subtypes of glia appear to be present throughout development and into the adult stage of Drosophila.
Midline glia are also present in the Drosophila embryonic CNS. These cells represent a unique class of mesectoderm-derived glia that have a critical role in axon pathfinding at the CNS midline and commissure formation in the VNC. Midline glia are a small subset of glia in the embryo (only about three per hemisegment at embryonic stage 17) and will not be discussed further here. Instead, we refer the reader to the excellent review by Jacobs (2000).
The earliest known marker for cells that will become glia in the embryonic nervous system is the novel transcription factor encoded by glial cells missing (gcm). Shortly after Gcm is activated, reversed polarity (repo) is expressed; this is a direct transcriptional target of Gcm that encodes a homeodomain transcription factor. Both of these (along with their enhancer traps and Gal4 driver lines) are extremely useful tools for labeling glia (except midline glia, which do not express Gcm or Repo) very early in their development (Table 1). The first Gcm-expressing glial cells appear in the developing embryonic VNC at late stage 10 or early stage 11, and Repo-positive glial cells become detectable shortly thereafter, at approximately stage 11 (Campbell et al. 1994; Halter et al. 1995). The majority of Gcm/Repo-positive embryonic VNC glia are likely present by early stage 14.
TABLE 1.
Useful tools in Drosophila glial research
| Markers/lines | CNS expression pattern | Type(s) of antibodies | Dilution(s) for embryo staining | Reference |
|---|---|---|---|---|
| Anti-Repo | All glia except midline | Mouse | 1:5, 1:10 | Campbell et al. (1994) |
| Anti-Gcm | All glia at early stages (except midline) | Rat | 1:400 | Alfonso and Jones (2002) |
| Anti-Moody-β | Subperineurial glia | Rabbit | 1:15 | Bainton et al. (2005) |
| Anti-Moody-α | Rat | 1:10 | ||
| Anti-Gs2 | Subset of longitudinal glia | Mouse | 1:10 | Thomas and van Meyel (2007) |
| Anti-NrxIV | Subperineurial glia, neurons, epithelia | Rabbit | 1:500 | Baumgartner et al. (1996) |
| nrxIV-GFP exon trap | Subperineurial glia, neurons, epithelia | NA | NA | Edenfeld et al. (2006) |
|
| ||||
| Driver line | CNS expression pattern | Comment | Reference(s) | |
|
| ||||
| repo-Gal4 | All glia except midline | Enhancer trap | Sepp et al. (2001) | |
| repo-Gal4-4.3 | All glia except midline (note that some variability in expression in different subsets of glia has been observed) |
Promoter fusion | Lee and Jones (2005) | |
| repo:LexA::GAD | All glia except midline | Promoter fusion | Lai and Lee (2006) | |
| gcm-Gal4 | All glia (except midline), apodemal cells, and macrophages | Enhancer trap | Paladi and Tepass (2004) | |
| htl-Gal4 | Longitudinal glia and other glia | Promoter fusion | Shishido et al. (1997) | |
| rl82-Gal4 (gliotactin-Gal4) | Pronounced in subperineurial glia, weaker in perineurial glia | Enhancer trap | Sepp and Auld (1999) | |
| Spg-Gal4 | Subperineurial glia | Promoter fusion | Stork et al. (2008), Mayer et al. (2009) | |
| alrm-Gal4 | Astrocyte-like glia | Promoter fusion | Doherty et al. (2009) | |
| deaat1-Gal4 | Astrocyte-like glia, some cortex, weak in neurons | Promoter fusion | Rival et al. (2004, 2006) | |
| Mz709-Gal4 | Ensheathing glia, neurons | Enhancer trap | Ito et al. (1995) | |
| nrv2-Gal4 | Cortex, subperineurial, ensheathing? astrocytes? | Promoter fusion | Sun et al. (1999) | |
| moody-Gal4 | Subperineurial glia | Promoter fusion | Schwabe et al. (2005) | |
| NP6293-Gal4 | Perineurial glia, subset neurons | Enhancer trap | Hayashi et al. (2002), Awasaki et al. (2008) | |
| NP2276-Gal4 | Subperineurial glia | Enhancer trap | Hayashi et al. (2002), Awasaki et al. (2008) | |
| NP577-Gal4 | Cortex glia | Enhancer trap | Hayashi et al. (2002), Awasaki et al. (2008) | |
| NP2222-Gal4 | Cortex glia | Enhancer trap | Hayashi et al. (2002), Awasaki et al. (2008) | |
| NP3233-Gal4 | Astrocyte-like glia | Enhancer trap | Hayashi et al. (2002), Awasaki et al. (2008) | |
| NP1243-Gal4 | Astrocyte-like glia and weaker in ensheathing glia and cortex glia | Enhancer trap | Hayashi et al. (2002), Awasaki et al. (2008) | |
| NP6520-Gal4 | Ensheathing glia and weaker in cortex glia | Enhancer trap | Awasaki et al. (2008) | |
This table gives a brief list of markers and tools that will be helpful for labeling and manipulating glial cells in Drosophila. Note that most markers, especially Gal4 driver lines, are often not exclusive to glia but show expression in other tissues as well. NA, not applicable.
Gcm is most useful for labeling or manipulating early embryonic glial lineages, but is not an appropriate glial-specific marker at later developmental stages. Gcm is expressed very early in glial development, but its expression fades by embryonic stage 14. Consequently, Gcm and gcm-Gal4 will not label late-stage embryonic glia. In addition, Gcm is expressed in cells outside the nervous system that are not glia (i.e., macrophages and apodemal cells). Finally, Gcm has recently been shown to be expressed in neural precursors in the larval/pupal brain and is required for the formation of many neurons that populate the adult brain (Chotard et al. 2005). These findings should be considered when attempting to label glia at larval or pupal stages.
Nevertheless, gcm-Gal4 is the earliest and strongest glial-specific driver in the embryonic CNS and it remains the best tool for experiments in which one wishes to ectopically express genes under UAS control at the earliest possible time point in embryonic glial development. Repo, in contrast to Gcm, appears to be expressed specifically in glia, and this holds true throughout the Drosophila life cycle (to our knowledge no Repo+ cells have been described that are not glia). Therefore, Repo antibodies remain the best marker to uniquely label glia at all developmental stages, and repo-Gal4 remains the best tool to specifically label and/or manipulate glia.
There is tremendous molecular heterogeneity in Drosophila embryonic glia, with probably 10–15 molecularly distinct subtypes of glia being present per hemisegment. One can assay this diversity in glial cell-fate specification in the embryo in exquisite detail by using a combination of cell position in the VNC along with an extensive list of available markers that label specific glial subtypes (e.g., Ito et al. 1995; Beckervordersandforth et al. 2008; von Hilchen et al. 2008). Here we will not delve deeply into this list of markers; rather we will briefly discuss the above-described broad categories of glia (surface, cortex, and neuropil) and show how they can be identified using simple combinations of antibody markers or Gal4 driver lines (e.g., Repo) in conjunction with position and cell morphology.
Embryonic glia can be successfully manipulated by a number of approaches including the Gal4/UAS system, “flip-out” clonal strategies, or through the functional analysis of mutations in specific glial genes. The embryo is also well suited to techniques such as immunohistochemistry, in situ hybridization, and live imaging because of preparation simplicity and tissue transparency. These techniques allow for incisive in vivo studies of the function of specific glia and glial-expressed genes in CNS development and physiology. Gene misexpression, or the expression of dominant negatives, via Gal4/UAS appears to be the best approach for assaying mosaic animals (e.g., altering the fate of a subset of glia with Gal4/UAS). Loss-of-function genetic mosaic methods, such as mosaic analysis with a repressible cell marker (MARCM; Lee and Luo 1999), have not proven highly successful in the embryo. It is thought that MARCM likely fails to reveal clones because of perdurance of the maternally contributed Gal80 repressor through late stages of embryogenesis. Likewise, although there are a few studies in the literature that report successful use of RNA interference (RNAi) in the embryonic CNS (both injected and transgenic RNAi), the vast majority of researchers report poor results with such methods. RNAi is generally slow to knock down target genes, and such a delayed knockdown, coupled with the speed with which the entire embryonic CNS develops (~18 h), may explain the lack of robust RNAi expression in the embryonic CNS. Attempts at transgenic RNAi (e.g., UAS-regulated RNAi constructs) likely exacerbate this issue because Gal4 itself will take additional time to activate high levels of the transgene encoding the double-stranded RNAi (dsRNAi).
Assaying Glial Cell-Fate Specification in the Embryonic Nervous System
Is the correct number of glia generated in the mutant background that I am studying? A simple experiment to determine whether the appropriate number of glia have been generated in an embryonic preparation would be to stain an embryo with an anti-Repo antibody (available from the Developmental Studies Hybridoma Bank at the University of Iowa) and simply count the number of glia per hemisegment. Segment boundaries can be delineated by costaining with anti-HRP (horseradish peroxidase), anti-Fasciclin II, or any other marker that has a segmentally reiterated pattern. Repo, being a nuclear transcription factor, will label all glial nuclei, thereby allowing for the identification of total glial number. Although the total number and subtypes of glia are nearly identical from embryo to embryo, the precise position of glial nuclei can be somewhat variable. As such, it is often difficult to identify exactly the same glial cell from animal to animal unless additional markers for specific glial subtypes are used (Beckervordersandforth et al. 2008), positions are scored very carefully in the CNS, or glial cell fate is assayed at very specific developmental stages. Simple examples are given below that use only Repo antibody and cell position at specific developmental stages.
NGB6-4T Glia
Glia produced by NGB6-4 can be uniquely identified by simply staining embryos with an anti-Repo antibody and scoring glial cell fates at embryonic stage 13. In the thorax, NGB6-4 (NGB6-4T) first produces three glia, and then only generates neurons. NGB6-4T-derived glia are generated by late stage 12, and include three glia per hemisegment, which appear to become either cortex or surface glia (Schmidt et al. 1997; Schmid 1999). By stage 13, two of these glia have migrated from the lateral position of NGB6-4T toward the midline and take on a stereotyped morphology when viewed from the ventral surface—they immediately flank the midline, positioned along the anteroposterior axis, and form a “box-like” structure with contralateral NGB6-4T-derived glia (Fig. 1A). To identify all NGB6-4T-derived glia in embryonic thoracic segments, one can also stain with Eagle antibody. NGB6-4 is one of only four NBs that express Eagle, and Eagle expression is also present in NGB6-4T progeny (Higashijima et al. 1996; Freeman and Doe 2001). The only cells in the embryo that costain with Repo and Eagle antibodies are the three NGB6-4T-derived glia.
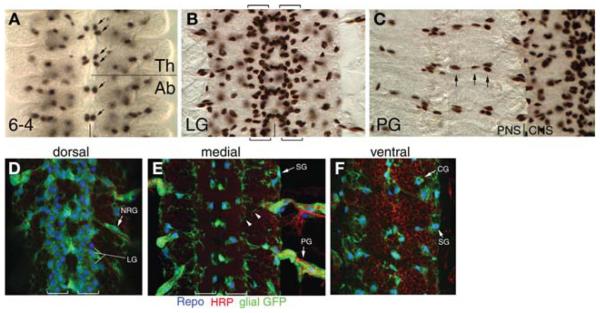
FIGURE 1.
Glial cell subtypes in the Drosophila embryonic CNS. Whole-mounted embryos processed by immunohistochemistry with anti-Repo (glial nuclei) and visualized with 3,3′-diaminobenzidine (DAB) histochemistry (A–C) or with anti-Repo, anti-HRP (neuropil), and anti-GFP (glia) and fluorescent secondary antibodies (blue, red, and green, respectively) (D–F). (A) Stage 13 embryo depicting NGB6-4 glial cell nuclei (arrows) in the thoracic (Th) and abdominal (Ab) hemisegments of the CNS. The lines mark the boundary between Th and Ab segments (horizontal) and location of the midline (vertical). (B) Longitudinal glia (LG) nuclei in an early stage 16 embryo. LG flank the midline (vertical line) and are present in columns along the length of the CNS (between brackets). (C) Peripheral glia (PG) nuclei labeled in an early stage 16 embryo. The boundary between the CNS and peripheral nervous system (PNS) is marked with a vertical line. PG migrate away from the CNS along motor neuron tracts (arrows). (D–F) Stage 16 transgenic embryos expressing UAS-cytoplasmic/membrane-bound-GFP driven by repo-Gal4. Each image represents a single slice from the same z-stack. Neuron processes are labeled in red to show their spatial relationship with glia. (D) Dorsal slice of the CNS showing examples of nerve root glia (NRG; arrow) and longitudinal glia (LG; lines). (E) Medial slice of the CNS with examples of surface glia (SG; arrow), peripheral glia (PG; arrow), and glial membrane processes adjacent to the neuropil and within the cortex (arrowheads). (F) Ventral slice of the CNS showing examples of cortex glia (CG; arrow) and surface glia (SG; arrowhead).
NGB6-4A Glia
NGB6-4 in abdominal segments (NGB6-4A) generates only two glia and no neurons. One of these glia migrates to a position adjacent to the midline, similar to that of the NGB6-4T-derived glia. Instead of appearing as a box of four cells per segment, NGB6-4A glia appear in pairs (Fig. 1A). Conveniently, both glia from NGB6-4A can be identified uniquely by using an eagle-Kinesin-LacZ reporter and scoring only abdominal segments. Although Eagle antibody does not label NGB6-4A-derived glia, the eagle-Kinesin-LacZ reporter does, perhaps because of the loss of a NGB6-4A-specific repressor for eagle when the construct was generated (Higashijima et al. 1996).
Longitudinal Glia
At late embryonic stages, the longitudinal glia (LG) reside at the dorsal surface of the CNS (Fig. 1B) and are thought to ensheath the longitudinal axon tracts. LG are derived exclusively from the glial precursor (GP), a stem cell that gives rise only to glia (in all segments), beginning at late stage 12. This precursor divides once, begins migrating medially, and continues to divide until it ultimately gives rise to seven to nine glia per hemisegment (Schmidt et al. 1997). These glia can be identified based on their dorsal position in the CNS, but there are a few additional glia that reside very close to the LG, and additional markers are essential to identify dorsal glial cells definitively as LG. Two extremely specific markers for LG are the enhancer trap line F236 (Jacobs et al. 1989) and the recently published Gal4 driver line alrm-Gal4 (Doherty et al. 2009; O Tasdemir, T Stork, and MR Freeman, unpubl.). LG can also be subdivided further at late embryonic stages by either of two available molecular markers: (1) a commercially available antibody to mammalian glutamine synthetase that cross-reacts with Drosophila glutamine synthetase 2 (GS2) and uniquely labels six LG per hemisegment, or (2) anti-Prospero antibody, which labels the same six anterior LG as GS2 (Thomas and van Meyel 2007). Thus, LG can be subdivided into Repo+/Pros+/GS2+ or Repo+/Pros−/GS2− subtypes.
Peripheral Glia
Embryonic peripheral glial are primarily derived from the CNS precursors NGB1-3 and NGB2-5, but quickly migrate out of the CNS along motor neuron tracts to the periphery in which they associate with motor neurons that project out of the CNS and sensory neurons that project into the CNS. Peripheral glia can be identified definitively based on their expression of Repo and position along the peripheral nerve (Fig. 1C), but they include a number of glial subtypes: The outermost layer is composed of perineurial glia, beneath these are subperineurial glia that form the BBB of the peripheral nerve, and the deepest cells are the wrapping glia that wrap axons (Stork et al. 2008). Leiserson et al. (2000) have also referred to wrapping glia as “ensheathing” glia. To avoid confusion we will refer to these as wrapping glia, which in the peripheral nerve refers specifically to their surrounding individual axons with membrane. In the adult, a morphologically distinct glial subtype that compartmentalizes brain regions has also been termed ensheathing glia (Awasaki et al. 2008; Doherty et al. 2009), although whether these glia wrap individual axons remains to be determined. In total, 12 glial cells per abdominal hemisegment are classified as peripheral glial cells. Of these, seven to nine are derived from CNS neuroblasts, and the remaining are derived from peripheral sensory organ precursor (SOP) lineages (Schmidt et al. 1997). A number of markers for further identification of specific peripheral glia subtypes have been described in detail elsewhere (von Hilchen et al. 2008).
Assaying Glial Cell Migration in the Embryonic Nervous System
Most glia are not generated at the site in which they will ultimately function within the nervous system. Instead, they have to migrate, sometimes significant distances, to their final positions within the developing nervous system. The vast majority of this migration occurs in the CNS/peripheral nervous system during embryonic stages 12–15. The lineages described above provide excellent examples of lineages that can be assayed for defects in glial migration in different mutant or misexpression backgrounds. For example, glia derived from NGB6-4T and NGB6-4A migrate medially along the ventral surface of the embryonic CNS; GP-derived LG migrate medially to the dorsalmost region of the CNS; and all peripheral glia migrate laterally out of the CNS to very specific positions along the peripheral nerve. Based on our knowledge of the positions in which these glia are generated, their migration pattern, and when they should arrive at their final destination, a specific mispositioning of selected subtypes of glia can be used to argue strongly for a defect in glial cell migration. For example, extensive screens for mutants with defects in peripheral glial migration along the nerve have led to the identification of key factors required to specify peripheral glial fates (Edenfeld et al. 2006, 2007). In addition, through the use of simple glial-specific drivers, one can force the expression of green fluorescent protein (GFP) and assay the dynamic movements of glia in live preparations by confocal microscopy (von Hilchen et al. 2008).
The analysis of stereotyped lineages such as those described above in combination with simple anti-Repo stains provides a good initial indicator of glial development in any mutant of interest. For example, if in a given mutant or misexpression background, NB6-4T glia are properly positioned near the midline at stage 13 and found in the appropriate numbers, then one can reasonably conclude that NB6-4T was formed, it generated glia, and those glia migrated medially and proliferated properly during development.
Visualizing Glial Cell Morphogenesis and Functions in the Embryonic Nervous System
After cell-fate specification and migration to the appropriate position within the nervous system are complete, glial cells elaborate fine membrane processes, which are presumably essential for functional maturation. During this developmental transition, glia undergo dramatic changes in shape that ultimately result in a striking array of specific glial morphological subtypes (Fig. 1D–F; Box 1). In this section, we outline a number of interesting changes in glial morphology or function that occur during embryonic neurogenesis, and describe simple assays for their visualization or functional dissection. However, owing to developmental and technical limitations, not all glial morphogenetic events are easily assayed in the embryo. For example, in some cases the elaboration of glial membranes does not occur until embryonic stage 17, which for technical reasons is difficult to study, or until larval stages, and some elaborations can be difficult to visualize because of the extremely small size of the embryonic CNS. As such, it is appropriate to study a number of neuron–glia associations specifically in the larva or adult, in which one can exploit the full array of genetic approaches available in Drosophila and glia can be considered to be functionally mature. These are discussed in the final section.
BOX 1. MAIN MORPHOLOGICAL CLASSES OF GLIAL CELLS IN DROSOPHILA.
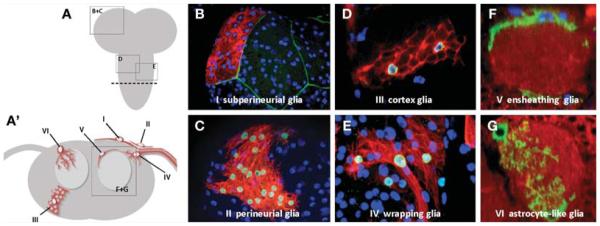
From embryonic to adult stages, three main classes of glial cells have been distinguished in Drosophila: surface-associated glia, cortex-associated glia, and neuropil-associated glia. These main classes can be further subdivided into morphologically distinct groups. The most common subtypes are exemplified in the third-instar larva (shown in Fig. 2) but can be found with very similar characteristics in the late embryo or in the adult.
Surface-Associated Glia
Perineurial glia: Perineurial glia form the outermost cellular sheath surrounding the nervous system. These Repopositive cells are believed to be of mesodermal origin (Edwards et al. 1993) and their function remains unknown. Although only very few perineurial cells cover the nervous system at the end of embryogenesis, these cells proliferate extensively to form a continuous monolayer in later larval stages and also in the adult stage (Awasaki et al. 2008; Stork et al. 2008).
Subperineurial glia
Underneath the perineurial sheath lies the thin monolayer of subperineurial glia. These extremely big, flat cells show epithelial characteristics. They ensheath the nervous system and seal their extensive cell–cell contacts with pleated septate junctions to form a paracellular diffusion barrier. This glial BBB is extremely important for neuronal physiology because it helps to exclude high potassium concentrations in the hemolymph from the nervous system and allows for proper action potential propagation (Auld et al. 1995; Baumgartner et al. 1996). Several structural components of pleated septate junctions and regulatory factors such as moody have been identified as being essential for BBB function, making subperineurial glia one of the best characterized glial subtypes in Drosophila.
Cortex-Associated Glia
This glial subtype surrounds and tightly ensheathes neuronal cell bodies in the CNS, forming a trophospongium (Dumstrei et al. 2003; Pereanu et al. 2005). A single cortex glial cell can ensheath dozens of neuronal cell bodies and typically contacts the subperineurial sheath, the neuropil associated glia, or both. Cortex glia are thought to supply trophic and metabolic support for neurons.
Neuropil-Associated Glia
Wrapping glia or nerve-associated glia
This subtype of Drosophila glia is closely associated with nerves and enwraps axonal profiles. In a mature state, a single wrapping glial cell is able to individually enwrap most axons in the diameter of a nerve simultaneously (Stork et al. 2008), resembling nonmyelinating Schwann cells in a Remak bundle in vertebrates (Nave and Salzer 2006; Nave and Trapp 2008). However, in the cortical regions of the CNS, wrapping glia also send out processes to ensheath neighboring neuronal cell bodies, as is seen in cortex glia.
Ensheathing glia
In contrast to wrapping glia, ensheathing glia are not associated with nerves but rather with the synaptic neuropil. Glial processes cover the synaptic neuropil (e.g., in the ventral nerve cord) and can ensheath and subdivide morphologically distinct substructures like the glomeruli of the adult antennal lobe (Awasaki et al. 2008; Doherty et al. 2009). In the adult, this subtype of glial cell (probably together with wrapping glia) has been shown to respond to axonal injury by clearing neuronal debris by phagocytosis (Doherty et al. 2009).
Astrocyte-like glia
These glia have their cell bodies closely associated with the synaptic neuropil. In contrast to ensheathing glia, the astocyte-like glial cell processes invade the neuropil and infiltrate the volume densely. Their tufted morphology and the close contact of glial membranes to synapses are reminiscent of vertebrate protoplasmic astrocyte morphology. Additionally, similar to the situation in vertebrates, Drosophila astrocyte-like glia cells express neurotransmitter transporters that are able to clear neurotransmitters, such as glutamate or GABA, from the synapse to ensure proper brain function (Rival et al. 2004; T Stork, unpubl.).
Formation of the BBB by Subperineurial Glia
Surface glia form the BBB that isolates and protects the embryonic CNS. A subset of glia, termed subperineurial glia (SPGs), position themselves along the surface of the CNS and by embryonic stage 15, these cells have flattened to cover the entire surface of the VNC and brain. Subsequently, they form pleated septate junctions (pSJs) with one another to seal off the CNS from the surrounding hemolymph (Auld et al. 1995; Baumgartner et al. 1996; Schwabe et al. 2005). SPGs are unique in their expression of moody, a seven-transmembrane-domain receptor required for the regulation of pSJs in SPGs (Bainton et al. 2005; Schwabe et al. 2005). moody-Gal4 or Spg-Gal4 can be used to drive markers for pSJs such as GFP::DlgS97 (Bachmann et al. 2004; Stork et al. 2008) and pSJ assembly can be visualized in live preparations (Bainton et al. 2005; Schwabe et al. 2005). Alternatively, GFP gene trap lines like Nrx-GFP (Edenfeld et al. 2006) can be used to visualize pSJs in vivo.
A number of groups have used a simple protocol initially designed to test the integrity of pSJs in the salivary gland epithelium (Lamb et al. 1998) to assay BBB integrity in vivo. Briefly, fluorescent dye-dextran conjugates (generally 10-kDa dextrans) are injected into the hemolymph of late-stage embryos, and after ~10–20 min, their CNS is visualized by confocal microscopy. If the BBB is intact, dye-dextran conjugates are excluded from the CNS. However, if the BBB is compromised, dyedextran conjugates accumulate in the CNS (around cell bodies and in the neuropil), indicating that the BBB is permeabilized. This simple assay has been used effectively to assay BBB integrity in a number of mutants affecting pSJs or their formation (Bainton et al. 2005; Schwabe et al. 2005; Banerjee et al. 2006; Stork et al. 2008).
Glial Engulfment of Neuronal Cell Corpses
Approximately 30% of CNS neurons generated during embryonic neurogenesis appear to undergo programmed cell death (Rogulja-Ortmann et al. 2007) and their cell corpses are engulfed and destroyed by glia (Sonnenfeld and Jacobs 1995; Freeman et al. 2003; Kurant et al. 2008). A simple approach to assay the efficiency of glial clearance of neuronal cell corpses is to stain the embryonic nervous system at a defined developmental stage (e.g., stage 15) with 7-aminoactinomycin-D (7-AAD; Franc et al. 1999). 7-AAD is a fluorescent nucleic acid intercalating agent that is used to visualize cellular nucleic acids. The vast majority of neuronal cell bodies in the CNS stain weakly with 7-AAD in fixed preparations, and neuronal cell corpses stain as bright puncta that are ~25% the size of a healthy neuronal cell (Freeman et al. 2003). The number of cell corpses per hemisegment can be quantified by costaining with a marker that delineates segment boundaries (e.g., Fasciclin II). It is important to note that this approach gives an overall assessment of the number of neuronal cell corpses present at a given developmental stage. Care must be taken in interpreting these results and distinguishing between glia function and neuronal survival. For example, a mutant may be suspected to affect engulfment by glia, but its actual role is to give trophic support to neurons. The increase in neuronal cell corpses may be a result of loss of trophic support rather than a decrease in engulfment by glia.
A more incisive method for exploring the dynamics of glial engulfment of neuronal cell corpses is to observe glial morphological changes directly during engulfment activity in live preparations. One can easily express GFP specifically in glia (e.g., repo-Gal4, UAS-mCD8-GFP) to visualize glial morphology and neuronal cell corpses can be visualized simultaneously using commercially available markers for apoptotic cells such as annexin V (Kurant et al. 2008). Levels of glial lysosomal activity can be assayed using LysoTracker probes (Watts et al. 2004; Kurant et al. 2008).
Axon Pathfinding and Trophic Support of Neurons by Glia
Based largely on cell ablation studies, embryonic CNS glial cells have been reported to play critical roles in axon pathfinding (Hidalgo and Booth 2000; Sepp et al. 2001; Sepp and Auld 2003) and trophic support of neurons (Booth et al. 2000), topics that are not discussed in detail here. It is thus expected that a number of glial-expressed genes will modulate axonal outgrowth and neuronal survival. With respect to axon pathfinding, we refer the reader to Sepp and Auld (2003) and Sepp et al. (2001) for discussions of the role of peripheral glia in entry of sensory neuronal processes into the embryonic CNS. A lack of glial trophic support of neurons would be expected to result in the apoptotic death of neurons that would normally survive, thereby generating an increase in overall neuronal cell death in the developing embryonic CNS. A simple initial method helpful for detecting increased neuronal cell death would be to combine the use of antibodies directed toward activated caspase-3 (Kurant et al. 2008) with markers for identifiable subsets of embryonic CNS neurons (i. e., do cells that normally survive inappropriately activate caspase-3?). Alternatively, one could use a battery of markers for uniquely identifiable neurons, count total numbers of neurons, and determine whether neuronal populations are reduced in genetic backgrounds of interest (i.e., are specific cells missing?).
GLIAL CELL DEVELOPMENT AND FUNCTION IN THE LARVAL AND ADULT NERVOUS SYSTEM
Over the last 20 years, the embryo has proven to be an excellent system in which to explore basic aspects of glial cell development. However, embryonic preparations also have their limitations. First, with standard staining procedures, only embryos that are at developmental stage 16 or earlier can be analyzed because the developing cuticle becomes increasingly impermeable to fixatives and antibodies by embryonic stage 17. At 25°C, neurogenesis begins ~3–4 h after egg lay (AEL) and continues until larval hatching. Stage 17 represents the final ~6 h of embryonic neurogenesis, or the last 33% of neural tissue development (Campos-Ortega and Hartenstein 1997; Prokop 1999). Therefore, the final stages of embryonic nervous system maturation are largely missed, and this may in fact be the time when some of the most interesting elaborations of glial processes take place to give rise to fully functional glia. Second, a number of additional interesting glial morphogenetic changes do not occur until larval stages. For example, at late embryonic stages, peripheral glia appear to wrap entire bundles of axons, but as animals progress through larval stages, these glia eventually separate and individually ensheath individual axons (Stork et al. 2008). Third, although it is generally difficult to manipulate embryos with powerful molecular-genetic tools like cell type-specific RNAi or MARCM, these approaches seem to work well in larval, pupal, and adult nervous system glia (Lee and Luo 1999; Aigouy et al. 2004; Dietzl et al. 2007; Doherty et al. 2009). The postembryonic CNS is an attractive system in which the full complement of sophisticated genetic tools available in Drosophila can be used to visualize and manipulate glia, even at the single-cell level. As discussed in Box 1, the major subtypes of glia established in the embryo are also found in the larva (Fig. 2A–G) and adult (not shown). Here we describe a number of genetic approaches for marking and manipulating specific subsets, or a very small number of glial cells, and discuss their advantages and limitations.
FIGURE 2.
Main morphological subclasses of glial cells in Drosophila. (A) Schematic overview of the CNS of a third-instar larva with the two brain lobes and the unpaired ventral nerve cord shown in gray. The dashed line indicates the plane of a schematic cross section shown in panel A′. (A′) Dark gray depicts the cortex, the region of cell bodies, and light gray depicts the synaptic neuropil. The different subtypes of glial cells described in the text are highlighted as single cells in red and numbered I–VI. The boxes refer to the approximate regions in the CNS in which the images shown in panels B–G were taken. (B–G) Confocal images of glial subtype morphology highlighted with different “flip-out” strategies. Glial nuclei are visualized with a-Repo staining (B–F; blue), and glial membranes are shown with the use of Gal4-driven mCD8-Cherry (B–E) or mCD8-GFP (F,G). Additional markers include a NrxIV-GFP exon trap, which labels the cell borders of subperineurial glia (B; green); nuclear β-galactosidase, which labels the cell nuclei of the glial cell clones (C–E; green); and α-HRP to label neuronal membranes (F,G; red). Note that in B a single subperineurial glial cell is labeled and that nearly all Repo-positive nuclei correspond to other, deeper located glial cells. The following genotypes were used: (B) act5c > CD2 > Gal4, repo-flp, and UAS mCD8-Cherry/nrxIV-GFP; (C–E) act5c > CD2 > Gal4, repo-flp, and UAS mCD8-Cherry/UAS lacZ::NLS; (F) UAS > CD2 > mCD8-GFP repo-flp and Mz709-Gal4; (G) UAS > CD2 > mCD8-GFP repo-flp and alrm-Gal4.
Molecular Markers for Postembryonic Stages
A number of molecular markers for stereotyped subpopulations of glial cells have been identified in Drosophila (e.g., Ito et al. 1995; Beckervordersandforth et al. 2008; von Hilchen et al. 2008). Most of these markers have been used primarily to study glial cell populations in the embryo, but remain poorly characterized at later developmental stages. An array of potential tools exist for studying glial cells at larval stages and beyond, but at the moment, we know surprisingly little about molecular identities and fine morphology of glial cells at these later time points. Nevertheless, a reasonable number of genetic tools for the visualization and manipulation of glial cells have been characterized in more detail in postembryonic stages (e.g., Awasaki et al. 2008; Stork et al. 2008; Doherty et al. 2009), and we have listed some of the tools we find particularly useful for the analysis of glia at different developmental stages (Table 1).
Analysis of Glial Gene Function by Cell Type-Specific dsRNAi
Cell type-specific dsRNAi can be accomplished through the use of Gal4-driven UAS constructs (Brand and Perrimon 1993) that carry inverted repeats of a gene of interest (Lee and Carthew 2003). On expression, these inverted repeats can fold back to form dsRNA, which is processed by Dicer (a ribonuclease), to give rise to the siRNAs (small interfering RNAs) (Fire et al. 1998). These siRNAs then lead to a sequence-specific knockdown of the corresponding gene of interest. By choosing glialspecific Gal4 driver lines, expression of genes can be manipulated in all glia, or even in small subsets of glial cells, without perturbing gene function in other tissues (Table 1). Although the Gal4 lines listed in Table 1 are predominantly expressed in glial cells, additional expression in other tissues is common, and expression patterns may dynamically change at different developmental stages. Therefore, care must be taken to show rigorously that any phenotype observed using UAS-dsRNAi with glial driver lines is in fact the result of glial gene expression manipulation rather than manipulation of gene expression in other tissues. Strong Gal4 driver lines and prolonged expression of the UAS-dsRNAi constructs are normally necessary for efficient knockdown of the targeted gene’s expression. Both of these issues are quite manageable in larval, pupal, and adult stages. Recent reports suggest that coexpression of UAS-dicer2 can enhance the effects of dsRNAi (Dietzl et al. 2007). The efficacy of dsRNAi-mediated knockdown may vary for different genes, UAS-dsRNAi constructs, and even different insertions of the same transgene, and therefore efficacy of dsRNAimediated knockdown should be empirically verified in each experiment (Dietzl et al. 2007; Ni et al. 2008). Furthermore, one needs to be aware that dsRNAi constructs may show off-target effects in which the resulting siRNAs are not entirely specific to the gene of interest (Kulkarni et al. 2006; Ma et al. 2006; Moffat et al. 2007). Ideally, data obtained by a UAS-dsRNAi approach should be complemented by additional genetic data, such as MARCM analysis, whenever possible (see below). Because UAS-dsRNAi constructs exist for ~90% of Drosophila genes and are readily available from public stock centers (Vienna Drosophila RNAi Center, Austria; Dietzl et al. 2007; National Institute of Genetics Fly Stock Center, Japan; Transgenic RNAi Project, USA; Ni et al. 2008, 2009), this approach has the potential to be extremely powerful in the analysis of glial cell biology from larval stages through adulthood.
Genetic Mosaic Approaches to Characterizing Glial Morphology and Function
“Flip-out” Techniques
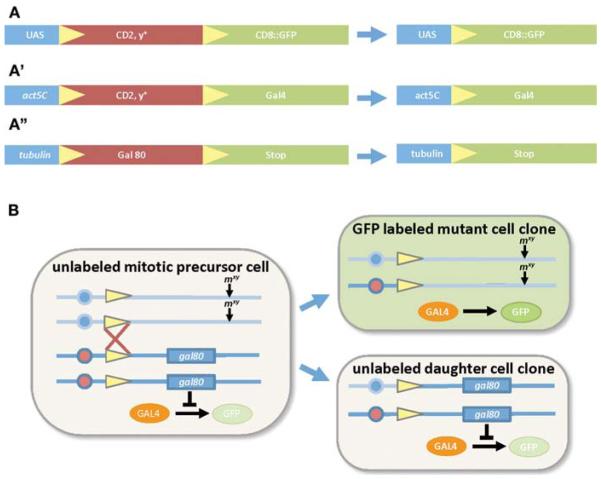
UAS Flip-Out Constructs
One way to restrict the expression patterns of Gal4 lines is by using a so-called “flip-out” approach (Basler and Struhl 1994). In this method, a promoter element is followed by a cassette, which is flanked by direct repeats of flippase recognition target (FRT) sites. These sites are recognized by the yeast site-specific recombinase, termed flippase (FLP), which leads to a circular excision of the FRT flanked sequences (Fig. 3A). One example of such a construct is UAS-FRT-CD2-y+-FRT-mCD8-GFP (or shorthand UAS > CD2 > mCD8-GFP; Wong et al. 2002). Without FLP activity, only the membrane marker CD2 is expressed by a Gal4 driver line, whereas the mCD8-GFP reporter remains silent. On activation of FLP, the CD2 flip-out cassette is excised and mCD8-GFP comes under direct Gal4 control (Fig. 3A). The excision of the FRT-flanked sequences is stochastic, depending on how strong the FLP activity is in a given cell, and can occur in either a mitotic or postmitotic cell. The latter point is important for studying glia during larval or adult stages because the vast majority of larval glia do not appear to proliferate. Several different FLP sources are available and their specific abilities are summarized in Box 2. Typically, FLP activity leads to clonal expression of the UAS-flip-out GFP reporter so that subtle changes in cell morphology can be visualized, rather than obscured by the full expression pattern normally generated by the Gal4 driver (Fig. 2A–G). Such approaches can lead to very clear images of the morphology of single glial cells. In addition, this labeling technique can be used in a mutant background, or coupled with a UAS-RNAi construct to assay changes in glial morphology in response to loss of a particular gene product.
FIGURE 3.
“Flip-out” and MARCM techniques. (A,A′,A′′) Schematic representations of flip-out events for three different constructs and (B) the genetic basis of the MARCM technique. (A,A′,A′′) The flip-out cassettes (red boxes) are flanked by directly repeated FRT sites (yellow triangles). Expression of genes contained in the flip-out cassette can be under the control of different promoter elements like Gal4 controllable UAS sequences (A) or specific promoter elements like the ubiquitously active actin5c or tubulin promoters (A′,A′′). Expression of Flp (not shown) mediates site-specific recombination between the FRT sites leading to the removal of the flip-out cassette and its respective marker expression. Instead, a second reporter element (green boxes) is now under the control of the promoter, here either CD8-GFP (A) or Gal4 (A′). For details, see text. (B) In a MARCM approach, the two homologous chromosome arms are carrying FRT sites (yellow triangles) near to their centrosomes (blue and red circles representing maternal and paternal copy of the chromosome). One of the chromosome arms is equipped with a tub-Gal80 insertion distal to the FRT site and on the other arm, a mutation of interest (mxy) can be placed. Gal80 suppresses Gal4-mediated GFP expression (orange and green oval, respectively; transgenic insertions for these proteins are not depicted and can be placed elsewhere in the genome). Flp-mediated site-specific recombination during the G2 phase of mitosis between the FRT sites of homologous chromosome arms (red cross; FLP not shown) leads to the separation of Gal80 and the mutation of interest after the mitotic division. This results in a homozygous mutant daughter cell that lost Gal80 and therefore starts to express the GFP reporter while the corresponding daughter cell and all other nonclonal cells keep expressing Gal80. GFP-labeled mutant clones can then be analyzed for morphological defects. For details, see text.
BOX 2. ADVANTAGES AND DISADVANTAGES OF DIFFERENT FLP SOURCES.
hs-flp is the workhorse for any flip-out or MARCM strategy. The expression of yeast recombinase FLP under the control of a heat shock-inducible promoter (Golic and Lindquist 1989; Struhl and Basler 1993) allows for high temporal control of clone induction. Additionally, strength of FLP activity can be adjusted by temperature, duration, and repetition of heat shocks. As a starting point, a typical heat shock would be conducted for 1 h at 37°C in a water bath at the desired developmental stage. Prolonged exposures or slightly higher temperatures are possible, but will likely lead to elevated levels of lethality. Instead, additional heat shocks can be given after a period of recovery on consecutive days. Furthermore, several hs-flp constructs and insertions are available that can differ in their abilities, like maximal FLP activity or basal level of expression, without heat shock induction (Theodosiou and Xu 1998).
repo-flp is a promoter fusion of the repo promoter (Lee and Jones 2005) with a FLP open reading frame (Silies et al. 2007). The repo promoter is active specifically in glia beginning at early developmental stages and remains on through adulthood, making it applicable for glial-specific flip-out as well as MARCM experiments. Because this construct is constitutively active in glia, FLP activity can only be modulated by copy number or choosing different repo-flp insertions, which can differ in FLP expression level quite substantially. Although repo-flp tends to work well for FLP-out approaches, activity can be low in early glial precursors, which may result in low clone frequency in some glial lineages when using MARCM. The clone frequency also depends on the efficiency of the FRT sites used (Rooke et al. 2000) and must be tested empirically for each glial MARCM setup. The main advantages of a repo-flp source are (1) mutant clones are generated specifically in glial cells and (2) clonal events can be induced without additional heat-shock protocols, which can be especially advantageous when large numbers of different genotypes have to be tested routinely.
UAS-flp/lexAop-flp
In combination with flip-out labeling approaches (e.g., UAS > CD2 > CD8GFP), UAS-flp and lexAop-flp constructs can be especially useful for characterizing expression patterns of Gal4 or LexA driver lines with high resolution because typically low numbers of cells will be labeled. For crossing schemes, including Gal80 constructs like MARCM or a tub > stop > Gal80 approach, these constructs are only useful if the corresponding Gal4 or LexA line is not repressible by Gal80 activity. This can be accomplished by use of the VP16 transactivation domain rather than that of Gal4 with Gal4 or LexA transgenes. Using such an approach, a glial LexA::VP16 driver can be used to drive lexAop-flp expression and generate Gal4-labeled clones in a MARCM background. At the moment, suitable glial VP16 fusions are not yet available but should be generated in the near future.
Flip-Out Gal4 Drivers
Without generating additional constructs, the approach described above is, at the moment, mainly restricted to mCD8::GFP as the clonal reporter. However, with other flip-out strategies it is possible to visualize and manipulate single glial cells with any available UAS line. This can be performed, for example, with an actin5C promoter-driven construct that conditionally activates Gal4 (act5c > CD2 > Gal4; Ito et al. 1997; Pignoni and Zipursky 1997). In this case, FLP activity leads to the expression of Gal4 under the control of the ubiquitiously expressed actin5C promoter, which then can be used to drive any UAS-regulated construct. Because this promoter is not specific to glial cells, one can increase its utility for studying glia by spatially restricting the FLP expression using either a repo promoter-driven version of FLP (repo-flp; Silies et al. 2007; Fig. 2) or through a LexA/lexA operon-controlled expression of FLP (Lai and Lee 2006; Shang et al. 2008). In general, it is also possible to exchange the ubiquitous promoters in the flip-out Gal4 constructs or the promoters controlling FLP expression for glial subset-specific promoters, as long as these sequences are identified.
Flip-Out Gal80 Constructs
Another more flexible approach that recently became available uses the clonal loss of the Gal4 repressor Gal80 as a way to activate Gal4 in subsets of cells. In this approach, a tubulin promoter drives the expression of a Gal80 gene that is flanked by FRT sites (tub > Gal80 > Stop) and ubiquitously represses Gal4 activity. On FLP-mediated clonal removal of the Gal80 repressor cassette, Gal4 is activated and any given UAS construct can be driven in the clonal cells (Shang et al. 2008; Gordon and Scott 2009). FLP activity can be induced by heatshock (hs-flp), or specifically targeted to glial cells by repo-flp or lexAop-flp under control of a glial LexA::VP16 driver (not repressible by Gal80). The advantage of this system is the high degree of experimental freedom because the expression can be controlled ad libitum by combining different Gal4 driver lines with any UAS effector line. Minor drawbacks of this approach might be the perdurance of the Gal80 protein after the cassette has been excised, which might lead to repressed Gal4 activity for another 24–48 h after excision (Lee and Luo 1999; Lee et al. 2000), or incomplete repression of Gal4 activity before the flip-out event of the Gal80 cassette, especially with strong Gal4 driver lines.
MARCM Analysis in Glial Cells
Another powerful method used to label and manipulate subsets of glial cells is MARCM. This approach allows positive genetic labeling of wild-type or mutant cell clones (Lee and Luo 1999) (Fig. 3B). Briefly, mitotic recombination is induced by FLP activity between homologous chromosome arms that carry FRT sites near to their centromeres. tub-Gal80 is present on one of the chromosome arms to repress activation of Gal4-regulated reporters (e.g., repo-Gal4, UAS-mCD8-GFP) in all heterozygous progenitor cells. The other chromosome arm either can be wild-type (control) or can carry a mutation of interest. Induction of mitotic recombination can result in production of two different daughter cell clones: one that is homozygous for tub-Gal80, and therefore Gal4/UAS remains repressed, and another that loses Gal80, which results in the derepression of Gal4/UAS and activation of UAS-driven reporter expression. By placing a mutation on the chromosome arm in trans to the tub-Gal80 (Fig. 3B), one can generate positively marked mutant glial cells that can be analyzed for cell-autonomous morphological defects. MARCM can also be used with any UAS-dsRNAi lines of interest to assay the autonomous effects of a gene in glia because the dsRNAi construct will only be driven in GFP-marked clones.
In contrast to the flip-out techniques described earlier, induction of MARCM clones requires mitotic recombination and therefore the presence of dividing glial precursor cells. Consequently, MARCM leads to lower clone frequencies, and the timing of clone production is restricted to specific developmental time points (Awasaki et al. 2008). For example, to make MARCM clones in larval astrocyte-like glia or subperineurial glia, one would have to induce clones in the embryo, when these populations of glia are still mitotically active. This presents a challenge for glial lineages with low proliferation rates because these will inherently generate fewer clones. Clone production can be enhanced by crossing in additional transgenes of the FLP source (Box 2), or in the case of hs-flp, through prolonged and repeated heat shock administrations during development. Finally, it is important to note that, depending on the FLP source used, MARCM can generate a substantial fraction of homozygous mutant cells in the nervous system (e.g., neurons) that are not labeled by the Gal4 reporter but could potentially contribute to any phenotypes observed. As with all analyses that involve the stochastic production of clones, these issues can be overcome by assaying for consistent phenotypes in many independently generated clonal animals. An additional possible pitfall of (glial) MARCM can be incomplete or inconsistent suppression of Gal4 activity by tub-Gal80. This can lead to reporter expression in small subsets of glial cells even in the absence of any FLP source, which could be mistaken as mutant clones. Awasaki et al. (2008) used an additional repo-Gal80 construct in their MARCM setup to circumvent this problem.
CONCLUSIONS
The embryonic nervous system has been and will continue to be a superb setting for assaying some of the earliest events in glial cell development including glial cell-fate specification, generation of molecularly and morphologically diverse glial subtypes, glial cell migration, and morphogenesis of the BBB. In addition, the embryo has been quite helpful for understanding glial control of neuronal development, including neuronal trophic support, axon pathfinding, nerve fasciculation, and clearance of dead cells from the CNS. Because we know so little about glial cell morphogenesis, there is clearly a wealth of interesting glial developmental biology to be uncovered through studies in the embryo, and our hope is that the methods outlined in this article will serve as a starting point for those types of analyses. At the same time, some of the most amazing features of glial cells, such as their massive expansion in size (e.g., along segmental nerves during the L1 to L3 growth), intimate association with highly organized brain structures (e.g., the lamina of the visual system), or fine elaboration of membranes (e.g., the tufted morphology of astrocyte vs. the flattened morphology of subperineurial glia), seem better addressed in the larval, pupal, and adult nervous system. In the second part of this article, we have attempted to outline a number of useful tools, genetic techniques, and glial markers that should facilitate the exploration of these later steps in glial cell morphogenesis. Finally, a major goal in the fleld of glial biology, and neuroscience in general, is to understand how glial cells control neural function to ultimately affect animal behavior. There are probably few synapses, neural circuits, or behaviors that are not regulated in some way by glia. The challenge before us now is to understand the cellular and molecular basis of these interactions. We believe the future is bright in this regard for Drosophila researchers, and hope that the information provided in this article will aid in this endeavor.
REFERENCES
- Abbott NJ. Dynamics of CNS barriers: Evolution, differentiation, and modulation. Cell Mol Neurobiol. 2005;25:5–23. doi: 10.1007/s10571-004-1374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigouy B, Van de Bor V, Boeglin M, Giangrande A. Time-lapse and cell ablation reveal the role of cell interactions in fly glia migration and proliferation. Development. 2004;131:5127–5138. doi: 10.1242/dev.01398. [DOI] [PubMed] [Google Scholar]
- Alfonso TB, Jones BW. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev Biol. 2002;248:369–383. doi: 10.1006/dbio.2002.0740. [DOI] [PubMed] [Google Scholar]
- Auld VJ, Fetter RD, Broadie K, Goodman CS. Gliotactin, a novel transmembrane protein on peripheral glia, is required to form the bloodnerve barrier in Drosophila. Cell. 1995;81:757–767. doi: 10.1016/0092-8674(95)90537-5. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol. 2004;14:668–677. doi: 10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann A, Timmer M, Sierralta J, Pietrini G, Gundelfinger ED, Knust E, Thomas U. Cell type-specific recruitment of Drosophila Lin-7 to distinct MAGUK-based protein complexes defines novel roles for Sdt and Dlg-S97. J Cell Sci. 2004;117:1899–1909. doi: 10.1242/jcs.01029. [DOI] [PubMed] [Google Scholar]
- Bainton RJ, Tsai LT, Schwabe T, DeSalvo M, Gaul U, Heberlein U. Moody encodes two GPCRs that regulate cocaine behaviors and bloodbrain barrier permeability in Drosophila. Cell. 2005;123:145–156. doi: 10.1016/j.cell.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Pillai AM, Paik R, Li J, Bhat MA. Axonal ensheathment and septate junction formation in the peripheral nervous system of Drosophila. J Neurosci. 2006;26:3319–3329. doi: 10.1523/JNEUROSCI.5383-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA. The mystery and magic of glia: A perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- Baumgartner S, Littleton JT, Broadie K, Bhat MA, Harbecke R, Lengyel JA, Chiquet-Ehrismann R, Prokop A, Bellen HJ. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 1996;87:1059–1068. doi: 10.1016/s0092-8674(00)81800-0. [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth RM, Rickert C, Altenhein B, Technau GM. Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mech Dev. 2008;125:542–557. doi: 10.1016/j.mod.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Booth GE, Kinrade EF, Hidalgo A. Glia maintain follower neuron survival during Drosophila CNS development. Development. 2000;127:237–244. doi: 10.1242/dev.127.2.237. [DOI] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe CQ, Tomlinson A. RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development. 1994;120:2957–2966. doi: 10.1242/dev.120.10.2957. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. Springer; Berlin: 1997. [Google Scholar]
- Chotard C, Leung W, Salecker I. Glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron. 2005;48:237–251. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Tasdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Hartenstein V. Role of DE-cadherin in neuroblast proliferation, neural morphogenesis, and axon tract formation in Drosophila larval brain development. J Neurosci. 2003;23:3325–3335. doi: 10.1523/JNEUROSCI.23-08-03325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebens AJ, Garren H, Cheyette BN, Zipursky SL. The Drosophila anachronism locus: A glycoprotein secreted by glia inhibits neuroblast proliferation. Cell. 1993;74:15–27. doi: 10.1016/0092-8674(93)90291-w. [DOI] [PubMed] [Google Scholar]
- Edenfeld G, Volohonsky G, Krukkert K, Naffin E, Lammel U, Grimm A, Engelen D, Reuveny A, Volk T, Klambt C. The splicing factor crooked neck associates with the RNA-binding protein HOW to control glial cell maturation in Drosophila. Neuron. 2006;52:969–980. doi: 10.1016/j.neuron.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Edenfeld G, Altenhein B, Zierau A, Cleppien D, Krukkert K, Technau G, Klambt C. Notch and Numb are required for normal migration of peripheral glia in Drosophila. Dev Biol. 2007;301:27–37. doi: 10.1016/j.ydbio.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Edwards JS, Swales LS, Bate M. The differentiation between neuroglia and connective tissue sheath in insect ganglia revisited: The neural lamella and perineurial sheath cells are absent in a mesodermless mutant of Drosophila. J Comp Neurol. 1993;333:301–308. doi: 10.1002/cne.903330214. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Franc NC, Heitzler P, Ezekowitz RA, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284:1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Doe CQ. Asymmetric Prospero localization is required to generate mixed neuronal/glial lineages in the Drosophila CNS. Development. 2001;128:4103–4112. doi: 10.1242/dev.128.20.4103. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- Gilmour DT, Maischein HM, Nusslein-Volhard C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron. 2002;34:577–588. doi: 10.1016/s0896-6273(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes sitespecific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, Technau GM. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development. 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, Aigaki T, Matsuzaki F, Nakagoshi H, Tanimura T, et al. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 2002;34:58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Booth GE. Glia dictate pioneer axon trajectories in the Drosophila embryonic CNS. Development. 2000;127:393–402. doi: 10.1242/dev.127.2.393. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Shishido E, Matsuzaki M, Saigo K. eagle, a member of the steroid receptor gene superfamily, is expressed in a subset of neuroblasts and regulates the fate of their putative progeny in the Drosophila CNS. Development. 1996;122:527–536. doi: 10.1242/dev.122.2.527. [DOI] [PubMed] [Google Scholar]
- Ito K, Urban J, Technau GM. Distribution, classification and development of Drosophila glial cells during late embryogenesis. Roux’s Arch Dev Biol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jacobs JR. The midline glia of Drosophila: A molecular genetic model for the developmental functions of glia. Prog Neurobiol. 2000;62:475–508. doi: 10.1016/s0301-0082(00)00016-2. [DOI] [PubMed] [Google Scholar]
- Jacobs JR, Hiromi Y, Patel NH, Goodman CS. Lineage, migration, and morphogenesis of longitudinal glia in the Drosophila CNS as revealed by a molecular lineage marker. Neuron. 1989;2:1625–1631. doi: 10.1016/0896-6273(89)90051-2. [DOI] [PubMed] [Google Scholar]
- Kimelberg HK. Astrocyte heterogeneity or homogeneity? In: Parpura V, Haydon PG, editors. In Astrocytes in (patho)physiology of the nervous system. Springer Science + Business Media, LLC; New York: 2009. pp. 1–26. [Google Scholar]
- Kofuji P, Newman EA. Regulation of potassium by glial cells in the central nervous system. In: Parpura V, Haydon PG, editors. In Astrocytes in (patho)physiology of the nervous system. Springer Science and Business Media, LLC; New York: 2009. pp. 151–176. [Google Scholar]
- Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- Kurant E, Axelrod S, Leaman D, Gaul U. Six-microns-under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell. 2008;133:498–509. doi: 10.1016/j.cell.2008.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- Lamb RS, Ward RE, Schweizer L, Fehon RG. Drosophila coracle, a member of the protein 4.1 superfamily, has essential structural functions in the septate junctions and developmental functions in embryonic and adult epithelial cells. Mol Biol Cell. 1998;9:3505–3519. doi: 10.1091/mbc.9.12.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Lee BP, Jones BW. Transcriptional regulation of the Drosophila glial gene repo. Mech Dev. 2005;122:849–862. doi: 10.1016/j.mod.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Leiserson WM, Harkins EW, Keshishian H. Fray, a Drosophila serine/threonine kinase homologous to mammalian PASK, is required for axonal ensheathment. Neuron. 2000;28:793–806. doi: 10.1016/s0896-6273(00)00154-9. [DOI] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- Mayer F, Mayer N, Chinn L, Pinsonneault RL, Kroetz D, Bainton RJ. Evolutionary conservation of vertebrate blood-brain barrier chemoprotective mechanisms in Drosophila. J Neurosci. 2009;29:3538–3550. doi: 10.1523/JNEUROSCI.5564-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Reiling JH, Sabatini DM. Off-target effects associated with long dsRNAs in Drosophila RNAi screens. Trends Pharmacol Sci. 2007;28:149–151. doi: 10.1016/j.tips.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Nave KA, Salzer JL. Axonal regulation of myelination by neuregulin 1. Curr Opin Neurobiol. 2006;16:492–500. doi: 10.1016/j.conb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Nave KA, Trapp BD. Axon-glial signaling and the glial support of axon function. Annu Rev Neurosci. 2008;31:535–561. doi: 10.1146/annurev.neuro.30.051606.094309. [DOI] [PubMed] [Google Scholar]
- Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, Booker M, Perkins L, Perrimon N. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, Villalta C, Laverty TR, Perkins LA, Perrimon N. A Drosophila resource of transgenic RNAi line for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oland LA, Tolbert LP. Key interactions between neurons and glial cells during neural development in insects. Annu Rev Entomol. 2003;48:89–110. doi: 10.1146/annurev.ento.48.091801.112654. [DOI] [PubMed] [Google Scholar]
- Paladi M, Tepass U. Function of Rho GTPases in embryonic blood cell migration in Drosophila. J Cell Sci. 2004;117:6313–6326. doi: 10.1242/jcs.01552. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Shy D, Hartenstein V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev Biol. 2005;283:191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by Decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Prokop A. Integrating bits and pieces: Synapse structure and formation in Drosophila embryos. Cell Tissue Res. 1999;297:169–186. doi: 10.1007/s004410051345. [DOI] [PubMed] [Google Scholar]
- Rival T, Soustelle L, Strambi C, Besson MT, Iche M, Birman S. Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr Biol. 2004;14:599–605. doi: 10.1016/j.cub.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Rival T, Soustelle L, Cattaert D, Strambi C, Iche M, Birman S. Physiological requirement for the glutamate transporter dEAAT1 at the adult Drosophila neuromuscular junction. J Neurobiol. 2006;66:1061–1074. doi: 10.1002/neu.20270. [DOI] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Luer K, Seibert J, Rickert C, Technau GM. Programmed cell death in the embryonic central nervous system of Drosophila melanogaster. Development. 2007;134:105–116. doi: 10.1242/dev.02707. [DOI] [PubMed] [Google Scholar]
- Rooke JE, Theodosiou NA, Xu T. Clonal analysis in the examination of gene function in Drosophila. Methods Mol Biol. 2000;137:15–22. doi: 10.1385/1-59259-066-7:15. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: Neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in Drosophila. Cell. 2005;123:133–144. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Auld VJ. Conversion of lacZ enhancer trap lines to Gal4 lines using targeted transposition in Drosophila melanogaster. Genetics. 1999;151:1093–1101. doi: 10.1093/genetics/151.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp KJ, Auld VJ. Reciprocal interactions between neurons and glia are required for Drosophila peripheral nervous system development. J Neurosci. 2003;23:8221–8230. doi: 10.1523/JNEUROSCI.23-23-08221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp KJ, Schulte J, Auld VJ. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev Biol. 2001;238:47–63. doi: 10.1006/dbio.2001.0411. [DOI] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido E, Ono N, Kojima T, Saigo K. Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development. 1997;124:2119–2128. doi: 10.1242/dev.124.11.2119. [DOI] [PubMed] [Google Scholar]
- Silies M, Yuva Y, Engelen D, Aho A, Stork T, Klambt C. Glial cell migration in the eye disc. J Neurosci. 2007;27:13130–13139. doi: 10.1523/JNEUROSCI.3583-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenfeld MJ, Jacobs JR. Macrophages and glia participate in the removal of apoptotic neurons from the Drosophila embryonic nervous system. J Comp Neurol. 1995;359:644–652. doi: 10.1002/cne.903590410. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ, Klambt C. Organization and function of the blood-brain barrier in Drosophila. J Neurosci. 2008;28:587–597. doi: 10.1523/JNEUROSCI.4367-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Sun B, Xu P, Salvaterra PM. Dynamic visualization of nervous system in live Drosophila. Proc Natl Acad Sci. 1999;96:10438–10443. doi: 10.1073/pnas.96.18.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodosiou NA, Xu T. Use of FLP/FRT system to study Drosophila development. Methods. 1998;14:355–365. doi: 10.1006/meth.1998.0591. [DOI] [PubMed] [Google Scholar]
- Thomas GB, van Meyel DJ. The glycosyltransferase Fringe promotes Delta-Notch signaling between neurons and glia, and is required for subtype-specific glial gene expression. Development. 2007;134:591–600. doi: 10.1242/dev.02754. [DOI] [PubMed] [Google Scholar]
- von Hilchen CM, Beckervordersandforth RM, Rickert C, Technau GM, Altenhein B. Identity, origin, and migration of peripheral glial cells in the Drosophila embryo. Mech Dev. 2008;125:337–352. doi: 10.1016/j.mod.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol. 2004;14:678–684. doi: 10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Montell C. Defective glia induce neuronal apoptosis in the repo visual system of Drosophila. Neuron. 1995;14:581–590. doi: 10.1016/0896-6273(95)90314-3. [DOI] [PubMed] [Google Scholar]