Abstract
Dynamics are thought to be important features of structure and signaling in the cytoplasmic domain of bacterial chemoreceptors. However, little is known about which structural features are dynamic. For this largely helical domain, comprising a four-helix bundle and an extended four-helix coiled coil, functionally important structural dynamics likely involves helical mobility and stability. To investigate, we used continuous wave EPR spectroscopy and site-specific spin labels that directly probed, in essentially physiological conditions, mobility of helical backbones in the cytoplasmic domain of intact chemoreceptor Tar homodimers inserted into lipid bilayers of Nanodiscs. We observed differences among functional regions, between companion helices in helical hairpins of the coiled coil and between receptor conformational states generated by adaptational modification. Increased adaptational modification decreased helical dynamics while preserving dynamics differences among functional regions and between companion helices. In contrast, receptor ligand occupancy did not have a discernable effect on dynamics to which our approach was sensitive, implying that the two sensory inputs alter different chemoreceptor features. Spectral fitting indicated that differences in helical dynamics we observed for ensemble spin-label mobility reflected differences in proportions of a minority receptor population in which the otherwise helical backbone was essentially disordered. We suggest that our measurements provided site-specific snapshots of equilibria between a majority state of well-ordered helix and a minority state of locally disordered polypeptide backbone. Thus, the proportion of polypeptide chain that is locally and presumably transiently disordered is a structural feature of cytoplasmic domain dynamics that varies with functional region and modification-induced signaling state.
Keywords: Bacterial chemotaxis, helical disorder, conformational signaling, EPR spectroscopy, Nanodiscs
Graphical Abstract
INTRODUCTION
Bacterial chemoreceptors can be viewed as allosteric proteins in equilibrium between two signaling states [1–3]. One state activates the chemotaxis histidine kinase (kinase-on); serves as substrate for one of the enzymes of sensory adaptation, the methylesterase, but not the other, the methyltransferase (methylation-off, demethylation-on); and has low affinity for its attractant ligand (low-affinity). The other state does not activate kinase (kinase-off), is a substrate for methyltransferase but not methylesterase (methylation-on, demethylation-off), and has high ligand affinity (high affinity). Two opposing allosteric effectors, chemoattractants and receptor adaptational covalent modification, shift the conformational equilibrium in opposite directions. Chemoattractants are recognized directly or as an attractant-occupied binding protein by sterospecific sites in the receptor periplasmic domain. The adaptational modification is methylation of four or five specific glutamyl residues in the receptor cytoplasmic domain. The kinase-on, methylation-off, demethylation-on, low-affinity state is favored by empty attractant-binding sites and methylation of the glutamyl residues. The kinase-off, methylation-on, demethylation-off, high affinity state is favored by attractant binding or demethylation that regenerates the glutamyl side chains. These opposing effects on receptor conformation create a feedback loop that mediates sensory adaptation, which in turn is central to the mechanism of sensitivity to temporal gradients and thus chemotactic migration in spatial gradients [2].
The conformational features that distinguish the chemoreceptor functional states are subjects of much current interest. Several lines of evidence suggest that structural dynamics could be an important difference between conformational states in the cytoplasmic domain of transmembrane chemoreceptors [4–9]. Thus, we investigated the influence of conformational state on polypeptide backbone dynamics in the cytoplasmic domain of the Escherichia coli aspartate chemoreceptor Tar. Our studies utilized Nanodisc technology [10–12], site-directed spin labeling and continuous-wave electron paramagnetic resonance (EPR) spectroscopy [13–17].
Chemoreceptor structure and function
The fundamental structural unit of a bacterial chemoreceptor is a homodimer [11]. Fig. 1 shows the rod-like helical structure of the homodimer of E. coli chemoreceptor Tar, a representative of a large family of transmembrane chemoreceptors present in many species [18]. Biophysical, biochemical and genetic studies have identified structural and functional features along the length of the largely helical transmembrane homodimer (Fig. 1) [1–3]. Ligand-binding sites are at the membrane-distal end of the periplasmic domain, in the dimer interface between the two protomer four-helix bundles. One helix from each bundle, α-4 and α-4′, begins at the ligand-binding site and extends the length of the cytoplasmic domain to become transmembrane helix 2 (TM2) and transmembrane helix 2′ (TM2′), respectively. α-4/TM2 and α-4′/TM2′ connect the ligand-binding site to a HAMP domain (histidine kinases, adenylyl cyclases, methyl-accepting chemoreceptors, and phosphatases [19]), a parallel four-helix bundle containing two helices from each protomer. HAMP appears to act as a conversion unit for conformational signals, receiving from TM2 or TM2′ a piston motion that began at the ligand-binding site and crossed the membrane to impinge on HAMP [20], and converting that motion into a different, as-yet-undefined conformational change in the cytoplasmic domain. Membrane-distal to HAMP, the chemoreceptor is an extended four-helix coiled coil of two helical hairpins, one from each subunit of the receptor dimer. The carboxyl-terminal HAMP helices AS-2 and AS-2′ become the amino-terminal helices (N- and N′-helices) of the two helical hairpins. The N-helices extend ~200 Å to a tight turn at the membrane-distal tip of the cytoplasmic domain that reverses the direction of the polypeptide chain and from which the companion C-helices extend as antiparallel partners to the N-helices in the four-helix bundle (Fig. 1). Functional regions have been identified along the length of the cytoplasmic domain. From HAMP to the membrane-distal tip these are the modification region, which contains the glutamyl residues of adaptational modification (four in Tar); the glycine hinge, a functionally important cluster of glycines at which the receptor can exhibit a modest bend [21]; and the protein-interaction region, where receptor dimers interact with each other to form trimers and trimers interact with the histidine kinase and a coupling protein to form core signaling complexes [22–24]. In a recent study of one conformational state of Tar, we found that the N-helix was more dynamic than the C-helix from AS-2 of the membrane-proximal HAMP to the membrane-distal beginning of the stably packed protein-interaction region [9]. The differential dynamics of the two helices suggests an organization in which the two C-helices serve as a stable scaffold for two more dynamic N-helices.
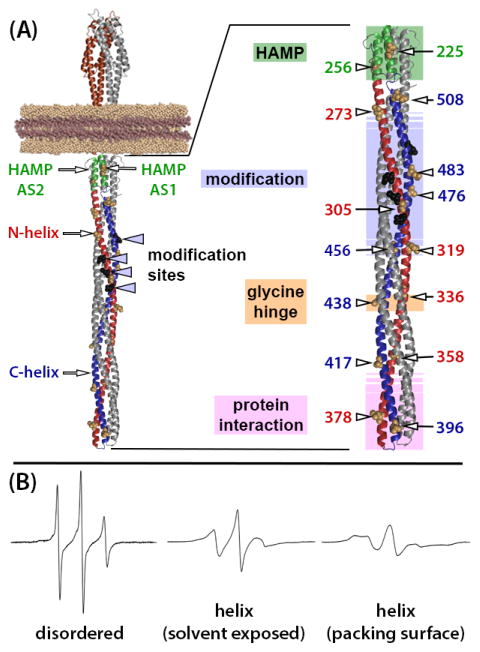
Fig. 1. Chemoreceptor structural organization and example EPR spectra.
(A) Ribbon diagrams of a molecular model of a chemoreceptor dimer based on X-ray and NMR structures of receptor fragments. The left-hand image is a model of an intact, single chemoreceptor dimer inserted into a Nanodisc with relevant structural features labeled (see text for explanations). The right-hand image is an enlargement of the receptor cytoplasmic domain with positions of mutagenically introduced cysteines indicated by residue number and an arrow pointing to the CPK representation of the respective native sidechains on the surface of the Tar helices. Functional regions are indicated by labels to the left of the cytoplasmic domain structure and by shaded colored boxes (HAMP pale green, modification region pale blue, glycine hinge pale orange and protein interaction region pale pink). Shaded boxes for the modification and interaction regions are shown with borders that fade out to symbolize a lack of definition for their precise positions. In both images, one receptor protomer is color coded: brown for the periplasmic, ligand-binding domain, green for HAMP, red for the N-helix in the coiled coil and blue for its C-helix. Sites of adaptational modification are indicated by black spheres, which are labeled with arrowheads in the left-hand diagram. (B) Example EPR spectra for a nitroxide spin label coupled by methanethiosulfonate chemistry to a cysteine introduced into chemoreceptor Tar (from left to right): in a disordered segment (position 549 in urea-denatured Tar [42]), on a solvent-exposed, non-interacting surface of a well-ordered alpha helix (Tar position 396, this study) or in a molecular interface between neighboring and interacting helices (Tar position 401 [9, 54]). Fig. 1 and a slightly modified version of its legend are from [9], with permission.
The chemoreceptor homodimer is the fundamental unit of both structure and function. Homodimer function has been documented by characterization of individual homodimers inserted into Nanodisc-contained bilayers of native E. coli lipid [25, 26]. Nanodiscs are small (~100 nm diameter) plugs of lipid bilayer surrounded by a protective belt of an amphipathic scaffold protein that renders water soluble the bilayer and membrane proteins inserted into it [10–12]. Individual, Nanodisc-inserted Tar dimers bind ligand, undergo adaptational modification (methylation and demethylation/deamidation) and perform conformational, transmembrane signaling, coupling ligand binding and adaptational modification to receptor conformation [25]. Thus we used Nanodisc-inserted Tar homodimers to investigate the effects of the allosteric effectors on chemoreceptor dynamics, comparing helical dynamics in the cytoplasmic domain of two signaling conformations of intact Tar inserted into a bilayer of native E. coli lipid and of those two conformations in the absence or presence of a saturating concentration of the Tar chemoattractant aspartate. One conformation was shifted strongly to the kinase-on, methylation-off, demethylation-on, low ligand affinity state by introduction of glutaminyl residues, functional mimics of methyl glutamyl residues [27–29], at all four positions of adaptational modification. The other form was shifted strongly to the kinase-off, methylation-on, demethylation-off, high ligand affinity state by glutamyl residues at those four positions. For Nanodisc-isolated chemoreceptor homodimers, functional differences for Tar-4E versus Tar-4Q have been documented for the activities of methylation and ligand affinity [25]. In addition, as components of chemotaxis signaling complexes Tar-4E and Tar-4Q generate low and high kinase activity, respectively [30, 31] and thus are considered to be in the “kinase-off” and “kinase-on” states. To relate our studies with isolated chemoreceptor dimers to this common terminology yet explicitly identify that the assayable differences between homodimers of Tar-4E and Tar-4Q are for adaptational modification, we will refer to the signaling states of Tar-4E and Tar-4Q as “kinase-off, methylation-on” and “kinase-on, methylation-off”, respectively.
RESULTS
Experimental strategy
We recently used site-directed spin labeling and EPR spectroscopy to characterize helical dynamics in the cytoplasmic domain of one signaling conformation of intact chemoreceptor Tar inserted into the lipid bilayer of a Nanodisc [9]. This protein, Tar-4Q, had a conformation shifted strongly toward the kinase-on, methylation-off state by glutaminyl residues at the four sites of adaptational modification. We have now extended our characterization of chemoreceptor helical dynamics to Tar-4E, a receptor form shifted strongly toward the kinase-off, methylation-on conformation by glutamyl residues at the modification sites. As in our characterization of Tar-4Q, cysteines were introduced individually into the cytoplasmic segment of Tar-4E by site-directed mutagenesis. Based on known structures of receptor fragments, the cysteines were introduced at fifteen of the same positions used for the characterization of Tar-4Q, on solvent-exposed helical surfaces of the cytoplasmic domain at positions where the side chain would not be able to interact with its neighbors. These cysteines were distributed along the length of the extended structure, on both the N- and C-helices and among the functional regions (Fig. 1). Each cysteine-substituted form of Tar-4E was solubilized in detergent, purified utilizing a six-histidine tag at its carboxyl terminus, coupled to a nitroxide spin label and reconstituted into a bilayer of native E. coli lipid contained in a Nanodisc [10–12, 32]. Spin label was introduced by reaction of the cysteine sulfhydryl with MTSL, 1-Oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl methanethiosulfonate spin label. The resulting spin-labeled protein was well suited for investigation of the dynamics of helical polypeptide backbones because nitroxides linked by this reagent to cysteines located on solvent-exposed helical surfaces away from contact with other side-chains or protein segments have EPR spectra that vary specifically as a function of nanosecond motions of the local polypeptide backbone [14, 16, 17, 33–35].
The functional state of cysteine-substituted, spin-labeled, Nanodisc-embedded Tar-4E
We tested for functionally native receptor structure by determining the extent to which each cysteine-substituted, spin-labeled Tar-4E was recognized and thus modified by the methyltransferase CheR. This modification enzyme recognizes its glutamyl substrate only when the residue and its surrounding consensus sequence are in an intact, natively structured chemoreceptor. For instance, receptors inserted in native membranes or Nanodisc-provided bilayers of native lipids are efficiently modified by this enzyme, but intact receptors perturbed by detergent solubilization are not modified at all [11], and those inserted into bilayers of non-native lipid compositions or ratios are inefficiently recognized by the enzyme [32]. For the fifteen cysteine-substituted, spin-labeled, Nanodisc-inserted forms of Tar-4E, all but four had methyl-accepting activity greater than half the activity of Tar-4E lacking a cysteine and those four had at least one-third of that the activity (Table 1).
Table 1.
Enzymatic methylation of cysteine-substituted, spin-labeled, Nanodisc-inserted Tar-4E
| Cysteine position | Methylationa | Cysteine position | Methylationa |
|---|---|---|---|
| None | 1.00 +/− 0.03 | 378 | 0.49 +/− 0.02 |
| 225 | 0.67 +/− 0.03 | 396 | 0.37 +/− 0.06 |
| 256 | 0.80 +/− 0.01 | 417 | 0.78 +/− 0.07 |
| 273 | 0.75 +/− 0.04 | 438 | 0.33 +/− 0.03 |
| 305 | 0.43 +/− 0.14 | 456 | 0.59 +/− 0.02 |
| 319 | 0.77 +/− 0.07 | 476 | 0.49 +/− 0.03 |
| 336 | 0.43 +/− 0.03 | 483 | 0.59 +/− 0.02 |
| 358 | 0.55 +/− 0.11 | 508 | 0.57 +/− 0.03 |
Relative to Nanodisc-embedded Tar without a cysteine and thus without a spin label. See Materials and Methods for details of assay.
Helical dynamics in the cytoplasmic domain of Tar-4E
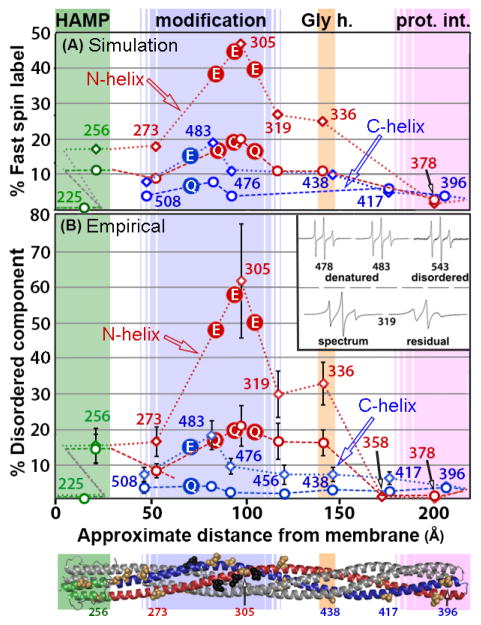
Continuous-wave EPR spectra for the fifteen spin-labeled forms of Tar-4E are shown in Fig. 2. Most spectra had features consistent with contributions from more than one component, each with a distinct spin-label mobility. Analysis of these components will be considered in a later section. As an aid to identifying patterns of differential helical dynamics along the receptor structure, we utilized two complementary mobility parameters that express spin-label mobility as a single numeric value. These parameters were h(+1)/h(0), the ratio of the amplitudes of the low field EPR spectrum line (h+1) and central line (h(0)) (inset Fig. 3A), and ΔHpp−1, the inverse of the central linewidth in Gauss (inset Fig. 3B). Each value has been used informatively as a parameter summarizing the ensemble mobility of spin labels generating an EPR spectrum [9, 13, 15, 33, 34, 36–43]. Fig. 3A shows h(+1)/h(0) and Fig. 3B shows ΔHpp−1 for the spectra in Fig. 2 plotted as a function of distance from the cytoplasmic membrane in the deduced three-dimensional structure of the Tar cytoplasmic segment.
Fig. 2. Continuous-wave EPR spectra of intact, spin-labeled Tar-4E inserted in the native lipid bilayer of a Nanodisc.
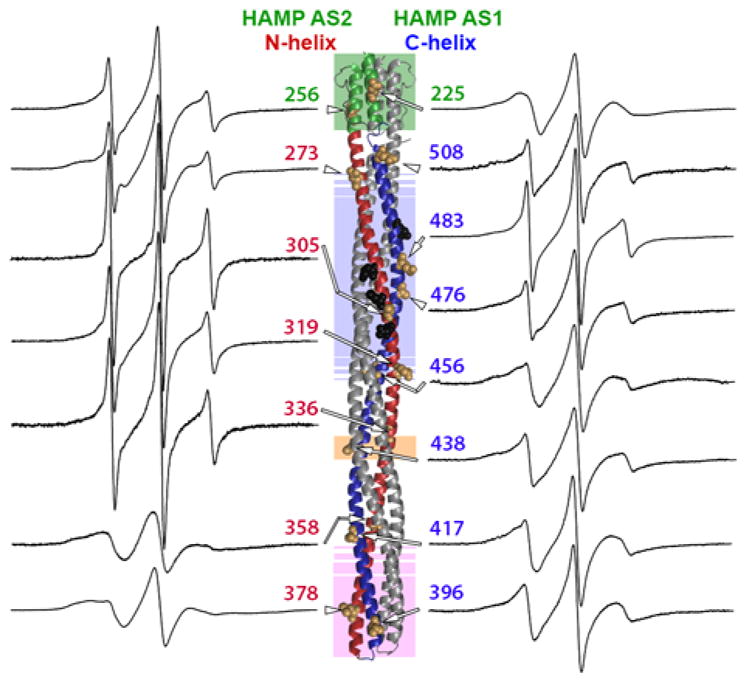
EPR spectra normalized to total spin for spin labels at 15 positions in the Tar cytoplasmic domain are displayed near the site of labeling on the ribbon diagram of the domain that is explained in Fig. 1A. Spectra are labeled with residue number and an arrow pointing to the residue position shown on the ribbon diagram as tan CPK models of the native sidechain. Sites of adaptational modification are black CPK models and functional regions identified by shading and helices as in Fig. 1A.
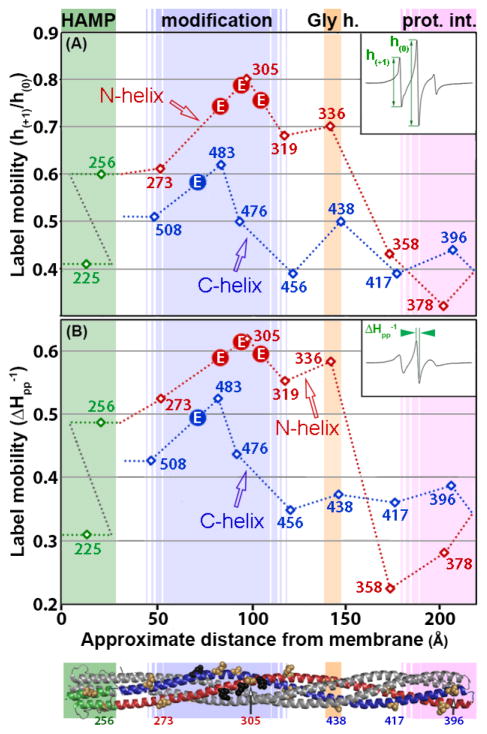
Fig. 3. Spin-label mobility as a function of position in the cytoplasmic of Tar-4E.
Mobility parameters h(+1)/h(0) (Fig. 3A) and ΔHpp−1 (Fig. 3B) (see insets and text) for the 15 spectra in Fig. 2 are plotted as a function of spin-label distance in Å from the membrane surface in the model of the Tar cytoplasmic domain (Fig. 1A). The ribbon diagram of the model is below the abscissa with some spin-label positions marked by tan CPK models of native side chains and functional regions identified by shading and helices as in Fig. 1A. Backbone connectivity is indicated on the plots by dotted lines. Lines and data points are color coded by structural element: HAMP green, N-helix red and C-helix blue. The N- and C-helices are labeled. Glutamyl sites of adaptational modification are indicated by circles containing an E.
A distinct pattern of differential helical dynamics in the cytoplasmic domain of the Tar-4E kinase-off, methylation-on conformation is evident in the series of spectra (Fig. 2) and is illustrated by the mobility parameters (Fig. 3A and 3B). Spectra and single-parameter mobilities characteristic of well-ordered alpha helices (see the middle spectrum in Fig. 1B and refs. [17, 33–35]) were observed for spin labels in or near the protein-interaction region (positions 358, 378, 396 and 417), in AS-1 of HAMP (225) and for the C-helix extending from the protein-interaction region through the glycine hinge region (438 and 456). In contrast, C-helix spin labels in the adaptation region (476 and 483) and near the end of the four-helix coiled-coil (508) had mobilities higher than typical of surface residues on helices, implying greater contributions from highly mobile components, i.e. a less consistently structured helix. Even greater mobility occurred in the N-helix. A region of substantial spin-label mobility and thus polypeptide backbone dynamics spanned the N-helix segment from AS-2 of the membrane-proximal HAMP (position 256) to the membrane-distal beginning of the protein-interaction region (position 336). The mobility parameters along this 80-residue segment of the N-helix were uniformly higher than those of labels on the corresponding portion of the partner C-helix. Thus for this major portion of the cytoplasmic domain of kinase-off, methylation-on Tar-4E, the N-helix was more dynamic than the C-helix. This is the same pattern observed for kinase-on, methylation-off Tar-4Q [9].
Similarities and differences in helical dynamics as a function of adaptational modification
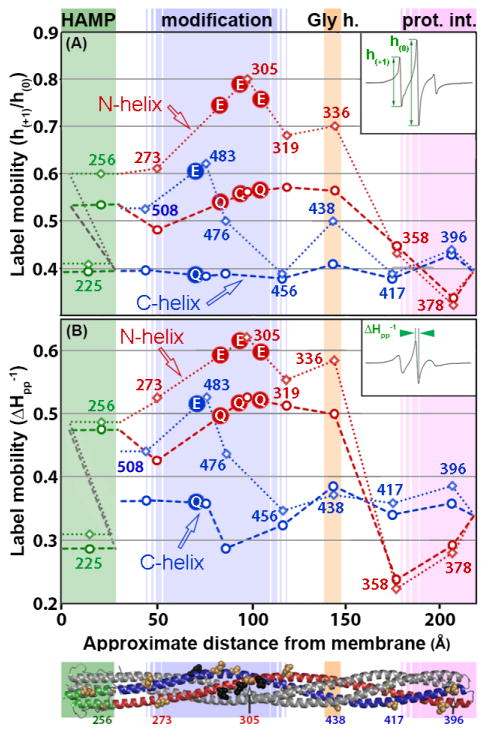
Comparisons of primary spectra (Fig. 2; [9]) or the mobility parameters h(+1)/h(0) (Fig. 4A) and ΔHpp−1 (Fig. 4B) of the unmodified, kinase-off, methylation-on receptor conformation of Tar-4E (Figs. 2 and 3) with those of the fully modified, kinase-on, methylation-off conformation of Tar-4Q [9] identified a consistent picture of similarities and differences in helical dynamics. The kinase-off, methylation-on conformation was more dynamic than the kinase-on, methylation-off conformation. For both conformations the N-helix was more dynamic than the C-helix. Shifting from the 4E kinase-off, methylation-on state to the 4Q kinase-on, methylation-off state reduced backbone mobility parameters for almost every position that had a value greater than typical of a consistently structured alpha helix (Fig. 4A and 4B). For the C-helix this produced a stable helix for the entire length of the polypeptide. For the multiple N-helix positions with mobility parameters in the 4E state greater than fully ordered helix, mobility was reduced in the 4Q state but remained consistently greater than the corresponding C-helix segment and consistently greater than the mobility of a well-ordered helix. Thus transition from the Tar-4E kinase-off, methylation-on conformational state to the Tar-4Q kinase-on, methylation-off state reduced helical dynamics over a considerable portion of the receptor cytoplasmic domain while preserving the distinct difference in dynamics between the N-helix and the C-helix. For positions near or in the protein-interaction region, (358 and 417; 378, 396, respectively) the shift from unmodified to fully modified resulted in only slight differences between the two signaling states (Fig. 5).
Fig. 4. Spin-label mobility as a function of position and modification state in the Tar cytoplasmic domain.
Mobility parameters h(+1)/h(0) (Fig. 4A) and ΔHpp−1 (Fig. 4B) (see insets and text) are shown for spectra of a spin-label at 15 positions on Tar with 4 glutamyl residues (Tar-4E, this study) and 4 glutaminyl residues (Tar-4Q, [9]) at the sites of adaptational modification. Data plots and structural model are as described for Fig. 3. Data for Tar-4E are open diamonds and connected by a dotted line. Data for Tar-4Q are open circles and connected by a dashed line. Lines and data points are color coded by structural element: HAMP green, N-helix red and C-helix blue. Sites of modification are marked with circles filled with E or Q according to the adaptation state.
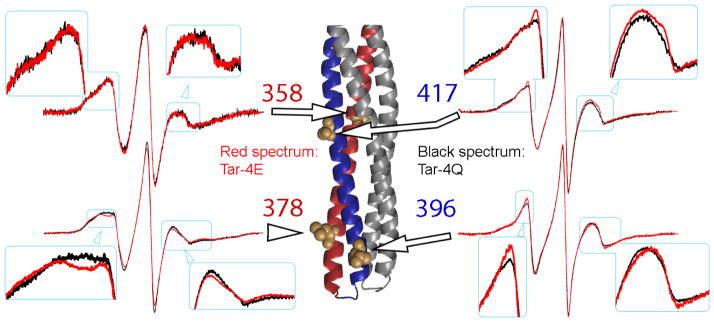
Fig. 5. Effects of adaptational modification on the protein interaction region.
Spectra normalized to total spin and plotted at same amplitude for Tar-4E (red, this study) and Tar-4Q (black, [9]) are shown for spin labels in the protein interaction region. Enlargements of regions of the spectra with spectral differences between the two modification states are provided above or below the respective spectra. Modest differences in spectral shape are seen for all but position 358 where there are no apparent differences between the noisy spectra.
Spectral deconvolution
EPR spectra of spin labels on proteins commonly have contributions from more than one mobility component and thus represent spectral ensembles. This is often the result of more than one conformational state of the spin label, each with distinct mobility characteristics. The spectra for spin labels at the positions characterized on Tar-4E and Tar-4Q had features consistent with contributions from more than one spin-label population. Thus we deconvoluted each spectrum using computational fitting coupled to simulated spectral parameters [44]. Three Tar-4Q and two Tar-4E spectra were too noisy to make meaningful estimates of contribution from a highly mobile population but the spectral shapes implied that such contributions were very minor. Simulating each of the other spectra with a multiple-component model generated satisfactory fits using two or three mobility states. For either class of fits, one mobility state had a short correlation time characteristic of a highly mobile component and the other state(s) had correlation time(s) corresponding to spin labels on a well-structured peptide backbone [17, 34]. The percent contributions of the short correlation-time, more mobile component to the respective fitted spectra are shown in Table 2. The highly mobile state was always in the minority and, with the exception of position 305 on Tar-4E, less than 30% of the spin-label population. The pattern of percent highly mobile component as a function of position and signaling state (Fig. 6A) strongly resembled the patterns observed for the mobility parameters (Fig. 4). Thus, three methods for assessing spin-label mobility indicated essentially the same pattern of differential backbone dynamics.
Table 2.
Percent disordered component as assessed by spectral fitting
| Pos. | Mod. | % Dis. | Tauc | Pos. | Mod. | % Dis. | Tauc | Pos. | Mod. | % Dis. | Tauc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 225 | 4E | 0 | - | 336 | 4E | 25 | 0.46 | 438 | 4E | 10 | 0.61 |
| QEQE | 0 | - | 4Q | 11 | 0.45 | 4Q | nd | nd | |||
|
| |||||||||||
| 256 | 4E | 17 | 0.52 | 358 | 4E | nd | nd | 456 | 4E | nd | nd |
| 4Q | 11 | 0.48 | 4Q | nd | nd | 4Q | nd | nd | |||
|
| |||||||||||
| 273 | 4E | 18 | 0.59 | 378 | 4E | 2 | 0.56 | 476 | 4E | 11 | 0.54 |
| 4Q | 9 | 0.54 | 4Q | 3 | 0.60 | 4Q | 4 | 0.52 | |||
|
| |||||||||||
| 305 | 4E | 47 | 0.38 | 396 | 4E | 4 | 0.55 | 483 | 4E | 19 | 0.52 |
| 4Q | 20 | 0.54 | 4Q | 4 | 0.39 | 4Q | 8 | 0.61 | |||
|
| |||||||||||
| 319 | 4E | 27 | 0.49 | 417 | 4E | 5 | 0.39 | 508 | 4E | 8 | 0.33 |
| 4Q | 11 | 0.44 | 4Q | 6 | 0.68 | 4Q | 4 | 0.43 | |||
The columns indicate the spin label position (Pos.), modification state (Mod.), percent disordered component (% Dis.), and motional correlation time of the disordered component in nanoseconds (Tauc) for each site probed by EPR. No Tauc value is shown for sites with no apparent fast motion component. Values for percent disorder and Tauc were not determined (nd) for spectra with insufficient signal-to-noise to estimate the fast motion component.
Fig. 6. Percent disordered component as a function of position and modification state in the Tar cytoplasmic domain.
Percent disordered component for spectra in Fig. 2 was determined by simulated fitting (Fig. 6A) and empirical (Fig. 6B) deconvolution of Tar (see text) with 4 glutamyl residues (Tar-4E, this study) or 4 glutaminyl residues (Tar-4Q, [9]) at the sites of adaptational modification. Data plots and structural model are as described for Fig. 3. Values for Tar-4E are indicated by open diamonds and connected by a dotted line. Values for Tar-4Q are indicated by open circles and connected by a dashed line. Line and data points are color coded by structural element as in Figure 1A. Sites of modification are marked with circles filled with E or Q, according to the adaptation state.
The highly mobile spin-label states identified by fitting to simulated spectra had subnanosecond motional correlation times, shorter than correlation times of well-ordered helices [17, 34]. Thus we compared those values to correlation times for spin labels on a disordered polypeptide backbone of Tar. Two of these correlation times were for spin labels on the cytoplasmic domain of Tar denatured in 4 M urea. The third was for a spin label near the end of the natively disordered, 35-residue flexible arm at the extreme carboxyl terminus of Tar [42]. Fitting these spectra computationally identified two components with different mobilities, a predominant (63 to 93%), highly mobile state and a minority (7 to 37%) less mobile state. The majority, highly mobile states had correlation times from ~0.4 to ~ 0.6 ns (mean 0.53 ± 0.09 ns). As seen in Table 2, these values were essentially the same as correlation times of the minority highly mobile state of spin labels on native Tar-4E and Tar-4Q, which had correlation times from ~0.4 to ~ 0.7 ns (mean 0.49 ± 0.09 ns and 0.52 ± 0.09 ns for Tar-4E and Tar-4Q, respectively). By these comparisons, it appeared that portions of the helical backbone of the cytoplasmic domain of intact, lipid-bilayer-inserted chemoreceptor Tar could assume a highly dynamic state indistinguishable in motional correlation time from a disordered polypeptide chain. For positions that had the features of well-ordered helices, the percent of the receptor population found in the apparently disordered state was small, generally 5% or less, which appears to be the limit of sensitivity of our deconvolution analysis and thus structural relevance. For positions on more dynamic segments of Tar helices, the proportion was significantly greater and likely structurally relevant.
To test the notion that the spectra for Tar-4E and Tar-4Q had contributions from a disordered helical backbone, we did empirical subtractions of spectra for spin labels on Tar polypeptide segments known to be disordered. As spectra from disordered backbones, we used those described in the previous paragraph, two for spin labels on urea-denatured Tar and one for a spin label on the disordered, Tar carboxyl-terminal flexible arm [42] (Fig. 6B, inset). Using a procedure similar to that routinely employed to subtract the contribution of a small proportion of free spin label from an experimental spectrum, we determined the hypothetical contribution of a “disordered spectrum” to each Tar-4E and Tar-4Q spectrum by subtracting one of our example spectra until the residual spectrum lacked features consistent with a high-mobility spin label. For spectra derived for more stable protein regions, very little could be subtracted without unreasonable spectral distortion, whereas for those containing significant mobile component, a more substantial portion could be removed. We did three independent subtractions, each using a different spectrum of a spin label on disordered Tar polypeptide. The resulting values of contributions by “disordered spectra” were averaged and plotted as a function of residue position (Fig. 6B). As for the results of fitting to simulated spectral parameters, values less than 5% are likely at the limit of structural relevance. The two deconvolution procedures, one to simulated spectral parameters and the other using spectra of spin labels on disordered polypeptide chains, generated similar values and similar patterns for percent disordered component as function of function of position and signaling state (Figs. 6A and 6B). Furthermore, the shared pattern strongly resembled those generated by spectral shapes (Fig. 2) and mobility parameters (Figs. 3 and 4).
Saturating ligand does not detectably modulate helical backbone stability
Ligand occupancy shifts the conformation of a chemoreceptor dimer to the methylation-on and thus kinase-off conformation [25]. Since Tar-4E is already strongly shifted toward that conformation, we first explored the effect of ligand occupancy using Tar-4Q, which is strongly shifted to the methylation-off, kinase-on state. We collected spectra for aspartate-saturated Tar-4Q with a spin label at all but a few of the positions we had tested for effects of adaptational modification (256, 305, 319, 336, 358, 378, 396, 417, 438, 456, 483, 508), including most that had exhibited substantial differences between the Tar-4E and Tar-4Q signaling state as well as a majority of those that had not exhibited such differences. For these eleven spin-label positions, there were no detectable differences between spectra collected in the presence or absence of saturating aspartate, yet assays of the influence of ligand occupancy on rates of chemoreceptor modification showed that ligand occupancy altered the conformation of the modification region. Fig. 7 shows representative spectra (Fig. 7A) for six of these eleven positions and documentation that each of these spin-labeled receptors coupled ligand occupancy via transmembrane conformational signaling to altered propensity for adaptational modification, specifically deamidation, the appropriate modification to be examined for Tar-4Q (Fig. 7B). Even though Tar-4E was already shifted strongly to the kinase-off, methylation-on state, we tested a few spin label positions (358, 378, 483, 508) on this form of the receptor. Again, no spectral differences were observed between the ligand-saturated and ligand-free receptors (data not shown). The implications of these observations are considered in the Discussion.
Fig. 7. No detectable effects of saturating ligand on helical dynamics in the cytoplasmic domain of Tar-4Q.
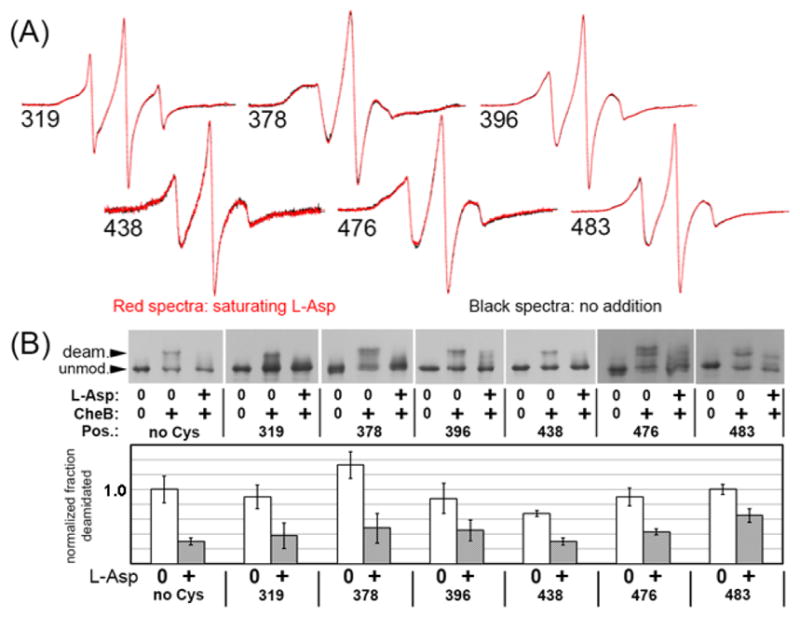
(A) Overlays of continuous-wave EPR spectra of cysteine-substituted (at the position indicated by the number), spin-labeled, Nanodisc-inserted Tar-4Q dimers in the absence (black) or presence (red) of saturating (1 mM) L-aspartate. Each pair of spectra were normalized to the same number of spins. (B) Effect of saturating ligand on CheB-mediated deamidation. The top panel shows immunoblots with anti-chemoreceptor serum of the forms of Tar-4Q dimers in (A) after 45 min in the absence or presence of methylesterase/deamidase CheB and the absence or presence of 1 mM L-aspartate. The bottom panel shows quantification of average percent deamidation and standard deviation (n=3) normalized to the percent deamidated of “no Cys” Tar-4Q. Positions of spin labeling are indicated by numbers and “no Cys” is Tar-4Q in the native state of no cysteine and thus no spin label that was submitted to the same labeling and reconstitution procedure as the cysteine-substituted receptors.
Discussion
We observed differences in helical dynamics between the two conformational states of chemoreceptor Tar generated by differences in adaptational modification. Notably, these dynamics differences were observed for intact, membrane-inserted receptor dimers at room temperature and in an essentially physiological solution. They were detected using continuous-wave EPR spectroscopy of site-specific spin labels placed on the receptor cytoplasmic domain at solvent-surrounded, non-interacting positions on helical surfaces, where they served as specific reporters of helical dynamics. The kinase-off, methylation-on state generated by negatively charged glutamyl residues at all four sites of adaptational modification was more dynamic than the kinase-on, methylation-off state generated by neutral glutaminyl residues at those sites. Although the two conformational states differed in extent of helical dynamics of the cytoplasmic domain, they shared a pattern of differential dynamics among structural and functional regions and of differential dynamics between the two helices of the paired helical hairpins of the four-helix coiled coil structure. Our analyses indicated that the differences in helical dynamics we observed corresponded to differential propensities of otherwise helical polypeptide backbones to be in an essentially disordered and thus highly mobile state. These observations and their implications are considered in more detail in the following sections.
Helical dynamics varies among functional regions and structural segments, independent of receptor conformational state
Independent of conformational state, the adaptation region was more dynamic than the protein-interaction region or the AS-1 helix of HAMP, each of which exhibited dynamics of a well-ordered helix. In both conformational states, the N-helix, from its origin as the HAMP AS-2 helix to the beginning of the well-ordered protein-interaction region, was more dynamic than a typical well-ordered helix and than its companion C-helix. These observations provide further support for the idea that the chemoreceptor cytoplasmic domain contains a stable structural scaffold, composed of the two well-ordered AS-1 helices of HAMP and the two well-ordered C-helices of the four-helix coiled coil, from which the more dynamic AS-2/N-helices fluctuate [9]. In addition, the stable-scaffold/dynamic helix asymmetry between companion helices is not a property of one conformational state, but instead is a more general feature of chemoreceptor structure. Throughout bacterial diversity, chemoreceptor cytoplasmic domains are organized as extended four-helix coiled coils [45, 46]. The asymmetric dynamics we observed for E. coli chemoreceptor Tar may be a general feature of these of chemoreceptor cytoplasmic domains.
Helical dynamics varies with modification-generated conformational state
For much of the rod-like cytoplasmic domain, helices of kinase-on, methylation-off Tar-4Q were less dynamic than corresponding helices of kinase-off, methylation-on Tar-4E. Specifically, completely modified Tar-4Q had reduced helical dynamics in all regions in which dynamics of the completely unmodified Tar-4E was greater than a well-ordered helix. These regions included over 60% of the N-helix, from its origin at AS-2 of HAMP up to the border with the protein-interaction region as well as the C-terminal 30% of the C-helix. The remaining regions or segments, the protein-interaction region, a portion of the C-helix adjacent to the protein-interaction region and HAMP AS-1, were well-ordered helices in both conformational states. Greater helical dynamics for kinase-off, methylation-on Tar-4E versus kinase-on, methylation-off Tar-4Q provides a basis for understanding the differences observed in the two states for rates of disulfide cross-linking between introduced cysteines in the HAMP AS-2 region and in the modification region [47, 48].
Differential receptor helical dynamics corresponds to differential propensities to be unstructured
Fitting of the multi-component Tar-4E and Tar-4Q spectra indicated that the differential helical dynamics we observed corresponded to differential proportions of a minority, very mobile spin-label population that had features of an unstructured protein backbone. This correspondence implies that differential dynamics among chemoreceptor segments and between conformational states reflected differential propensities of otherwise well-structured chemoreceptor helical backbones to be in an unstructured state in which mobility of the polypeptide backbone is no longer restricted by the hydrogen bonds that hold a helix in place. Is the unstructured polypeptide backbone irreversibly in that state, and thus a subpopulation in which the helix is locally denatured permanently, or is the unstructured state reversible and thus the relevant helical segment is in a dynamic equilibrium between a majority state of well-ordered helix and a minority state of locally disordered polypeptide backbone? Our EPR data cannot distinguish between the two possibilities, but we favor the notion of a dynamic equilibrium. This is because the spectral shapes and mobility parameters we observed are reproducible for different preparations of the same spin-labeled protein, in different solution conditions, and for measurements of the same preparation made after months or years of storage (data not shown). If differences we observed in extents of spin-label mobility were the result of differential propensities to assume an irreversibly unstructured, highly mobile state, mobility would likely vary with condition, preparation and storage time unless one assumes an upper limit for the proportion of the receptor population which can become unstructured and that limit has been reached for all conditions, preparations and at the earliest time of assay. In contrast, a dynamic equilibrium between structured and unstructured directly accounts for consistent extents of mobility over different preparations, solution conditions, and storage times.
The notion that the helices of the chemoreceptor cytoplasmic domain of all receptor homodimers have propensities to transiently assume an essentially unstructured state is strongly supported by the strikingly promiscuous oxidative cross-linking between symmetrical cysteines placed in the two protomers of the cytoplasmic domain of functional, membrane-inserted Tar dimers [49–53]. In these studies, cysteines on opposite sides of the four-helix coiled coil could be cross-linked to essentially 100% by mild oxidation conditions [49–53]. Essentially complete cross-linking indicates that the polypeptide backbone of every receptor dimer, not just a subpopulation, assumed the unstructured state that allowed otherwise distant cysteine pairs to achieve reactive proximity. Yet prior to cross-linking these receptors were in large part fully functional, implying that the unstructured state was transient. In addition, forays into an unstructured state provide a way to understand high temperature factors in the modification region of X-ray structures of chemoreceptor cytoplasmic domain fragments [54, 55], notably high hydrogen/deuterium exchange rates for amide hydrogens of the N-helix modification region in a cytoplasmic domain fragment [7], and distances significantly greater than the width of a stable, four-helix coiled coil determined by pulsed EPR techniques between spin labels on opposite sides of the cytoplasmic domain [8].
Helical dynamics unaffected by ligand occupancy
Ligand occupancy and increased adaptational modification have opposite effects on methylation rates of Nanodisc-inserted Tar homodimers [25] and fully modified Tar has reduced helical dynamics relative to the unmodified receptor (this study). Yet saturation of Nanodisc-inserted Tar dimers with the attractant ligand aspartate had no effect on helical dynamics detectable by our measurements for any of the many cytoplasmic domain sites we tested in Tar-4Q or Tar-4E, including ones that exhibited substantial differences in helical dynamics as a function of receptor modification (Fig. 7A). Absence of changes in dynamics as a function of attractant ligand was not the result of inactivated ligand binding or disrupted coupling for ligand binding to cytoplasmic domain conformation since the many spin-labeled, Nanodisc-inserted forms of Tar we tested exhibited functionally relevant changes in cytoplasmic domain conformation, detected as reduced ability to serve as substrate for the chemotaxis deamidase/methylesterase (Fig. 7B). It could be that Tar dimers behave differently as components of trimers of receptor dimers in signaling complexes with CheA and CheW. Whether or not this is the case, our data demonstrate that receptor modification but not ligand binding alters the helical backbone dynamics detected by EPR of spin-labels on solvent-facing helical faces of isolated chemoreceptor dimers. Thus our data indicate that ligand occupancy and adaptational modification alter different features of chemoreceptor structure while still providing opposing influences on the receptor activities of adaptational modification and kinase activation, an idea already advanced in the literature [47, 56]. This suggests that adaptational modification alters helical dynamics of the chemoreceptor cytoplasmic domain, as detected by EPR spectroscopy and particularly for the substantially more mobile AS-2/N-helix component, whereas ligand occupancy does not alter the same aspects of helical dynamics but instead some other feature of chemoreceptor structure and dynamics. These issues will be investigated in subsequent work.
Helical dynamics and chemoreceptor conformational signaling
Current models of conformational signaling in the chemoreceptor cytoplasmic domain suggest that neighboring segments along the extended length of that domain have different stabilities, e.g. helical packing and/or dynamics, between adjoining domain segments, and that sensory signals are conveyed from HAMP to the tip of the extended four-helix coiled coil by inverting the order of greater versus lesser helical packing/dynamics [2–5]. Thus in one signaling state HAMP, the modification region and the protein-interaction region would be in dynamic-static-dynamic states and in the other would be in static-dynamic-static states. The observations described here are relevant to several aspects of these models. Consistent with the notion of differential dynamics in adjoining regions, the relative helical dynamics of HAMP AS-1, the modification region and the protein-interaction region was low-high-low. Consistent with the notion that dynamics changes with signaling state, the dynamics of modification region was high in the kinase-off, methylation-on state of Tar-4E and lower in the kinase-on, methylation-off state of Tar-4Q. However, HAMP and the protein-interaction region did not exhibit an opposite shift from less to more dynamic. Instead, the shift from kinase-off, methylation-on Tar-4E to kinase-on, methylation-off Tar-4Q reduced helical dynamics in all domain segments that were more dynamic than a well-ordered helix. Thus, by an assay sensitive to certain aspects of helical mobility, the difference between kinase-off, methylation-on Tar-4E and kinase-on, methylation-off Tar-4Q was not a reversal of the relative helical dynamics of adjoining domain segments but rather a shift over much of the domain from a more dynamic to a less dynamic state.
In the protein-interaction region, spin-label mobility was characteristic of a well-ordered helix. There were only modest differences in spectral shape as a function of modification-generated conformational state (Fig. 5). This might mean that signaling changes in the protein-interaction region occur only in complete signaling complexes, in which individual receptor dimers interact with two other dimers in that region in trimers as well as with CheA and CheW. However, pulsed EPR studies of a soluble fusion protein of the extended coiled coil of the Tar cytoplasmic domain with HAMP domains from a heterologous species indicated that individual chemoreceptor homodimers couple inputs from HAMP and from adaptational modification to changes in the protein-interaction region [8]. Thus alternative explanations are needed. For instance, the protein-interaction region might be altered by sensory adaptational modification in ways other than the disorder-linked changes in helical mobility we observed for other segments of the cytoplasmic domain. Such changes might alter helical super-coiling, inter-helical packing, inter-helical distances or relative positions of nearby residues, changes to which our experimental approach would be insensitive. In fact, differential inter-helical distances as a function of signaling state have been detected by pulsed EPR analysis of spin-labeled fragments of the Tar cytoplasmic domain [8].
Concluding remarks
Characterization of helical dynamics in the cytoplasmic domain of intact, lipid-bilayer-inserted chemoreceptor Tar provided new observations about chemoreceptor structural dynamics. These are: 1) There is differential helical dynamics among structural regions and between signaling states in the chemoreceptor cytoplasmic domain that appear to reflect differential propensities for excursions of otherwise helical polypeptide chains into an unstructured state, 2) Chemoreceptor conformational states generated by the two extremes of adaptational modification differ in extent of helical stability but exhibit similar patterns of alternating stability in receptor segments, and 3) Ligand occupancy and adaptational modification have opposing influences on receptor activity by mechanisms other than simple reversals of helical backbone stability. Each of these observations merits further investigation and we hope this study encourages those investigations.
MATERIALS AND METHODS
Strains, plasmids and proteins
Tar proteins were produced in E. coli K-12 strain RP3098 [57], which contains a deletion from flhA to flhD that eliminates the presence or expression of all chemoreceptor and che genes, harboring a derivative of the Tar-encoding plasmid pAL67. This plasmid, which carries tar under control of a modified lac promoter as well as lacIq and codes for Tar with six histidines (Tar-6H) added to its carboxyl terminus [58], was altered by site-specific mutagenesis to produce pAL529, which codes for Tar-6H 4E with glutamyl residues (E in the single letter code) at the four sites of adaptational modification. Site-specific mutagenesis of pAL529 created plasmids coding for Tar-6H 4E with cysteines substituted at the indicated residues (native Tar contains no cysteines): pAL766, H256C; pAL727, D273C; pAL780, A305C; pAL769, D319C; pAL826, Q336C; pAL763, K358C; pNB12, L378C; pAL827, V396C; pNB14, A417C; pAL764, E438C; pAL828, E456C; pAL768, S476C; pAL770, Q483C; and pNB16, Q508C. Tar proteins bearing 4Q or QEQE at the sites of modification were described previously [9]. pAL plasmids were constructed by Angela Lilly and pNB plasmids by Mutagenex, Inc. (Piscataway, NJ)
Receptor purification, spin labeling, Nanodisc reconstitution and EPR spectroscopy
Receptor purification, spin labeling, Nanodisc reconstitution, and EPR spectroscopy of Tar were performed as previously described [9]. In ligand addition experiments, L-aspartate was added to a saturating concentration of 0.5 to 8 mM relative to 25 to 50 μM Tar in 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 50 mM KCl, 5 mM MgCl2.
Assays of Tar adaptational modification
The proportion of each cysteine-substituted, spin labeled Tar-4E that was sufficiently native to be recognized and thus methylated by the chemotaxis methyltransferease was determined by incubating 5 μM Nanodisc-inserted Tar with 5 μM CheR and 80 μM S-adenosylmethionine in 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 50 mM KCl, 5 mM MgCl2 for 4 hours at room temperature, separating methylated from not methylated receptor by SDS gel electrophoresis using a gel with a low-percentage of acrylamide, immunoblotting with anti-Tar serum, and densitometric quantification of the methylated and not methylated receptor forms.
Ability to couple ligand occupancy to conformational changes in the chemoreceptor cytoplasmic domain was assessed for cysteine-substituted, spin-labeled forms of Tar-4Q inserted into Nanodiscs by determining the extent of CheB-mediated deamidation after 45 minutes at room temperature in 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 50 mM KCl, 5 mM MgCl2, at 5 μM Tar, 5 μM CheB, 50 μM phosphoramidate in the presence or absence of 500 μM L-aspartate. Extent of modification was determined by quantification of modified and unmodified forms of the receptor on immunoblots using anti-receptor serum.
Spectral deconvolution
Computational fitting and simulation were performed using the microscopic order with macroscopic disorder (MOMD) model [44] implemented with Labview-based MultiComponent software (Version 810) (Christian Altenbach, University of California – Los Angeles). We employed the Levenberg-Marquardt algorithm, using a minimum number of parameters to achieve reasonable fits of each spectrum. For tensor parameters we used the modified spherical form, initial default values of A1 = 16.4; A2 = 31.65; A3 = −0.30; g1, 2.00530 = g2, −0.00450; g3 = 0.00200, an isotropic linewidth tensor (W1) of 0.05, and an isotropic rate tensor (R1) of 8.00. To fit, we varied A1 within a limited range (+/− 1.0) and allowed the isotropic rate tensor to vary as necessary. All other tensors remained at their default values. Very mobile components characteristic of fast isotropic motion were fit without additional motional constraints. Slow motion spectral components generally required introduction of additional parameters characteristic of anisotropic motion. This was accomplished by applying orienting potentials (C20, C22, C40, C42, or C44), adding additional rate tensor components (R2, R3), or both. Deconvolution of the final fit, generated values for the relative contribution of highly mobile and less mobile spin labels. Spectra with poor signal to noise could not be reliably fit to provide an estimate of a highly mobile component.
For determinations of the proportion of a spectrum attributable to contributions from the spin label on the disordered Tar polypeptide backbone, we utilized EPR130_NoDAQ as described [9] to subtract the spectrum of a spin label on a Tar polypeptide backbone known to be disordered from the spectrum of interest. These spectra were for Tar carrying a spin-labeled cysteine at position 478 or 483 denatured in 4M urea, and Tar spin labeled at position 543 in the 35-residue disordered flexible arm at the carboxyl terminus of Tar [42].
HIGHLIGHTS.
Helical dynamics in cytoplasmic domain of intact, bilayer-inserted chemoreceptors
Receptor helical dynamics varies with functional segment and structural element
Variable chemoreceptor dynamics correlates with proportion of helical disorder
Adaptational modification alters chemoreceptor helical dynamics/percent disordered
Ligand binding and adaptational modification alter different structural features
Acknowledgments
We thank Angela Lilly for constructing many plasmids, Wing-Cheung Lai for preparation of membrane scaffold protein and Christian Altenbach for software and “hands-on” training in spectral simulation and deconvolution. This work was supported by National Institute of General Medical Sciences Grant GM29963
ABBREVIATIONS
- EPR
electron paramagnetic resonance spectroscopy
- HAMP
histidine kinases, adenylyl cyclases, methyl-accepting chemoreceptors, and phosphatases
- AS-1 and AS-2
amphipathic sequence 1 and 2 respectively
- Tar
chemoreceptor mediating taxis to aspartate and repellents
- Tar-4E
Tar with 4 glutamyl residues at the sites of adaptational modification
- Tar-4Q
Tar with 4 glutaminyl residues at the sites of adaptational modification
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hazelbauer GL, Lai W-C. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr Opin Microbiol. 2010;13:124–32. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkinson JS, Hazelbauer GL, Falke JJ. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015;23:257–66. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–77. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol Microbiol. 2009;73:801–14. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koshy SS, Li X, Eyles SJ, Weis RM, Thompson LK. Hydrogen exchange differences between chemoreceptor signaling complexes localize to functionally important subdomains. Biochemistry. 2014;53:7755–64. doi: 10.1021/bi500657v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samanta D, Borbat PP, Dzikovski B, Freed JH, Crane BR. Bacterial chemoreceptor dynamics correlate with activity state and are coupled over long distances. Proc Natl Acad Sci USA. 2015;112:2455–60. doi: 10.1073/pnas.1414155112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartelli NL, Hazelbauer GL. Differential backbone dynamics of companion helices in the extended helical coiled-coil domain of a bacterial chemoreceptor. Protein Sci. 2015;24:1764–76. doi: 10.1002/pro.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12:2476–81. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci USA. 2006;103:11509–14. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boldog T, Li M, Hazelbauer GL. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–35. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- 13.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Mol Biol. 2000;7:735–9. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 14.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem Sci. 2002;27:288–95. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 15.Klug CS, Feix JB. Methods and applications of site-directed spin labeling EPR spectroscopy. In: Correia JJ, Detrich WH III, editors. Methods Cell Biol. Academic Press; 2008. pp. 617–58. [DOI] [PubMed] [Google Scholar]
- 16.Hubbell WL, López CJ, Altenbach C, Yang Z. Technological advances in site-directed spin labeling of proteins. Current Opinion in Structural Biology. 2013;23:725–33. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altenbach C, López CJ, Hideg K, Hubbell WL. Chapter three - Exploring structure, dynamics, and topology of nitroxide spin-labeled proteins using continuous-wave electron paramagnetic resonance spectroscopy. In: Peter ZQ, Kurt W, editors. Methods Enzymol. Academic Press; 2015. pp. 59–100. [DOI] [PubMed] [Google Scholar]
- 18.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci USA. 2007;104:2885–90. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–6. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 20.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–65. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman MD, Bass RB, Mehan RS, Falke JJ. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry. 2005;44:7687–95. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci USA. 2011;108:9390–5. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci USA. 2012;109:3766–71. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci USA. 2012;109:E1481–E8. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amin DN, Hazelbauer GL. The chemoreceptor dimer is the unit of conformational coupling and transmembrane signaling. J Bacteriol. 2010;192:1193–200. doi: 10.1128/JB.01391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, Khursigara CM, Subramaniam S, Hazelbauer GL. Chemotaxis kinase CheA is activated by three neighbouring chemoreceptor dimers as effectively as by receptor clusters. Mol Microbiol. 2011;79:677–85. doi: 10.1111/j.1365-2958.2010.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park C, Dutton DP, Hazelbauer GL. Effects of glutamines and glutamates at sites of covalent modification of a methyl-accepting transducer. J Bacteriol. 1990;172:7179–87. doi: 10.1128/jb.172.12.7179-7187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunten P, Koshland DE., Jr Tuning the responsiveness of a sensory receptor via covalent modification. J Biol Chem. 1991;266:1491–6. [PubMed] [Google Scholar]
- 29.Borkovich KA, Alex LA, Simon MI. Attenuation of sensory receptor signaling by covalent modification. Proc Natl Acad Sci USA. 1992;89:6756–60. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bornhorst JA, Falke JJ. Evidence that both ligand binding and covalent adaptation drive a two-state equilibrium in the aspartate receptor signaling complex. J Gen Physiol. 2001;118:693–710. doi: 10.1085/jgp.118.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amin DN, Hazelbauer GL. Chemoreceptors in signalling complexes: shifted conformation and asymmetric coupling. Mol Microbiol. 2010;78:1313–23. doi: 10.1111/j.1365-2958.2010.07408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amin DN, Hazelbauer GL. Influence of membrane lipid composition on a transmembrane bacterial chemoreceptor. J Biol Chem. 2012;287:41697–705. doi: 10.1074/jbc.M112.415588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 34.Columbus L, Hubbell WL. Mapping backbone dynamics in solution with site-directed spin labeling: GCN4–58 bZip free and bound to DNA. Biochemistry. 2004;43:7273–87. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 35.Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18:893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langen R, Cai K, Altenbach C, Khorana HG, Hubbell WL. Structural features of the C-terminal domain of bovine rhodopsin: a site-directed spin-labeling study. Biochemistry. 1999;38:7918–24. doi: 10.1021/bi990010g. [DOI] [PubMed] [Google Scholar]
- 37.Barnakov A, Altenbach C, Barnakova L, Hubbell WL, Hazelbauer GL. Site-directed spin labeling of a bacterial chemoreceptor reveals a dynamic, loosely packed transmembrane domain. Protein Sci. 2002;11:1472–81. doi: 10.1110/ps.0202502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin B, Bourhis J-M, Belle V, Woudstra M, Carrière F, Guigliarelli B, et al. Assessing induced folding of an intrinsically disordered protein by site-directed spin-labeling electron paramagnetic resonance spectroscopy. J Phys Chem B. 2006;110:20596–608. doi: 10.1021/jp063708u. [DOI] [PubMed] [Google Scholar]
- 39.Belle V, Rouger S, Costanzo S, Liquière E, Strancar J, Guigliarelli B, et al. Mapping α-helical induced folding within the intrinsically disordered C-terminal domain of the measles virus nucleoprotein by site-directed spin-labeling EPR spectroscopy. Proteins. 2008;73:973–88. doi: 10.1002/prot.22125. [DOI] [PubMed] [Google Scholar]
- 40.López CJ, Fleissner MR, Guo Z, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18:1637–52. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pirman NL, Milshteyn E, Galiano L, Hewlett JC, Fanucci GE. Characterization of the disordered-to-α-helical transition of IA3 by SDSL-EPR spectroscopy. Protein Sci. 2011;20:150–9. doi: 10.1002/pro.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartelli NL, Hazelbauer GL. Direct evidence that the carboxyl-terminal sequence of a bacterial chemoreceptor is an unstructured linker and enzyme tether. Protein Sci. 2011;20:1856–66. doi: 10.1002/pro.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López CJ, Oga S, Hubbell WL. Mapping molecular flexibility of proteins with site-directed spin labeling: a case study of myoglobin. Biochemistry. 2012;51:6568–83. doi: 10.1021/bi3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg–Marquardt algorithm. Journal of Magnetic Resonance, Series A. 1996;120:155–89. [Google Scholar]
- 45.Lai W-C, Barnakova LA, Barnakov AN, Hazelbauer GL. Similarities and differences in interactions of the activity-enhancing chemoreceptor pentapeptide with the two enzymes of adaptational modification. J Bacteriol. 2006;188:5646–9. doi: 10.1128/JB.00497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422:3–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Moual H, Quang T, Koshland DE., Jr Conformational changes in the cytoplasmic domain of the Escherichia coli aspartate receptor upon adaptive methylation. Biochemistry. 1998;37:14852–9. doi: 10.1021/bi980343y. [DOI] [PubMed] [Google Scholar]
- 48.Starrett DJ, Falke JJ. Adaptation mechanism of the aspartate receptor: Electrostatics of the adaptation subdomain play a key role in modulating kinase activity. Biochemistry. 2005;44:1550–60. doi: 10.1021/bi048089z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danielson MA, Bass RB, Falke JJ. Cysteine and disulfide scanning reveals a regulatory alpha-helix in the cytoplasmic domain of the aspartate receptor. J Biol Chem. 1997;272:32878–88. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winston SE, Mehan R, Falke JJ. Evidence that the adaptation region of the aspartate receptor is a dynamic four-helix bundle: cysteine and disulfide scanning studies. Biochemistry. 2005;44:12655–66. doi: 10.1021/bi0507884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998;37:10746–56. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bass RB, Falke JJ. Detection of a conserved alpha-helix in the kinase-docking region of the aspartate receptor by cysteine and disulfide scanning. J Biol Chem. 1998;273:25006–14. doi: 10.1074/jbc.273.39.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bass RB, Coleman MD, Falke JJ. Signaling domain of the aspartate receptor is a helical hairpin with a localized kinase docking surface: cysteine and disulfide scanning studies. Biochemistry. 1999;38:9317–27. doi: 10.1021/bi9908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim KK, Yokota H, Kim S-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–92. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 55.Ferris HU, Zeth K, Hulko M, Dunin-Horkawicz S, Lupas AN. Axial helix rotation as a mechanism for signal regulation inferred from the crystallographic analysis of the E. coli serine chemoreceptor. J Struct Biol. 2014;186:349–56. doi: 10.1016/j.jsb.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 56.Murphy OJ, Yi X, Weis RM, Thompson LK. Hydrogen exchange reveals a stable and expandable core within the aspartate receptor cytoplasmic domain. J Biol Chem. 2001;276:43262–9. doi: 10.1074/jbc.M105585200. [DOI] [PubMed] [Google Scholar]
- 57.Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–13. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai W-C, Hazelbauer GL. Carboxyl-terminal extensions beyond the conserved pentapeptide reduce rates of chemoreceptor adaptational modification. J Bacteriol. 2005;187:5115–21. doi: 10.1128/JB.187.15.5115-5121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]