Abstract
Versatility of CRISPR/Cas9-based platforms makes them promising tools for the correction of diverse genetic/epigenetic disorders. Here we contrast the use of these genome editing tools in two myopathies with very different molecular origins: Duchenne muscular dystrophy, a monogenetic disease, and facioscapulohumeral muscular dystrophy, an epigenetic disorder with unique therapeutic challenges.
Keywords: CRISPR, Cas9, genome editing, muscular dystrophy, FSHD, DMD
CRISPR/Cas9 genome editing tools are being widely investigated for the treatment of many disorders [1], with one neuromuscular disease at the forefront. Duchenne Muscular Dystrophy (DMD) is one of the most prevalent fatal genetic diseases, with no approved therapies currently available. DMD is caused by any of a large spectrum of mutations in the Dystrophin gene that lead to loss of functional protein. Although many therapeutic approaches for DMD have been attempted over the years, success has been limited, in part by the large size of Dystrophin and the difficulty of achieving long-term rescue. Since 62% of DMD patients have mutations in exons 45-55 of Dystrophin, targeting this non-essential region to restore the open reading frame (ORF) has been a compelling strategy. In fact, deleting or otherwise editing this mutation hotspot is amenable to the CRISPR/Cas9 approach in patient cells [2] (Figure 1). Following proof-of-principle studies, several groups recently reported Cas9-mediated gene editing in vivo using the mdx mouse model of DMD, which contains a natural mutation in exon 23 of Dystrophin [3–5]. Using AAV delivery, all three groups targeted Cas9 to the exon 23 splice junctions in Dystrophin, taking advantage of repair by non-homologous end joining (NHEJ) to delete the mutated exon and restore the ORF. In all three reports, Dystrophin expression was recovered to therapeutic levels in the affected muscles and the dystrophic phenotype was improved.
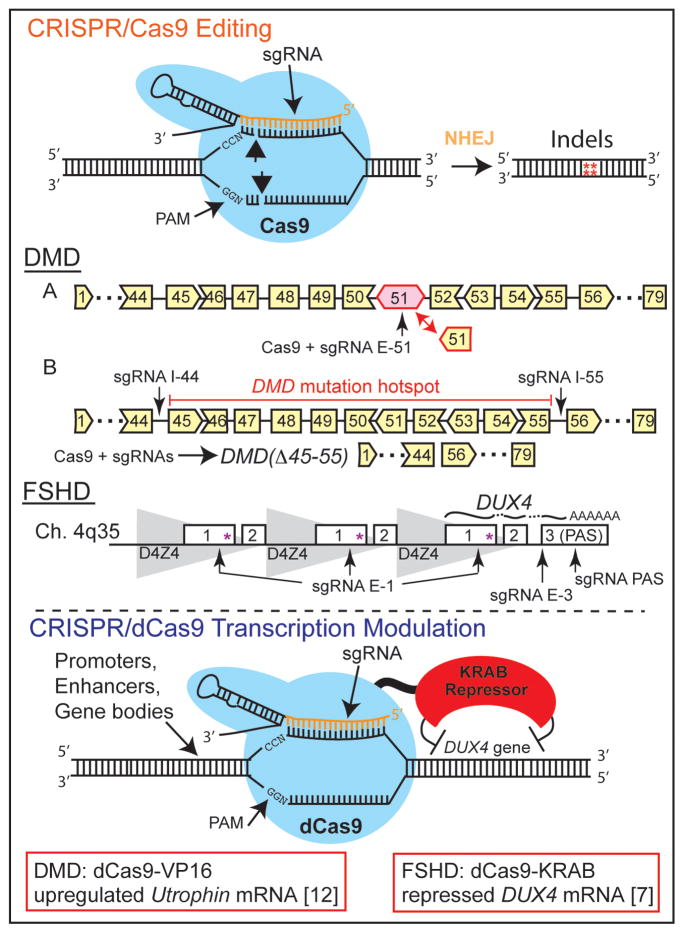
Figure 1. CRISPR/Cas9 tools for genome editing or transcription modulation in skeletal myocytes.
Upper panel: The Cas9 enzyme forms a complex with a sequence-specific single guide RNA (sgRNA), which guides binding to a genomic target. Cas9 cuts DNA, generating double-stranded breaks which are repaired by non-homologous end joining (NHEJ) to produce various insertions and deletions (indels) at the target sequence. Cas9 guided by sgRNAs flanking a target sequence can be used to generate a larger deletion, albeit at a lower frequency. In DMD patient myoblasts harboring an out-of-frame deletion of exons 48-50 in Dystrophin, Cas9 guided by an sgRNA targeting exon 51 (A) was used to create frameshifts in this exon and restore the open reading frame (ORF) [2]. In a strategy designed to correct a majority of DMD lesions, Cas9 guided by sgRNAs flanking a mutation hotspot comprising exons 45-55 of Dystrophin (B) was used to delete these exons and restore the ORF [2]. A similar approach can be used in FSHD myocytes, targeting Cas9 to various locations in DUX4 to decrease DUX4-fl mRNA from the pathogenic D4Z4 repeat (e.g., exon 1 to disrupt the ORF, splice junctions to prevent splicing, or flanking the polyadenylation signal to delete this sequence). Lower panel: The catalytically inactive dCas9 is incapable of cutting DNA, but can still be recruited via sgRNAs to specific genomic targets. When fused to transcriptional modulators (e.g., the KRAB repressor domain), this platform can alter gene expression at defined loci. In DMD patient myoblasts, dCas9 fused to the VP16 transcriptional activator was used to upregulate expression of the compensatory Utrophin gene [12]. In FSHD patient myocytes, dCas9-KRAB was used to repress DUX4-fl mRNA from the pathogenic D4Z4 repeat [7].
If only all diseases were so straightforward.
Consider Facioscapulohumeral Muscular Dystrophy (FSHD), the most prevalent myopathy affecting males and females of all ages (reviewed in [6]). In contrast to DMD, which is caused by the loss of a functional protein, FSHD is caused by the aberrant expression of a protein that is normally silent due to its extreme toxicity. In healthy individuals, the DUX4 gene—embedded within each repeat of the D4Z4 macrosatellite array—is under strong epigenetic repression in somatic cells. In FSHD, contraction of the D4Z4 array on chromosome 4 or mutations in proteins required to maintain epigenetic silencing leads to de-repression of the pathogenic allele and two array-proximal enhancers help to drive aberrant DUX4 expression in skeletal muscle. Once stably expressed, DUX4 protein acts as a transcription factor, activating immune mediators, retroelements, and germline genes with pathological consequences.
DMD is caused by mutations in a single gene that lies in a typical region of the genome, making it a prime candidate for editing by CRISPR/Cas9. By contrast, FSHD is caused by the pathogenic expression of one copy of a gene that lies in every repeat unit of a macrosatellite array. To complicate matters, this array occurs not only on Chromosome 4, but also on Chromosome 10, with other polymorphic homologs scattered throughout the genome. With hundreds of inert copies of DUX4 present in a human cell, would a gene-editing platform be able to target the single pathogenic copy? In a recent proof-of-principle study [7], we showed that a CRISPR platform can, in fact, be targeted to the pathogenic D4Z4 repeat in FSHD muscle cells (Figure 1). Using the nuclease-dead version of Cas9 fused to a transcriptional repressor (dCas9-KRAB), we returned the chromatin at the disease locus to a more normal heterochromatic state, repressing expression of DUX4 and its target genes.
Why not simply use Cas9 to disrupt the DUX4 ORF, alternative splicing (Box 1), or polyadenylation signal (Figure 1)? In principle, such a strategy is feasible. As with exon-skipping for DMD, disrupting the DUX4 locus can be accomplished with indels created by the imprecise, but efficient NHEJ pathway, which—unlike homology-directed repair—is highly active in post-mitotic cells. While disrupting the DUX4 locus with Cas9 is a viable therapeutic avenue, the use of a dCas9 platform with transcriptional effectors or epigenetic modifiers has the distinct advantage of not cutting the genome at hundreds of unintended places. Additionally, an epigenetic “straitjacket” can potentially be removed, whereas the effects of cutting with the Cas9 molecular scalpel are permanent. Importantly, since other D4Z4 arrays throughout the genome are normally silent, targeting a transcriptional repressor to these off-target locations shouldn’t cause undue harm.
Box 1. Challenges and perspectives for genome targeting in FSHD (200 words).
DUX4, the causal gene in FSHD, exists in hundreds of copies throughout the genome, but only one copy is stably expressed, leading to disease [6]. Amongst hundreds of non-pathogenic copies, the dCas9-KRAB repressor was successfully used to decrease expression of the single pathogenic copy of DUX4 [7].
The repeat harboring DUX4 contains long stretches of low-complexity sequence, limiting selection of sgRNA targets.
In normal cells, DUX4 produces a short mRNA isoform that is translated into a non-toxic protein. In FSHD myocytes, there is a shift in mRNA splicing to generate the full-length pathogenic DUX4 isoform (DUX4-fl) [6]. Using Cas9 or a dCas9 modifier to affect splicing represents an alternate approach for decreasing levels of DUX4-fl.
DUX4 is primate-specific; no natural animal models of FSHD exist. Although attempts to model the disease in mice have been problematic [6], generation of a valid mouse model is key for testing any therapeutic approaches in vivo. In an FSHD-like model, DUX4 would be expressed in sporadic bursts in rare myocytes, with all muscle cells poised for expression. With this in mind, gene targeting strategies may need to correct a majority of myocytes—and possibly muscle satellite cells—in order to provide therapeutic benefit.
Although our study is the first reported use of a dCas9 platform for a muscular dystrophy, many questions and issues remain (Box 1). Whereas the phenotype in DMD can be rescued with only a small fraction of wild-type dystrophin levels, the therapeutic threshold for FSHD is not so clear. How many myofibers will need to be corrected, and how much of a reduction in DUX4 expression will be required to provide a functional benefit? Although DUX4 mRNA and protein are only rarely expressed in muscle nuclei at any given time, DUX4 appears to be poised for expression in a majority of FSHD myocytes [6, 8]. Thus, any gene-targeting approach for this disease may need to correct a majority of myofibers—and possibly the muscle stem cells that give rise to them. With this in mind, it is encouraging that following muscle-tropic AAV9 delivery of Cas9 components in the mdx mouse, a small percentage of muscle satellite cells displayed evidence of gene editing [4], and numbers of dystrophin-positive myofibers increased over time [3], consistent with editing of satellite cells.
Developing Cas9-based technologies into a therapeutic approach for DMD or FSHD will require advances on several fronts, including tissue delivery. Thanks to improvements in stem cell therapy, correction of a patient’s iPSC-derived myoblasts followed by transplantation into dystrophic muscle is feasible, in principle. Although correction would be limited to injected muscles, such a strategy would allow for selection and expansion of gene-edited/modified cells. Systemic delivery of muscle-tropic AAVs (e.g., AAV9) can reach a wider range of anatomical muscles, but AAV vectors have a limited packaging capacity—a challenge that is already being addressed by minimizing the AAV regulatory cassette [3], trans-splicing the large Cas9 enzyme from S. pyogenes [9], or using smaller Cas9 orthologs [4, 5, 10].
A successful Cas9-based therapy for FSHD and many other disorders will surely require increased efficiency of genome editing/modification. Technical improvements in the stability, specificity, and delivery of Cas9 components are rapidly evolving to meet this need. Additionally, greater versatility in Cas9 platforms make multiplexing—targeting multiple genomic regions with one or more enzymes or chromatin effectors—strategically possible [11]. For example, one could envision a combination therapy that targets either DUX4 in FSHD muscles or Dystrophin in DMD muscles, while upregulating expression of a compensatory gene such as Utrophin in the same cells [12].
Safety has always been a paramount concern for gene therapy, and the prospect of creating permanent changes in the genome has put Cas9-based approaches under a great deal of scrutiny. Alongside the need for more sensitive and comprehensive assessment of off-target effects is the need to determine immunogenicity of Cas9 components and the long-term effects—both intended and unintended—of genome modification. Fortunately for DMD, FSHD, and a host of other disorders, the need to correct somatic—not germline—cells makes the prospect of genome targeting both ethically and technically feasible.
Ultimately, CRISPR/Cas9 platforms should be broadly applicable, from the treatment of conceptually simple monogenic diseases to complex multigene disorders. It is a testament to the synergistic nature of science that advances in the treatment of any disease are really advances for most, if not all, diseases. Even for one caused by a toxic, sporadically expressed gene lurking in a repeat array amongst hundreds of decoy copies. With emerging functions for repetitive sequences that comprise nearly half the human genome, who knows what other disorders wait to be targeted?
Acknowledgments
We wish to thank NIAMS/NIH, the Chris Carrino Foundation for FSHD, the FSH Society, and the Muscular Dystrophy Association for funding our FSHD research program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maeder ML, Gersbach CA. Genome Editing Technologies for Gene and Cell Therapy. Mol Ther. 2016 doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ousterout DG, et al. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nature communications. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long C, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabebordbar M, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Himeda CL, et al. Facioscapulohumeral muscular dystrophy as a model for epigenetic regulation and disease. Antioxidants & redox signaling. 2015;22:1463–1482. doi: 10.1089/ars.2014.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Himeda CL, et al. CRISPR/dCas9-mediated Transcriptional Inhibition Ameliorates the Epigenetic Dysregulation at D4Z4 and Represses DUX4-fl in FSH Muscular Dystrophy. Mol Ther. 2015 doi: 10.1038/mt.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones TI, et al. Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clinical epigenetics. 2015;7:37. doi: 10.1186/s13148-015-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fine EJ, et al. Trans-spliced Cas9 allows cleavage of HBB and CCR5 genes in human cells using compact expression cassettes. Sci Rep. 2015;5:10777. doi: 10.1038/srep10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wade M. High-Throughput Silencing Using the CRISPR-Cas9 System: A Review of the Benefits and Challenges. Journal of biomolecular screening. 2015;20:1027–1039. doi: 10.1177/1087057115587916. [DOI] [PubMed] [Google Scholar]
- 12.Wojtal D, et al. Spell Checking Nature: Versatility of CRISPR/Cas9 for Developing Treatments for Inherited Disorders. Am J Hum Genet. 2016;98:90–101. doi: 10.1016/j.ajhg.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]