SUMMARY
Generic spinal motor neuron identity is established by cooperative binding of programming transcription factors (TFs), Isl1 and Lhx3, to motor-neuron-specific enhancers. How expression of effector genes is maintained following downregulation of programming TFs in maturing neurons remains unknown. High-resolution exonuclease (ChIP-exo) mapping revealed that the majority of enhancers established by programming TFs are rapidly deactivated following Lhx3 downregulation in stem-cell-derived hypaxial motor neurons. Isl1 is released from nascent motor neuron enhancers and recruited to new enhancers bound by clusters of Onecut1 in maturing neurons. Synthetic enhancer reporter assays revealed that Isl1 operates as an integrator factor, translating the density of Lhx3 or Onecut1 binding sites into transient enhancer activity. Importantly, independent Isl1/Lhx3- and Isl1/Onecut1-bound enhancers contribute to sustained expression of motor neuron effector genes, demonstrating that outwardly stable expression of terminal effector genes in postmitotic neurons is controlled by a dynamic relay of stage-specific enhancers.
In Brief
Rhee et al. performed high-resolution genome-wide analysis of TF binding in nascent and maturing ESC-derived motor neurons. The study revealed mechanisms by which Isl1 integrates stage-specific TFs at dynamic enhancers to maintain neuronal cell identity.
INTRODUCTION
The vertebrate central nervous system is a complex organ composed of many thousands of neuronal subtypes organized in a highly stereotypical pattern. Construction of such a sophisticated system relies on the precise and reproducible establishment of cell-type-specific gene expression programs that define the morphology, connectivity, and functionality of individual neuronal subtypes.
The acquisition of neuronal identity is driven by a developmental transcriptional program executed in neural progenitors that culminates in the activation of a unique combination of transcription factors (TFs) in nascent postmitotic neurons (Bertrand et al., 2002; Shirasaki and Pfaff, 2002). These TFs are often referred to as master regulators or programming factors, as they are both necessary and sufficient to activate expression of cell-type-specific combinations of effector genes that define neuronal identity and functionality (Hobert, 2008; Vierbuchen et al., 2010). How programming TFs selectively recognize their genomic binding sites and which of the bound sites will get activated and regulate target genes have been the topic of intense scrutiny; yet, our understanding of the process remains rather limited. Furthermore, many programming TFs are expressed only transiently in the developing mammalian nervous system and are rapidly downregulated in maturing postmitotic neurons (Mong et al., 2014; Son et al., 2011; Wapinski et al., 2013), raising the question of how expression of the cell-type-specific effector genes is maintained in maturing neurons.
Recent studies established that enhancers in progenitor cells transitioning from one developmental state to another are highly dynamic (Hong et al., 2013; Johnson et al., 2011; Velasco et al., 2016). In contrast to dividing cells, many genes induced in postmitotic neurons (axon guidance receptors, neurotransmitters and receptors, ion channels, etc.) operate as terminal effector genes and remain stably expressed despite downregulation of TFs that initiated their expression. Whether enhancers established in nascent postmitotic neurons become stabilized by the recruitment of secondary TFs or whether the continuity of expression of postmitotic neuronal effector genes depends on a transcriptional cascade engaging independent stage-specific enhancers in maturing motor neurons remain unknown.
Generic vertebrate spinal motor neuron identity is established by three TFs—proneural basic helix-loop-helix (bHLH) TF Neurogenin 2 (Ngn2), and two LIM homeodomain (HD) TFs, Lhx3 and Islet 1 (Isl1) (Briscoe et al., 2000; Jessell, 2000; Novitch et al., 2001). The three factors function as programming TFs, sufficient to reprogram embryonic stem cells (ESCs) or neural progenitors into spinal motor neurons (Hester et al., 2011; Lee et al., 2012; Lee and Pfaff, 2003; Mazzoni et al., 2013). Ngn2 is expressed only transiently in motor neuron progenitors as they initiate their terminal differentiation. Isl1 and Lhx3 are initially co-expressed in nascent postmitotic motor neurons, but Lhx3 is rapidly downregulated in all hypaxial and limb muscle-innervating motor neurons (Bonanomi and Pfaff, 2010; Sharma et al., 1998). Mapping TF binding sites during transcriptional reprogramming of ESCs to motor neurons revealed that Isl1 and Lhx3 co-occupy more than 10,000 genomic sites (Mazzoni et al., 2013). However, the precise syntax of cis-regulatory elements underlying cooperative recruitment of Isl1 and Lhx3 to their preferred binding sites remained occluded by the poor spatial resolution of chromatin immunoprecipitation sequencing (ChIP-seq) assays.
ESC differentiation into spinal hypaxial motor neurons undergoes a progressive transition from Ngn2-positive progenitors into Isl1/Lhx3-positive nascent postmitotic motor neurons and, ultimately, into Isl1-positive/Lhx3-negative maturing motor neurons (Tan et al., 2016). Temporal downregulation of two of the three motor neuron programming TFs (Ngn2 and Lhx3) under this differentiation protocol provides an opportunity to study gene regulatory mechanisms contributing to the stable expression of motor neuron effector genes in the context of a dynamic transcriptional environment. Our study demonstrates that downregulation of Lhx3 results in the release of Isl1 from the Isl1/Lhx3 co-bound sites followed by the decommissioning of nascent motor neuron enhancers. Isl1 is then recruited to a new set of enhancers activated in maturing motor neurons. By using high-resolution mapping of TF binding sites (Rhee and Pugh, 2011), we discovered that the dynamic behavior of Isl1 can be in part explained by its indirect recruitment to enhancers through protein-protein interactions with clusters of Lhx3 and Onecut1 TFs in nascent and maturing motor neurons, respectively. Gene expression analysis revealed that the majority of motor neuron effector genes expressed in postmitotic neurons are associated with transiently active stage-specific enhancers. These results indicate that outwardly stable expression of effector genes in terminally differentiated cells is controlled by transient enhancers established and activated by stage-specific combinations of TFs.
RESULTS
Dynamic Changes in the Genomic Regulatory Landscape in Postmitotic Motor Neurons
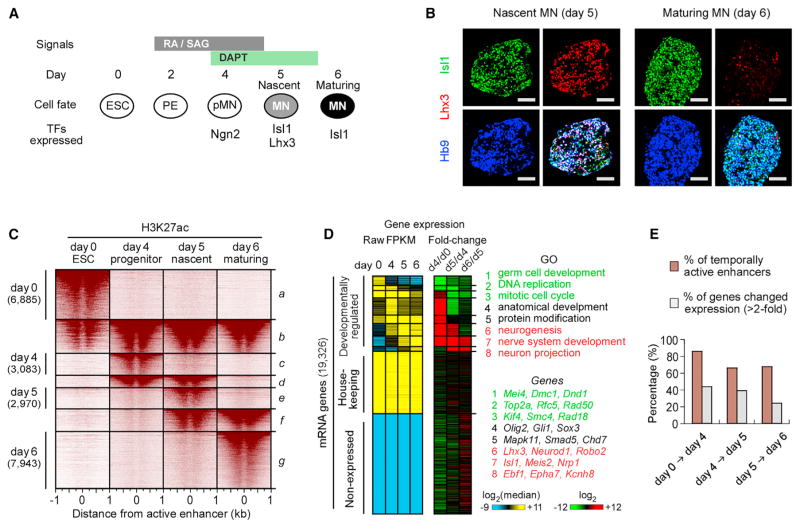
The progressive differentiation of pluripotent cells into their terminal identity is controlled by developmentally regulated TFs that dynamically establish, activate, silence, and decommission enhancers regulating lineage-specific gene expression programs (Dixon et al., 2015; Shlyueva et al., 2014; Whyte et al., 2012). Mouse ESCs induced on day 2 of differentiation with patterning factors, retinoic acid (RA), and smoothened agonist (SAG) acquire motor neuron progenitor identity characterized by expression of Olig2 on day 4 of differentiation (Wichterle et al., 2002; Wichterle and Peljto, 2008). Treatment of motor neuron progenitors with γ-secretase inhibitor (DAPT) transiently activates expression of Ngn2, followed by cell cycle exit and terminal differentiation of progenitors into postmitotic motor neurons by day 5 (Figure 1A) (Tan et al., 2016). Under this protocol, there are virtually no dividing progenitors remaining on day 5 of differentiation, with ~80% of cells becoming postmitotic motor neurons. Motor neuron maturation under this protocol is temporally well synchronized, as 63% of day 5 cells co-express nascent motor neuron markers Isl1 and Lhx3, but 1 day later, 91% of maturing motor neurons downregulate Lhx3 and acquire an expression profile of hypaxial motor neurons (Figure 1B; Figure S1).
Figure 1. Dynamic Changes in Enhancer Organization and Gene Expression during Motor Neuron Differentiation.
(A) Overview of mouse ESC-directed differentiation to postmitotic hypaxial motor neuron (MN). Differentiating cells become nascent postmitotic MNs on day 5 and maturing MNs on day 6. PE, primitive ectoderm; pMN, MN progenitor.
(B) Expression of Isl1, Lhx3, and Hb9 in embryoid bodies. Lhx3 is transiently expressed in nascent MNs on day 5 and rapidly downregulated in maturing MNs on day 6 (Figure S1). Scale bars represent 50 μm.
(C) H3K27ac intensity relative to active enhancer midpoints (30,648 rows, Table S1) during MN differentiation. Active enhancers were grouped and sorted by stage-specific H3K27ac intensity, demonstrating that the majority of enhancers are transient during MN differentiation.
(D) Expression levels of 19,326 annotated RefSeq genes grouped by fold changes in expression between individual MN differentiation stages. Left: FPKM (fragments per kilobase of exon per million fragments mapped) values were median normalized and log2 transformed. Middle: Log2 RNA fold change, ordered as shown in the left panel. Right: representative GO terms and examples in each group of genes.
(E) Percentage of enhancers and genes that dynamically change more than 2-fold between indicated time points. Enhancers remain dynamic between day 5 and day 6 MNs, while gene expression becomes more stable.
See also Figure S1.
Developmentally active enhancers are commonly associated with enriched histone H3 lysine 27 acetylation (H3K27ac) and increased chromatin accessibility (Calo and Wysocka, 2013; Heintzman and Ren, 2009). To examine genome-wide changes in active enhancer signatures in differentiating motor neurons, we performed ChIP-seq of H3K27ac and an assay for transposase-accessible chromatin with sequencing (ATAC-seq) (Buenrostro et al., 2013) in mouse ESCs (day 0), motor neuron progenitors (day 4), nascent postmitotic motor neurons (day 5), and maturing motor neurons (day 6). We identified 30,648 unique distal genomic regions (referred to as “active enhancers”), having significant increase of both H3K27ac and ATAC-seq intensity, across the four stages of motor neuron differentiation. We observed that a significant fraction of active enhancers is not developmentally stable (Figure 1C). Approximately 66% of active enhancers are reorganized during a 24 hr period when cells undergo a transition from dividing motor neuron progenitors to nascent postmitotic motor neurons (10,404 of 15,802 identified active enhancers). Surprisingly, comparably dynamic enhancer behavior is detected in maturing postmitotic motor neurons. More than 67% of enhancers are temporally reorganized (13,628 of 20,237 active enhancers) during the first 24 hr of motor neuron maturation. The high enhancer turnover in postmitotic motor neurons contrasts with the drop in the total number of developmentally regulated genes (Figures 1D and 1E). RNA sequencing (RNA-seq) analysis revealed that 39% of all expressed genes (4,256/11,049) significantly change their expression between day 4 progenitors and day 5 nascent postmitotic motor neurons (>2-fold; Supplemental Experimental Procedures), while only 24% of all expressed genes (2,607/11,049) change expression between nascent and maturing motor neurons. These results suggest that the genomic organization of active enhancers remains highly dynamic even in postmitotic neurons exhibiting fewer changes in gene expression.
Divergent Binding of Ngn2 and Isl1 to Stage-Specific Enhancers
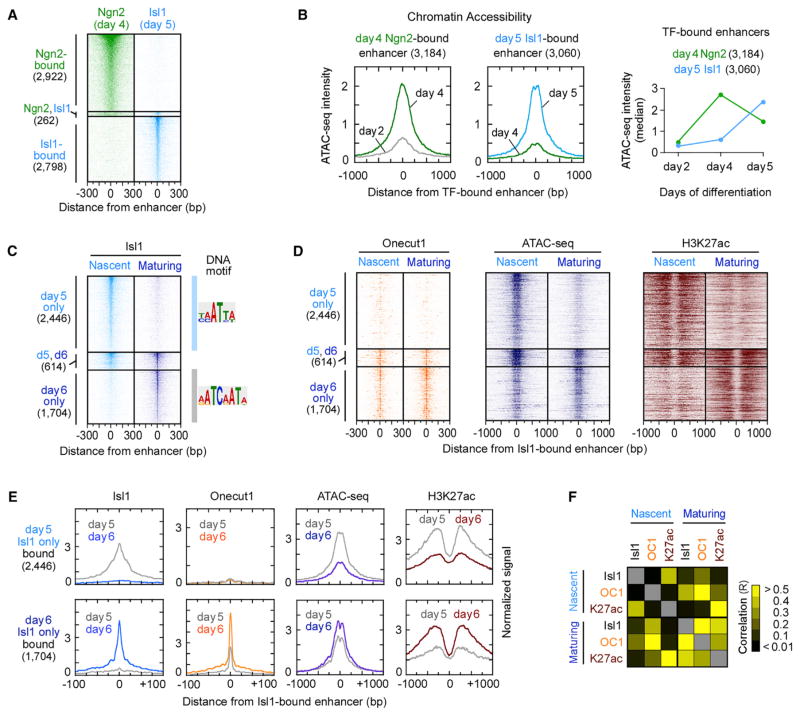
Considering the dynamic behavior of active enhancers in postmitotic motor neurons, we examined whether programming TFs controlling terminal effector gene expression preferentially bind to the small subset of stably maintained enhancers in postmitotic motor neurons. Previously, it has been shown that during neuronal reprogramming of fibroblasts, the proneural bHLH TF Ascl1 establishes accessible chromatin regions to which the programming TF Brn2 is recruited (Wapinski et al., 2013). To examine whether the proneural bHLH TF Ngn2 functions similarly to recruit the programming TF Isl1 in motor neuron differentiation, we performed a series of ChIP-seq experiments to identify Ngn2-bound enhancers (1 kb region around Ngn2-bound site) in day 4 motor neuron progenitors and Isl1-bound enhancers in day 5 nascent motor neurons. Surprisingly, we found that only 4% of bound enhancers (262/5,982) were co-occupied by both TFs (Figure 2A), suggesting that Ngn2 expressed in progenitors and Isl1 expressed in postmitotic neurons control gene expression programs through distinct sets of stage-specific enhancers.
Figure 2. Ephemeral Nature of Enhancers Established by Motor Neuron Programming TFs.
(A) ChIP mapping of Ngn2-bound enhancers in day 4 progenitors and Isl1-bound enhancers in day 5 postmitotic MNs, sorted by TF occupancy. Ngn2 and Isl1 TFs were found within 1 kb genomic region only in 262 locations.
(B) Composite (average read counts) plots of days 2 and 4 ATAC-seq intensity for Ngn2-bound enhancers (left) and days 4 and 5 ATAC-seq intensity for Isl1-bound enhancers (middle) (Figure S2A). Right panel depicts a transient increase in median ATAC-seq intensity of Ngn2-bound enhancers on day 4 and a gain of chromatin accessibility of Isl1-bound enhancers on day 5, coincident with the time of TF expression.
(C and D) ChIP intensity of Isl1 (C), Onecut1, and H3K27ac and ATAC-seq (D) measured in nascent (day 5) and maturing (day 6) postmitotic MNs. All data are plotted relative to the Isl1-bound enhancer midpoints (Table S2), sorted and ordered by Isl1 occupancy. If the midpoints between day 5 and day 6 Isl1-bound enhancers reside within 1 kb, they were defined as maintained enhancers (n = 614). DNA motif represents the most enriched sequence within ±14 bp from the midpoint of Isl1-bound site (Figure S2B).
(E) Composite plots of (C) and (D).
(F) A heatmap of Pearson correlation coefficients (R) for pairwise combinations of TFs and H3K27ac intensity, shown in (C) and (D) (Table S3).
See also Figure S2.
Next, we asked whether Ngn2 and Isl1 are preferentially recruited to existing accessible chromatin regions. Mapping of chromatin accessibility by ATAC-seq revealed that neither Ngn2- nor Isl1-bound enhancers were accessible in primitive ectodermal cells on day 2 of differentiation (Figure 2B; Figure S2A). Ngn2-bound enhancers became accessible in progenitors, coincident with the onset of Ngn2 expression, but rapidly lost their chromatin accessibility 1 day later when Ngn2 is downregulated and cells differentiate into postmitotic motor neurons. Similarly, we found that Isl1-bound enhancers were largely inaccessible in day 4 progenitors prior to Isl1 expression but exhibited increased chromatin accessibility in day 5 nascent postmitotic motor neurons. These findings indicate that Ngn2 and Isl1 contribute to the highly dynamic genomic regulatory landscape by establishing largely non-overlapping sets of stage-specific accessible enhancers during the transition from progenitors to nascent postmitotic motor neurons.
Isl1 Rapidly Relocates to Onecut1-Bound Sites Following Lhx3 Downregulation
During motor neuron specification, Isl1 and Lhx3 form a heterodimer complex that binds distal enhancers of many effector genes expressed in nascent postmitotic motor neurons (Lee et al., 2013; Mazzoni et al., 2013; Thaler et al., 2002). However, the fate of Isl1/Lhx3-bound enhancers following Lhx3 downregulation in maturing postmitotic motor neurons has not been previously examined. High-resolution exonuclease (ChIP-exo) mapping of Isl1 binding before and after Lhx3 downregulation identified 4,128 genomic sites (22–28 bp protected regions, referred to as Isl1-bound sites) in nascent (Lhx3-positive) motor neurons and 3,153 Isl1-bound sites in maturing (Lhx3-negative) motor neurons. Remarkably, only 6% of Isl1-bound sites in nascent neurons (250/4,128) were occupied by Isl1 in maturing motor neurons (Figure S2B). Analysis of Isl1-bound enhancers (1 kb region around Isl1-bound site) revealed that 80% of enhancers bound by Isl1 in nascent motor neurons (2,446/3,060) exhibited a significant depletion of Isl1 binding in maturing motor neurons and vice versa; 74% of mature Isl1-bound enhancers lacked Isl1 binding in nascent motor neurons (1,704/2,318) (Figure 2C; Figure S2C). Most Isl1-bound sites were far from annotated transcription start sites (TSSs), and the median distance between the closest nascent and maturing motor neuron Isl1-bound sites was 109 kb (Figure S2D), demonstrating that Isl1 does not simply move to a new site within the same genomic territory. The rapid displacement of Isl1 from nascent enhancers depends on Lhx3 downregulation, as Isl1 remained bound to nascent enhancers in maturing median motor neurons that maintain Lhx3 expression (Figures S2E and S2F).
Considering that Isl1 tends to form heterodimers with other TFs, we examined DNA sequences at the new Isl1-bound sites in maturing motor neurons using de novo motif discovery (MEME) (Bailey et al., 2015). We identified an unexpected primary motif, AATCAATA, annotated as the Onecut/HNF6 DNA binding motif (Figure 2C). Onecut1 and Onecut2 members of the Cut-HD TF family are induced 6- to 12-fold in postmitotic motor neurons relative to motor neuron progenitors. Loss of function analysis established that Onecut TFs are necessary for motor neuron maturation, specification of motor neuron subtype identity, and formation of functional neuromuscular synapses (Audouard et al., 2012; Francius and Clotman, 2010; Roy et al., 2012). To determine whether Onecut TFs co-occupy genomic regions bound by Isl1 in maturing motor neurons, we performed ChIP-exo analysis for Onecut1. We observed that more than 60% of all Isl1-bound sites (1,918/3,153) in maturing motor neurons were located within 50 bp of Onecut1-bound sites (Figure 2D). Interestingly, ChIP-exo and ATAC-seq analyses revealed that many Onecut/Isl1-bound enhancers are already accessible and bound by Onecut1 in nascent motor neurons prior to Isl1 recruitment (Figures 2D and 2E), indicating that Isl1 presence is not a prerequisite for the binding of Onecut1 to maturing motor neuron enhancers.
As Isl1 does not appear to be necessary for the establishment of Onecut1-bound sites, we wondered whether it might be involved in functional regulation of these enhancers. ChIP-seq profiling of histone modifications revealed a correlation between Isl1 binding and accumulation of H3K27ac (Figures 2C–2F; Figure S2C). In nascent motor neurons, Isl1-bound enhancers exhibited high levels of H3K27ac, but histone acetylation was significantly decreased in maturing motor neurons, suggesting that Isl1-bound enhancers become rapidly deactivated following the displacement of Isl1. In contrast, Onecut1-bound enhancers in nascent neurons exhibited low levels of H3K27ac, but their acetylation levels significantly increased following Isl1 recruitment in maturing motor neurons. The close correlation between Isl1 binding and H3K27ac intensity raises the possibility that Isl1 is instrumental in the productive recruitment of a histone acetyltransferase (HAT) and the activation of motor neuron enhancers.
Stage-Specific Enhancers Maintain Expression of Postmitotic Motor Neuron Genes
Neuronal progenitors undergo a major transition from highly proliferative cells with a simple bipolar morphology to postmitotic motor neurons extending synaptic processes, acquiring mature electrophysiological properties, and synthesizing appropriate neurotransmitters. This transition is accompanied by the activation of a battery of postmitotic effector genes that are expressed throughout the lifespan of motor neurons. Our observation that motor neuron programming TFs bind to transient stage-specific enhancers raises the important question of how expression of terminal effector genes is maintained in postmitotic neurons.
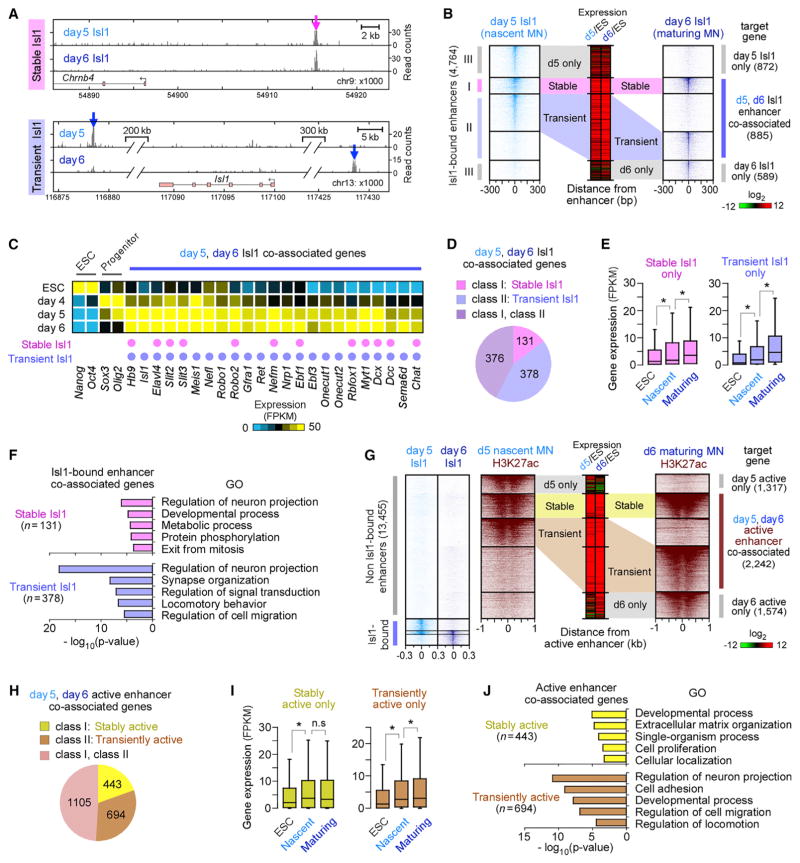
To correlate enhancers with changes in gene expression, we assigned distal enhancers to the nearest TSS (Creyghton et al., 2010; Rada-Iglesias et al., 2011; Wang et al., 2015; Whyte et al., 2013). We defined three classes of Isl1-bound enhancers (Figures 3A and 3B): (1) enhancers bound by Isl1 in both nascent and maturing motor neurons (referred to as “stable” Isl1 enhancers) (class I), (2) enhancers differentially bound by Isl1 in nascent or maturing motor neurons but co-associated with the same gene (referred to as “transient” Isl1 enhancers) (class II), and (3) enhancers associated with genes only in nascent or maturing motor neurons (day 5 or day 6 only) (class III). Approximately 62% of all Isl1-bound enhancers (2,932/4,764) belonged to class I or II. RNA-seq analysis revealed that day 5 and day 6 Isl1 co-associated genes (class I or II) were significantly induced in postmitotic motor neurons compared to other Isl1-associated genes (Figure S3A). Moreover, Isl1 co-associated genes included many genes previously known as postmitotic motor neuron effector genes (e.g., Hb9, Isl1, Slit2, Lhx3, Chat, and Nrp1), and these genes were highly expressed in both nascent and maturing hypaxial motor neurons (Figure 3C). Importantly, over 85% of these Isl1 co-associated genes (754/885) contained transient Isl1 enhancers (Figure 3D).
Figure 3. Motor Neuron Effector Genes Are Associated with Stage-Specific Isl1-Bound Enhancers.
(A) Examples of a stable Isl1-bound enhancer (class I, magenta) proximal to Chrnb4 and transient enhancers (class II, blue) proximal to Isl1 gene in nascent (day 5) and maturing (day 6) MNs. ChIP mapping of Isl1 is shown.
(B) Isl1 occupancy relative to Isl1-bound enhancers, sorted and ordered by Isl1 occupancy. These enhancers were grouped by the presence of stable (I), transient (II), day 5 only (III), and day 6 only (III) Isl1-bound enhancers. Middle panel shows relative changes in associated gene expression in day 5 or day 6 MNs relative to ESCs (Log2 FPKM).
(C) Expression profiles of selected genes developmentally regulated in ESCs, progenitors, and postmitotic MNs. Association of a gene with stable and/or transient enhancers is marked by a pink and purple dot, respectively.
(D) Number of genes associated only with stable, transient, or both classes of Isl1-bound enhancers, shown in (B).
(E) Boxplots of RNA expression on days 0, 5, and 6 for genes associated only with stable enhancers (n = 131) and only with transient enhancers (n = 378), shown in (D) (Table S4). Boxplots show the median (line), second to third quartiles (box), and 1.5× the interquartile range (whiskers). *p < 1 × 10−8, n.s. non-significant (p > 0.05); Wilcoxon rank-sum test.
(F) Top five non-redundant GO terms of genes shown in (D) (Table S5).
(G) H3K27ac intensity relative to active enhancers (n = 13,455), which were enriched for both H3K27ac and ATAC-seq intensity in the absence of Isl1 binding (subset of groups d, e, f, and g in Figure 1C), sorted and ordered by H3K27ac intensity. These enhancers were grouped by the presence of stable, transient, day 5 only, and day 6 only active enhancers.
(H) Number of genes associated only with stable, transient, or both classes of active enhancers, shown in (G).
(I) Same as in (E), except for genes shown in (H).
(J) Top five non-redundant GO terms of genes shown in (H).
See also Figure S3.
To investigate the regulatory effects of transient and stable Isl1 enhancers, we separated Isl1 co-associated genes into two groups: genes associated only with stable Isl1 enhancers (131 genes; Figure 3D) and genes associated only with transient Isl1 enhancers (378 genes). Both groups of genes were highly induced during motor neuron maturation (Figure 3E). Interestingly, genes associated only with transient Isl1 enhancers (e.g., Isl1, Robo1, Nrp1, Onecut1/2, and Ebf3) were significantly overrepresented in gene ontology (GO) terms for neuron-specific categories (neurite projection, synaptogenesis, and cell migration, p < 1 × 10−18), compared to stable Isl1-only-associated genes (p > 1 × 10−6; Figure 3F). These results suggest that genes playing an important role in neuronal maturation preferentially rely on stage-specific transient Isl1 enhancers instead of stable enhancers. We further refined the set of genes associated with transient Isl1 enhancers to a subset associated also with stage-specific enhancers transiently bound by Ngn2 in progenitors. Within this subset of genes, we observed even more significant enrichment for neuronal and developmental GO categories (p < 1 × 10−20; Figure S3B), indicating that genes highly relevant to neuronal development are associated with transient enhancers occupied by stage-specific combinations of programming TFs from late progenitor to postmitotic stages.
To determine whether this is a more general principle, we extended the analysis to active enhancers that are not bound by Isl1. We identified over 13,400 active enhancers in nascent and maturing motor neurons, which lacked Isl1 binding (Figure 3G). By assigning these enhancers to the nearest TSS, we obtained 443 genes associated only with stable enhancers (Figure 3H) and 694 genes associated only with transient enhancers. Both groups of putative target genes were highly induced in postmitotic motor neurons (Figure 3I). However, as observed above, genes associated only with transiently active enhancers were more significantly overrepresented in GO terms for neuron projection and cell migration (p < 1 × 10−11) and included many important motor neuron effector genes (e.g., Robo2, Dcc, Nrcam, etc.) compared to the group of genes associated only with stable active enhancers (p > 1 × 10−3; Figure 3J). Altogether, these results indicate that transiently active enhancers are broadly utilized regulatory features contributing to stable expression of developmentally regulated effector genes in postmitotic motor neurons.
Transient Isl1-Bound Enhancers Control Stable Motor Neuron Gene Expression
To test the ability of transient Isl1 enhancers to control gene expression in the context of motor neuron maturation in vivo, we utilized an electroporation system in the developing chick embryos. A nascent motor-neuron-specific enhancer (274 bp) at a neuropilin receptor gene Nrp2 (43 kb from the TSS) and a maturing motor-neuron-specific enhancer (143 bp) proximal to a glutamate-receptor-associated gene Gripap1 (32 kb from the TSS) were cloned upstream of a minimal promoter driving expression of a destabilized GFP reporter (Figures 4A and 4B). Electroporation of the construct carrying the nascent Isl1-specific enhancer resulted in strong GFP expression in nascent spinal motor neurons 24 hr post-electroporation, followed by rapid loss of GFP expression by 48 hr (Figure 4C, top). In contrast, the maturing Isl1-specific enhancer failed to activate reporter expression in nascent motor neurons at 24 hr post-electroporation (Figure 4C, middle). However, by 48 hr, we observed robust induction of the reporter in postmitotic neurons within the ventral spinal cord, recapitulating the in vitro inferred temporal pattern of gene regulation. Similar results were obtained with three out of four tested nascent Isl1-bound enhancers and two out of three tested maturing Isl1-bound enhancers, indicating that transient Isl1-bound enhancers regulate gene expression with high temporal resolution. Since we found that many stably expressed genes associate with independent stage-specific enhancers, we also tested regulatory activity of an artificial enhancer that combined one nascent and one maturing Isl1-bound enhancer (Figure 4B). Upon electroporation of this construct, we observed robust GFP expression that persisted from nascent to maturing motor neurons (Figure 4C, bottom), demonstrating that a combination of stage-specific enhancers is sufficient to maintain stable reporter gene expression in the chick spinal cord in vivo.
Figure 4. Spatiotemporal Specificity of Transient Isl1-Bound Enhancers in the Developing Neural Tube.
(A) Examples of a nascent MN-specific Isl1-bound enhancer (274 bp, cyan arrow) at Nrp2 and a maturing MN-specific Isl1-bound enhancer (143 bp, blue arrow) proximal to Gripap1. ChIP mapping of Isl1, H3K27ac, and Onecut1 is shown.
(B) Stage-specific enhancers were cloned individually or in combination into a destabilized GFP reporter plasmid containing a minimal promoter (minP).
(C) Analysis of the reporter gene expression 24 and 48 hr after electroporation of the plasmids into the Hamburger Hamilton stage 13 chick neural tube. A CMV-mCherry reporter (red) was co-electroporated with GFP plasmids to assess the efficiency of electroporation. Embryos were fixed at 24 hr (stage 18) and 48 hr later (stage 23) and stained with MN marker Hb9 (blue) and GFP (green) antibodies (n = 2). Scale bars represent 50 μm.
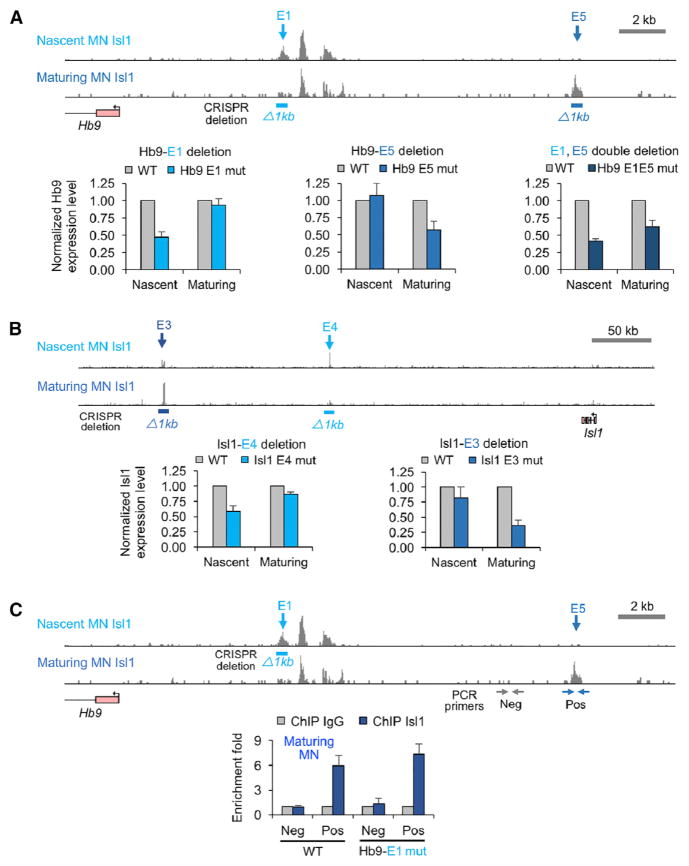
To test the requirement for stage-specific enhancers during motor neuron differentiation we generated a series of ESC lines carrying homozygous deletions of nascent and/or maturing motor neuron enhancers. We used the CRISPR/Cas9 system (Ran et al., 2013) to excise ~1 kb regions harboring nascent and maturing Isl1-bound enhancers associated with motor neuron genes Hb9 (enhancers E1 and E5, respectively) and Isl1 (enhancers E4 and E3, respectively) (Figures 5A and 5B). After differentiation of control and mutant ESC lines to nascent and maturing motor neurons, we examined transcription of Isl1 and Hb9 genes using qPCR. We determined that the deletion of transient enhancers resulted in a stage-specific decrease in Hb9 and Isl1 expression. Furthermore, double deletion of nascent and maturing motor-neuron-specific enhancers resulted in a persistent decrease in Hb9 expression (Figure 5A), indicating that individual nascent and maturing motor neuron enhancers are necessary for maintained expression of associated genes. Importantly, deletion of nascent Isl1-bound enhancers did not compromise Isl1 recruitment to the maturing enhancers, demonstrating functional independence of stage-specific enhancers (Figure 5C; Figure S4). Together, these results demonstrate that transient stage-specific Isl1-bound enhancers operate independently and that sequential activation of these enhancers in maturing neurons is sufficient and, in many instances, necessary for sustained expression of terminal effector genes during motor neuron maturation.
Figure 5. Transient Isl1-Bound Enhancers Are Necessary for a Stable Expression of Motor Neuron Genes.
(A) Top: the location of Isl1-bound enhancers proximal to Hb9 gene in wild-type (WT) MNs. 629 bp genomic DNA of a nascent MN-specific Isl1-bound enhancer (Hb9-E1) and 847 bp of a maturing MN-specific Isl1-bound enhancer (Hb9-E5) were deleted using CRISPR genome editing (Ran et al., 2013). Bottom: Hb9 expression levels measured by quantitative RT-PCR in nascent and maturing MNs (normalized to WT expression levels of Hb9). Error bars represent SD, n = 4, two independent differentiations.
(B) Same as (A) except transient enhancers proximal to Isl1 gene (1,413 bp of the maturing MN-specific enhancer [Isl1-E3] and 1,519 bp of nascent MN-specific enhancer [Isl1-E4]) were deleted. Error bars represent SD. n = 4, two independent differentiations.
(C) ChIP-PCR analysis of Isl1 binding to maturing MN enhancers in an ESC line containing the deletion of a nascent MN-specific enhancer (Hb9-E1). Horizontal arrows mark the position of positive (blue; Pos) and negative (gray; Neg) PCR primer sets. Error bars represent SD, n = 3, two independent differentiations.
See also Figure S4.
Isl1 Is Recruited to Stage-Specific Enhancers through Interactions with Onecut1 or Lhx3
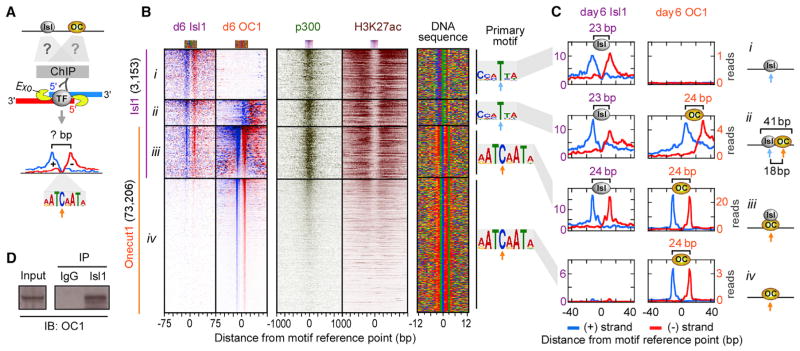
The highly dynamic changes in Isl1 binding patterns prompted us to examine mechanisms underlying the rapid relocation of Isl1- to Onecut1-bound sites upon Lhx3 downregulation. We hypothesized that, in maturing motor neurons, Isl1 might form heterodimers with Onecut1 and bind to sites containing adjacent Isl1 and Onecut1 binding motifs. To test this, we mapped Isl1 and Onecut1 binding sites with ChIP-exo at near single-base resolution (Rhee and Pugh, 2011) (Figure 6A). Indeed, at 33% of Isl1/Onecut1 co-bound regions (629/1,918) in maturing motor neurons, the two TFs occupied adjacent sites, containing appropriate Isl and Onecut binding motifs, respectively (subset ii in Figures 6B and 6C). Each of these sites also exhibited a unique 23 bp (Isl1) and 24 bp (Onecut1) ChIP-exo footprint, centered over the expected motifs. Interestingly, at two-thirds of co-bound regions (1,289/1,918), Isl1 and Onecut1 bound the same genomic site (subset iii). Closer examination of the ChIP-exo footprints revealed that both Isl1 and Onecut1 binding profiles at these sites were similar and matched the footprint and motif found at sites bound by Onecut1 alone (subset iv). These observations indicate that Isl1 might be recruited to a subset of maturing motor neuron enhancers through protein-protein interactions with Onecut1. Indeed, biochemical analysis revealed that Onecut1 co-immunoprecipitates from lysates of maturing motor neurons with Isl1-containing protein complexes (Figure 6D).
Figure 6. High-Resolution Mapping of Isl1 and Onecut1 Binding Sites in Maturing Motor Neuron Enhancers.
(A) ChIP-exo relies on the treatment of immunoprecipitated DNA with a 5′ to 3′ exonuclease. The 5′ ends of the digested DNA on the + and − strands are enriched at a fixed distance from the TF-DNA crosslinking sites, demarcating TF footprint regions protected from the exonuclease.
(B) ChIP-exo for Isl1 and Onecut1 and ChIP-seq for p300 and H3K27ac plotted relative to the TF motif reference point in day 6 maturing MNs, sorted and ordered by Isl1 and Onecut1 occupancy (Table S6). TF occupancy on the + strand (blue, left border) and − strand (red, right border) is shown. We classified four subsets: (1) 1,235 Isl1-only-bound sites (subset i), (2) 629 Isl1-bound sites next to Onecut1 (plots were reoriented to keep Onecut1 to the right side of Isl1) (subset ii), (3) 1,289 Isl1/Onecut1 co-bound sites (subset iii), and (4) 71,917 Onecut1-only-bound sites (10,426 sites on chromosomes 1 and 2 are shown) (subset iv). Right panel shows a color chart representation of the DNA sequence located ±12 bp from the motif reference point (cyan arrow for Isl and orange arrow for Onecut motif), which is the midpoint between the left and right border of TF.
(C) Composite plots of TF ChIP-exo profiles on the + and − strand (TF footprints) shown in (B). Right panel shows models of TF binding sites. The distance between adjacent Isl1 and Onecut1 motif reference points of heterodimers (subset ii) is 18 bp.
(D) Biochemical demonstration of Isl1 and Onecut1 interactions in maturing MNs. Immunoprecipitation (IP) of day 6 MN lysates with Isl1 antibody followed by a western blot (IB) with Onecut1 antibody. The detected band is ~51 kD, consistent with Onecut1 protein. Input is 5% of whole cell extract; IgG is a negative control. Shown is a representative blot (n = 2).
See also Figure S5.
Isl1 has been previously shown to form a complex with Lhx3 and a LIM domain-binding protein, Ldb1 (Lee et al., 2013; Mazzoni et al., 2013; Thaler et al., 2002), raising the possibility that, even in nascent motor neurons, Isl1 might be recruited to a subset of enhancers through protein-protein interactions with Lhx3. Examination of ChIP-exo profiles of Isl1 and Lhx3 in nascent motor neurons recovered adjacent 23 bp (Isl1) and 28 bp (Lhx3) footprints at 31% of co-bound regions (subset ii in Figures S5A and S5B). Interestingly, in the remaining co-bound regions, both TFs co-occupied the same 28 bp footprint centered over a 15 bp HD motif (subset iii in Figure S5B), which is identical to the footprint and motif recovered from sites bound by Lhx3 alone (subsets iv in Figure S5B). Our data indicate that protein-protein interactions of Isl1 with stage-specific TFs, Lhx3 and Onecut1, play an important role in the recruitment of Isl1 to transient motor neuron enhancers.
Previous studies established that Isl1 and Lhx3 engage in heterodimer complexes relying on interaction between Isl1 LIM-binding domain (LBD) and Lhx3 LIM domain (Lee et al., 2013; Mazzoni et al., 2013; Thaler et al., 2002). Since we observed mostly mutually exclusive recruitment of Isl1 to either Lhx3- or Onecut1-bound enhancers, we wondered whether Onecut1 and Lhx3 compete for the same Isl1 protein-binding domain. To address this question, we transfected ESCs with Onecut1- and Isl1-expressing vectors, together with increasing amount of Lhx3, and performed co-immunoprecipitation assays. We observed that Lhx3 is not co-immunoprecipitated with Onecut1 (Figure S5C), indicating that the two factors are not found in the same protein complexes. We also observed that Isl1 co-immunoprecipitation with Onecut1 is disrupted by increasing concentration of Lhx3, supporting the competitive binding of the two TFs to Isl1. Moreover, we functionally tested the competitive interactions among the TFs using luciferase reporter assays. We observed that activity of Isl1/Lhx3 or Isl1/Onecut1 enhancers was significantly diminished by overexpression of Onecut1 or Lhx3, respectively, presumably due to their ability to titrate Isl1 away from the enhancers (Figure S5D). Together, these results indicate that Onecut1 and Lhx3 interactions with Isl1 are mutually exclusive and that Onecut1 likely competes for the same Isl1 LDB domain that Lhx3 binds.
Isl1 and Histone Acetyltransferase Are Selectively Recruited to Clusters of Onecut1 or Lhx3
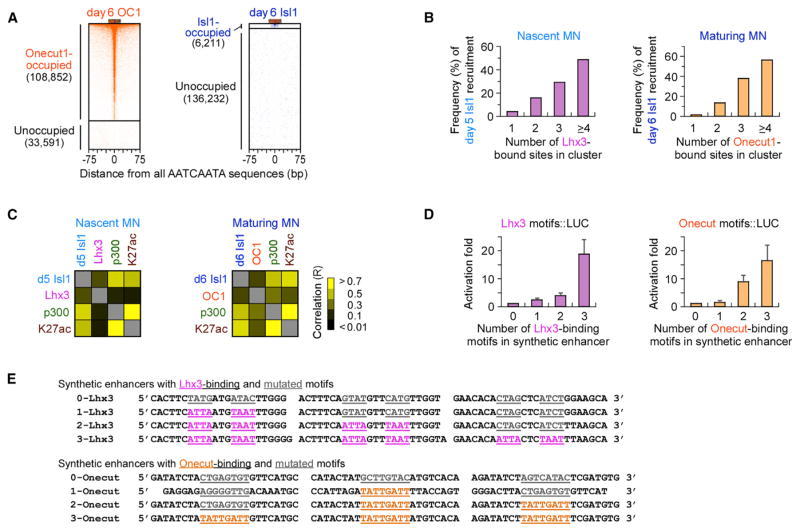
Global analysis of TF-bound sites in nascent and maturing motor neurons revealed a discrepancy between the large number of Lhx3- or Onecut1-bound sites and the relatively small number of sites bound by Isl1 (Figure 6B; Figure S5A). Throughout the mouse genome, there are over 142,000 occurrences of the 8 bp Onecut consensus motif (AATCAATA). Remarkably, ChIP-exo analysis revealed that more than 75% of these sites (>108,800 sites) were occupied by Onecut1 in maturing motor neurons (Figure 7A). In contrast, Isl1 was present at only 5% of Onecut1-occupied sites, prompting us to investigate what additional features contribute to the productive recruitment of Isl1 to a small subset of Onecut1 sites. Notably, we found that Isl1 is preferentially recruited to closely spaced clusters of Onecut1 TFs (defined as Onecut1 binding sites within a 200 bp window). The majority of genomic regions (228/413) containing four or more Onecut1-bound sites effectively recruited Isl1 in maturing motor neurons, whereas only 1% of regions containing a single Onecut1-bound site (657/61,248) recruited Isl1 (Figure 7B). Similarly, Isl1 was preferentially recruited to regions containing multimeric Lhx3 binding sites in nascent motor neurons.
Figure 7. Isl1 Is Preferentially Recruited to Enhancers Containing Clusters of Onecut1 or Lhx3 TFs.
(A) Onecut1 and Isl1 occupancy in 142,443 genomic sites containing canonical Onecut binding motif (AATCAATA), sorted by Onecut1 (left) and Isl1 (right) occupancy.
(B) Percentage of Lhx3-bound (in nascent iNIL MNs) and Onecut1-bound (in maturing MNs) enhancers that successfully recruited Isl1 in nascent and maturing MNs, respectively. The enhancers were subdivided based on the number of clustered Lhx3 or Onecut1 binding sites (number of identified binding sites within a 200 bp window; Table S7).
(C) Heatmaps of the Pearson correlation coefficients (R) for pairwise comparisons of TFs and H3K27ac intensity in nascent MNs shown in Figure S5A and in maturing MNs shown in Figure 6B (Table S3).
(D) Luciferase reporter assays with synthetic enhancers containing increasing number of Lhx3 or Onecut1 binding motifs in ESCs, transfected with Lhx3- and Isl1-expressing or Onecut1- and Isl1-expressing vectors (fold activation relative to a synthetic enhancer containing no Lhx3 or Onecut1 binding motifs). Error bars represent SD, n = 3.
(E) Synthetic enhancer sequences used for luciferase assays in (D). The linker sequences between the motifs were randomly synthesized.
See also Figure S6.
Next, we examined whether recruitment of Isl1 to a subset of enhancers correlates with enhancer activity. Above we described a strong correlation between Isl1 recruitment and accumulation of H3K27ac at transiently bound nascent motor neuron enhancers (Figures 2D–2F). Analysis of the histone acetyl transferase p300 and H3K27ac enrichment revealed that p300 and H3K27ac intensities correlated most closely with Isl1-binding occupancy, irrespective of whether these regions contained Lhx3 or Onecut1 binding (Figure 7C; Figure S6A). In contrast, genomic regions that were occupied by Lhx3 alone or by Onecut1 alone exhibited little or no p300 or H3K27ac enrichment. Together, these results indicate that Isl1 is preferentially recruited to enhancers containing clusters of Lhx3 or Onecut1 and that histone acetyl transferase p300 is selectively recruited to enhancers containing Isl1 TF.
To experimentally probe whether effective transcriptional activation depends on clustered Lhx3 or Onecut1 TFs, we performed luciferase reporter assays with synthetic enhancers containing increasing numbers of Lhx3 or Onecut binding motifs. Quantification of luciferase expression revealed a super-linear relationship between the number of binding sites within a clustered enhancer and the reporter gene expression, supporting our conclusion that efficient recruitment of trans-activator complexes to motor neuron enhancers depends on clustered multimeric Lhx3 or Onecut1 binding sites (Figures 7D and 7E). Importantly, efficient activation of enhancers containing clustered multimeric Lhx3 or Onecut1 binding sites depends on the presence of Isl1 TF (Figure S6B). Together, these results support a model in which Isl1 functions as an integrator of Lhx3 and Onecut1 binding patterns, functionally differentiating between singular and clustered binding sites in the genome.
Isl1 and Isl2 and Lhx3 and Lhx4 are paralogous pairs of TFs containing LIM and HD domains, whose expression pattern and function largely overlap in developing motor neurons (Briscoe et al., 2000; Sharma et al., 1998). Onecut1 and Onecut2 are also a paralogous pair of TFs containing CUT and HD domains, and their expression patterns and functions are partially overlapping during neural development (Francius and Clotman, 2010; Roy et al., 2012). We examined whether the behavior of Isl1, Lhx3, and Onecut1 can be extended to their paralogs by performing combinatorial luciferase reporter assays. While Isl2 and Onecut2 functioned in this simplified assay analogously to Isl1 and Onecut1, respectively (Figures S6C and S6D), Lhx4 turned out to be sufficient to activate reporter gene expression even in the absence of Isl1/2 TFs, suggesting that Lhx4 can employ alternative strategies for co-activator recruitment.
DISCUSSION
Outwardly Stable Gene Expression in a Dynamic Regulatory Environment
Embryonic development can be viewed as a multi-step process during which individual cells acquire unique and cell-type-specific gene expression profiles. Programming TFs play a pivotal role in this process by regulating the transition from a multipotent progenitor state to a terminally differentiated cell. The ability of programming TFs to induce expression of cell-type-specific effector genes and to establish a defined cell identity has been successfully exploited in cell reprogramming experiments (Mazzoni et al., 2013; Mong et al., 2014; Son et al., 2011; Vierbuchen et al., 2010; Wapinski et al., 2013). Recent convergence of biochemical, genomic, and computational approaches yielded maps of genomic regions bound by programming TFs (Lodato et al., 2013; Mazzoni et al., 2013; Stampfel et al., 2015; Wapinski et al., 2013). However, these maps remain relatively coarse to reveal the fine-grain organization of cis-regulatory elements controlling the combinatorial transcriptional inputs into cell-type-specific gene expression programs. Moreover, many mammalian programming TFs are expressed only transiently at the time when cell identity is being specified (Briscoe et al., 2000; Jessell, 2000; Novitch et al., 2001; Sharma et al., 1998), leaving open the question of what ensures continuous expression of cell-type-specific effector genes in the context of such a dynamic gene regulatory environment.
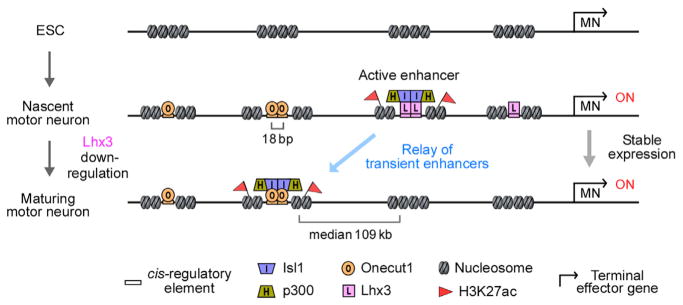
In theory, enhancers established by transiently expressed programming TFs might be maintained in postmitotic neurons by secondary TFs recruited to these accessible chromatin regions. Our high-resolution mapping analysis of TF binding sites, chromatin accessibility, and histone modifications in ESC-derived hypaxial motor neurons supports an alternative model (Figure 8). We report that many enhancers established by Isl1 and Lhx3 TFs in nascent motor neurons are truly ephemeral, in existence for less than 24 hr, losing not only TF binding, but also histone acetylation and chromatin accessibility. At the same time, Isl1 is recruited to a distinct set of enhancers bound by clustered Onecut1 TFs that become rapidly activated. We propose that the maintenance of motor neuron gene expression relies on a dynamic handover (relay) of regulatory control from one stage-specific enhancer to another.
Figure 8. Model of a Transient Enhancer Relay Controlling the Stable Motor Neuron Gene Expression Program.
We propose that expression of MN effector genes is maintained by transient enhancers bound by Isl1/Lhx3 in nascent MNs and Isl1/Onecut1 in maturing hypaxial MNs. The median distance between the closest day 5 and day 6 Isl1-bound sites is 109 kb (Figure S2D), indicating engagement of a large genomic territory in the regulation of MN gene expression. Isl1 is preferentially recruited to enhancers containing clusters of stage-specific TFs. We propose that stage-specific Isl1 binding leads to the recruitment of the histone acetyl transferase p300, acetylation of H3K27, and transient activation of enhancers. Following Isl1 release, enhancers are rapidly deactivated and decommissioned as manifested by the loss of p300, H3K27ac, and chromatin accessibility.
Indirect Recruitment of Isl1 to Enhancers through Protein-Protein Interactions
The rapidity with which Isl1 is displaced from its initial binding sites following Lhx3 downregulation and recruited to the new sites occupied by Onecut1 is notable. We speculated that this instability might be due to a strictly cooperative binding of Isl1/Lhx3 and Isl1/Onecut1 heterodimers to genomic sites containing pairs of heterodimer-specific motifs (LIM-LIM; LIM-Onecut) (Lee et al., 2013; Mazzoni et al., 2013). Indeed, we identified such binding pattern in one-third of Isl1/Lhx3 and Isl1/Onecut1 co-bound regions. Surprisingly, we discovered a significant fraction of sites at which Isl1 does not bind to its cognate motif; instead, Isl1 appears to be recruited to these sites through protein-protein interactions with Lhx3 or Onecut1. We propose that the observed tendency of Isl1 to engage in protein-protein interactions contributes to the rapid displacement of Isl1 from nascent motor neuron enhancers following Lhx3 downregulation and to its relocation to a new set of Onecut1-bound enhancers in maturing motor neurons.
While surprising, indirect recruitment of proteins traditionally recognized as DNA-binding TFs is not without precedent. An analogous mechanism has been reported for the recruitment of MEC-3, a LIM HD TF controlling postmitotic mechanosensory neuronal identity in C. elegans (German et al., 1992; Gordon and Hobert, 2015; Lichtsteiner and Tjian, 1995; Xue et al., 1993). Biochemical and genetic studies demonstrated that the HD DNA-binding domain of MEC-3 is dispensable for its normal function and that MEC-3 is recruited to relevant enhancers primarily through a protein-protein interaction with a POU TF, UNC-86. Besides enhancers to which Isl1 is recruited indirectly, we identified a number of Isl1-bound sites with the preferred Isl HD binding motif. However, since the motif is short and degenerate, we favor a model in which additional, yet-to-be-identified TFs interact with Isl1 to stabilize its binding to DNA. Ultimately, it will be interesting to systematically probe TFs with short and degenerate binding motifs to determine whether there is a larger group of TFs preferentially relying on protein-protein interactions for their recruitment to DNA.
Isl1 Operates as an Integrator TF, Functionally Integrating Binding Patterns of Anchor TFs
In contrast to Isl1, Onecut1 and Lhx3 TFs appear to bind many genomic sites independently of other TFs (75% of all preferred motifs in the genome are bound by Onecut1), but their recruitment is not sufficient to activate enhancers. We propose referring to such factors as anchor TFs—they strongly bind to DNA, yet possess limited inherent regulatory capacity, and their function therefore depends on combinatorial interaction with additional TFs or co-factors. We reason that Isl1 functions as a transcriptional co-activator for Lhx3 and Onecut1 that might directly or indirectly recruit HATs or other histone-modifying enzymes (Wang et al., 2016) to activate subsets of anchor TF-bound enhancers. It is of interest that Velasco et al. (2016) reported a subset of enhancers in transcriptionally programmed motor neurons co-occupied by Onecut2, Lhx3, and Isl1 TFs, raising a possibility that Isl1 might be effectively recruited not only by pure Lhx3 or Onecut1 clusters, but also to hybrid clusters composed of Lhx3and Onecut binding sites. Isl1 is a key TF involved in the specification of many cell types within the brain, pancreas, retina, heart, gut, and spinal cord. Previous biochemical studies revealed that Isl1 interacts with several cell-type-specific TFs to regulate expression of cell identity genes. Isl1 has been shown to partner with a bHLH TF Beta2 in pancreatic β cells (Zhang et al., 2009), a POU-HD TF Brn3b in retinal ganglion cells (Audouard et al., 2012), and with Phox2a in cranial motor neurons (Mazzoni et al., 2013). Ultimately, it will be interesting to determine whether Isl1 functions primarily as a co-activator of enhancers in all of these cell types and whether its partner TFs function primarily as anchor TFs.
As effective Isl1 recruitment positively correlates with the number of clustered anchor TFs in postmitotic spinal motor neurons, we propose that Isl1 operates as an integrator TF, reading and translating the density of anchor TFs into enhancer activity. Integrator TFs can add a layer of complexity to transcriptional networks and contribute to higher-order combinatorial regulation of gene expression. Thus, the observed “division of labor” between anchor TFs strongly interacting with their cognate DNA motifs and integrator TFs that functionally stratify anchor TF-bound regions might constitute an important logical component of transcriptional regulatory networks, contributing to specification of cell diversity in highly complex organs and organisms.
Our findings suggest that the process of neuronal maturation is controlled by cell-type-specific dynamic gene regulatory networks, engaging stage-specific enhancers associated with terminal effector genes. Mapping and understanding these regulatory networks might yield new strategies for the direct reprogramming of stem cells into mature or aging neurons, a critical step toward the development of more realistic models of late-onset neurodegenerative diseases.
EXPERIMENTAL PROCEDURES
Cell Culture
Motor neuron differentiation of mouse ESCs was performed as described previously (Tan et al., 2016; Wichterle et al., 2002; Wichterle and Peljto, 2008) with some modifications. Briefly, ESCs were seeded in ADFNK medium (day 0). Medium was supplemented on day 2 with 1 μM all-trans retinoic acid and 0.25 μM smoothened agonist (SAG) (Millipore, 566660). DAPT (5 μM, Selleckchem, S2215) was added to the culture medium on day 4 and day 5. Inducible iNgn2 and iNIL lines (Mazzoni et al., 2013) were differentiated using the same conditions and induced by adding 3 μg/mL of doxycycline (Clontech, NC0424034) on day 3 and day 2, respectively.
Immunocytochemistry
Embryoid bodies (EBs) were fixed with 4% paraformaldehyde, embedded in OCT, sectioned, and immunostained (antibodies against Isl1, RRID: AB_2126323; Hb9, RRID: AB_2145209). For quantification, an average of eight randomly selected EBs were analyzed per time point (average of 380 cells per EB) and scored for expression of motor neuron markers.
ChIP-Exo and ChIP-Seq
Cells were fixed with 1% formaldehyde and then processed as described previously (Rhee and Pugh, 2011). Briefly, cells were lysed, and chromatin pellets were isolated and fragmented by sonication, then subjected to immunoprecipitation using antibodies (Isl1, RRID: AB_1157901; Onecut1, RRID: AB_2251852; H3K27ac, RRID: AB_2118291; p300, RRID: AB_2293429; V5, RRID: AB_2556564). After washing, ChIP-seq samples were eluted. While ChIP-exo samples were still on the beads, the samples were ligated to a sequencing adaptor and digested by lambda exonuclease. Single-stranded DNA was eluted and converted to double-stranded DNA. A second sequencing adaptor was ligated to exonuclease-treated DNA ends, PCR amplified, gel purified, and sequenced by Illumina HiSeq 2000.
Data Analysis
All sequencing datasets were aligned to mm9 mouse genome assembly. We used the GEM (Guo et al., 2012) and the GeneTrack peak calling algorithms to identify regions enriched by TFs over background without DNA motif information from ChIP-seq and ChIP-exo datasets. If multiple peaks resided within 1 kb to each other, we merged these peaks into a single enhancer, and a midpoint of a peak with highest occupancy was considered an enhancer midpoint. Enhancer intensity (read counts) for TFs, histone modification mark, and ATAC-seq signal was plotted relative to the enhancer midpoint, and summed within ±50 bp for ChIP-exo and ±500 bp for ChIP-seq or ATAC-seq datasets. TF occupancies were normalized across time points using a median occupancy at the shared enhancers as a normalizer. See Supplemental Experimental Procedures for detailed experimental procedures and data analysis.
Supplementary Material
Highlights.
Enhancer landscape in stem-cell-derived motor neurons remains highly dynamic
Motor neuron effector gene expression is maintained by stage-specific enhancers
Isl1 is recruited to transient enhancers via stage-specific programming TFs
Isl1 translates the density of clustered TFs into enhancer activity
Acknowledgments
We thank Kristina Krebs for experimental support and members of the H.W. laboratory, Oliver Hobert, Stavros Lomvardas (Columbia University), and Shaun Mahony (Penn State University) for valuable discussions and comments. We are grateful to Martin Jacko for sharing a modified Cas9 expression plasmid, Phuong Hoang for sharing a PGK-Lhx3 expression plasmid, Susan Morton and Thomas Jessell (Columbia University) for sharing Isl1 and Lhx3 monoclonal antibodies, Richard Sherwood (Brigham and Women’s Hospital) for sharing Onecut1 cDNA, Samuel Pfaff (Salk Institute) for sharing Isl2 and Lhx4 cDNA, and Esteban Mazzoni (New York University) for sharing Onecut2 cDNA and for valuable discussions. Sequencing was performed at the MIT BioMicro Center. H.S.R. is the Howard Hughes Medical Institute Fellow of The Helen Hay Whitney Foundation. This work was supported by funding from the Project ALS foundation and by National Institutes of Health NINDS grants NS078097 and NS092043.
Footnotes
ACCESSION NUMBERS
The accession number for all sequencing data reported in this paper is GEO: GSE79561.
Supplemental Information includes Supplemental Experimental Procedures, six figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2016.11.037.
AUTHOR CONTRIBUTIONS
H.S.R., M.C., and G.C.T. performed ChIP-seq experiments. G.C.T. generated the inducible Ngn2 cell line. M.C. and E.V.B. performed ATAC-seq and DNase-seq experiments. H.S.R. and M.C. performed RNA-seq experiments. Y.G. performed computational analyses of deep sequencing data. H.S.R., M.C., D.K.G., and H.W. conceived the experiments and analyses. H.S.R., M.C., and H.W. co-wrote the manuscript.
References
- Audouard E, Schakman O, René F, Huettl RE, Huber AB, Loeffler JP, Gailly P, Clotman F. The Onecut transcription factor HNF-6 regulates in motor neurons the formation of the neuromuscular junctions. PLoS ONE. 2012;7:e50509. doi: 10.1371/journal.pone.0050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Res. 2015;43(W1):W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bonanomi D, Pfaff SL. Motor axon pathfinding. Cold Spring Harb Perspect Biol. 2010;2:a001735. doi: 10.1101/cshperspect.a001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francius C, Clotman F. Dynamic expression of the Onecut transcription factors HNF-6, OC-2 and OC-3 during spinal motor neuron development. Neuroscience. 2010;165:116–129. doi: 10.1016/j.neuroscience.2009.09.076. [DOI] [PubMed] [Google Scholar]
- German MS, Wang J, Chadwick RB, Rutter WJ. Synergistic activation of the insulin gene by a LIM-homeo domain protein and a basic helix-loop-helix protein: building a functional insulin minienhancer complex. Genes Dev. 1992;6:2165–2176. doi: 10.1101/gad.6.11.2165. [DOI] [PubMed] [Google Scholar]
- Gordon PM, Hobert O. A competition mechanism for a homeotic neuron identity transformation in C. elegans. Dev Cell. 2015;34:206–219. doi: 10.1016/j.devcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester ME, Murtha MJ, Song S, Rao M, Miranda CJ, Meyer K, Tian J, Boulting G, Schaffer DV, Zhu MX, et al. Rapid and efficient generation of functional motor neurons from human pluripotent stem cells using gene delivered transcription factor codes. Mol Ther. 2011;19:1905–1912. doi: 10.1038/mt.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci USA. 2008;105:20067–20071. doi: 10.1073/pnas.0806070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Park KW, Levine MS. Temporal regulation of single-minded target genes in the ventral midline of the Drosophila central nervous system. Dev Biol. 2013;380:335–343. doi: 10.1016/j.ydbio.2013.05.015. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Johnson SA, Harmon KJ, Smiley SG, Still FM, Kavaler J. Discrete regulatory regions control early and late expression of D-Pax2 during external sensory organ development. Dev Dyn. 2011;240:1769–1778. doi: 10.1002/dvdy.22672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Lee S, Cuvillier JM, Lee B, Shen R, Lee JW, Lee SK. Fusion protein Isl1-Lhx3 specifies motor neuron fate by inducing motor neuron genes and concomitantly suppressing the interneuron programs. Proc Natl Acad Sci USA. 2012;109:3383–3388. doi: 10.1073/pnas.1114515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Shen R, Cho HH, Kwon RJ, Seo SY, Lee JW, Lee SK. STAT3 promotes motor neuron differentiation by collaborating with motor neuron-specific LIM complex. Proc Natl Acad Sci USA. 2013;110:11445–11450. doi: 10.1073/pnas.1302676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtsteiner S, Tjian R. Synergistic activation of transcription by UNC-86 and MEC-3 in Caenorhabditis elegans embryo extracts. EMBO J. 1995;14:3937–3945. doi: 10.1002/j.1460-2075.1995.tb00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato MA, Ng CW, Wamstad JA, Cheng AW, Thai KK, Fraenkel E, Jaenisch R, Boyer LA. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS Genet. 2013;9:e1003288. doi: 10.1371/journal.pgen.1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Closser M, Morrison CA, Nedelec S, Williams DJ, An D, Gifford DK, Wichterle H. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat Neurosci. 2013;16:1219–1227. doi: 10.1038/nn.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mong J, Panman L, Alekseenko Z, Kee N, Stanton LW, Ericson J, Perlmann T. Transcription factor-induced lineage programming of noradrenaline and motor neurons from embryonic stem cells. Stem Cells. 2014;32:609–622. doi: 10.1002/stem.1585. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Francius C, Rousso DL, Seuntjens E, Debruyn J, Luxenhofer G, Huber AB, Huylebroeck D, Novitch BG, Clotman F. Onecut transcription factors act upstream of Isl1 to regulate spinal motoneuron diversification. Development. 2012;139:3109–3119. doi: 10.1242/dev.078501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampfel G, Kazmar T, Frank O, Wienerroither S, Reiter F, Stark A. Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature. 2015;528:147–151. doi: 10.1038/nature15545. [DOI] [PubMed] [Google Scholar]
- Tan GC, Mazzoni EO, Wichterle H. Iterative role of notch signaling in spinal motor neuron diversification. Cell Rep. 2016;16:907–916. doi: 10.1016/j.celrep.2016.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Velasco S, Ibrahim MM, Kakumanu A, Garipler G, Aydin B, Al-Sayegh MA, Hirsekorn A, Abdul-Rahman F, Satija R, Ohler U, et al. A multi-step transcriptional and chromatin state cascade underlies motor neuron programming from embryonic stem cells. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.11.006. Published online December 8, 2016 http://dx.doi.org/10.1016/j.stem.2016.11.006. [DOI] [PMC free article] [PubMed]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA, Muth K, Palmer J, Qiu Y, Wang J, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16:386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Shi Q, Guo T, Yang Z, Jia Z, Chen P, Zhou C. PDX1 and ISL1 differentially coordinate with epigenetic modifications to regulate insulin gene expression in varied glucose concentrations. Mol Cell Endocrinol. 2016;428:38–48. doi: 10.1016/j.mce.2016.03.019. [DOI] [PubMed] [Google Scholar]
- Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Peljto M. Differentiation of mouse embryonic stem cells to spinal motor neurons. In: Schlaeger T, Schamback A, Snyder EY, Wu J, editors. Current Protocols in Stem Cell Biology. John Wiley & Sons; 2008. pp. Unit 1H.1.1–1H.1.9. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Xue D, Tu Y, Chalfie M. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science. 1993;261:1324–1328. doi: 10.1126/science.8103239. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang WP, Guo T, Yang JC, Chen P, Ma KT, Guan YF, Zhou CY. The LIM-homeodomain protein ISL1 activates insulin gene promoter directly through synergy with BETA2. J Mol Biol. 2009;392:566–577. doi: 10.1016/j.jmb.2009.07.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.