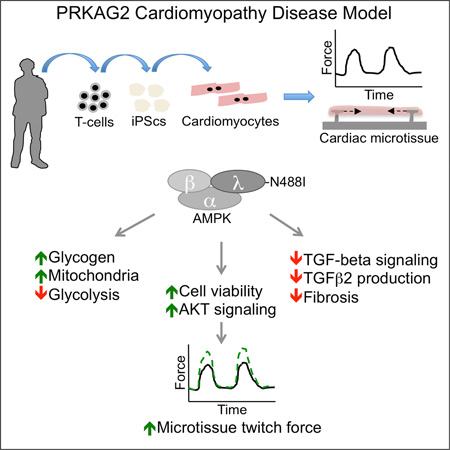
Summary
AMP-activated protein kinase (AMPK) is a metabolic enzyme that can be activated by nutrient stress or genetic mutations. Missense mutations in the regulatory subunit, PRKAG2, activate AMPK and cause left ventricular hypertrophy, glycogen accumulation and ventricular pre-excitation. Using human iPS cell models combined with three-dimensional cardiac microtissues, we show that activating PRKAG2 mutations increase microtissue twitch force by enhancing myocyte survival. Integrating RNA sequencing with metabolomics, PRKAG2 mutations that activate AMPK remodeled global metabolism by regulating RNA transcripts to favor glycogen storage and oxidative metabolism instead of glycolysis. Like patients with PRKAG2 cardiomyopathy, iPS cell and mouse models are protected from cardiac fibrosis, and we define a crosstalk between AMPK and post-transcriptional regulation of TGFβ isoform signaling that has implications in fibrotic forms of cardiomyopathy. Our results establish critical connections between metabolic sensing, myocyte survival and TGFβ signaling.
Graphical Abstract
Introduction
PRKAG2 is one of three regulatory subunits of the AMP-activated protein kinase (AMPK), and is highly expressed in the heart (Lang et al., 2000). The activity of AMPK is determined physiologically by energy status. Changes in AMPK activity have been observed in acquired forms of cardiac remodelling such as pressure overload (Tian et al., 2001) and inherited as autosomal-dominant left ventricular hypertrophy (LVH) caused by PRKAG2 missense mutations (Gollob et al., 2001). In vitro studies indicate that PRKAG2 mutations decrease the nucleotide-dependence of AMPK catalytic activity (Scott et al., 2004), resulting in gain of function. Once activated, AMPK regulates multiple metabolic pathways including increased glucose uptake by GLUT4 translocation (Kurth-Kraczek et al., 1999) and glycolysis by phosphofructokinase-2 regulation (Marsin et al., 2000). In addition to its metabolic effects, AMPK regulates diverse energy-dependent cellular functions including protein synthesis, autophagy, cytoskeletal dynamics and cell polarity (Hardie et al., 2012).
PRKAG2 mutations are identified in about 1% of patients with unexplained LVH (Murphy et al., 2005). PRKAG2 cardiomyopathy mimics some features of hypertrophic cardiomyopathy (HCM), a genetic disorder caused by mutations in contractile components of the sarcomere, but with notable differences. HCM but not PRKAG2 mutations exhibit myocyte disarray and markedly increased fibrosis (Ho et al., 2010). By contrast, PRKAG2 mutations cause electrophysiologic abnormalities such as atrioventricular conduction disease and mal-development of the annulus fibrosus that predisposes to ventricular pre-excitation (Arad et al., 2002). Some features of the PRKAG2 cardiomyopathy can be explained by alterations in glucose handling (Kim et al., 2014), which leads to increased glycogen accumulation in myocytes and LVH (Arad et al., 2002). Mechanisms for the paucity of myocardial fibrosis in PRKAG2 cardiomyopathy prior to end-stage disease (Poyhonen et al., 2015), remains an enigma.
We developed two human in vitro models of PRKAG2 cardiomyopathy to study AMPK function using myocytes (iPS-CMs) differentiated from induced pluripotent stem cells (iPScs) from patients and by TALEN genome engineering. We analyzed function in myocytes and cardiac microtissue (CMT) assays that better recapitulate cardiac architecture and myocyte maturation (Boudou et al., 2012; Hinson et al., 2015). We combined these in vitro analyses with mouse models to further probe the mechanisms that distinguish PRKAG2 from HCM mutations.
Results
PRKAG2 mutations increase AMPK activity, glycogen accumulation and AKT signaling resulting in iPS-CM hypertrophy
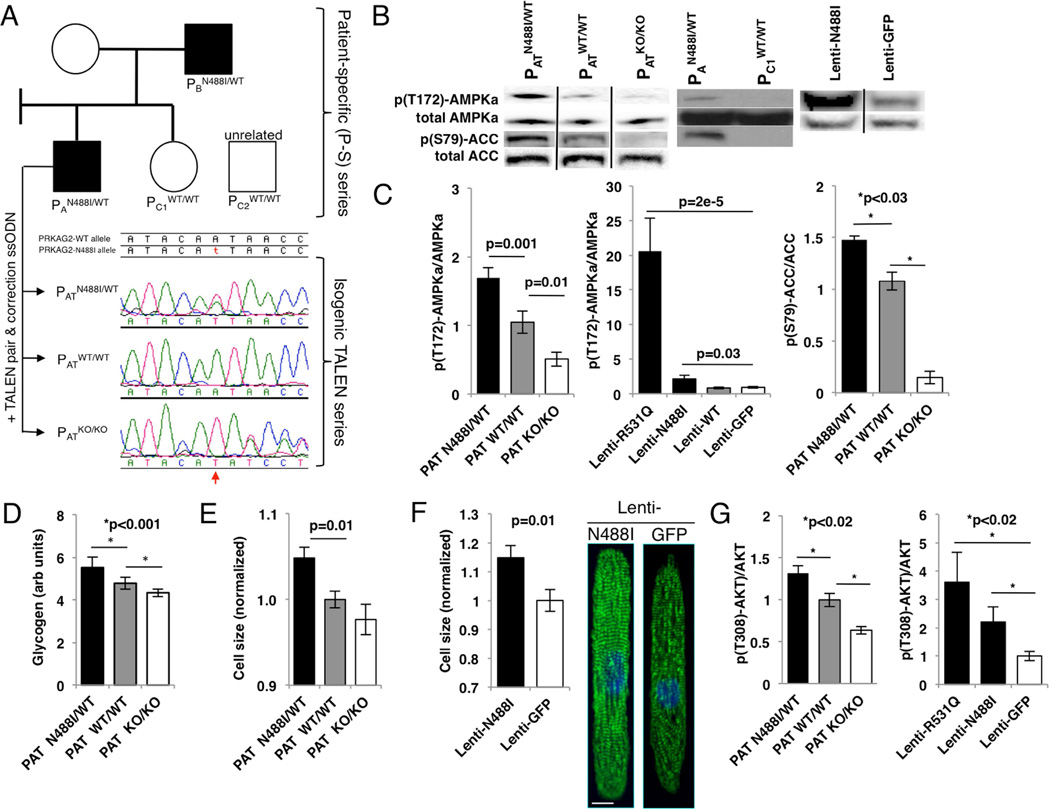
A patient-specific (P-S) iPSc model was engineered from members of a large family (Arad et al., 2002) with a heterozygous, missense mutation in PRKAG2 substituting asparagine for isoleucine at residue 488 (N488I). To create P-S iPScs (fig. 1A), we reprogrammed T-cells from two affected family members (PAN488I/WT and PBN488I/WT), one unaffected relative (PC1WT/WT) and one unaffected and unrelated control (PC2WT/WT). In parallel, we engineered a series of scarless, isogenic iPSc lines derived from PAN488I/WT by electroporation of TALE-nucleases (TALENs) (Ding et al., 2013) with wildtype single-stranded donor oligonucleotide that target sequences flanking the N488I mutation (figs. 1A, S1A, B). The TALEN isogenic series included an unmodified N488I mutation (PATN488I/WT), wildtype-corrected PRKAG2 (PATWT/WT) and homozygous null alleles in PRKAG2 (PATKO/KO).
Figure 1. PRKAG2 cardiomyopathy iPS-CMs recapitulate hypertrophy and glycogen accumulation due to AMPK activation.
(A) IPScs were engineered from two affected individuals (PAN488I/WT and PBN488I/WT), and a related (PC1WT/WT) and unrelated control (PC2WT/WT) (circle=female, square=male; shaded=PRKAG2 cardiomyopathy, unshaded=normal heart). PAN488I/WT iPScs were genome-edited with TALENs and a wildtype PRKAG2 oligonucleotide to create an isogenic series at the N488I locus (PATN488I/WT, PATWT/WT and PATKO/KO). Sanger tracings of PRKAG2 amplicons derived from the isogenic TALEN series (red arrow=A/T substitution). (B) Representative immunoblots probed with anti-p(T172)-AMPKα subunit, p(S79)-ACC and total AMPKα and ACC, and (C) quantified by densitometric analysis (n≥3). (D) Quantification of intracellular glycogen in iPS-CMs (n≥3). (E) iPS-CM size measured by normalized forward scatter (FSC) by flow cytometry (n≥15 differentiations) and by (F) pixel area on fibronectin lines (n≥20 myocytes); representative myocytes stained with anti-cardiac actin-in A (green) and DAPI (blue; scale bar=10 microns). (G) Quantification of anti-p(T308)-AKT by normalized densitometry (n≥3 lanes each) of immunoblots from lysates derived from iPS-CMs. Significance assessed by Student’s t-test (C–G) and error bars are mean +/− SEM (C–G).
IPScs were then differentiated to iPS-CMs and purified by metabolic selection. Since prior publications reported conflicting effects of N488I on AMPK activity in vivo (Arad et al., 2003; Sidhu et al., 2005), we initially measured phosphorylation of AMPKα at threonine 172. Both PAN488I/WT and PATN488I/WT iPS-CMs had similarly increased basal AMPKα phosphorylation compared to controls, while PATKO/KO has the lowest AMPKα phosphorylation (figs. 1B, C). We deduced that the TALEN isogenic series is a model of gain- and loss-of-function in AMPK activity. We extended these studies to characterize a second AMPK missense mutation, R531Q, which causes profound neonatal PRKAG2 cardiomyopathy (Burwinkel et al., 2005). Using a lentiviral system we expressed N488I and R531Q in iPS-CMs. While lenti-N488I increased AMPKα phosphorylation 2× compared to lenti-wildtype (WT) and green fluorescent protein (GFP) controls, lenti-R531Q increased AMPKα phosphorylation by over 20× (fig. 1C, middle panel). N488I also increased acetyl-CoA carboxylase (ACC) phosphorylation at the AMPK target site serine 79 (fig. 1C, right panel). These results confirm that N488I and R531Q mutations that cause PRKAG2 cardiomyopathy increase AMPK activity in proportion to the degree of cardiomyopathy severity.
Next, we used the TALEN isogenic series to model the consequences of gain- and loss-of-function in AMPK activity. Glycogen content in PATN488I/WT iPS-CMs was 17% higher than in PATWT/WT iPS-CMs, while the glycogen content in PATKO/KO iPS-CMs was the lowest (fig. 1D). We analyzed iPS-CM size by flow cytometry and after patterning iPS-CMs onto fibronectin lines to align sarcomeres to more closely resemble in vivo sarcomere structure (figs. 1E, F and S1J.). By either method, N488I iPS-CMs were larger. Mutant iPS-CMs also had increased insulin signaling, a recognized hypertrophic signal, as supported by increased AKT phosphorylation at threonine 308 (fig. 1G). These data confirm that LVH associated with PRKAG2 cardiomyopathy correlates with both glycogen accumulation and myocyte hypertrophy that is associated with AKT phosphorylation.
AMPK increases microtissue twitch force by enhancing myocyte survival
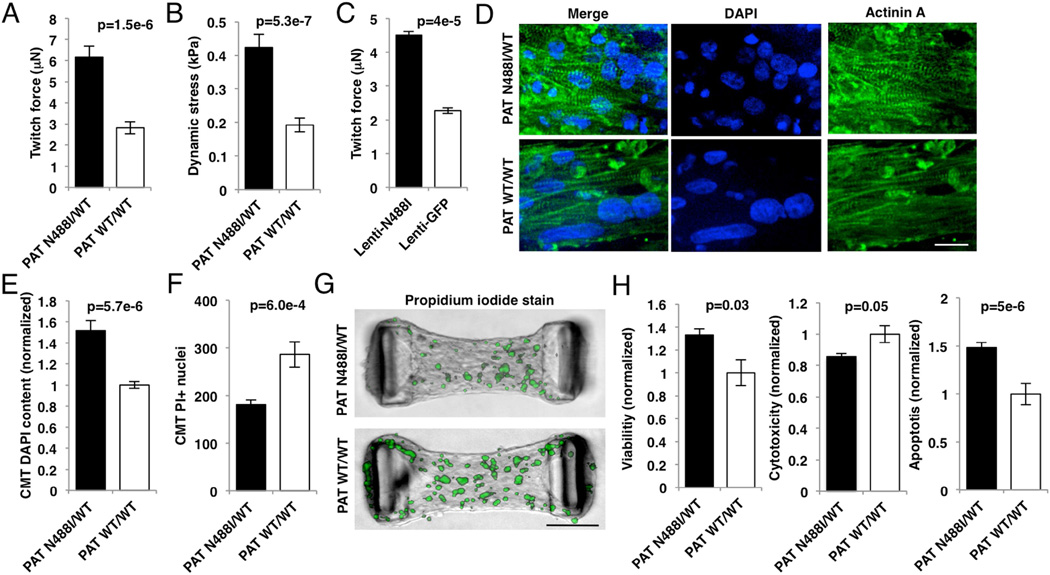
Unlike mutations in beta-myosin heavy chain that cause HCM by altering properties of contractile components (Debold et al., 2007), whether AMPK regulates cardiac force production remains unknown. To address this, we measured twitch force in cardiac microtissues (CMTs) that are composed of iPS-CMs (movies S1, 2). PATN488I/WT CMTs generated 6.16 µN of twitch force compared to 2.81 µN by PATWT/WT CMTs (p=1.5e-6; fig. 2A), an increase that remained after normalization for CMT width (fig. 2B). As twitch force in CMTs is dependent on cell composition and maturity, we expressed N488I or GFP by lentiviral transduction into iPS-CMs with identical iPS-CM content and made CMTs. Lenti-N488I similarly increased twitch force by 98% (p=4e-5; fig. 2C). Next, iPS-CMTs were stained with the sarcomeric isoform of actinin A and nuclear stain 4’,6-diamidino-2-phenylindole (DAPI) (fig. 2D) to identify structural changes that may explain increased CMT twitch force. Analysis of stained PATN488I//WT CMTs identified a 51% increase (fig. 2E; p=5.7e-6) in iPS-CM number despite controlling for iPS-CM seeding density. Since single cell traction force assays were not different in iPS-CMs with N488I (fig. S2A), and expression of maturity and chamber-specific transcript markers were also not regulated by N488I (figs.S2B–E), we conclude that PATN488I/WT CMTs have increased iPS-CM number per CMT as the major mechanism for increased CMT twitch force.
Figure 2. AMPK increases twitch force by enhancing viability in microtissues.
(A) Twitch force (µN) measured by cantilever displacement by CMTs generated from iPS-CMs and paced at 1Hz (n≥5 CMTs). (B) Tissue dynamic stress measured by twitch force normalized to CMT cross-sectional area (n≥5 CMTs). (C) Twitch force (µN) from IPS-CMs transduced with lentivirus expressing N488I or GFP (n≥5 CMTs). (D) Representative CMTs fixed and immunostained with anti-cardiac actinin A (green) to highlight sarcomeres, and DAPI (blue) to identify nuclei (scale bar=20 microns). (E) Normalized nuclear content in CMTs by DAPI staining (n≥10 CMTs). (F) Non-viable cells in live CMTs labelled with propidium iodide (PI; n≥10 CMTs). (G) Representative CMTs stained with PI (green) to identify dead cells (scale bar=100 microns). (H) IPS-CMs were cultured on tissue culture plates and analyzed for viability (left panel), cytotoxicity (middle panel), and apoptosis (right panel) (n≥6 replicates). Significance assessed by Student’s t-test (A–C, E–F and H) and error bars are mean +/− SEM (A–C, E–F and H).
To consider whether the increase in iPS-CM number was due to increased iPS-CM survival or proliferation, we stained live CMTs with propidium iodide (PI), which penetrates and binds DNA only in non-viable cells. As PATN488I/WT CMTs had 37% fewer PI-positive nuclei (p=6e-4; figs. 2F, G), we deduced that N488I increased iPS-CM survival in CMT assays, but did not alter proliferation rates since BRDU+ and cyclin B1 expression were not increased in PATN488I/WT iPS-CMs (figs.S2F, G). Consistent with increased survival, PATN488I/WT iPS-CMs cultured routinely in standard tissue culture were 33% more viable at baseline (p=0.03; fig. 2H, left panel) and after exposure to the cardiotoxic agent doxorubicin (p=0.02; fig. S2H). To determine whether the enhanced viability was due to inhibition of apoptosis, we measured caspase-3/7 cleavage in iPS-CMs. While overall cytotoxicity was decreased by 14% (p=0.05; fig. 2H, middle panel) in PATN488I/WT iPS-CMs consistent with PI staining, cell death by apoptosis was increased by 48% (p=5e-6; fig. 2H, right panel). These results indicate that AMPK enhances twitch force in CMTs by inhibiting non-apoptotic cell death.
AMPK regulates metabolism by transcript regulation
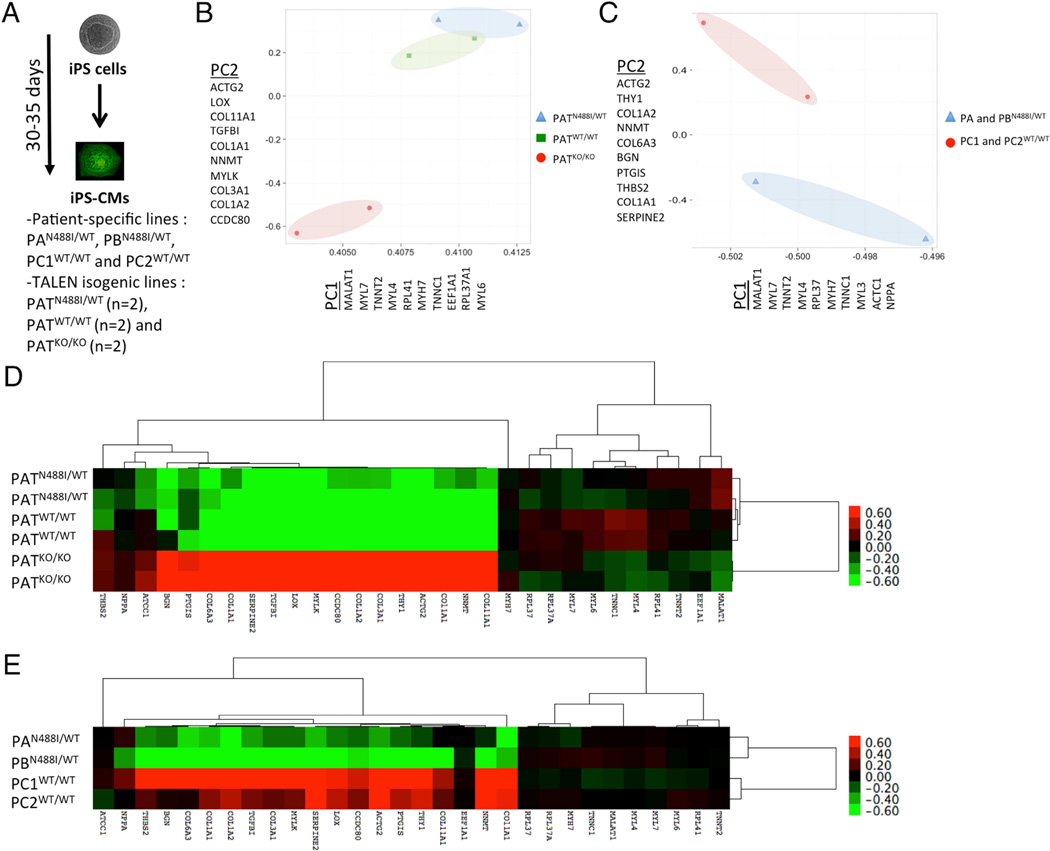
Since PRKAG2 cardiomyopathy is associated with life-long AMPK changes, we speculated that transcript regulation would reflect mechanisms of the genetic disorder. We analyzed gene transcripts by RNA sequencing (RNAseq) of iPS-CMs derived from P-S and TALEN isogenic cohorts (fig. 3A; tables S3, 4). We then performed unsupervised principle component analysis (PCA) of expression patterns to identify transcripts that separate cells within P-S and TALEN isogenic models (figs. 3B, C and table S5). In both datasets, iPS-CMs with N488I were separated from controls by the first two principle components. Principle component 1 (PC1) included components of the cardiac sarcomere including myosin heavy chains (MYH6 and 7), myosin light chains (MYL3, 4 and 7) and thin filament components (TNNT2, TNNC1 and ACTC1). Moreover, PC1 contained genes associated with hypertrophy such as ribosomal and translational transcripts (RPL41, EEF1A1, RPL37A1 and RPL37) and atrial-type natriuretic peptide (NPPA). Principle component 2 (PC2) contained gene transcripts involved in extracellular matrix (ECM) including collagens (COL11A1, COL1A1, COL3A1, COL1A2 and COL6A3) and ECM regulators (THBS2, LOX, BGN and SERPINE2). PC2 also contained gene transcripts involved in cytoskeletal dynamics (ACTG2 and MYLK). Analysis of combined PC1 and PC2 transcripts by hierarchical clustering and illustrated in a heatmap (figs. 3D, E) confirmed shared gene expression patterns between iPS-CMs with PRKAG2-N488I.
Figure 3. Gene expression analysis by RNAseq of TALEN and P-S iPS-CM models.
(A) Experimental design of RNA sequencing for purified P-S and TALEN isogenic iPS-CMs (pooled triplicates for P-S, and duplicates of pooled triplicates for TALEN isogenic). (B) Unsupervised principle components analysis (PCA) of all TALEN isogenic and (C) P-S iPS-CM gene transcripts separates cell populations by genotype by principle components 1 (PC1) and 2 (PC2). Gene components of PC1 and PC2 are identified by official gene symbol. A heatmap displays 30 gene transcripts from all PC1 and PC2 components for (D) TALEN isogenic (n=6 pooled triplicates) and (E) P-S iPS-CMs (n=4 pooled triplicates). Gene transcripts and iPS-CMs were organized by hierarchical clustering.
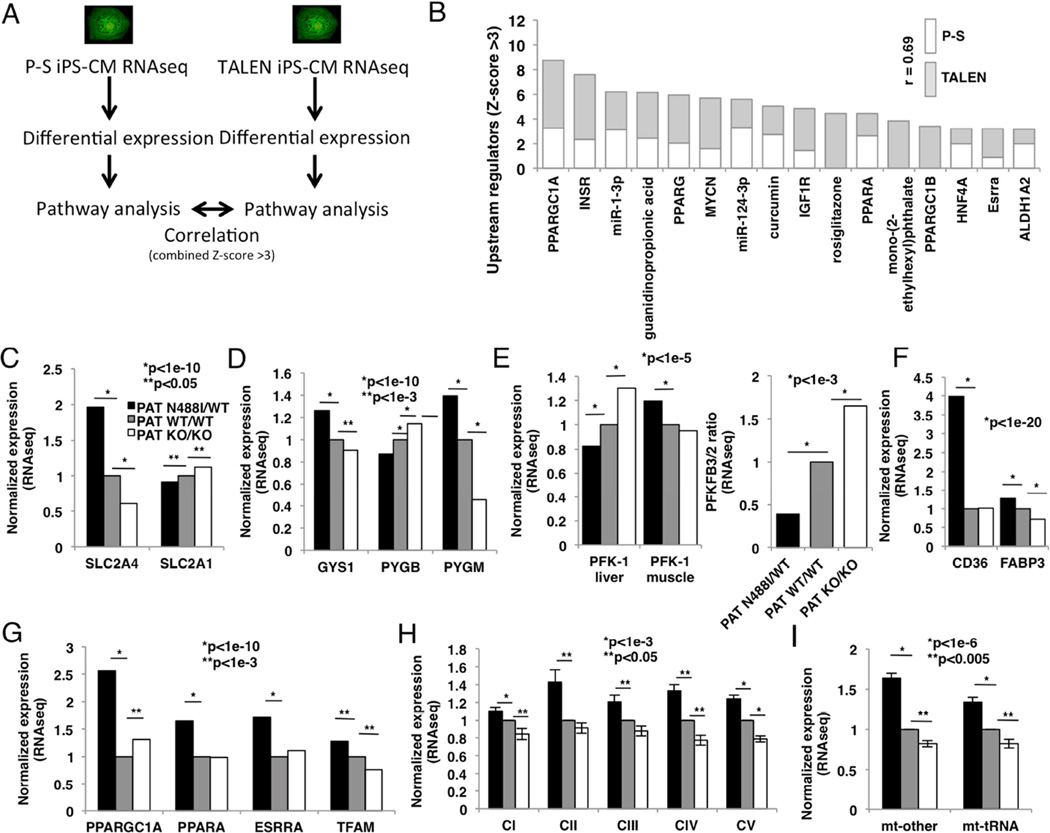
We proceeded to analyze differentially regulated gene transcripts from TALEN isogenic and P-S iPS-CM cohorts (fig. 4A). We identified 623 differentially regulated transcripts in the P-S cohort, and 1660 in the TALEN isogenic cohort (tables S3, S4). Differentially regulated transcripts were then analyzed by pathway analysis using Ingenuity Pathway Analysis (IPA) and ranked by Z-score of enrichment. Like PCA, analysis of pathways enriched in both iPS-CM cohorts identified highly correlated (r=0.69) pathways increased in both N488I iPS-CM models (fig. 4B). Key metabolic factors were enriched in iPS-CMs with PRKAG2-N488I including regulators of mitochondrial biogenesis and oxidative metabolism such as PGC-1α, PPARγ, PPARα, HNF-4α and estrogen-related receptor α. Chemical agonists of these pathways were also identified, including guanidinopropionic acid (Reznick et al., 2007), rosiglitazone (Lehmann et al., 1995), mono-(2-ethylhexyl) phthalate (Lovekamp-Swan et al., 2003). The N488I mutation increased RNA transcripts associated with increased microRNA activity that regulated myocyte differentiation (miR-124) (Cai et al., 2012) and pathologic cardiac hypertrophy (miR-1) (Ikeda et al., 2009) as well as transcripts downstream of signaling by the insulin receptor family (INSR and IGF1R).
Figure 4. Pathway analysis of RNAseq transcripts identifies metabolic and signaling pathways that regulate glucose handling and oxidative metabolism in iPS-CMs with PRKAG2-N488I.
(A) Experimental overview to identify transcript pathways regulated by N488I in P-S and TALEN cohorts. (B) Transcript pathways increased (Z-score >3) by N488I (P-S – no shade; TALEN isogenic – grey) associate with metabolic and growth factor signaling and are positively correlated (r=0.69). Transcript networks include PGC-1α/β (PPARGC1A and PPARGC1B), insulin receptor (INSR), PPARα/γ (PPARA and PPARG), IGF1R, hepatocyte nuclear factor 4α (HNF4A), estrogen receptor related α (ESRRA). Pathways regulated by microRNAs-1 and -124 and activators of mitochondrial biogenesis like guanidinopropionic acid, circumin, rosiglitazone and mono-(2-ethylhexyl) phthalate. (C) Transcripts of glucose transporters GLUT1 (SLC2A1) and the insulin-sensitive GLUT4 (SLC2A4), (D) glycogen synthase-1 (GYS1), isoforms of glycogen phosphorylase (PYGM (muscle) and PYGB (brain)), (E) glycolytic enzymes phosphofructokinase-1 (PFK-1) and the bifunctional glycolysis regulator 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases PFKFB2 and PFKFB3, and (F) fatty acid transporters CD36 and FABP3. (G) Regulators of mitochondrial biogenesis PGC-1α, PPARα, estrogen-related receptor α and mitochondrial transcription factor A (TFAM). Transcripts of (H) nuclear-encoded (merged) and (I) mitochondrial DNA-encoded (merged) genes that are components of respiratory chain complexes I–V (CI–V), tRNAs (mt-tRNAs) and all other genes (mt-other) encoded by the mitochondrial DNA. Data are normalized fragments per kilobase of transcript per million (FPKM) (C–I) and means +/− SEM (H, I). Significance assessed by Z-score of enrichment (B), Bayesian p-values (C–G) or Students t-test (H, I)
Because of increased glycogen storage, we analyzed glucose transporters and the rate-limiting enzymes that regulate glycogen content. Transcript data indicated that N488I mutation favored glycogen accumulation by coordinated regulation of key glucose handling transcripts. PATN488I/WT iPS-CMs have increased insulin-dependent GLUT4 (SLC2A4, fig. 4C) transcripts that are responsible for the majority of glucose transport in myocytes (Kraegen et al., 1993), but reduced levels of GLUT1 (SLC2A1). In parallel, transcripts encoding glycogen synthase (GYS1) were increased in PATN488I/WT iPS-CMs, while glycogen phosphorylase, the rate–limiting glycogen degradation enzyme, shifted from the more AMP-sensitive brain isoform (PYGB) to the less AMP-sensitive muscle isoform (PYGM) (Lehmann et al., 1995) (fig. 4D).
To explore how AMPK regulates glycolysis and fatty acid oxidation, we analyzed transcripts in these pathways. PATN488I/WT iPS-CMs exhibited an isoform switch in phosphofructokinase-1 (PFK-1), the rate-limiting step in glycolysis, to the less active muscle isoform from the liver isoform (fig. 4E, left panel), and favored expression of PFK-2/FBPase PFKFB2 instead of PFKFB3. These changes implied that glycolysis would be less active in PATN488I/WT iPS-CMs (fig. 4E, right panel). Both CD36 and FABP3, genes that regulate fatty acid uptake into myocytes, were increased in PATN488I/WT iPS-CMs (fig. 4F), a finding that is consistent with increased transcripts of regulators of mitochondrial biogenesis and oxidative phosphorylation such as PGC1-1α itself (fig. 4G). Both mitochondrial transcripts encoded by nuclear DNA and in this organelle were also increased (figs. 4H, I).
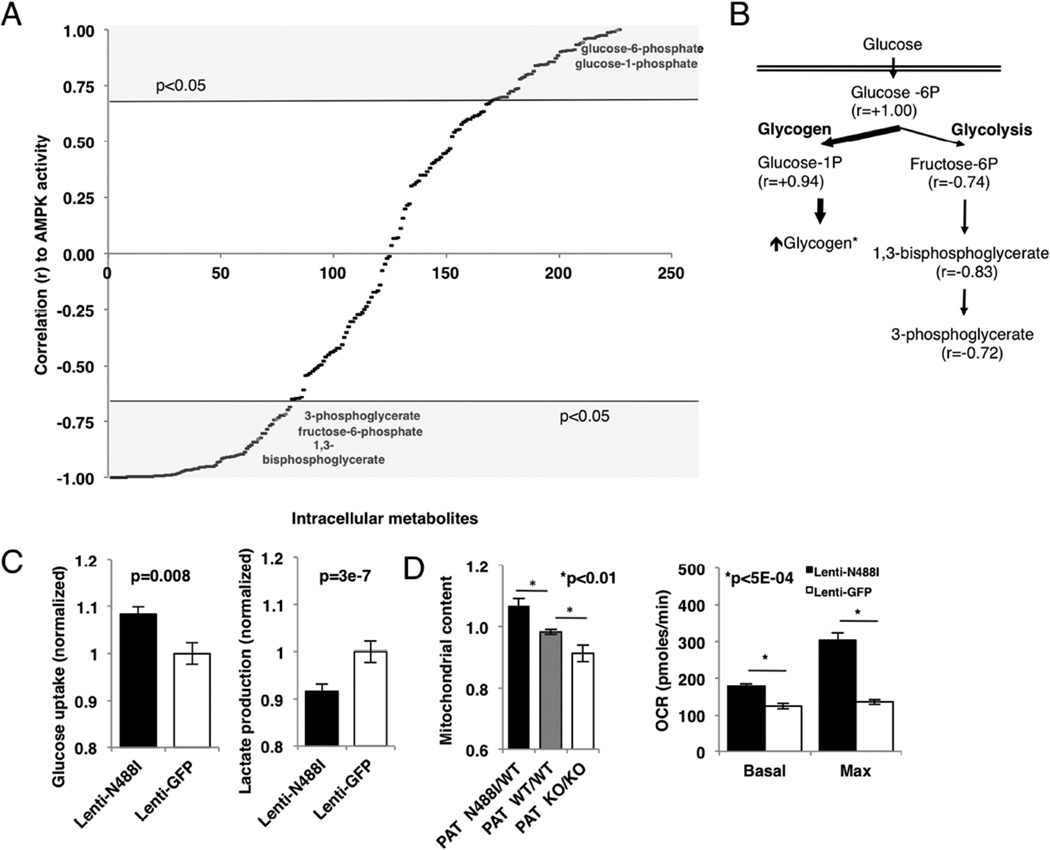
To determine the functional relevance of transcript changes, we measured steady-state levels of intracellular metabolites by LC-MS/MS, mitochondrial content and respiration, and glucose uptake and lactate production in conditioned media from TALEN isogenic iPS-CMs. We focused on pathways involved in glucose handling and oxidative metabolism, and identified metabolites that correlated with AMPK activity, as determined by the level of p(T172)-AMPKα (fig. 1C). Among 224 metabolites detected (fig. 5A and table S6), 70 were significantly increased (r>0.67) and 78 were significantly decreased (r<−0.67) in PATN488I/WT iPS-CMs. We analyzed metabolites associated with glucose handling first. Of four measured metabolites associated with glycolysis with significant differences (p<0.05), only glucose-6 phosphate was increased (r=1.00) in PATN488I/WT iPS-CMs. By contrast, the downstream glycolytic metabolites fructose-6-phosphate (r=−0.74), 1,3-bisphosphoglycerate (r=−0.83) and 3-phosphoglycerate (r=−0.72) were significantly decreased in PATN488I/WT iPS-CMs (fig. 5B). Consistent with this mismatch between glucose uptake and glycolysis, the glycogen precursor glucose-1-phosphate was similarly increased (r=0.94). To determine whether these steady state levels reflected changes in the kinetics of glucose handling, we measured glucose uptake in conditioned media from PATN488I/WT iPS-CMs compared to controls. Glucose uptake was increased by 8.3% (p=0.008; fig. 5C, left panel) in parallel to glucose-6-phosphate, and lactate production was decreased by 8.3% (p=3e7; fig. 5C, right panel) in parallel to reduction in three downstream glycolytic intermediates. Activation of AMPK by A769662 similarly regulated glucose and lactate metabolism in iPS-CMs (fig. S3B).
Figure 5. Metabolic assays by LC-MS/MS, glucose handling and mitochondrial function.
(A) 224 intracellular metabolites quantified by LC-MS/MS at steady state in iPS-CMs (n=3) and correlated (r) with AMPK activity. Metabolites shaded in grey satisfy p<0.05. (B) Schematic showing metabolites involved in glucose handling that are significantly correlated with AMPK activity. (C) Normalized glucose uptake and lactate production by iPS-CMs (n>3). (D) Normalized mitochondrial content measured by FACS analysis of Mitotracker-stained iPS-CMs, and mitochondrial function measured by basal and maximum oxygen consumption rate (pmoles/min). Data are means +/− SEM (C–D). Significance assessed by Pearson correlation (A) or Student’s t-test (C, D).
Metabolic intermediates associated with fatty acid oxidation identified by LC-MS/MS were increased, including carnitine (r=0.91), C2-, C5- and C8-long chain acylcarnitines (r=0.99, 0.69 and 0.70, respectively) and long chain acyl-CoA (r=0.77). Based on the increased transcripts encoding regulators of mitochondrial biogenesis (e.g., PGC-1α and PPARα), we suggest that increased mitochondrial content and function may account for increased steady-state levels of fatty acid intermediates. Indeed, both mitochondrial content and oxygen consumption were increased in PATN488I/WT iPS-CMs compared to isogenic controls (figs. 5D, S3A).
AMPK activation inhibits TGFβ signaling by inhibition of TGFβ–2 production in vitro
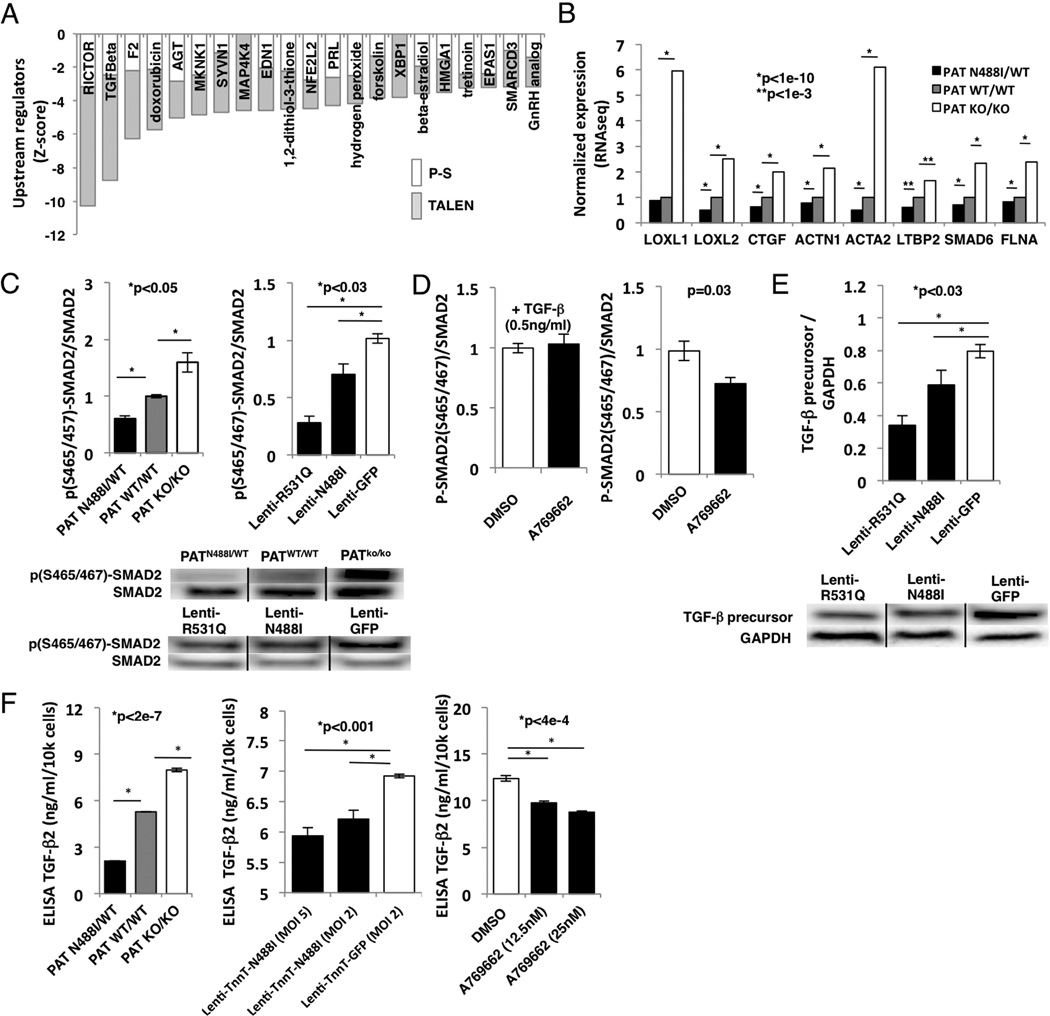
RNAseq data revealed changes in gene expression that predicted inhibition of distinct signaling networks (fig. 6A). Among these, we noted that N488I mutations reduced expression of transcript targets of TGFβ signaling and other pathways implicated in cardiac fibrosis, including rictor (RICTOR) (Li et al., 2015), thrombin (F2) (Carney et al., 1992), angiotensinogen (AGT) (Ruperez et al., 2003), endothelin-1 (EDN1) (Widyantoro et al., 2010) and the known cardiotoxic agent doxorubicin. Also, pathway regulators with functions downstream of non-canonical TGFβ signaling were predicted to be inhibited such as MAP kinase kinase kinase kinase 4 (MAP4K4) and MAP kinase signal-integrating kinase 1 (MKNK1). In addition, specific TGFβ transcriptional targets including genes that regulate collagen cross-linking (LOXL1 and LOXL2), growth factor (CTGF), cytoskeleton (ACTN1, ACTA2 and FLNA), and signaling (LTBP2 and SMAD6) were reduced in PATN488I/WT iPS-CMs (fig. 6B).
Figure 6. AMPK inhibits transcripts associated with cardiac fibrosis in vitro.
(A) Transcript pathways depleted (Z-score<-3) in PRKAG2-N488I iPS-CMs (P-S – no shade; TALEN isogenic – grey) include RICTOR, transforming growth factor-beta (TGFβ), thrombin (F2), doxorubicin, angiotensinogen (AGT), MAP kinase interacting kinase 1 (MKNK1) and endothelin-1 (EDN1). (B) Transcripts of TGFβ-regulated genes: lysyl oxidase-1 and -2 (LOXL1 and LOXL2), connective tissue growth factor (CTGF), smooth muscle alpha actinin (ACTN1), smooth muscle actin (ACTA2), latent TGFβ binding protein-2 (LTBP2), SMAD6 and filamin A (FLNA). (C) (left panel) Immunoblots probed with anti-p(S465/457)-SMAD2 and total SMAD2 and quantified by densitometry (n>3 per genotype) with (bottom panel) representative blots. (D) Immunoblots analyzed from IPS-CMs pre-treated with AMPK agonist A769662, and (left panel) stimulated with 0.5 ng/ml exogenous TGFβ for 30 mins or (right panel) no stimulation (n>3 per treatment). (E) Lysates of iPS-CMs transduced with lentivirus and probed with anti-TGFβ precursor or glyceraldehyde-3-phosphate dehydrogenase, GAPDH (n>3 per genotype), with (bottom panel) representative blots. (F) ELISA for TGF-β2 from conditioned media of iPS-CMs transduced with N488I expressed by troponin T promoter (MOI 2 and MOI 5), and by A769662 (12.5 and 25nM)(n>3 per condition). Data are normalized FPKM (B) and means +/− SEM (C–F). Significance assessed by Bayesian p-value (B) or Student’s t-test (C–F).
As reduced activation of TGFβ pathways could account for the unusual lack of fibrosis in PRKAG2 cardiomyopathy and the loss of integrity in the annulus fibrosis, we probed canonical TGFβ signaling pathways by measuring SMAD2 phosphorylation at serine 465/457 in iPS-CMs. SMAD2 phosphorylation was inhibited in proportion to AMPK activation in both the TALEN isogenic iPS-CMs and with lenti-N488I and -R531Q to control for maturation and purity (fig. 6C). As decreased SMAD2 phosphorylation could reflect mechanisms upstream or downstream of the TGFβ receptor, we re-measured SMAD2 phosphorylation after pre-treatment with the AMPK agonist A769662, in iPS-CMs treated with or without exogenous TGFβ. Only A769662 pre-treatment without exogenous TGFβ reduced SMAD2 phosphorylation by 28% (p=0.03; fig. 6D, right panel), which suggested a mechanism upstream of the receptor. Furthermore, TGFβ precursor protein was reduced in iPS-CMs expressing either the N488I or R531Q mutation (p<0.03; fig. 6E).
We next determined the TGFβ isoform that was regulated by AMPK by ELISA assays of conditioned culture media. PATN488I/WT iPS-CMs, and iPS-CMs with myocyte-specific lentiviral transduction of N488I or treated with A769662 dose-dependently reduced levels of TGFβ-2 (fig. 6F), but not TGFβ-1 (fig. S4A). By contrast, we did not observe a significant relationship between AMPK activity and TGFβ1 or TGFβ2 transcript levels (fig. S4B). TGFβ-3 was not expressed highly in iPS-CMs (fig. S4B). In summary, PRKAG2 mutations or A769662 that activate AMPK inhibit the production of TGFβ precursor protein and leads to reduced TGFβ-2 produced in iPS-CMs by post-transcriptional regulation.
AMPK activation inhibits TGFβ-regulated transcripts in vivo
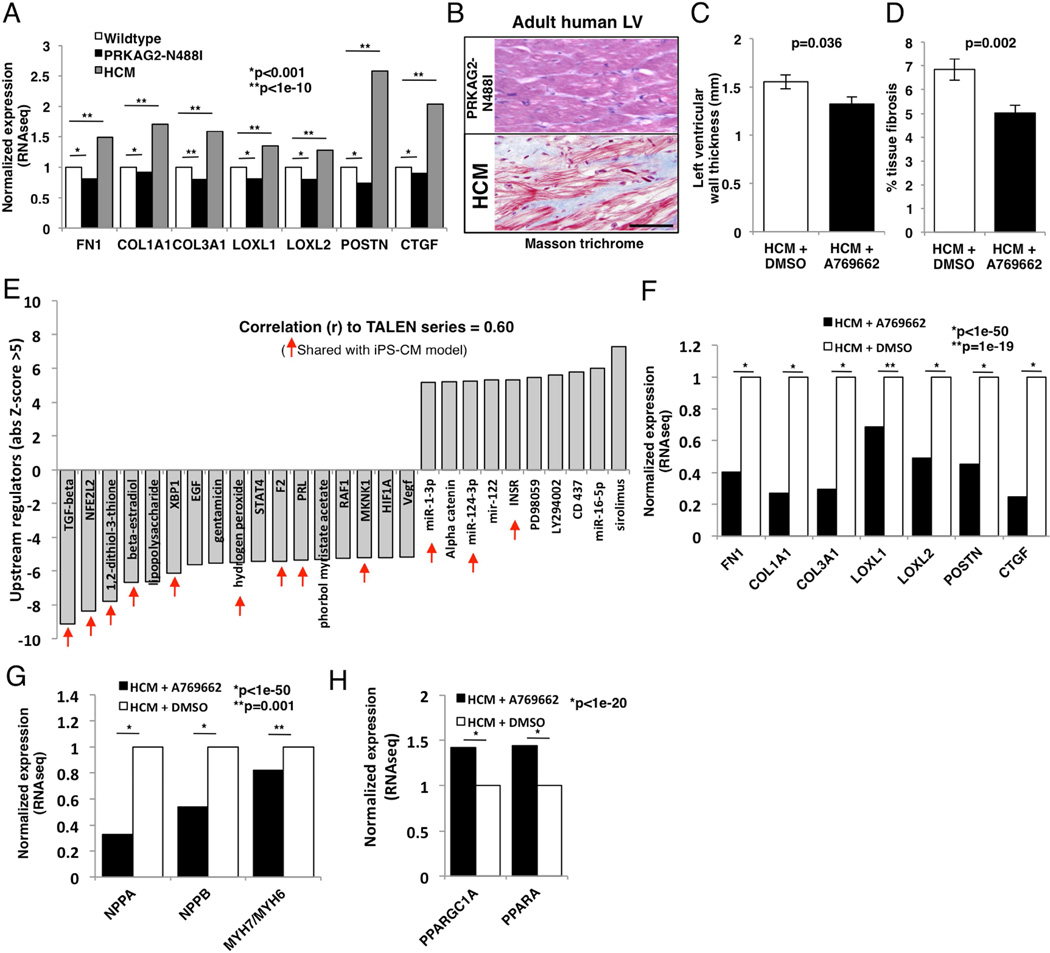
We hypothesized that AMPK activation might provide a therapeutic strategy to inhibit pathological forms of cardiac remodeling that are associated with fibrosis such as in HCM where TGFβ signaling is increased (Teekakirikul et al., 2010). We initially analyzed transcripts associated with fibrosis in RNAseq data from pre-hypertrophic mice with PRKAG2 cardiomyopathy (Arad et al., 2003) with RNAseq from wildtype and pre-hypertrophic HCM mice (MHCR403Q/+) (Geisterfer-Lowrance et al., 1996). Similar to iPS-CM models, N488I mice had reduced expression of TGFβ targets that are associated with fibrosis including extracellular matrix components and regulators (fig. 7A), which is in contrast to HCM mice. Human histopathology (fig. 7B) was consistent with these data. LV sections stained with Mason trichrome showed little fibrosis in a patient with PRKAG2 (N488I) cardiomyopathy compared to HCM (MYBPC3+/−). Therefore, while lifelong, constitutive AMPK activation leads to the PRKAG2 cardiomyopathy, a method to provide a tunable AMPK activation could achieve reduction in gene transcripts involved in fibrosis that are regulated by TGFβ in vivo.
Figure 7. AMPK prevents cardiac fibrosis and pathologic hypertrophy in vivo.
(A) Transcripts by RNAseq of TGFβ targets from mouse left ventricular tissue with pre-hypertrophic PRKAG2 cardiomyopathy (N488I; black bars), pre-hypertrophic HCM (grey bars) and wildtype controls (white bars). B) Representative formalin-fixed sections stained with Masson trichrome (fibrosis=blue) in adult human LV tissue from patient PAN488I/WT compared to HCM (MYBPC3 mutation). Scale bar=100 microns. (C) Pre-hypertrophic HCM mice were treated with A769662, and hypertrophy was measured by LV wall thickness (n>15 mice per condition). (D) Tissue fibrosis (%) measured by Masson trichrome staining (N>35 sections from >3 mice per condition). (E) Transcript pathway analysis of transcripts from HCM mice treated with A769662 or carrier were identified and compared to transcript pathways identified from iPS-CM models (shared =red arrows). (F) Transcript levels of TGFβ targets, (G) pathological hypertrophy genes and (H) mitochondrial biogenesis factors from HCM mice treated with A769662. Data are normalized FPKM (A, F–H) and mean +/− SEM (C, D). Significance assessed by Bayesian p-value (A, F–H) or Student’s t-test (C, D).
Next, we tested whether the AMPK agonist A769962 could prevent the development of hypertrophy and fibrosis in HCM mice. We treated six week-old pre-hypertrophic HCM mice with A769662, using a once instead of twice daily dose that did not change weight, blood pressure, or glucose (Cool et al., 2006). A769662-treated HCM mice developed less hypertrophy (1.32 mm versus 1.55 mm, p=0.04; fig. 7C) and tissue fibrosis (5.02% compared to 6.85%, p=0.002; fig. 7D) compared to carrier-treated HCM mice. LV transcriptional analyses of A769662-treated HCM mice identified 2768 differentially expressed genes compared to controls (table S7). Comparison of pathway analyses (IPA) of differentially expressed genes from the in vitro TALEN isogenic iPS-CM cohort with in vivo A769662-treated HCM mice, were highly correlated (r=0.60). The activation states of multiple pathways were common to both including TGFβ that was the most negatively regulated pathway in vivo (Z-score of −9.1; fig. 7E). Moreover, A769662-treated HCM mice had markedly reduced cardiac expression of the same TGFβ targets, including extracellular matrix components and regulators reduced in pre-hypertrophic N488I mice (fig. 7F). In addition, A769662-treatment reduced hypertrophic gene expression (NPPA, NPPB), normalized myosin isoform expression (MYH7/MYH6 ratios), and increased oxidative transcripts (e.g. PGC-1α and PPARα) in HCM hearts (figs. 7G, H) and in control hearts (fig. S5).
Discussion
AMPK coordinates metabolic sensing with a diverse group of energy-dependent cellular functions. The integration of biochemical, transcriptional, and functional datasets using human iPS-CMs, microtissues and mouse models allowed us to deconvolute how mutations produce the phenotypes observed in PRKAG2 mutations. Our analyses of human iPS-CMs provide evidence that PRKAG2 mutations increase myocyte size in correlation with increased glycogen content and by activating AKT signaling, which is consistent with findings in PRKAG2 mouse models (Kim et al., 2014). We further demonstrate that regulation of glucose metabolism that results in glycogen accumulation by PRKAG2 mutations is due to coordinated changes in transcript abundance of key regulators of glucose handling. While other studies have defined the acute or chronic effects of AMPK activity on specific factors (Bultot et al., 2012; McGee et al., 2008), this study is the first to define the transcriptional network that drives glycogen accumulation. For example, we find increased transcripts encoding glycogen synthase, and isoform shifts in glycogen phosphorylase, phosphofructokinase, and glucose transporters that parallel changes in steady-state metabolomics and glucose handling kinetics. On the other hand, we identify mitochondrial biogenesis factors increased such as PGC-1α and PPARα, which parallel increased mitochondrial content and respiration. This pattern of metabolic remodelling is opposite to the changes associated with heart failure (Doenst et al., 2013), which is reflected in the iPS-CMs by reduced transcripts associated with heart failure such as natriuretic peptides.
Unexpectedly, the metabolic and transcriptional changes induced by mutational activation of AMPK were associated with increased viability leading to increased twitch force in microtissues. These observations are consistent with in vivo studies that indicate increased AMPK activation can protect the heart from ischemic stress (Ofir et al., 2008). Several mechanisms could account for the improved stress-response, including increased AKT signaling observed here, glycogen content that would provide a ready supply of glucose, that is the preferred energy substrate in stressed myocytes (Ofir et al., 2008), increased mitochondrial biogenesis and activation of autophagy (Baskin and Taegtmeyer, 2011). We recognize that the heightened viability and twitch force of mutant myocytes contrasts with the poor clinical courses of adult patients and mice with PRKAG2 cardiomyopathy (Murphy et al., 2005). We suggest several observations that account for these differences. First, cardiac conduction system cells that were not modelled in these studies with PRKAG2 mutations progressively develop dysfunction and contribute substantially to in vivo phenotypes. Second, late-onset cardiac deficits of PRKAG2 mutations may not be well modelled in short-term tissue culture studies. Finally, as both embryonic myocytes and immature iPS-CMs routinely metabolize glucose for energy, these cells may have multiple adaptations to accommodate excess glucose and glycogen.
Unlike most other forms of LVH including HCM, PRKAG2 cardiomyopathy is distinguished by a remarkable absence of myocardial fibrosis. Attenuated TGFβ signaling, that we identified in both iPS-CMs and mouse models of PRKAG2 cardiomyopathy, appears to account for the absence in fibrosis. While others have implicated AMPK in TGFβ in other tissues (Lee et al., 2013; Lim et al., 2012), we demonstrate that AMPK provides post-transcriptional regulation of TGFβ-2, the most abundant TGFβ isoform produced by iPS-CMs. We suggest that reduced TGFβ-2 is also a contributing mechanism for ventricular pre-excitation arrhythmias that patients with PRKAG2 mutations. The annulus fibrosus, a band of connective tissue that insulates and prevents direct electrophysiologic connections between the atria and ventricles, develops from the embryonic cardiac cushions in response to TGFβ-2 signals (Azhar et al., 2009). Consistent with this TGFβ-2 dependence, the annulus fibrosus is hypomorphic in PRKAG2 cardiomyopathy mouse models (Arad et al., 2003) and exhibits ventricular pre-excitation and arrhythmias. By activating AMPK and reducing TGFβ-2 production, we propose that PRKAG2 mutations that are selectively expressed in the heart may cause these developmental abnormalities.
Finally, we show that the potential for harnessing AMPK-dependent changes in TGFβ signaling. We demonstrate that A769662 prevented fibrosis and cardiac hypertrophy in HCM mice. Consistent with these observations, others have reported that AMPK inhibition exacerbates cardiac remodelling and dysfunction after thoracic-aortic banding (Shimano et al., 2010) while exercise promotes a reduction in cardiac fibrosis through AMPK-dependent mechanisms (Ma et al., 2015). AMPK activation also increased expression of PGC-1α and PPARα potentially providing an improved metabolic profile to HCM hearts. We suggest that pulsatile AMPK activation by direct agonists, such as A769662, has the potential to reduce myocardial fibrosis that accompanies adverse remodelling and promotes arrhythmias in HCM and other cardiomyopathies.
In conclusion, the critical linkages between metabolism and transcript regulation that we report provide novel insights into AMPK signaling and the pathophysiologic mechanisms for PRKAG2 cardiomyopathy. The global transcriptional remodelling of metabolism that favors glycogen accumulation instead of glycolysis and crosstalk between AMPK activity and TGFβ signaling point to the complex roles by which altered AMPK activity impacts cardiac function. Small molecules that target AMPK activation are under development for several conditions, albeit with concerns that these may incite cardiomyopathy, similar to PRKAG2 mutations (Zaha and Young, 2012). Our findings indicate the considerable potential for molecules that provide tailored and intermittent AMPK activation to attenuate fibrosis and adverse remodelling in cardiomyopathies and many other forms of heart disease.
Experimental Procedures
IPSc production, differentiation and PRKAG2 lentivirus
Patient samples were obtained after informed written consent using protocols approved by the Institutional Review Board of Partners HealthCare. Patient-specific iPScs were produced from T-cells (Loh et al., 2009) using STEMCCA lentivirus. IPScs were screened for pluripotency markers (figs.S1F–H), copy number variants, karyotyping (fig. S1G) or virtual karyotyping using Illumina HumanOmniExpress-12v1 arrays. iPScs were maintained in feeder-free conditions, and were differentiated to the CM lineage by small molecules (Lian et al., 2012). IPS-CMs were purified using metabolic selection (Tohyama et al., 2013) and studied on day 30–40 after initiation of differentiation. Following differentiation, iPS-CM purity was determined by FACS analysis (>90% TnnT+) or by morphology. We amplified human PRKAG2 from a human iPS-CM cDNA library, and inserted N488I and R531Q by site-directed mutagenesis (Agilent). We inserted sequence-verified PRKAG2 into the third generation lentivirus backbone pLenti (Addgene) and transduced iPS-CMs with MOI of 2.
TALEN genome editing
TALE nucleases were designed to flank the targeted mutation at N488I in human PRKAG2 (fig. S1A) as previously described (Ding et al., 2013). Briefly, TALEN genomic binding sites 15bp in length (table S1A) were chosen within 30 bp of the nucleotide change required during homologous recombination. The TALEN pair along with a correction, 50-mer ssODN (table S1A) that encoded the wildtype PRKAG2 sequence, were electroporated into PAN488I/WT iPScs. GFP/RFP double-positive iPScs were sorted (fig. S1C), expanded and Sanger sequenced (tables S1B, S2) to confirm either scarless correction to wildtype (PATN488I/WT) or in/dels that created a homozygous loss-of-function line (PATKO/KO). Genotypes were validated with RFLP analysis using restriction enzyme digest with MSE1 (NEB) (figs.S1D, E). Experimental overview summarized in fig. S1B.
RNA sequencing and computational methods
Total RNA was isolated from day 30–40 iPS-CMs or 3-week old mouse left ventricular tissue using Trizol (Invitrogen). CDNA libraries were constructed using Superscript III First-Strand synthesis (Invitrogen). To reduce biologic variation among samples, cDNAs were pooled from at least biological triplicates for each sample and cDNA libraries were constructed using Nextera XT DNA sample prep (Illumina). Libraries were prepared and sequenced as described previously (Christodoulou et al., 2011) and were aligned with STAR (Engstrom et al., 2013). Differentially regulated transcripts were analysed, and transcript network analysis was generated through the use of Ingenuity Pathway Analysis (IPA, QIAGEN). For hierarchical cluster analysis, we used Cluster 3.0.
Cardiac microtissues and microcontact printing
CMTs were prepared as previously described (Boudou et al., 2012). Micropatterned substrates were prepared as previously described (Tan et al., 2004).
Cell size and flow cytometry assays
Cell size was measured by determining cell area on micropatterned surface using ImageJ (National Institutes of Health). IPS-CMs were disassociated and analyzed by flow cytometry using a LSR Fortessa analyzer (BD). Cell size was determined by analyzing forward scatter (FSC). Mitochondrial content was measured by staining iPS-CMs with Mitotracker Green FM (Invitrogen) and analyzed using flow cytometry according to manufacturer’s protocol. All FACS assays were done with at least biological triplicates.
LC-MS/MS metabolomics and other metabolic assays
Intracellular metabolites were extracted from day 30–40 iPS-CMs differentiated from TALEN isogenic iPScs. All assays were done in biological triplicates. We performed metabolomic profiling using liquid chromatography-tandem mass spectrometry (LC-MS/MS). For extended methods, see supplemental methods. Seahorse XF24 (Seahorse Bioscience) was utilized to measure mitochondrial oxygen consumption according to manufacturers protocol. Glucose uptake was measured by biochemical methods from conditioned media using the glucose oxidase assay (Sigma), while lactate production was measured using a lactate dehydrogenase assay (Cayman). All metabolic assays were done with at least biological triplicates.
iPS-CM viability and cytotoxicity assays
Day 30–40 iPS-CMs were cultured in two dimensional tissue culture formats and were stained for viability with ApoTox-Glo assay (Promega). For doxorubicin (Tocris) experiments, iPS-CMs were exposed to doxorubicin (6µM) for 12 hours prior to viability assay. For CMT viability experiments, propidium iodide (Invitrogen) was used according to manufacturer’s protocol. PI+ nuclei were counted manually using ImageJ (NIH).
Western blotting, tissue staining and ELISA
IPS-CM lysates were solubilised in RIPA buffer followed by western blotting using Cell Signaling antibodies: AMPKα duet, ACC duet, AKT Duet, SMAD2 duet, cardiac actinin A (Abcam), TGFβ and GAPDH. ImageJ (NIH) was used to quantify western blot densitometries with at least three lanes per condition. For imaging CMTs, we fixed CMTs with 4% PFA followed by staining with cardiac actinin A and DAPI. To measure conditioned media TGF-beta isoform production, we used Quantikine ELISA (R&D).
Mouse protocols, cardiac imaging, and fibrosis quantification
All mice were maintained and studied using protocols approved by the Animal Care and Use Committee of Harvard Medical School. Studies used male heterozygous HCM (MHCR403Q/+) mice that were in 129SvEv background, and transgenic N488I that were in the FVB background. 6-week old pre-hypertrophic HCM mice (n=15 mice in each arm) were treated with either DMSO or A769662 (Cayman) subcutaneously once daily for three weeks at a dose of 30 mg/kg (Cool et al., 2006). To accelerate hypertrophy, mice were treated with 1mg/g Cyclosporine A (CsA; Novartis), which was administered via oral chow. We studied male mice that more consistently develop HCM than do female littermates. Fibrosis was quantified using analysis of Masson trichrome stained sections. Cardiac hypertrophy was measured using a Vevo 770 Micro-Imager (VisualSonics). For extended methods, see supplemental methods.
Supplementary Material
Acknowledgments
We thank Barbara McDonough, Kaoru Ito and Gregory Fishbein for their contributions. C.S.C. is the scientific founder of Innolign Biological, which is developing CMTs for commercial applications. This work was supported in part by grants from NIH (HL125807(J.T.H.), AR062128(G.R.), UH3EB017103(C.S.C), EB001046(C.S.C.), HL080494(J.G.S., C.S.C.), HL128810(R.M.G.), HL084553(J.G.S.), John S. Ladue foundation(J.T.H), Leducq Foundation(J.G.S, C.E.S.), Sarnoff Foundation(C.C.S.), and the Howard Hughes Medical Institute (C.E.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, J.T.H. and C.E.S.; Methodology, J.T.H., A.C., A.L., K.Z., R.M.G., R.K., J.O., G.R., J.G., K.M., R.G., S.M.W., C.S.C., J.G.S. and C.E.S.; Investigation, J.T.H., A.C., K.Z., C.C.S., A.L., H.W., and J.O.; Writing-Original Draft, J.T.H.; Writing – Review & Editing, all authors; Funding acquisition, J.T.H., C.S.C., J.G.S., and C.E.S..
References
- Arad M, Benson DW, Perez-Atayde AR, McKenna WJ, Sparks EA, Kanter RJ, McGarry K, Seidman JG, Seidman CE. Constitutively active AMP kinase mutations cause glycogen storage disease mimicking hypertrophic cardiomyopathy. J Clin Invest. 2002;109:357–362. doi: 10.1172/JCI14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arad M, Moskowitz IP, Patel VV, Ahmad F, Perez-Atayde AR, Sawyer DB, Walter M, Li GH, Burgon PG, Maguire CT, et al. Transgenic mice overexpressing mutant PRKAG2 define the cause of Wolff-Parkinson-White syndrome in glycogen storage cardiomyopathy. Circulation. 2003;107:2850–2856. doi: 10.1161/01.CIR.0000075270.13497.2B. [DOI] [PubMed] [Google Scholar]
- Azhar M, Runyan RB, Gard C, Sanford LP, Miller ML, Andringa A, Pawlowski S, Rajan S, Doetschman T. Ligand-specific function of transforming growth factor beta in epithelial-mesenchymal transition in heart development. Dev Dyn. 2009;238:431–442. doi: 10.1002/dvdy.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin KK, Taegtmeyer H. AMP-activated protein kinase regulates E3 ligases in rodent heart. Circ Res. 2011;109:1153–1161. doi: 10.1161/CIRCRESAHA.111.252742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudou T, Legant WR, Mu A, Borochin MA, Thavandiran N, Radisic M, Zandstra PW, Epstein JA, Margulies KB, Chen CS. A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue Eng Part A. 2012;18:910–919. doi: 10.1089/ten.tea.2011.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultot L, Guigas B, Von Wilamowitz-Moellendorff A, Maisin L, Vertommen D, Hussain N, Beullens M, Guinovart JJ, Foretz M, Viollet B, et al. AMP-activated protein kinase phosphorylates and inactivates liver glycogen synthase. Biochem J. 2012;443:193–203. doi: 10.1042/BJ20112026. [DOI] [PubMed] [Google Scholar]
- Burwinkel B, Scott JW, Buhrer C, van Landeghem FK, Cox GF, Wilson CJ, Grahame Hardie D, Kilimann MW. Fatal congenital heart glycogenosis caused by a recurrent activating R531Q mutation in the gamma 2-subunit of AMP-activated protein kinase (PRKAG2), not by phosphorylase kinase deficiency. Am J Hum Genet. 2005;76:1034–1049. doi: 10.1086/430840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B, Li J, Wang J, Luo X, Ai J, Liu Y, Wang N, Liang H, Zhang M, Chen N, et al. microRNA-124 regulates cardiomyocyte differentiation of bone marrow-derived mesenchymal stem cells via targeting STAT3 signaling. Stem Cells. 2012;30:1746–1755. doi: 10.1002/stem.1154. [DOI] [PubMed] [Google Scholar]
- Carney DH, Mann R, Redin WR, Pernia SD, Berry D, Heggers JP, Hayward PG, Robson MC, Christie J, Annable C, et al. Enhancement of incisional wound healing and neovascularization in normal rats by thrombin and synthetic thrombin receptor-activating peptides. J Clin Invest. 1992;89:1469–1477. doi: 10.1172/JCI115737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou DC, Gorham JM, Herman DS, Seidman JG. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Curr Protoc Mol Biol. 2011;Chapter 4(Unit4):12. doi: 10.1002/0471142727.mb0412s94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool B, Zinker B, Chiou W, Kifle L, Cao N, Perham M, Dickinson R, Adler A, Gagne G, Iyengar R, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 2006;3:403–416. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Debold EP, Schmitt JP, Patlak JB, Beck SE, Moore JR, Seidman JG, Seidman C, Warshaw DM. Hypertrophic and dilated cardiomyopathy mutations differentially affect the molecular force generation of mouse alpha-cardiac myosin in the laser trap assay. Am J Physiol Heart Circ Physiol. 2007;293:H284–H291. doi: 10.1152/ajpheart.00128.2007. [DOI] [PubMed] [Google Scholar]
- Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doenst T, Nguyen TD, Abel ED. Cardiac metabolism in heart failure: implications beyond ATP production. Circ Res. 2013;113:709–724. doi: 10.1161/CIRCRESAHA.113.300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom PG, Steijger T, Sipos B, Grant GR, Kahles A, Ratsch G, Goldman N, Hubbard TJ, Harrow J, Guigo R, et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisterfer-Lowrance AA, Christe M, Conner DA, Ingwall JS, Schoen FJ, Seidman CE, Seidman JG. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272:731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- Gollob MH, Green MS, Tang AS, Gollob T, Karibe A, Ali Hassan AS, Ahmad F, Lozado R, Shah G, Fananapazir L, et al. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med. 2001;344:1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Lopez B, Coelho-Filho OR, Lakdawala NK, Cirino AL, Jarolim P, Kwong R, Gonzalez A, Colan SD, Seidman JG, et al. Myocardial fibrosis as an early manifestation of hypertrophic cardiomyopathy. N Engl J Med. 2010;363:552–563. doi: 10.1056/NEJMoa1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N, Lee KH, Ma Q, Kang PM, Golub TR, et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193–2204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Hunter RW, Garcia-Menendez L, Gong G, Yang YY, Kolwicz SC, Jr, Xu J, Sakamoto K, Wang W, Tian R. Mutation in the gamma2-subunit of AMP-activated protein kinase stimulates cardiomyocyte proliferation and hypertrophy independent of glycogen storage. Circ Res. 2014;114:966–975. doi: 10.1161/CIRCRESAHA.114.302364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraegen EW, Sowden JA, Halstead MB, Clark PW, Rodnick KJ, Chisholm DJ, James DE. Glucose transporters and in vivo glucose uptake in skeletal and cardiac muscle: fasting, insulin stimulation and immunoisolation studies of GLUT1 and GLUT4. Biochem J. 1993;295(Pt 1):287–293. doi: 10.1042/bj2950287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth-Kraczek EJ, Hirshman MF, Goodyear LJ, Winder WW. 5' AMP-activated protein kinase activation causes GLUT4 translocation in skeletal muscle. Diabetes. 1999;48:1667–1671. doi: 10.2337/diabetes.48.8.1667. [DOI] [PubMed] [Google Scholar]
- Lang T, Yu L, Tu Q, Jiang J, Chen Z, Xin Y, Liu G, Zhao S. Molecular cloning, genomic organization, and mapping of PRKAG2, a heart abundant gamma2 subunit of 5'-AMP-activated protein kinase, to human chromosome 7q36. Genomics. 2000;70:258–263. doi: 10.1006/geno.2000.6376. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim JH, Kim JS, Chang JW, Kim SB, Park JS, Lee SK. AMP-activated protein kinase inhibits TGF-beta-, angiotensin II-, aldosterone-, high glucose-, and albumin-induced epithelial-mesenchymal transition. Am J Physiol Renal Physiol. 2013;304:F686–F697. doi: 10.1152/ajprenal.00148.2012. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- Li J, Ren J, Liu X, Jiang L, He W, Yuan W, Yang J, Dai C. Rictor/mTORC2 signaling mediates TGFbeta1-induced fibroblast activation and kidney fibrosis. Kidney Int. 2015;88:515–527. doi: 10.1038/ki.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2012;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JY, Oh MA, Kim WH, Sohn HY, Park SI. AMP-activated protein kinase inhibits TGF-beta-induced fibrogenic responses of hepatic stellate cells by targeting transcriptional coactivator p300. J Cell Physiol. 2012;227:1081–1089. doi: 10.1002/jcp.22824. [DOI] [PubMed] [Google Scholar]
- Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC, Kim K, Miller JD, Ng K, Daley GQ. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovekamp-Swan T, Jetten AM, Davis BJ. Dual activation of PPARalpha and PPARgamma by mono-(2-ethylhexyl) phthalate in rat ovarian granulosa cells. Mol Cell Endocrinol. 2003;201:133–141. doi: 10.1016/s0303-7207(02)00423-9. [DOI] [PubMed] [Google Scholar]
- Ma X, Fu Y, Xiao H, Song Y, Chen R, Shen J, An X, Shen Q, Li Z, Zhang Y. Cardiac Fibrosis Alleviated by Exercise Training Is AMPK-Dependent. PLoS One. 2015;10:e0129971. doi: 10.1371/journal.pone.0129971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsin AS, Bertrand L, Rider MH, Deprez J, Beauloye C, Vincent MF, Van den Berghe G, Carling D, Hue L. Phosphorylation and activation of heart PFK-2 by AMPK has a role in the stimulation of glycolysis during ischaemia. Curr Biol. 2000;10:1247–1255. doi: 10.1016/s0960-9822(00)00742-9. [DOI] [PubMed] [Google Scholar]
- McGee SL, van Denderen BJ, Howlett KF, Mollica J, Schertzer JD, Kemp BE, Hargreaves M. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- Murphy RT, Mogensen J, McGarry K, Bahl A, Evans A, Osman E, Syrris P, Gorman G, Farrell M, Holton JL, et al. Adenosine monophosphate-activated protein kinase disease mimicks hypertrophic cardiomyopathy and Wolff-Parkinson-White syndrome: natural history. J Am Coll Cardiol. 2005;45:922–930. doi: 10.1016/j.jacc.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Ofir M, Arad M, Porat E, Freimark D, Chepurko Y, Vidne BA, Seidman CE, Seidman JG, Kemp BE, Hochhauser E. Increased glycogen stores due to gamma-AMPK overexpression protects against ischemia and reperfusion damage. Biochem Pharmacol. 2008;75:1482–1491. doi: 10.1016/j.bcp.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Poyhonen P, Hiippala A, Ollila L, Kaasalainen T, Hanninen H, Helio T, Tallila J, Vasilescu C, Kivisto S, Ojala T, et al. Cardiovascular magnetic resonance findings in patients with PRKAG2 gene mutations. J Cardiovasc Magn Reson. 2015;17:89. doi: 10.1186/s12968-015-0192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick RM, Zong H, Li J, Morino K, Moore IK, Yu HJ, Liu ZX, Dong J, Mustard KJ, Hawley SA, et al. Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell Metab. 2007;5:151–156. doi: 10.1016/j.cmet.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruperez M, Lorenzo O, Blanco-Colio LM, Esteban V, Egido J, Ruiz-Ortega M. Connective tissue growth factor is a mediator of angiotensin II-induced fibrosis. Circulation. 2003;108:1499–1505. doi: 10.1161/01.CIR.0000089129.51288.BA. [DOI] [PubMed] [Google Scholar]
- Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano M, Ouchi N, Shibata R, Ohashi K, Pimentel DR, Murohara T, Walsh K. Adiponectin deficiency exacerbates cardiac dysfunction following pressure overload through disruption of an AMPK-dependent angiogenic response. J Mol Cell Cardiol. 2010;49:210–220. doi: 10.1016/j.yjmcc.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu JS, Rajawat YS, Rami TG, Gollob MH, Wang Z, Yuan R, Marian AJ, DeMayo FJ, Weilbacher D, Taffet GE, et al. Transgenic mouse model of ventricular preexcitation and atrioventricular reentrant tachycardia induced by an AMP-activated protein kinase loss-of-function mutation responsible for Wolff-Parkinson-White syndrome. Circulation. 2005;111:21–29. doi: 10.1161/01.CIR.0000151291.32974.D5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JL, Liu W, Nelson CM, Raghavan S, Chen CS. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 2004;10:865–872. doi: 10.1089/1076327041348365. [DOI] [PubMed] [Google Scholar]
- Teekakirikul P, Eminaga S, Toka O, Alcalai R, Wang L, Wakimoto H, Nayor M, Konno T, Gorham JM, Wolf CM, et al. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-beta. J Clin Invest. 2010;120:3520–3529. doi: 10.1172/JCI42028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R, Musi N, D'Agostino J, Hirshman MF, Goodyear LJ. Increased adenosine monophosphate-activated protein kinase activity in rat hearts with pressure-overload hypertrophy. Circulation. 2001;104:1664–1669. doi: 10.1161/hc4001.097183. [DOI] [PubMed] [Google Scholar]
- Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Widyantoro B, Emoto N, Nakayama K, Anggrahini DW, Adiarto S, Iwasa N, Yagi K, Miyagawa K, Rikitake Y, Suzuki T, et al. Endothelial cell-derived endothelin-1 promotes cardiac fibrosis in diabetic hearts through stimulation of endothelial-to-mesenchymal transition. Circulation. 2010;121:2407–2418. doi: 10.1161/CIRCULATIONAHA.110.938217. [DOI] [PubMed] [Google Scholar]
- Zaha VG, Young LH. AMP-activated protein kinase regulation and biological actions in the heart. Circ Res. 2012;111:800–814. doi: 10.1161/CIRCRESAHA.111.255505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.