Abstract
Since emerging in Saint Martin in 2013, chikungunya virus (CHIKV), an alphavirus transmitted by the Aedes aegypti mosquito, has infected approximately two million individuals in the Americas, with over 500,000 reported cases in the Dominican Republic (DR). CHIKV-infected patients typically present with a febrile syndrome including polyarthritis/polyarthralgia, and a macropapular rash, similar to those infected with dengue and Zika viruses, and malaria. Nevertheless, many Dominican cases are unconfirmed due to the unavailability and high cost of laboratory testing and the absence of specific treatment for CHIKV infection. To obtain a more accurate representation of chikungunya fever (CHIKF) clinical signs and symptoms, and confirm the viral lineage responsible for the DR CHIKV outbreak, we tested 194 serum samples for CHIKV RNA and IgM antibodies from patients seen in a hospital in La Romana, DR using quantitative RT-PCR and IgM capture ELISA, and performed retrospective chart reviews. RNA and antibodies were detected in 49% and 24.7% of participants, respectively. Sequencing revealed that the CHIKV strain responsible for the La Romana outbreak belonged to the Asian/American lineage and grouped phylogenetically with recent Mexican and Trinidadian isolates. Our study shows that, while CHIKV-infected individuals were infrequently diagnosed with CHIKF, uninfected patients were never falsely diagnosed with CHIKF. Participants testing positive for CHIKV RNA were more likely to present with arthralgia, although it was reported in just 20.0% of CHIKF+ individuals. High percentages of respiratory (19.6%) signs and symptoms, especially among children, were noted, though it was not possible to determine whether individuals infected with CHIKV were co-infected with other pathogens. These results suggest that CHIKV may have been underdiagnosed during this outbreak, and that CHIKF should be included in differential diagnoses of diverse undifferentiated febrile syndromes in the Americas.
Author Summary
Chikungunya virus (CHIKV) is known for its ability to cause explosive outbreaks of flu-like illness followed by severe joint pain and swelling, which can persist for months to several years. We tested patient serum of both suspected and unsuspected chikungunya fever (CHIKF) cases collected at an emergency clinic in La Romana, Dominican Republic for markers of current or recent CHIKV infection, and showed through next generation sequencing that the causative CHIKV strain is closely related to other isolates from the Americas. After matching clinical outcomes with diagnostic data, we found that relatively few CHIKF patients presented with the joint symptoms typically associated with CHIKV infection in the past, which likely contributed to a notable frequency of CHIKF misdiagnosis. Other deviations from Old World CHIKF outbreaks were also discovered, including shifts in affected age groups and in the frequency of respiratory signs and symptoms. These data are particularly important for improving future surveillance endeavors, as well as the diagnosis and treatment of CHIKF in the Americas.
Introduction
Chikungunya virus (CHIKV), a mosquito-borne virus in the family Togaviridae, genus Alphavirus, was first isolated in 1952 from a patient on the Makonde plateau that is now part of Tanzania. It derives its name from the Makonde term kungunyala, meaning roughly, “that which bends up” [1]. Urban chikungunya fever (CHIKF) outbreaks were first described in the late 1950s in Asia, transmitted by the mosquito vector Aedes aegypti. In 2005, CHIKV emerged again from enzootic circulation in Africa and spread to the Indian Ocean Basin, India and other Asian countries and also made incursions into Italy and France, which reported local transmission for the first time in Europe [2–6]. This unprecedented spread was partly credited to the ability of the CHIKV strains within the new Indian Ocean lineage (IOL) to adapt through E1 and E2 envelope glycoprotein substitutions to a new vector species, Ae. albopictus, which itself spread from Asia to many parts of the world since 1985 [7–9]. Then, in 2013, a CHIKV strain belonging to the same lineage that had been circulating in Asia at least since the 1950s (Asian lineage) was introduced into the Americas, first detected on the Caribbean island of St. Martin before spreading through South and Central America, including Mexico [10, 11]. Local transmission was reported in the U.S. in Florida and Texas after the arrival of infected travelers [12, 13]. To date, this New World introduction has resulted in over 1.9 million suspected cases of CHIKF in North, Central, and South America as well as the Caribbean Islands [Pan American Health Organization (PAHO) data]. An independent introduction of an East, Central, and South Africa (ECSA)-derived enzootic CHIKV strain from Africa was also reported in Brazil in 2014 [14].
Strains from all CHIKV lineages can cause explosive outbreaks of CHIKF, which is characterized by a generalized febrile syndrome typically including high fever, maculopapular rash, muscle and joint aches, headache, and malaise, which progresses to severe polyarthritis/polyarthralgia. Although CHIKF is generally self-limiting and sometimes resolves within a month of onset, up to 30–70% of CHIKF patients experience chronic and/or recurring arthritis lasting months to years [4].
Polyarthritis and polyarthralgia are hallmarks of CHIKF, and can range between 80% and 97% of patients reporting these symptoms during past epidemics [2, 3, 15–18]. However, data from New World CHIKV infections remain limited with few reports examining in detail patient signs and symptoms. Some studies suggest that infection with New World/Asian CHIKV lineage strains results in arthritic outcomes similar to those observed in previous epidemics, but with only limited data available for long-term joint sequelae [19].
Among New World CHIKF outbreaks, the Dominican Republic has had the most suspected cases, with over 500,000 reported since 2014 [PAHO data] (Fig 1). Here, we describe an outbreak in the southeastern city of La Romana. In February, 2014, the Dominican Republic’s first suspected cases were reported in Nigua, a town in San Cristóbal Province, southwest of the capital. Suspected cases were defined by the Dominican Ministerio de Salud Pública (MSP) by acute febrile syndrome and polyarthralgia, though many patients also presented cervical, supraclavicular, and inguinal adenomegalies and facial, vulva and scrotal edema. It was hypothesized that CHIKV entered the country through Bajos de Haina, a port city located 2 km from Nigua. On April 3, 2014, the U.S. Centers for Disease Control and Prevention (CDC) confirmed CHIKV infection by detecting CHIKV-specific IgM antibodies in a patient’s blood [10]. Nationally, cases continued to rise, peaking between mid-July and mid-August with up to 45,000 new cases each week. After administering brief questionnaires in major cities, the MSP estimated attack rates ranging from 40% of the population-at-risk in Higüey to 81% in Azua de Compostela. In La Romana, up to 89% of households interviewed were suspected to have been affected by CHIKV as of August, 2014 [20]. Clinicians and patients reported a high fever and arthralgia in the wrists and ankles, the latter which lasted up to six months in middle-aged and elderly female patients. Nevertheless, little published information describes the rate of CHIKV seropositivity or CHIKF symptoms in the La Romana population.
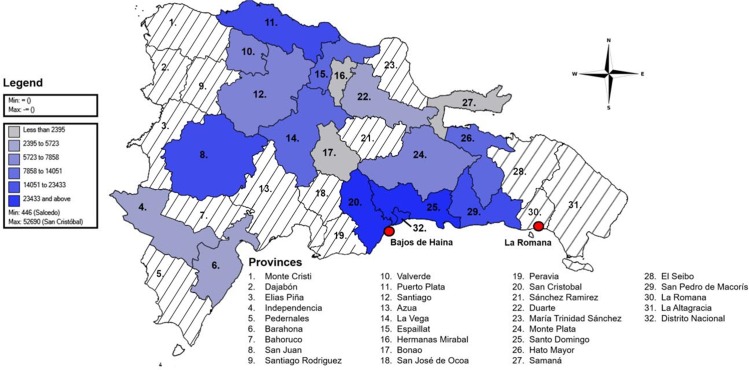
Fig 1. Distribution of total suspected cases of CHIKF reported in the Dominican Republic by province.
A suspected case was defined by the presence of sudden fever and arthralgia. The color scheme classifies provinces by number of total suspected CHIKF cases, determined by summing the number of cases reported in MSP DIGEPI weekly bulletins and Chikungunya Outbreak Bulletins for each epidemiological weeks between February 16, 2014 and June 6, 2015. No exact case numbers were reported to MSP DIGEPI for provinces shaded in white. Number 32, Distrito Nacional, represents the national district, which does not pertain to a province. The city of La Romana and the port of Bajos de Haina, where the outbreak is suspected to have started, are highlighted in red. Map created using Epi InfoTM 7.1.5 software licensed by the Centers for Disease Control and Prevention (http://wwwn.cdc.gov/epiinfo/7/). MSP DIGEPI weekly bulletins publicly available through Minesterio de Salud Publica (http://digepisalud.gob.do/).
Here, we report a case series of CHIKF patients in La Romana from June-August, 2014 during the CHIKF outbreak, following diagnostic tests of patient sera and retrospective chart reviews of signs and symptoms as well as clinical data associated with these patients, and genetic characterization of CHIKV isolates from patients.
Methods
Patient data and serum collection
Between June and August, 2014, serum collected for complete blood counts (CBC) was collected from discarded diagnostic blood samples of patients attended by the department of emergency medicine at Hospital el Buen Samaritano (HBS). The criteria for serum collection in HBS included patients for whom a complete blood count (CBC) was performed. A retrospective chart review was performed to collect patient data. Patient age, gender, symptoms, and CBC results were collected retrospectively wherever possible. For patients who were admitted into the hospital, a physical examination and medical history were recorded, including a history of the present illness and clinical and familial antecedents. All patient samples and data were assigned institution-specific identification numbers to ensure patient anonymity. Neither patient names nor clinic-assigned laboratory numbers were used, and patient age was used in lieu of birthdate, eliminating all potential identifying information from the study without compromising data integrity.
All patient data were deidentified and handled under University of Texas Medical Branch Institutional Review Board Protocol #15–0265.
Quantitative RT-PCR
RNA was first isolated from serum samples using the ZR-96 Viral RNA Kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s protocol. Quantitative reverse transcription PCR (RT-qPCR) was then performed using the TaqMan RNA-Ct 1-step Kit (Applied Biosystems, San Francisco, CA, USA) and previously described primers [21].
CHIKV IgM ELISA
Serum samples were screened for anti-CHIKV IgM antibodies by enzyme-linked immunosorbent assay (ELISA) as previously described [22] using the CHIKjj Detect MAC-ELISA kit (InBios, Inc., Seattle, WA), which was validated by the CDC [23]. All samples were tested in duplicate and any inconclusive samples were retested.
Plaque assay
African green monkey kidney cells (Vero; American Type Cell Culture, Bethesda, MD) were grown to 90–100% confluency in 12-well plates. Serum was diluted in Dulbecco's mimimal essential medium (DMEM) (supplemented with 5% FBS and from a series of 10-fold dilutions, medium was removed from plates, and 100uL of serum dilution was plated per well. After 60 minutes incubation, an overlay composed of DMEM and 0.2% agarose was added and incubated for 24–48 hours. Plates were then fixed with 10% formaldehyde for one hour before staining with crystal violet, and plaques counted.
Sequencing and phylogenetic analysis
Ten serum samples obtained during the CHIKV RT-qPCRs with low RT-qPCR Ct values were submitted for Illumina HiSeq sequencing, without passaging, as previously described [19]. Viral genomes were assembled using the Abyss software [24]. Assembled contigs were checked using bowtie2 to align reads to the contigs followed by visualization using the integrative genomics viewer [25]. Genomic sequences were manually aligned with those representing all three genotypes downloaded from Genbank using Se-Al (http://tree.bio.ed.ac.uk/software/seal/), and non-coding sequences were removed from the alignment, resulting in a common length of 11,241 nt. The final data set comprised of 85 complete open reading frame sequences from 26 countries isolated during 1953–2014. A Bayesian maximum clade credibility (MCC) phylogeny was inferred using the GTR+G4 nucleotide substitution model with BEAST version 1.8.2 [26].
Statistics
This study is primarily observational as it followed a series of CHIKF cases, and thus most data were reported in raw form. Contingency tables for admitted vs. outpatients were constructed with outpatients in the first row and hospitalized in the second, and those with symptoms/parameter in the first column and without in the second; relative risk analysis was then performed using Medcalc (MedCalc Software, Ostend, Belgium). Continuous variables were first tested for normalcy, normalized as needed, and then means were analyzed by either T-test or one-way ANOVA using SPSS software. Regression analyses were employed to ascertain correlation between CT value and symptom frequency using SPSS software.
Results
Finalized sample pool
A total of 194 serum samples, collected between July and August, 2014, were included in the study; at the time of sample receipt, scientists were blinded to disease state of the patients sampled. Of those, 95 (49%) were positive for CHIKV RNA by RT-qPCR, 48 (24.7%) were positive for acute CHIKV antibodies by IgM ELISA, and 2 samples were positive for both (1%; Table 1). In total, 145/194 (74.7%) sera tested positive for recent CHIKV infection. CT values for RNA-positive samples ranged between 34 and 17.39 (S1 Fig), and plaque assays on RNA-positive samples revealed infectious titers ranging between 2.9x105 plaque forming units/mL to below the limit of detection of 1x102 PFU/mL. Sequencing and phylogenetic analysis confirmed that the CHIKV isolates obtained from the DR were closely related to other isolates collected from Caribbean outbreaks, stemming from an Asian lineage strain (S2 Fig).
Table 1. Diagnostic outcomes for serum samples collected from patients in La Romana, Dominican Republic between July 2014 and August 2014.
| Parameter | n(percent) |
|---|---|
| Total samples | 194 |
| Samples CHIKV-positive | 145(74.7) |
| by RT-qPCR | 95(49.0) |
| by IgM ELISA | 48(24.7) |
| By both RTq-PCR and IgM ELISA | 2(1.0) |
Demographic data
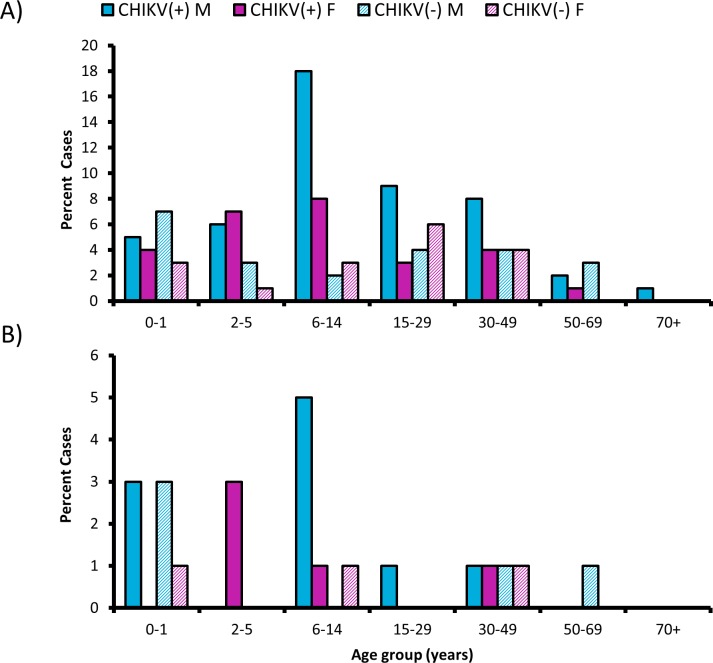
After samples underwent diagnostic testing, de-identified patient data were analyzed for demographic characteristics such as gender, age, and hospital admission (Table 2). As inclusion criteria were not used to regulate sample submission, these data are strictly observational and are presented to describe the general clinical picture associated with this particular sample set. Only RNA-positive samples were included in these analyses because IgM can persist up to several months after infection, thus not necessarily indicating active infection. In total, demographic data were available for 75 CHIKF(+) (CHIKV RNA-positive) and 41 CHIKF(-) (both RNA- and IgM-negative) patients. More male samples were represented for both CHIKF(+) and CHIKF(-) patient groups. A broad age range of both CHIKF(+) and CHIKF(-) patients was represented, between 13 days and 85 years, with the majority of cases involving children between the ages of 6 and 14 years (Fig 2). The mean time from fever onset to clinic visit ranged from 2–7 days, with an average of 3.8 days for CHIKF(+) patients and 4.0 days for CHIKF(-) patients. Viremia was detectable up to 7 days post-onset of illness in CHIKF(+) patients. Patients in the CHIKF(-) group had a higher rate of hospitalization, with 18.7% of CHIKF(+) patients and 26.8% of CHIKF(-) patients requiring admission. The mean age and time of symptom onset (before visiting the clinic) were not significantly different between admitted and outpatient CHIKF(+) and CHIKF(-) groups (ANOVA with Tukey’s post-hoc, 0.27<p<0.93; Table A in S1 Tables), nor did gender significantly affect the relative risk (RR) of being hospitalized for either group.
Table 2. Demographic data for CHIKF(+) and CHIKF(-) patient serum samples collected from patients in La Romana, Dominican Republic July-August 2014, and associated relative risk.
| Characteristic | CHIKV(+) value | CHIKV(-) value |
|---|---|---|
| Male: Female (ratio) | 2 | 1.3 |
| Mean years of age, ± STD | 16.2±16.9 | 19.6±19.1 |
| Time from onset to hospital visit, mean days ± STD | 3.8±1.8 | 4±1.4 |
| Hospitalized n(%) | 15 (18.7) | 8 (26.8) |
Fig 2. Age and gender distribution of patient sample pool.
Serum from patients visiting an emergency clinic in the DR was tested for CHIKV RNA and IgM, and results were retrospectively matched to demographic data. A) The age and gender distribution of CHIKV-RNA positive [CHIKF(+)] and CHIKF(-) (both IgM and RNA negative) patients. The majority of patients were under the age of 50 for both CHIKF(+) and CHIKF(-) groups. B) The age and gender distribution of patients who were hospitalized.
Symptoms, clinical diagnoses, and blood findings
In addition to demographic data, specific sign and symptom data were matched to samples (Table 3). These data were available for 48 CHIKF(+) patients and 19 CHIKF(-) patients. The average ages for patients with available symptom data were 16.5±16.4 years for CHIKF(+) patients and 29.6±23.3 years for CHIKF(-) patients. More symptom data were available for admitted patients, as hospital admission rates for patients with available symptom data were 31.2% for CHIKF(+) and 57.9% for CHIKF(-) patients; however, similar to demographic data, no demographic trait significantly affected the risk of hospitalization (Table B in S1 Tables). The most common feature of both CHIKF(+) and CHIKF(-) patients was fever (91.7% and 63.2%, respectively) averaging 39.4°C for CHIKF(+) patients, while the average fever for CHIKF(-) patients was slightly lower at 38.8°C. Complaints of arthralgia and myalgia were surprisingly low for CHIKF(+) patients at 20.0% and 13.3%, respectively. Despite the low frequency of these particular symptoms, they may still be considered of diagnostic value, as arthralgia and myalgia appeared to be CHIKV-specific, with no patients in the CHIKF(-) group exhibiting these symptoms. Several preexisting conditions were noted, the most prominent being pregnancy of greater than 12 weeks and hypertension However, because these occurred so infrequently, the effect of risk on hospital admission was not assessed.
Table 3. Signs and symptoms recorded for CHIKF-positive (and CHIKF-negative) patients for whom complete blood counts were ordered between July 2014 and August 2014 at Hospital Buen Samaritano, La Romana, Dominican Republic.
| Symptom | CHIK(+) | CHIKV(-) |
|---|---|---|
| TOTAL (no. patients) | 46 | 19 |
| Fever | 42 (91.7) | 12 (63.2) |
| Average (°C ± STD) | 39.4±0.5 | 38.8±0.3 |
| Arthralgia* | 6 (20) | 0 (0) |
| Myalgia* | 4 (13.3) | 0 (0) |
| Headache* | 7 (23.3) | 5 (38.46) |
| Enophthalmos | 5 (14.6) | 1 (5) |
| Malaise/fatigue | 4 (8.7) | 2 (10.5) |
| Rash | 0 (0) | 0 (0) |
| Dehydration | 14 (30.5) | 5 (26.3) |
| GastrointestinalA | 7 (15.2) | 7 (36.8) |
| RespiratoryB | 9 (19.6) | 6 (31.6) |
| PREXISTING CONDITIONS | ||
| Pregnancy >12 weeks† | 2 (16.6) | 1 (20.0) |
| Hypertension | 1 (2.3) | 1 (5.3) |
*Patient data for children under 3 years of age not included
†Percent calculated from number of female patients of child-bearing age (9–45)
ANausea, diarrhea, vomiting
Bpneumonia, dyspnea, rhonchus, difficulty breathing, rhinorrhea
Values in n(percent) unless otherwise noted.
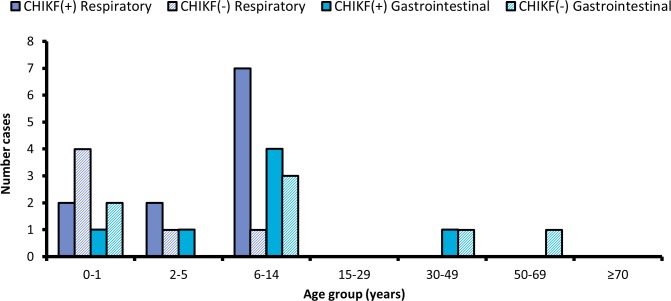
Other signs and symptoms less commonly associated with CHIKV infection can be broadly categorized as gastrointestinal, respiratory, and neurological. About 15% of CHIKF(+) patients presented with gastrointestinal signs and symptoms, including diarrhea, nausea, and gastroenteritis. A surprising 19.6% of CHIKF(+) patients presented with respiratory signs and symptoms. Most CHIKF(+) patients exhibiting gastrointestinal signs and symptoms were below the age of 15; patients exhibiting respiratory signs and symptoms were also largely under the age of 15 (Fig 3). No sign or symptom correlated to level of viremia, as determined by regression analysis of CT bin against patient data (0.32<p<0.98; S3 Fig).
Fig 3. Distribution of respiratory and gastrointestinal symptoms by age.
Serum from patients visiting an emergency clinic in the DR was tested for CHIKV RNA and IgM, and results were retrospectively matched to clinical features documented by physicians. Children and young adults below the age of 15 were the primary demographic groups presenting with respiratory and gastrointestinal symptoms for both CHIKF(+) (RNA-positive) and CHIKF(-) (both IgM and RNA negative) patients.
Given the high rate of hospitalization among CHIKF(+) patients relative to past outbreaks, the relative risk of presenting with specific signs and symptoms among admitted patients was assessed (Table C in S1 Tables). Firstly, while the mean age of admitted CHIKF(+) patients was lower than CHIKF(+) outpatients, this difference was not significant (ANOVA with Tukey’s post-hoc, p = 0.55); a similar observation was made for CHIKF(-) patients (ANOVA with Tukey’s post-hoc, p = 0.74). While fever and myalgia were not specifically associated with either admitted nor outpatient CHIKF(+) patients, admitted CHIKF(+) patients were 4.7 times more likely to present with arthralgia (95%CI = 1.03,21.06), 12.7 times more likely to present with headache (95%CI = 1.78, 90.18), 6.3 times more likely to present with dehydration (95%CI = 2.42, 16.67), 6.9 times more likely to present with gastrointestinal signs and symptoms (95%CI = 1.53, 30.84), and 9.6 times more likely to present with respiratory signs or symptoms (95%CI = 2.31, 40.05). Temperature was not significantly different between admitted and outpatient CHIKF(+) patients (ANOVA with Tukey’s post-hoc, p = 0.94), nor was time of symptom onset (ANOVA with Tukey’s post-hoc, p = 0.99). While no signs or symptoms imparted a significant risk of hospitalization among CHIKF(-) individuals, this finding may reflect the small number of patients in the CHIKF(-) group (n = 11 and n = 8 for admitted and outpatient groups, respectively) rather than the biological significance of respiratory and gastrointestinal features. Time of symptom onset was not significantly different between admitted and outpatient CHIKF(-) patients (p = 0.99).
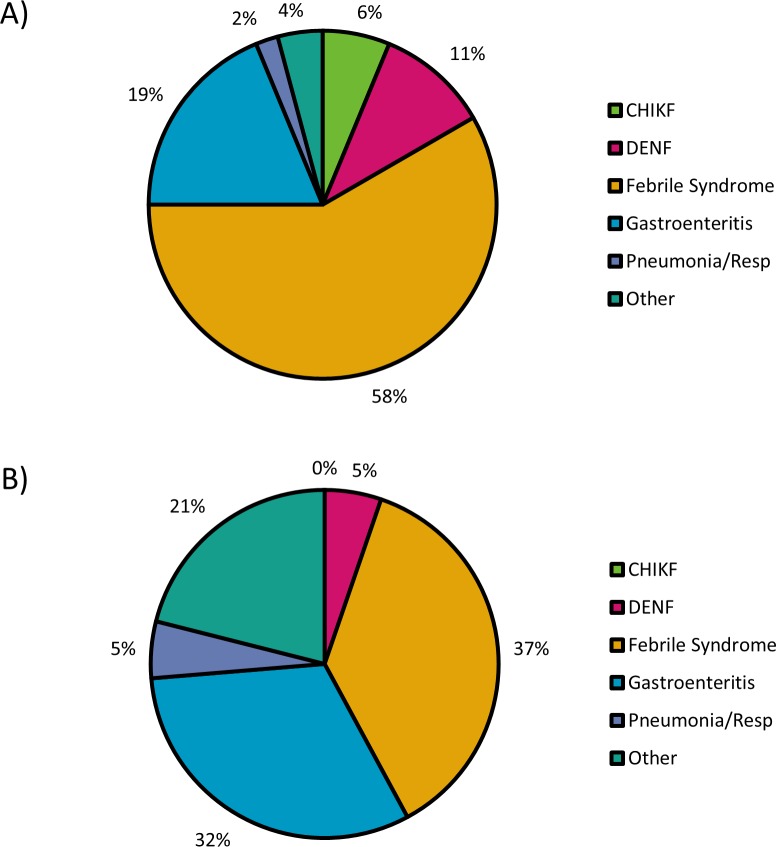
Initial diagnoses were made based on clinical findings (Fig 4). Only 6% of CHIKF(+) cases were clinically diagnosed as such, likely due to the absence of joint symptoms normally associated with CHIKF and possibly a lack of knowledge about CHIKF by some health care providers. CHIKF(+) patients were more likely to be diagnosed with idiopathic febrile syndrome, dengue fever, or pneumonia. No CHIKF(-) patients were misdiagnosed with CHIKF. Diagnoses classified under “other” included meningitis, pregnancy, asthma complications, and trauma. Some diagnoses for “generalized febrile syndrome” were inferred from the prescription of fever reducing agents, namely metamizole.
Fig 4. Clinical diagnoses made for CHIKF-positive and CHIKF-negative patients.
Serum from patients visiting an emergency clinic in the DR was tested for CHIKV RNA and IgM, and results were retrospectively matched to initial diagnoses based on clinical presentation. The most common diagnosis for patients in both groups was undifferentiated febrile illness. Most notably, while CHIKF-positive (RNA-positive) patients (A) were more likely to be diagnosed with something other than CHIKF, no CHIKF-negative (RNA and IgM negative) patients (B) were misdiagnosed with CHIKF.
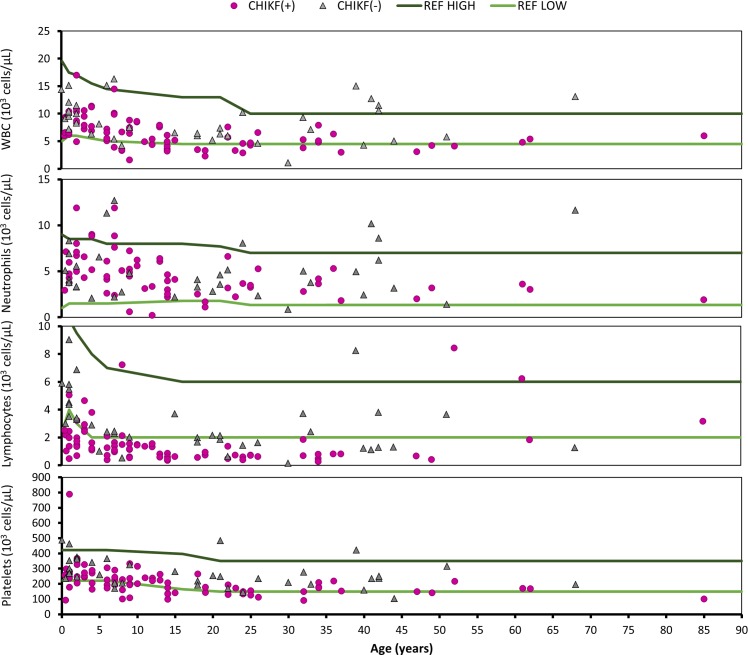
Pediatric (age<21) blood cell reference values were derived from standard hematology references [27], while adult reference values were provided by the laboratory at the Fundación Hospital General el Buen Samaritano. White blood cell (WBC) counts were generally unremarkable for both CHIKF(+) and CHIKF(-) groups, with the average for most age groups falling within normal ranges albeit with large variation; this also made statistical comparison of means impractical. Median values for complete WBC and lymphocytes were generally lower in CHIKF(+) patients than CHIKV(-) patients (Table 4). For CHIKF(+) patients: complete WBC ranged between 1.6–16.3x 103 cells/μL; neutrophils ranged between 0.2–11.9x103 cells/μL; lymphocytes ranged between 0.3–8.4x103 cells/μL; and platelets ranged between 91-789x103 cells/μL. CHIKF(-) patients showed similar ranges to CHIKF(+) patients: complete WBC ranged between 1.1–16.3x103 cells/μL; neutrophils ranged between 0.8–12.7x103 cells/μL; lymphocytes ranged between 0.2–9.0x103 cells/μL; and platelets ranged between 103-487x103 cells/μL. While most values fell within normal ranges for whole WBC, neutrophils, and platelets, many CHIKF(+) patients presented with varying degrees of lymphopenia when compared to reference values (Fig 5). Lymphopenia was significantly associated with relative level of viremia in CHIKF(+) patients, with the percent of patients presenting with lymphopenia decreasing with decreasing level of viral RNA; thrombocytopenia was not significantly associated with the level of viremia (linear regression with ANOVA, p = 0.006 and p = 0.081, respectively;S3 Fig), However, basal lymphocyte counts are highly specific to individuals, so it is possible that some of the deviant values fell within normal limits for some patients.
Table 4. Median values and interquartile range in parentheses for white blood cell (WBC) and differential counts for complete WBC, neutrophils, lymphocytes, and platelets by CHIKF positivity and age group.
| Age | CHIKV+/- | WBC | Neutrophil | Lymphocyte | Platelets |
|---|---|---|---|---|---|
| ALL | + | 6.2 (4.5–1.6) | 4.2 (3.0, 6.1) | 1.2 (0.7, 1.8) | 205 (153.5, 249.5) |
| - | 8.2 (6.0, 11.2) | 4.1 (3.0, 6.4) | 2.4 (1.5, 3.7) | 250 (205, 325) | |
| 0–1 | + | 6.6 (6.4,7.5) | 4.28 (3.7, 5.4) | 2.0 (1.3, 2.5) | 265.5 (223.3, 286) |
| - | 10.2 (9.4, 12.7) | 5.1 (4.1, 6.9) | 4.5 (3.5, 5.8) | 300 (274, 407) | |
| 2–5 | + | 9.1 (7.8, 10.8) | 6.9 (5.5, 8.6) | 1.9 (1.4, 2.9) | 260.5 (244.5, 317.5) |
| - | 8.3 (8.1, 10) | 3.3 (3.3, 5.6) | 3.3 (2.9, 3.4) | 341 (259, 364) | |
| 6–14 | + | 6.3 (4.3, 7.7) | 4.6 (3.0, 6.1) | 1.22 (0.8, 1.5) | 213.5 (173.8, 238.5) |
| - | 7.5 (5.4, 15.1) | 4.8 (2.7, 11.3) | 2.3 (2.0, 2.4) | 205 (205, 325) | |
| 15–29 | + | 4.5 (3.3, 5.3) | 3.4 (2.3, 4.0) | 0.7 (0.6, 0.7) | 146.5 (132.3, 173.8) |
| - | 6.3 (6, 6.5) | 3.6 (2.8, 4.6) | 1.8 (1.6, 2.1) | 234 (191, 253) | |
| 30–49 | + | 4.8 (3.8, 5.3) | 3.2 (2.6, 3.8) | 0.7 (0.5, 0.8) | 152 (150, 179) |
| - | 9.3 (5, 11.5) | 5.0 (3.2, 6.2) | 1.3 (1.2, 3.7) | 234 (198, 250) | |
| 50–69 | + | 4.8, (4.5, 5.1) | 3.3 (3.2, 3.5) | 6.24 (4.0, 7.3) | 171 (169.5, 194) |
| - | 9.45 (7.6, 11.3) | 6.5 (4.0, 9.1) | 2.5 (1.9, 3.1) | 255.5 (225.8, 285.3) |
Fig 5. White blood cell values for CHIKF(+) and CHIKF(-) patients by age.
Complete blood count (CBC) panel data for patients visiting an emergency clinic in La Romana, Dominican Republic, which were matched to Chikungunya virus (CHIKV) diagnostic results from discarded samples. Patients positive for CHIKV RNA by RT-qPCR are represented by solid purple circles, and patients negative for both CHIKV RNA and IgM antibodies [CHIKF(-)] are represented by grey triangles; green lines indicate suggested low and high reference values.
Discussion
CHIKV infection was detected in 145 patients in La Romana, Dominican Republic by either RT-qPCR or IgM ELISA, and CHIKF-associated disease was described for patients; while only IgM antibodies were detected in 48 samples, CHIKV RNA was detected in 95 samples, providing the opportunity to isolate and sequence CHIKV. It is important to adequately characterize the molecular and clinical aspects of the CHIKV strain responsible for CHIKF outbreaks, which can cause financial harm to affected populations and severely affect quality of life. For example, it is estimated that the 2005–2006 CHIKF epidemic on La Réunion island cost €43.9 million (approx. $60.4 million USD at the time) for healthcare costs associated with infection [28]. This estimate excludes the cost of long-term treatment, typically for persistent arthralgia. Another study following a cohort of patients from the Réunion CHIKF epidemic from 2006 found that a significant portion of those infected with CHIKV continued to seek medical care up to 30 months after the acute infection resolved [29]. Similarly high costs are well documented through India, where medical expenses are out-of-pocket and thus present a significant financial challenge to affected families [30–32]. In Tolima, Colombia, 44.3% of patients with laboratory-confirmed CHIKV infection continued to suffer from polyarthralgia 7 months after their initial diagnosis, indicating that New World CHIKV infection may also result in chronic joint symptoms [33]. Although our study includes potential sources of bias, most importantly an interview bias inherent to the retrospective nature of the medical chart review, the results provide useful preliminary data for evaluating the clinical phenotype of Asian CHIKV genotypes circulating in the Americas.
Reports describing CHIKF outbreaks in the Caribbean and South and Central America caused by strains derived from the Asian lineage introduced in the Caribbean suggest that arthritis and arthralgia/joint pain are major symptoms. In Trinidad and Tobago, 83.3% of confirmed patients complained of joint pain, while arthralgia was reported in 96% of patients in Colombia [19, 34]. In our study, although arthritis was not reported by any physician, there was relatively little arthralgia associated with CHIKV infection, with only 20.0% of patients reporting joint pain. Furthermore, other symptoms heretofore considered typical of CHIKF—rash, headache, and myalgia—were nearly absent in La Romana. Colombia reported that 64% of CHIKF cases exhibited rash, 57% headache, and 24% myalgia, in contrast La Romana, where none of the patients evaluated exhibited rash, only 20% reported headache, and only 13.3% reported myalgia. These discrepancies may reflect sampling practices, as previous Caribbean outbreak studies have included only suspected cases of CHIKF and dengue while our study broadly evaluated all patients for whom a CBC test was ordered, regardless of clinical diagnosis. As such, cases of CHIKF were described in our study, which would not have otherwise been recognized as such without the characteristic joint symptoms and rash. In fact, many of the patients we studied visited the clinics for other indications ranging from gastrointestinal or respiratory infections to pregnancy-related issues, and CHIKF was only confirmed incidentally. Interestingly, in a prospective cohort study reported by Yoon et al, a CHIKF outbreak in the Philippines caused by a strain closely related to the Asian strains circulating in the Americas resulted in only 45% of patients reporting arthralgia [35]. The study by Yoon et al broadly evaluated febrile patients, supporting our symptomology findings in La Romana.
Historically, joint symptoms have been a hallmark of CHIKF. Outbreak descriptions from La Réunion and Italy, for example, found that 96.1% and 97% of laboratory-confirmed cases reported joint pain. Similarly, evaluation of an outbreak in Singapore caused by an Asian-lineage CHIKV strain found that 87.6% of patients reported arthralgia. Furthermore, rash, headache, and myalgia were also more prominent than in our study, with 38–52% of patients reporting skin rash, 40–51% reporting headache, and 46–60% reporting myalgia in Old World CHIKF outbreaks [15, 18, 36]. The reasons behind these discrepancies are unknown, but may include environmental factors such as diet, human genetic factors, as well as CHIKV lineage and strain-specific variation in pathogenesis. The diagnostic value of differential CBC analysis has been a focus of debate among CHIKV researchers, as some epidemics include a large majority of patients presenting with profound lymphopenia [15] while other studies show that changes in lymphocyte counts may not be CHIKV-specific compared to other indications, namely dengue hemorrhagic fever [37]. In La Romana, CHIKV infection greatly increased the risk of lymphopenia, given reference values derived from a combination of sources, when compared to CHIKF(-) patients. This suggests that, in La Romana, lymphopenia may potentially be used to distinguish CHIKF from other febrile illnesses. A more specific study comparing WBC results in dengue virus- and CHIKV-infected individuals would be needed to further substantiate this claim. However, in La Romana a larger percentage of children under the age of 14 were diagnosed as CHIKF(+) than previously reported. Over 62% of the cases we detected were children below the age of 14 and 50% were 10 or below. This is in stark contrast to past outbreaks, such as in La Réunion where only 5–14% of patients were 0–9 years of age and in Italy where only 6% of patients were 0–19 years old [15, 36, 38]. It is unclear whether this demographic difference reflects cultural differences, such financial constraints forcing the decision to treat a child instead an adult from a family with multiple febrile members, or a higher risk of CHIKV infection in children in La Romana due to differences in exposure to mosquitoes. As previously described, gastrointestinal CHIKF signs and symptoms such as nausea, vomiting, and diarrhea were more common among our younger patients. However, an overall larger percentage of patients presented with respiratory symptoms in La Romana compared to most CHIKF outbreaks. This increase in respiratory illness seen with CHIKF cases in the Americas is not restricted to La Romana—respiratory signs, namely cough, were also reported in 23% of cases in Trinidad [19]. Further, the CHIKF outbreak in the Philippines resulted in up to 40% of patients experiencing respiratory symptoms [35]. Although CHIKV is known to affect the cardiopulmonary system, causing heart palpitations, dyspnea, chest pain, and rarely death due to cardiac complications, it has not been shown to directly cause overt respiratory pathology [39–41]. Most likely, the respiratory illness associated with CHIKF in La Romana was either an exacerbation of existing conditions such as asthma, or co-infection with a respiratory pathogen. Respiratory co-infection in CHIKF cases confirmed by pathogen isolation has been documented in past outbreaks. In La Réunion Island during 2005–2006, Lemant and colleagues reported two patients over the age of 60 with laboratory-confirmed CHIKF who were initially diagnosed with pneumonia caused by Streptococcus pneumoniae and Candida albicans, respectively [39]. Additionally, in a sample of cases from a 2006–2007 outbreak in Pondicherry and Karaikal, India, 87% of laboratory-confirmed CHIKF patients were co-infected with respiratory syncytial virus (RSV) [42], and 9% were also infected with adenoviruses; just 4% of CHIKF cases were mono-infected with CHIKV. Curiously, only CHIKV(+) cases were co-infected with multiple respiratory viruses. In our study, five patients received a diagnosis of bronchopneumonia, two of pneumonia, and two of acute respiratory infection. However, it is unclear whether these diagnoses were confirmed with x-ray imaging. The relatively small number of CHIKF(+) patients with respiratory signs and symptoms, paired with the concurrently higher percentage of CHIKF(-) patients diagnosed with respiratory findings, suggests that CHIKV and respiratory co-infection is likely. Respiratory infections inflict a high burden of disease among Dominican children, and in 2002 accounted for 5% of all pediatric deaths [43]. Our retrospective chart review did not permit us to track the morbidity and mortality of patients following initial diagnosis and hospital admission. Nevertheless, respiratory failure and death in laboratory-confirmed CHIKF cases was a feature in the outbreaks in La Réunion, France, and in southern India [39, 42, 44, 45], where co-infection with respiratory pathogens was hypothesized to contribute to the elevated morbidity and mortality. Understanding how respiratory co-infections and underlying respiratory conditions may have contributed to morbidity and mortality in the 2014 Dominican Republic outbreak and in potential future outbreaks should be a research priority, and may be applicable to other areas in the Americas.
Other researchers have suggested that an attenuated disease phenotype may be associated with some Asian lineage CHIKV strains. Yoon et al. reported an unprecedented rate of subclinical CHIKV infections in the Philippines, with as many as 2–12 subclinical infections for every symptomatic infection, depending on the age group [35]. Similarly, Simmons et al. examined blood donation pools in Puerto Rico and found an alarming number of CHIKV RNA(+) samples, indicating that many pre-symptomatic or asymptomatic CHIKV-infected individuals donated blood [46]. Our study supports to these findings, indicating that typical markers of CHIKV infection, i.e. joint symptoms, were not as common among CHIKF(+) patients in La Romana. Further, we observed many more infections in children and adolescents, consistent with the finding that fewer asymptomatic cases were associated with younger patients in the study by Yoon et al [35]. This may also explain the higher rate of respiratory signs and symptoms observed in La Romana and Trinidad, as children are naturally predisposed to respiratory infections. Genetically, CHIKV isolates from all of these outbreaks fall within the Asian lineage now circulating in the Americas [19, 35, 46]. While Zika virus RNA has been detected in the blood of a pregnant patient 10 weeks after symptom resolution [47], CHIKV RNA and/or antigen has only been shown to persist in joint and muscle tissue of humans (although no replicating virus has ever been isolated from individuals with chronic symptoms)[48, 49]. Therefore, rather than persistent viremia in CHIKV-infected patients biasing our symptom data, there could be an attenuation in the disease phenotype associated with American/Asian CHIKV strains compared to IOL strains.
Taken together, our data suggest that traditional symptom-based methods of diagnosing CHIKF may be insufficient in the Americas and other areas affected by recently emerging Asian lineage CHIKV strains; updating diagnostic approaches could greatly enhance surveillance in the Americas, an important consideration for designing vaccine and therapeutic clinical trials.
In summary, this study provided valuable clinical insights into the CHIKF outbreak in La Romana, DR with potential relevance to other areas in the New World where the Asian lineage of CHIKV is now circulating. Although limitations in our study design limit the ability to extrapolate these data to the Americas at large, the data the need to reevaluate current CHIKV surveillance practices and symptom-based diagnosis criteria.
Limitations of our study
Our study succeeded in identifying unique clinical and laboratory factors present during the outbreak of CHIKF in the Dominican Republic in 2014. These data demonstrate new clinical characteristics that may be valuable for diagnostic and surveillance considerations in other areas affected by CHIKV strains closely related to the Asian strain introduced to the Caribbean. However, as an outbreak investigation, our study had several limitations including an inherent selection bias in that patients came from a single hospital in La Romana, which is not necessarily representative of a random sample of the Dominican population. The selection was further biased by the fact that RT-qPCR and IGM ELISA for CHIKV were run on all patients subjected to a CBC test. Reporting data, therefore, were sparse and inconsistent, and included patients who may or may not have experienced febrile symptoms. Since historical and physical exam data were not standardized, extracting them from clinical records created an information bias that included the possibility of various measurement errors (vital signs, interview information, etc.).
Finally, there appeared to be an information bias toward hospitalized patients, particularly in the CHIKV(-) group, as percent hospitalization increased with increasing level of information (i.e., symptoms were more likely to be recorded and/or records were more likely to be stored appropriately if a patient was admitted to the hospital). This is also a bias inherent to utilizing emergency clinic samples, as patients with severe illness will more likely visit the emergency clinic than those with minor (or no) symptoms, and thus a greater rate of hospital admissions will be detected in this particular patient population regardless of disease state. While this complicated our ability to interpret risk analyses when comparing CHIKF(+) to CHIKF(-) individuals, it did not necessarily affect our ability to detect symptoms in CHIKF(+) patients. Roughly 30% of the CHIKF(+) patients with symptom data were admitted to HBS; therefore roughly 30% of our population exhibited the most severe CHIKF symptoms (while 70% exhibited slight to moderate symptoms which did not necessitate hospital admission); even among the most severe CHIKF manifestations, only 30% of CHIKF(+) patients presented with arthralgia (compared to just 12% among the CHIKF(+) outpatient group). Furthermore, the failure to associate any sign or symptom (except lymphopenia) with the CT value indicates that viremia was not an accurate predictor of disease severity. Along with the absence of fever in some patients with low to moderate CT values, this suggests viremia in the absence of signs and symptoms as reported by Simmons et al, who found many blood donation pools to be strongly CHIKV RNA(+)[46]. Thus, information bias toward admitted patients and its complicating effect on comparing CHIKF(+) patients to the general CHIKF(-) population probably contributes to an overestimation of CHIKF symptoms among CHIKF(+) individuals.
Supporting Information
Serum collected from discarded blood samples of patients visiting an emergency clinic in the DR was tested for CHIKV RNA. CT values from positive results are displayed as bins (e.g., bin 17 represents 17.0≤CT<18.0)
(TIF)
Using an Illumina HiSeq platform, complete genome sequences were determined directly from RNA isolated from the sera of 10 individuals. The overall alignment rate of the reads varied widely among samples, ranging from 7–88%, with a mean of approximately 20%. Nucleotide and amino acid identity among the consensus sequences from these individuals was >99.9%. The MCC phylogeny showed that the Dominican Republic sequences clustered within the Asian lineage together with other Caribbean sequences isolated in 2014 [11, 16]. The most closely related Asian sequences circulated between 2012 and 2013 in Micronesia, the Philippines and China. The three major CHIKV genotypes are labeled. Nodes with clade credibility’s ≥ 95% are labeled accordingly.
(TIF)
Serum collected from discarded blood samples of patients visiting an emergency clinic in the DR was tested for CHIKV RNA and IgM, and samples testing positive for RNA were retroactively matched to clinical signs and symptoms documented by physicians (arthralgia, myalgia, enophthalmos, respiratory signs and symptoms, and gastrointestinal signs and symptoms) or results from complete blood count analysis (lymphopenia and thrombocytopenia). Results were organized by CT bins defined by approximate log change from positive control (e.g., bin 20–22 represents samples within a 10-fold decrease from control, bin 23–25 within a 100-fold decrease, etc.). Only percent patients presenting with lymphopenia was significantly associated with CT bin by linear regression analysis (p = 0.006), suggesting a positive correlation between percent patients presenting with lymphopenia and relative viremia.
(TIF)
(DOCX)
Acknowledgments
We thank UTMB Global Health Track medical students Johnae D. Snell, Carlos Chavez, Aidan Deleon, and Russel Purpura, who established the cold chain for sample transport and helped with various laboratory tasks, as well as Carmen Dilia de la Cruz de Noble, Jennifer Jorge, Maribel Vicioso, Carlos de Mateo Valdez, and Geisel Mojica Acevedo in the DR, who assisted with sample and patient data acquisition. We also thank Netanya Utay and Michael Goodman for their thoughtful advice on data formatting and statistical analysis. Finally, we thank everyone at the Direccion General de Epidemiologia (DIGEPI) for their comments on the initial data, and thank Rhina Reyes and Jose Rodriguez Abreu from the Directorate of the Provincial and Regional Ministerio de Salud Publica for their support for this project.
Data Availability
Genbank accession numbers: BankIt1972278 Seq1 KY272961, BankIt1972278 Seq2 KY272962, BankIt1972278 Seq3 KY272963, BankIt1972278 Seq4 KY272964, BankIt1972278 Seq5 KY272965, BankIt1972278 Seq6 KY272966, BankIt1972278 Seq7 KY272967, BankIt1972278 Seq8 KY272968, BankIt1972278 Seq9 KY272969, BankIt1972278 Seq10 KY272970. All other relevant data are within the paper and its supporting data.
Funding Statement
This work was funded by the Institute for Human Infections and Immunity at the University of Texas Medical Branch (http://www.utmb.edu/ihii/), National Institutes of Health grant AI120942, and the Institute for Collaboration in Health at the University of Texas Medical Branch (http://www.utmb.edu/ccgh/education/). RML and JHE were supported by the National Institutes of Health NCATS CTSA UL1 TR001439. The genomes referenced in this publication were sequenced through funding provided by Department of Homeland Security through Contract No. HSHQDC-13-C-B0009 "Capturing Global Biodiversity of Pathogens by Whole Genome Sequencing." The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tesh RB. Arthritides caused by mosquito-borne viruses. Annu Rev Med. 1982;33:31–40. 10.1146/annurev.me.33.020182.000335 [DOI] [PubMed] [Google Scholar]
- 2.Angelini R, Finarelli AC, Angelini P, Po C, Petropulacos K, Silvi G, et al. Chikungunya in north-eastern Italy: a summing up of the outbreak. Euro Surveill. 2007;12(11):E071122.2. [DOI] [PubMed] [Google Scholar]
- 3.Josseran L, Paquet C, Zehgnoun A, Caillere N, Le Tertre A, Solet JL, et al. Chikungunya disease outbreak, Reunion Island. Emerg Infect Dis. 2006;12(12):1994–5. PubMed Central PMCID: PMCPMC3291364 10.3201/eid1212.060710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schilte C, Staikowsky F, Staikovsky F, Couderc T, Madec Y, Carpentier F, et al. Chikungunya virus-associated long-term arthralgia: a 36-month prospective longitudinal study. PLoS Negl Trop Dis. 2013;7(3):e2137 PubMed Central PMCID: PMCPMC3605278 10.1371/journal.pntd.0002137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roiz D, Boussès P, Simard F, Paupy C, Fontenille D. Autochthonous Chikungunya Transmission and Extreme Climate Events in Southern France. PLoS Negl Trop Dis. 2015;9(6):e0003854 PubMed Central PMCID: PMCPMC4469319 10.1371/journal.pntd.0003854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weaver SC, Lecuit M. Chikungunya virus and the global spread of a mosquito-borne disease. The New England journal of medicine. 2015;372(13):1231–9. Epub 2015/03/26 10.1056/NEJMra1406035 [DOI] [PubMed] [Google Scholar]
- 7.Tsetsarkin KA, Weaver SC. Sequential adaptive mutations enhance efficient vector switching by Chikungunya virus and its epidemic emergence. PLoS Pathog. 2011;7(12):e1002412 PubMed Central PMCID: PMCPMC3234230 10.1371/journal.ppat.1002412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraemer MU, Sinka ME, Duda KA, Mylne AQ, Shearer FM, Barker CM, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife. 2015;4:e08347 PubMed Central PMCID: PMCPMC4493616 10.7554/eLife.08347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsetsarkin KA, Chen R, Sherman MB, Weaver SC. Chikungunya virus: evolution and genetic determinants of emergence. Curr Opin Virol. 2011;1(4):310–7. PubMed Central PMCID: PMCPMC3182774 10.1016/j.coviro.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer M, Staples JE, Arboviral Diseases Branch NtCfEaZID, C. D. C. Notes from the field: chikungunya virus spreads in the Americas—Caribbean and South America, 2013–2014. MMWR Morb Mortal Wkly Rep. 2014;63(22):500–1. [PMC free article] [PubMed] [Google Scholar]
- 11.Díaz-Quiñonez JA, Ortiz-Alcántara J, Fragoso-Fonseca DE, Garcés-Ayala F, Escobar-Escamilla N, Vázquez-Pichardo M, et al. Complete genome sequences of chikungunya virus strains isolated in Mexico: first detection of imported and autochthonous cases. Genome Announc. 2015;3(3). PubMed Central PMCID: PMCPMC4424286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kendrick K, Stanek D, Blackmore C, (CDC) CfDCaP. Notes from the field: Transmission of chikungunya virus in the continental United States—Florida, 2014. MMWR Morb Mortal Wkly Rep. 2014;63(48):1137 [PMC free article] [PubMed] [Google Scholar]
- 13.CDC. Table. Laboratory-confirmed chikungunya virus disease cases reported to ArboNET by state or territory—United States, 2015. In: 2015table-062216-finalpdf, editor Atlanta: Centers for Disease Control and Prevention; 2016. [Google Scholar]
- 14.Teixeira MG, Andrade AM, Costa MaC, Castro JN, Oliveira FL, Goes CS, et al. East/Central/South African genotype chikungunya virus, Brazil, 2014. Emerg Infect Dis. 2015;21(5):906–7. PubMed Central PMCID: PMCPMC4412231 10.3201/eid2105.141727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borgherini G, Poubeau P, Staikowsky F, Lory M, Le Moullec N, Becquart JP, et al. Outbreak of chikungunya on Reunion Island: early clinical and laboratory features in 157 adult patients. Clin Infect Dis. 2007;44(11):1401–7. 10.1086/517537 [DOI] [PubMed] [Google Scholar]
- 16.Horwood PF, Reimer LJ, Dagina R, Susapu M, Bande G, Katusele M, et al. Outbreak of chikungunya virus infection, Vanimo, Papua New Guinea. Emerg Infect Dis. 2013;19(9):1535–8. PubMed Central PMCID: PMCPMC3810919 10.3201/eid1909.130130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manimunda SP, Sugunan AP, Rai SK, Vijayachari P, Shriram AN, Sharma S, et al. Outbreak of chikungunya fever, Dakshina Kannada District, South India, 2008. Am J Trop Med Hyg. 2010;83(4):751–4. PubMed Central PMCID: PMCPMC2946737 10.4269/ajtmh.2010.09-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Win MK, Chow A, Dimatatac F, Go CJ, Leo YS. Chikungunya fever in Singapore: acute clinical and laboratory features, and factors associated with persistent arthralgia. J Clin Virol. 2010;49(2):111–4. 10.1016/j.jcv.2010.07.004 [DOI] [PubMed] [Google Scholar]
- 19.Sahadeo N, Mohammed H, Allicock OM, Auguste AJ, Widen SG, Badal K, et al. Molecular Characterisation of Chikungunya Virus Infections in Trinidad and Comparison of Clinical and Laboratory Features with Dengue and Other Acute Febrile Cases. PLoS Negl Trop Dis. 2015;9(11):e0004199 PubMed Central PMCID: PMCPMC4651505 10.1371/journal.pntd.0004199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pimentel R, Skewes-Ramm R, Moya J. [Chikungunya in the Dominican Republic: lessons learned in the first six months]. Rev Panam Salud Publica. 2014;36(5):336–41. [PubMed] [Google Scholar]
- 21.Powers AM, Roehrig JT. Alphaviruses. Methods Mol Biol. 2011;665:17–38. Epub 2010/12/01 10.1007/978-1-60761-817-1_2 [DOI] [PubMed] [Google Scholar]
- 22.Erasmus JH, Needham J, Raychaudhuri S, Diamond MS, Beasley DW, Morkowski S, et al. Utilization of an Eilat Virus-Based Chimera for Serological Detection of Chikungunya Infection. PLoS Negl Trop Dis. 2015;9(10):e0004119 PubMed Central PMCID: PMCPMC4619601 10.1371/journal.pntd.0004119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson BW, Goodman CH, Holloway K, de Salazar PM, Valadere AM, Drebot M. Evaluation of Commercially Available Chikungunya Virus Immunoglobulin M Detection Assays. Am J Trop Med Hyg. 2016; 95(1):182–92. PubMed Central PMCID: PMC4944686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19(6):1117–23. PubMed Central PMCID: PMCPMC2694472 10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. PubMed Central PMCID: PMCPMC3322381 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–73. PubMed Central PMCID: PMCPMC3408070 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheehan AM, Yee DL. Resources for the Hematologist In: Hoffman R, editor Hematology: Basic Principles and Practice. 6th ed. Philadelpha, PA: Elsevier Saunders; 2013. p. e1–e32. [Google Scholar]
- 28.Soumahoro MK, Boelle PY, Gaüzere BA, Atsou K, Pelat C, Lambert B, et al. The Chikungunya epidemic on La Réunion Island in 2005–2006: a cost-of-illness study. PLoS Negl Trop Dis. 2011;5(6):e1197 PubMed Central PMCID: PMCPMC3114750 10.1371/journal.pntd.0001197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sissoko D, Malvy D, Ezzedine K, Renault P, Moscetti F, Ledrans M, et al. Post-epidemic Chikungunya disease on Reunion Island: course of rheumatic manifestations and associated factors over a 15-month period. PLoS Negl Trop Dis. 2009;3(3):e389 PubMed Central PMCID: PMCPMC2647734 10.1371/journal.pntd.0000389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishnamoorthy K, Harichandrakumar KT, Krishna Kumari A, Das LK. Burden of chikungunya in India: estimates of disability adjusted life years (DALY) lost in 2006 epidemic. J Vector Borne Dis. 2009;46(1):26–35. [PubMed] [Google Scholar]
- 31.Nandha B, Krishnamoorthy K. Cost of illness due to Chikungunya during 2006 outbreak in a rural area in Tamil Nadu. Indian J Public Health. 2009;53(4):209–13. [PubMed] [Google Scholar]
- 32.Seyler T, Hutin Y, Ramanchandran V, Ramakrishnan R, Manickam P, Murhekar M. Estimating the burden of disease and the economic cost attributable to chikungunya, Andhra Pradesh, India, 2005–2006. Trans R Soc Trop Med Hyg. 2010;104(2):133–8. 10.1016/j.trstmh.2009.07.014 [DOI] [PubMed] [Google Scholar]
- 33.Rodríguez-Morales AJ, Calvache-Benavides CE, Giraldo-Gómez J, Hurtado-Hurtado N, Yepes-Echeverri MC, García-Loaiza CJ, et al. Post-chikungunya chronic arthralgia: Results from a retrospective follow-up study of 131 cases in Tolima, Colombia. Travel Med Infect Dis. 2016;14(1):58–9. 10.1016/j.tmaid.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 34.Mattar S, Miranda J, Pinzon H, Tique V, Bolanos A, Aponte J, et al. Outbreak of Chikungunya virus in the north Caribbean area of Colombia: clinical presentation and phylogenetic analysis. J Infect Dev Ctries. 2015;9(10):1126–32. 10.3855/jidc.6670 [DOI] [PubMed] [Google Scholar]
- 35.Yoon IK, Alera MT, Lago CB, Tac-An IA, Villa D, Fernandez S, et al. High rate of subclinical chikungunya virus infection and association of neutralizing antibody with protection in a prospective cohort in the Philippines. PLoS Negl Trop Dis. 2015;9(5):e0003764 PubMed Central PMCID: PMCPMC4423927 10.1371/journal.pntd.0003764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, et al. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370(9602):1840–6. 10.1016/S0140-6736(07)61779-6 [DOI] [PubMed] [Google Scholar]
- 37.Lee VJ, Chow A, Zheng X, Carrasco LR, Cook AR, Lye DC, et al. Simple clinical and laboratory predictors of Chikungunya versus dengue infections in adults. PLoS Negl Trop Dis. 2012;6(9):e1786 PubMed Central PMCID: PMCPMC3459852 10.1371/journal.pntd.0001786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renault P, Solet JL, Sissoko D, Balleydier E, Larrieu S, Filleul L, et al. A major epidemic of chikungunya virus infection on Reunion Island, France, 2005–2006. Am J Trop Med Hyg. 2007;77(4):727–31. [PubMed] [Google Scholar]
- 39.Lemant J, Boisson V, Winer A, Thibault L, André H, Tixier F, et al. Serious acute chikungunya virus infection requiring intensive care during the Reunion Island outbreak in 2005–2006. Crit Care Med. 2008;36(9):2536–41. 10.1097/CCM.0b013e318183f2d2 [DOI] [PubMed] [Google Scholar]
- 40.Mirabel M, Vignaux O, Lebon P, Legmann P, Weber S, Meune C. Acute myocarditis due to Chikungunya virus assessed by contrast-enhanced MRI. Int J Cardiol. 2007;121(1):e7–8. 10.1016/j.ijcard.2007.04.153 [DOI] [PubMed] [Google Scholar]
- 41.Obeyesekere I, Hermon Y. Myocarditis and cardiomyopathy after arbovirus infections (dengue and chikungunya fever). Br Heart J. 1972;34(8):821–7. PubMed Central PMCID: PMCPMC486987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sankari T, Hoti SL, Govindaraj V, Das PK. Chikungunya and respiratory viral infections. Lancet Infect Dis. 2008;8(1):3–4. 10.1016/S1473-3099(07)70295-5 [DOI] [PubMed] [Google Scholar]
- 43.Cashat-Cruz M, Morales-Aguirre JJ, Mendoza-Azpiri M. Respiratory tract infections in children in developing countries. Semin Pediatr Infect Dis. 2005;16(2):84–92. [DOI] [PubMed] [Google Scholar]
- 44.Renault P, Josseran L, Pierre V. Chikungunya-related fatality rates, Mauritius, India, and Reunion Island. Emerg Infect Dis. 2008;14(8):1327 PubMed Central PMCID: PMCPMC2600376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Economopoulou A, Dominguez M, Helynck B, Sissoko D, Wichmann O, Quenel P, et al. Atypical Chikungunya virus infections: clinical manifestations, mortality and risk factors for severe disease during the 2005–2006 outbreak on Réunion. Epidemiol Infect. 2009;137(4):534–41. 10.1017/S0950268808001167 [DOI] [PubMed] [Google Scholar]
- 46.Simmons G, Brès V, Lu K, Liss NM, Brambilla DJ, Ryff KR, et al. High Incidence of Chikungunya Virus and Frequency of Viremic Blood Donations during Epidemic, Puerto Rico, USA, 2014. Emerg Infect Dis. 2016;22(7):1221–8. PubMed Central PMCID: PMCPMC4918147 10.3201/eid2207.160116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Driggers RW, Ho CY, Korhonen EM, Kuivanen S, Jääskeläinen AJ, Smura T, et al. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N Engl J Med. 2016;374(22):2142–51. 10.1056/NEJMoa1601824 [DOI] [PubMed] [Google Scholar]
- 48.Hoarau JJ, Jaffar Bandjee MC, Krejbich Trotot P, Das T, Li-Pat-Yuen G, Dassa B, et al. Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol. 2010;184(10):5914–27. 10.4049/jimmunol.0900255 [DOI] [PubMed] [Google Scholar]
- 49.Ozden S, Huerre M, Riviere JP, Coffey LL, Afonso PV, Mouly V, et al. Human muscle satellite cells as targets of Chikungunya virus infection. PLoS One. 2007;2(6):e527 PubMed Central PMCID: PMCPMC1885285 10.1371/journal.pone.0000527 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Serum collected from discarded blood samples of patients visiting an emergency clinic in the DR was tested for CHIKV RNA. CT values from positive results are displayed as bins (e.g., bin 17 represents 17.0≤CT<18.0)
(TIF)
Using an Illumina HiSeq platform, complete genome sequences were determined directly from RNA isolated from the sera of 10 individuals. The overall alignment rate of the reads varied widely among samples, ranging from 7–88%, with a mean of approximately 20%. Nucleotide and amino acid identity among the consensus sequences from these individuals was >99.9%. The MCC phylogeny showed that the Dominican Republic sequences clustered within the Asian lineage together with other Caribbean sequences isolated in 2014 [11, 16]. The most closely related Asian sequences circulated between 2012 and 2013 in Micronesia, the Philippines and China. The three major CHIKV genotypes are labeled. Nodes with clade credibility’s ≥ 95% are labeled accordingly.
(TIF)
Serum collected from discarded blood samples of patients visiting an emergency clinic in the DR was tested for CHIKV RNA and IgM, and samples testing positive for RNA were retroactively matched to clinical signs and symptoms documented by physicians (arthralgia, myalgia, enophthalmos, respiratory signs and symptoms, and gastrointestinal signs and symptoms) or results from complete blood count analysis (lymphopenia and thrombocytopenia). Results were organized by CT bins defined by approximate log change from positive control (e.g., bin 20–22 represents samples within a 10-fold decrease from control, bin 23–25 within a 100-fold decrease, etc.). Only percent patients presenting with lymphopenia was significantly associated with CT bin by linear regression analysis (p = 0.006), suggesting a positive correlation between percent patients presenting with lymphopenia and relative viremia.
(TIF)
(DOCX)
Data Availability Statement
Genbank accession numbers: BankIt1972278 Seq1 KY272961, BankIt1972278 Seq2 KY272962, BankIt1972278 Seq3 KY272963, BankIt1972278 Seq4 KY272964, BankIt1972278 Seq5 KY272965, BankIt1972278 Seq6 KY272966, BankIt1972278 Seq7 KY272967, BankIt1972278 Seq8 KY272968, BankIt1972278 Seq9 KY272969, BankIt1972278 Seq10 KY272970. All other relevant data are within the paper and its supporting data.