Abstract
Borrelia mayonii, a Borrelia burgdorferi sensu lato (Bbsl) genospecies, was recently identified as a cause of Lyme borreliosis (LB) among patients from the upper midwestern United States. By microscopy and PCR, spirochete/genome loads in infected patients were estimated at 105 to 106 per milliliter of blood. Here, we present the full chromosome and plasmid sequences of two B. mayonii isolates, MN14-1420 and MN14-1539, cultured from blood of two of these patients. Whole genome sequencing and assembly was conducted using PacBio long read sequencing (Pacific Biosciences RSII instrument) followed by hierarchical genome-assembly process (HGAP). The B. mayonii genome is ~1.31 Mbp in size (26.9% average GC content) and is comprised of a linear chromosome, 8 linear and 7 circular plasmids. Consistent with its taxonomic designation as a new Bbsl genospecies, the B. mayonii linear chromosome shares only 93.83% average nucleotide identity with other genospecies. Both B. mayonii genomes contain plasmids similar to B. burgdorferi sensu stricto lp54, lp36, lp28-3, lp28-4, lp25, lp17, lp5, 5 cp32s, cp26, and cp9. The vls locus present on lp28-10 of B. mayonii MN14-1420 is remarkably long, being comprised of 24 silent vls cassettes. Genetic differences between the two B. mayonii genomes are limited and include 15 single nucleotide variations as well as 7 fewer silent vls cassettes and a lack of the lp5 plasmid in MN14-1539. Notably, 68 homologs to proteins present in B. burgdorferi sensu stricto appear to be lacking from the B. mayonii genomes. These include the complement inhibitor, CspZ (BB_H06), the fibronectin binding protein, BB_K32, as well as multiple lipoproteins and proteins of unknown function. This study shows the utility of long read sequencing for full genome assembly of Bbsl genomes, identifies putative genome regions of B. mayonii that may be linked to clinical manifestation or tissue tropism, and provides a valuable resource for pathogenicity, diagnostic and vaccine studies.
Introduction
Members of the Borrelia genus group into two major disease causing clades, the Lyme borreliosis (LB) group and the relapsing fever (RF) group [1, 2]. Genospecies of the Borrelia burgdorferi sensu lato (Bbsl) complex are the causative agents of LB, the most common tick-borne disease in the northern hemisphere [3–6]. Humans acquire Bbsl infection by the bite of a hard tick, Ixodes spp., and in nature, small mammals or birds serve as hosts for Bbsl genospecies [4, 7, 8]. Although 20 Bbsl genospecies have been described worldwide, B. afzelii, B. burgdorferi sensu stricto (hereafter called B. burgdorferi), B. bavariensis and B. garinii, most commonly cause LB in Europe and Asia, whereas B. burgdorferi causes the majority of cases in North America [3]. Clinical features of LB are broad, variable, and disease manifestations seem to be linked with distinct tissue tropisms of specific Bbsl genospecies [9]. Early infection is typically localized and in the absence of treatment, the spirochete can disseminate to the nervous system (neuroborreliosis), joints (arthritis), heart (carditis), or other regions of the skin [10, 11]. B. burgdorferi is more often associated with Lyme arthritis as compared to other genospecies [9].
A novel Bbsl genospecies, Borrelia mayonii, was recently described as a cause of LB among six patients in the upper midwestern United States [12, 13]. Spirochetes isolated from two infected patients and taxonomically characterized using an eight gene multi-locus sequencing analysis (MLSA) demonstrated that the pairwise similarity to Bbsl genospecies fell below the system threshold for delineating Bbsl genospecies, confirming it as a novel genospecies [12]. For the six patients described, somewhat differing clinical presentations as compared to infection with B. burgdorferi were noted including, nausea or vomiting, symptoms potentially consistent with neurological effects, and a diffuse macular rash. Two of the six patients were hospitalized. By microscopy and quantitative PCR, B. mayonii infected patients were shown to have elevated numbers of spirochetes/genomes in their blood (105 to 106 spirochetes/genomes per milliliter) as compared to loads previously reported for B. burgdorferi (102 to 103 genomes per milliliter; 0.1 spirochete per milliliter) [14–16]. This elevated spirochetemia coupled with blood based PCR detection for five patients led to the suggestion that B. mayonii may have a distinct tissue tropism as compared to B. burgdorferi [11]. Patients’ infections were likely acquired by the bite of I. scapularis, which transmits B. burgdorferi in the United States. I. scapularis positive for B. mayonii were identified in several counties in Wisconsin and Minnesota [12, 13].
Bbsl genospecies possess one of the most unique and complex prokaryotic genomes [17, 18]. Their genomes are comprised of a long linear chromosome (~1Mb) and multiple (up to 21) linear and circular plasmids (collectively ~600 kbp), ranging in size from 5 to 56 kbp, of which several encode highly redundant sequence [17, 18]. The main chromosome of Bbsl genospecies is highly syntenic with low sequence variation across nearly the entire length [19]. This is in contrast to the extrachromosomal plasmids which are more structurally and genetically variable and encode proteins necessary for infection of vertebrate hosts and tick vectors, pathogenesis and immune cell invasion [19, 20]. The nomenclature developed for Bbsl plasmids is based on the reference strain B. burgdorferi B31. Plasmids from other Bbsl strains which show ≥95% percent identity to the PF32 plasmid partitioning proteins from B31 plasmids are named according to the B31 nomenclature [20]. The plasmids lp54 and cp26, have been observed in all characterized strains [21]. Species specific sequence differences in these two plasmids occur primarily in surface exposed genes [20]. Other plasmids are found in fewer than 10% of the analyzed Bbsl strains, while some plasmids are nearly always present but vary by size, gene content and copy number [21]. Cross species genomic comparisons suggest gene duplication/loss and differential expression patterns in addition to sequence variation in conserved lipoproteins are the primary drivers of differing tissue tropisms rather than gain of virulence genes [21–23].
Because of the similar sizes and highly paralogous nature of Bbsl plasmid sequences, next generation sequencing platforms have often yielded unfinished genome assemblies of Bbsl genospecies [22]. Short read platforms are typically unable to span the large number of repetitive regions present in Borrelia strains which in turn means the origin of the read cannot be precisely determined. PacBio® sequencing offers the potential to overcome these limitations for Bbsl genospecies allowing for de novo assembly of full genomes. The long, multikilobase read lengths combined with high read coverage, can allow for accurate, non-hybrid, de novo assemblies that span problematic repetitive regions [24].
To investigate the genome structure of B. mayonii as well as genetic differences between B. mayonii and other Bbsl genospecies, we performed whole genome sequencing on two B. mayonii strains isolated from the blood of two acutely ill patients. Genomes were sequenced using the Pacific Biosciences RSII SMRT platform. By generating long reads (~6-15kb), with an average of >200X coverage, full genomes could be assembled and highly redundant plasmid sequences readily differentiated. These results demonstrate that the B. mayonii genome has a unique repertoire of plasmids and genetic features that distinguish it from other Bbsl genomes. Full chromosome and plasmid sequences for the B. mayonii MN14-1420 (type strain) and MN14-1539 genomes have been deposited in the NCBI database under BioProject # PRJNA321302.
Materials and Methods
Bacterial culture and DNA isolation
Low passage (P3) frozen stocks of two B. mayonii strains, MN14-1420 and MN14-1539, were grown in modified BSK at 34°C under microaerophilic conditions [13]. Spirochetes were pelleted by centrifugation at 8000 X g for 10 minutes. DNA was isolated from the pellet using the Qiagen DNA mini kit tissue protocol (Qiagen, Valencia, CA). Purified DNA was eluted in dH20 and quantified using the Qubit fluorimeter (Life Technologies, Grand Island, NY).
Library preparation and whole genome sequencing
Whole-genome sequencing was carried out using the PacBio RSII (Pacific Biosciences, Menlo Park, CA) platform. 5 μg of input genomic DNA was used for 10-kb fragment library preparation. DNA was sheared with g-TUBE® microcentrifuge tubes (Covaris, Woburn, MA) and SMRTbell libraries prepared with DNA Template Kit 2.0 (Pacific Biosciences, Menlo Park, CA). Sequencing primers were annealed to the SMRTbell template and samples were sequenced using C3 chemistry, Polymerase version 5, SMRTAnalysis software version 2.3.0 and a single SMRT cell (Pacific Biosciences, Menlo Park, CA). Raw read quality and average reference consensus were determined using SMRTAnalysis software version 2.3.0.
Genome assembly
De novo genome assembly of PacBio reads for MN14-1420 (49,701 reads totaling 343,091,509 bases, mean length 6,903 bp, max 34,633) and MN14-1539 (87,914 reads totaling 474,379,679 bases, mean length 5,395 bp, max 22,928) was conducted using the hierarchical genome-assembly process (HGAP3, SmrtAnalysis 2.3.0) with default parameters (expected genome size 2 Mbp, minimum target coverage 15X), which included consensus polishing with Quiver [24]. Briefly, raw reads were converted into high-quality pre-assembled reads which were assembled into contigs followed by a final consensus calling step to polish the assembled contigs. The mean quality score for the pre-assembled genomic reads was 85% for both strains. The assembly resulted in 15 and 14 polished contigs for MN14-1420 and MN15-1539 respectively. Each polished contig was quality assessed for topology by determining end to end overlap (circular) or long read-through of palindromic telomere sequence at the 5’ and 3’ ends of the contig (linear) using gepard 1.30 [25]. All contig assemblies were also subjected to re-sequencing analysis to validate trimmed regions and circular versus linear topology. Suspect circular contigs were trimmed to remove end to end overlap and re-oriented and stitched so that the circularized portion was located at the center of the contig. Reads were re-mapped to the trimmed re-oriented contig to ensure consistent coverage across the stitched (circularized) portion of the contig. Suspect linear contigs were trimmed to remove 5’ and 3’ sequence resulting from telomere read through. Reads were re-mapped to trimmed contigs to ensure consistent raw read coverage across the assembled genomes. The initial MN14-1539 assembly was missing two small contigs as compared to MN14-1420, a circular ~8 kbp contig and linear ~5 kbp contig. Reads from the MN14-1539 dataset were therefore mapped to the two contigs from MN14-1420. Reads corresponding to the circular ~8 kbp contig were present in MN14-1539 (92X coverage), while no trace reads mapped to the ~5 kbp linear contig. Sequence trimming and stitching was performed using CLC Genomics Workbench 8.0 (Qiagen, Valencia, CA). Re-sequencing analysis was conducted using SMRTAnalysis 2.3.0 (maxHits = 1, minAccuracy = 0.75, minLength = 50).
Genome annotation
Genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline, to take advantage of the ability to combine annotation methods based on alignment with other Bbsl genospecies along with annotation methods predicting functional elements directly from sequence [26]. The annotation was compared between the two isolates to identify any major differences and overall percentages of gene specific ontology determined, i.e. hypothetical proteins, lipoproteins, pseudogenes, and plasmid partitioning genes. Additional lipoprotein prediction was performed using the spirochete specific (SpLip) algorithm with the B. burgdorferi prediction matrix [27].
Nomenclature for B. mayonii plasmids roughly followed the previously established criteria of naming plasmids according to the B. burgdorferi B31 nomenclature if PF32 plasmid partitioning proteins display ≥95% average amino acid identity (AAI) [20]. Additionally, if B. mayonii plasmids shared ≤95% AAI to B31 PF32 proteins, but ≥ 95% to PF32 proteins in other B. burgdorferi strains or genospecies, they were named thusly. For example, cp32-13 does not exist in B31, however the B. mayonii PF32 protein from cp32-13 shares 100% AAI to cp32-13 PF32 proteins in 3 B. burgdorferi strains (CA-11.2A, 72a, 118a). cp9 does not encode a PF32 protein, however the B. mayonii cp9 molecule contained all of the obvious genetic elements of cp9 from other Bbsl genospecies. The PF32 proteins from B mayonii lp28-4 and lp36 share 95% and 99% with lp-28-4 and lp36 PF32s from B. bissettii and B. burgdorferi strain 29805, respectively. B. mayonii lp28-10 was given a unique plasmid designation as its PF32 protein shares ≤ 89% AAI with PF32 proteins in the NCBI non-redundant protein database.
Chromosomal synteny was examined on a gene by gene basis by comparing the B. burgdorferi B31 chromosome and two B. mayonii chromosomes using a progressive Mauve 2.4.0 alignment [28]. The Mauve alignment was compared to the Core Genome comparison tool available at www.borreliabase.org [29]. Ideograms of each annotated plasmid were generated using CViT 1.2.1 (linear) and Circos 0.69 (circular) [30, 31]. Silent vls cassettes were determined using BLAST 2.2.30+ [32] by querying the lp28-10 plasmid DNA sequence with the DNA sequence from the predicted vlsE open reading frame (ORF) from the template strand at the 3’ terminus of the plasmid. Comparisons of silent vls cassette copy number between MN14-1420 and MN14-1539 were determined by whole plasmid alignments generated by STRETCHER [33]. Silent vls cassettes and PF54 array comparisons were visualized using EasyFig 2.1 [34].
Comparative genomics
Two way average nucleotide identity (ANI) analysis was calculated using best hit and reciprocal best hit analysis (window size 1000 bp, step size 200 bp) as previously described (http://enve-omics.ce.gatech.edu/ani/) [35]. BLAST analyses between B. mayonii and B. burgdorferi B31 chromosomal and plasmid proteins were conducted using standalone BLAST 2.2.30+ and default blastp parameters. Chromosomal and plasmid proteins were compared separately. B. burgdorferi B31 homologs were identified using the “max_target_seqs 1” argument and an e-value ≥10−2. Each BLAST alignment with an e-value ≥10−2 was manually inspected. B. burgdorferi B31 plasmid proteins identified as absent were blasted against the NCBI non-redundant protein database to confirm they were present in at least one other B. burgdoferi sensu stricto strain and not unique to the B. burgdorferi B31 genome. Nucleotide sequence for B. burgdorferi genes considered missing from the B. mayonii genome were also queried against all unmapped reads from the two assemblies.
For comparison of proteins present on B. mayonii plasmids to other Bbsl genospecies, all individual protein sequence data was downloaded from NCBI (April 2016) for B. burgdorferi (n = 41,177), B. garinii (n = 17,334), B. afzelii (n = 11,502), B. bissettii (n = 2,946), and B. miyamotoi (n = 5,264). The downloaded proteins were converted to BLAST databases using the makeblastdb functionality in BLAST 2.2.30+. Each database was queried with a multi-fasta file comprised of each predicted protein from the individual B. mayonii plasmids and an average AAI determined for proteins greater than or equal to 100 amino acids in length. Each BLAST hit was inspected to ensure equivalent query coverage and alignment length. Phylogenetic analysis of a ~6.5 kbp region of cp26 from Bbsl genospecies was performed using MEGA6 [36]. Briefly, DNA sequences were aligned using MUSCLE, maximum likelihood trees generated using 4 discrete gamma categories and rooted at the midpoint based on Qiu et al 2014 [22], followed by 1000 bootstrap replicates. Sequences included in the phylogenetic analysis were retrieved from GenBank. Multi-alignment of the 26 amino acid C6 peptide was performed using Clustal Omega with default parameters [37].
GenBank accession numbers
The two B. mayonii genome assemblies have been deposited in the GenBank public database under BioProject # PRJNA321302. BioSample, and individual Accession numbers for each DNA molecule are shown in Table 1.
Table 1. GenBank accession details for B. mayonii MN1420 and MN14-1539.
| BioProject | BioSample | Strain | Plasmid name | Accession |
|---|---|---|---|---|
| PRJNA321302 | SAMN04979181 | MN14-1420 | Chromosome | CP015780 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | cp26 | CP015781 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | cp32-1 | CP015782 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | cp32-13 | CP015783 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | cp32-3 | CP015784 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | cp32-4 | CP015785 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | cp32-6 | CP015786 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | cp9 | CP015787 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | lp17 | CP015788 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | lp25 | CP015789 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | lp28-10 | CP015790 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | lp28-3 | CP015791 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | lp28-4 | CP015792 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | lp36 | CP015793 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | lp5 | CP015794 |
| PRJNA321302 | SAMN04979181 | MN14-1420 | lp54 | CP015795 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | Chromosome | CP015796 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | cp26 | CP015797 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | cp32-1 | CP015798 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | cp32-13 | CP015799 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | cp32-3 | CP015800 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | cp32-4 | CP015801 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | cp32-6 | CP015802 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | cp9 | CP015962 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | lp17 | CP015803 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | lp25 | CP015804 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | lp28-10 | CP015805 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | lp28-3 | CP015806 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | lp28-4 | CP015807 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | lp36 | CP015808 |
| PRJNA321302 | SAMN04979182 | MN14-1539 | lp54 | CP015809 |
Results
Genome sequencing and assembly of B. mayonii
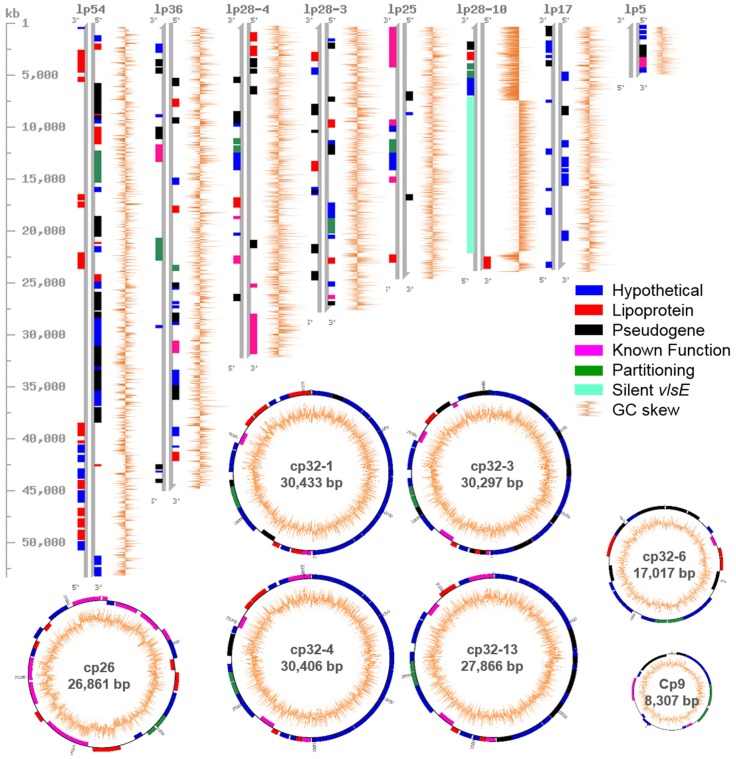
For the two B. mayonii genomes, MN14-1420 and MN14-1539, the mean read length was 6149 bp and the average nucleotide coverage was 202X and 232X, respectively (range: 98X – 593X). The overall raw read quality was 0.85 with an average reference consensus of 99.99%. Complete contigs were generated for the chromosome and each of the extrachromosomal plasmids. The MN14-1420 genome is 1,311,545 bp in size (26.9% average GC content) and is comprised of a 904,387 bp linear chromosome, 8 linear plasmids and 7 circular plasmids (Table 2, Fig 1). It is predicted to encode 850 chromosomal genes and 411 plasmid-borne genes. The MN14-1539 genome is 1,293,788 bp and comprised of an identical sized linear chromosome, 7 linear and 7 circular plasmids. The MN14-1539 genome is predicted to encode 850 chromosomal genes and 405 plasmid-borne genes. Overall, the MN14-1420 and MN14-1539 genome sequences were highly similar and will therefore be discussed jointly, with detected differences noted. Size, average nucleotide coverage, GC content, and number of predicted open reading frames (ORF) is shown in Table 2 for the B. mayonii linear chromosome and each of the plasmids.
Table 2. Whole genome sequencing and annotation statistics for B. mayonii MN14-1420 and MN14-1539.
| Contig | Size (bp) | Coverage (Avg) | Predicted Genes | Coding | rRNA | tRNA | GC% |
|---|---|---|---|---|---|---|---|
| Chromosome | 904,387 | 208.4X | 850 | 788 | 5 | 33 | 28.7 |
| cp32-1 | 30,433 | 217.1X | 42 | 39 | 0 | 0 | 29.3 |
| cp32-3 | 30,297/30,362* | 157.2X | 43 | 31 | 0 | 0 | 28.7 |
| cp32-4 | 30,406 | 160.4X | 43 | 42 | 0 | 0 | 28.9 |
| cp32-6 | 17,017 | 153.1X | 22 | 15 | 0 | 0 | 27.2 |
| cp32-13 | 27,866 | 174.8X | 39 | 34 | 0 | 0 | 28.9 |
| cp26 | 26,861/26,835* | 145.5X | 26 | 26 | 0 | 0 | 25.8 |
| cp9 | 8,307 | 98.2X | 8 | 6 | 0 | 0 | 24.2 |
| lp54 | 53,359/53,343* | 593.5X | 62 | 45 | 0 | 0 | 27.0 |
| lp36 | 44,035 | 218.8X | 35 | 23 | 0 | 0 | 23.6 |
| lp28-3 | 27,809 | 578.7X | 27 | 18 | 0 | 0 | 24.2 |
| lp28-4 | 32,294/32,269* | 502.9X | 20 | 13 | 0 | 0 | 24.2 |
| lp28-10 | 23,800/19,566* | 235.8X | 8 | 7 | 0 | 0 | 39.4 |
| lp25 | 24,702/24696* | 139.1X | 12 | 10 | 0 | 0 | 23.1 |
| lp17 | 23,735/23,770* | 403.9X | 18 | 15 | 0 | 0 | 23.5 |
| lp5 | 5,225** | 309.7X | 6 | 5 | 0 | 0 | 23.6 |
*Plasmid size differences displayed as MN14-1420/MN14-1539
**lp5 was not detected in the MN14-1539 genome
Fig 1. Full plasmid content of B. mayonii.
Visual depiction and annotation of the linear and circular plasmids identified in B. mayonii. Linear plasmid (lp) ideograms were drawn with CViT 1.2.1 and are shown in relation to the size marker in kilobases (kb). Circular plasmid (cp) ideograms were drawn with Circos 0.69, circular plasmids are not drawn to scale. Annotation data was derived from the NCBI Prokaryotic Genome Annotation Pipeline. Proteins annotated as hypothetical (blue), lipoprotein (red), known function (pink), or plasmid partitioning (green) are indicated. Also shown are pseudogenes (black), the silent vls cassettes (light blue) and GC skew (orange lines), lp5 was only present in MN14-1420. lp28-10 is ~4 kbp shorter in MN14-1539.
B. mayonii chromosome
Sequence comparison of the linear chromosomes of MN14-1420 and MN14-1539 revealed an average nucleotide identity (ANI) of >99.99%, with no whole gene differences and only 9 single nucleotide polymorphisms (SNPs), two of which were non-synonymous (Table 3). The coding density of the B. mayonii chromosome is 95.2%. Analysis of the structural arrangement of the B. mayonii chromosome showed it is highly syntenic with chromosomes from other Bbsl genospecies (data not shown). When the sequence of the B. mayonii chromosome was compared with chromosomes available for 8 other Bbsl genospecies, the ANI was less than 94%. The highest ANI was to B. burgdorferi B31 at 93.83% followed by B. bissetii DN127 at 93.52% (S1 Table). The B. mayonii chromosome is predicted to contain 32 tRNAs, 5 rRNAs (16S, and two copies of 23 and 5S), 3 ncRNAs, 22 pseudogenes and 788 protein encoding genes. Whole chromosome BLAST analysis between B. mayonii and B. burgdorferi B31 revealed 763 (97%) proteins with greater than 80% amino acid identity (AAI), consistent with a highly conserved gene repertoire for the B. mayonii and B. burgdorferi chromosomes.
Table 3. Nucleotide differences detected between B. mayonii strains MN14-1420 and MN14-1539*.
| Molecule | Position | Difference | MN14-1420 | MN14-1539 | Result |
|---|---|---|---|---|---|
| Chromosome | 153,205 | SNP | C | A | Synonymous |
| Chromosome | 208,477 | SNP | A | C | Non-synonymous |
| Chromosome | 258,586 | SNP | A | G | Synonymous |
| Chromosome | 410,389 | SNP | T | C | Synonymous |
| Chromosome | 459,470 | SNP | G | T | Intergenic |
| Chromosome | 506,116 | SNP | G | A | Synonymous |
| Chromosome | 524,398 | SNP | C | T | Synonymous |
| Chromosome | 589,450 | SNP | T | C | Synonymous |
| Chromosome | 680,050 | SNP | C | T | Non-synonymous |
| lp54 | 9,877 | VNTR | CTAAACTTAATTAAAA | - | Intergenic |
| lp54 | 28456/28440** | SNP | A | G | Intergenic |
| lp54 | 33165/33149** | SNP | T | C | Synonymous |
| cp26 | 10,001 | VNTR | TATGGAACTAACAGTTCCA | - | Intergenic |
| cp26 | 12204/12185** | VNTR | AAAACAT | - | Intergenic |
| lp25 | 24612 | VNTR | TTAAAG | - | Intergenic |
| lp17 | 19980 | VNTR | - | TAATTAATATATGATATAAATAAA | Intergenic |
| cp32-3 | 206 | SNP | C | T | Synonymous |
| cp32-3 | 3926 | SNP | T | G | Non-synonymous |
| cp32-3 | 22155 | VNTR | - | CACTAAAATAGACAATGTTGAAAAGAATTTAAA | Intergenic |
| lp28-4 | 17602 | VNTR | TAGTTCATCTTCTTCTTGTTTTTTCTT | - | Intergenic |
| lp28-4 | 23,645/23619** | INDEL | - | AT | Intergenic |
| lp28-3 | 3637 | SNP | C | A | Non-synonymous |
| lp36 | 31962–3 | SNP | AA | TT | Intergenic |
*Excluding nucleotide differences in the vls locus of lp28-10
**Due to VNTR based differences in sequence length respective base position is displayed as MN14-1420/MN14-1539
SNP: Single Nucleotide Polymorphism, VNTR: Variable Number Tandem Repeat, INDEL: Insertion/Deletion
Plasmid content of B. mayonii
Like other Bbsl genospecies, B. mayonii harbors a wide range of both linear and circular extrachromosomal plasmids totaling 407,158 bp, with 329 predicted protein coding genes and an average GC content of 26.7%. The overall coding density of the B. mayonii plasmids is 71.9%. Analogous Bbsl plasmids identified in B. mayonii include lp54, lp36, lp28-3, lp28-4, lp25, lp17, lp5, 5 cp32s, cp26, and cp9 (Fig 1). Sequence conservation and overall gene content and structural organization were noted for lp54, lp25, lp5, cp9, cp26, and the cp32s as compared to the respective plasmids in other Bbsl genospecies. The B. mayonii plasmids lp36, lp28-3, lp28-4, lp28-10, and lp17 show the greatest diversity in structural organization and gene content as compared to the respective plasmids from other Bbsl genospecies. One plasmid, lp5, is present in the MN14-1420 genome and absent from the MN14-1539 genome. Overall, the plasmid gene content of B. mayonii is characterized by a large number of predicted hypothetical proteins (59.3%), pseudogenes (19.9%), and lipoproteins (13.1%) (Fig 1). Only 6 SNPs, with the exception of the vls locus, were observed between the plasmid sequences of MN14-1420 and MN14-1539 (Table 3). Size differences noted between the cp32-3, cp26, lp54, lp28-4, lp25 and lp17 plasmids in MN14-1420 and MN14-1539 were due to either insertion/deletion (INDEL) or variable number of tandem repeats (VNTR), all of which were intergenic (Table 3).
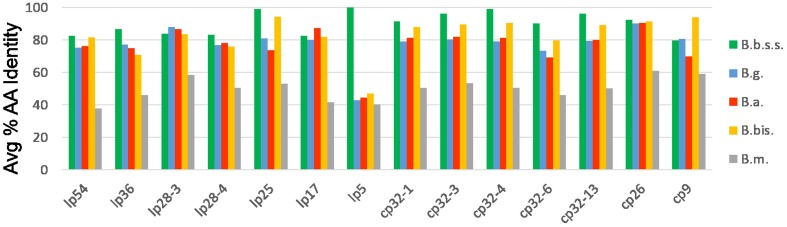
Amino acid sequences for all proteins encoded on the B. mayonii plasmids were compared to protein databases constructed for 4 Bbsl genospecies, B. burgdorferi, B. bissettii, B. garinii and B. afzelii, as well as the RF-like Borrelia, B. miyamotoi. Fig 2 shows the mean AAI of all proteins encoded on each of the 14 B. mayonii plasmids as compared to plasmid located proteins from the other Borrelia species. Lp28-10 was left out of the analysis as ORFs are predicted throughout the silent vls cassettes. Proteins present on 11 B. mayonii plasmids, lp54, lp36, lp28-4, lp25, lp5, cp32-1, cp32-3, cp32-4, cp32-6, cp32-13, and cp26, show the highest AAI at 82.6%, 86.6%, 83.2%, 99.3%, 100%, 91.7%, 96.3, 99.3%, 90.2%, 96.4%, and 92.6% respectively, to homologs present in B. burgdorferi.
Fig 2. Plasmid based protein composition of B. mayonii in comparison to 4 other Bbsl genospecies and 1 RF Borrelia species.
The mean amino acid identity is indicated for proteins present on each of the 14 B. mayonii plasmids. Comparisons are to protein databases constructed for B. burgdorferi (B.b.s.s.; green), B. garinii (B.g.; blue), B. afzelii (B. a.; red), B. bissettii (B.b.; yellow), and B. miyamotoi (B.m.; grey).
Proteins present on three B. mayonii plasmids, lp28-3, lp17 and cp9, display slightly higher AAI to Bbsl genospecies other than B. burgdorferi. B. mayonii cp9 encoded proteins share the highest AAI (94.2%) to cp9 proteins from B. bissettii, whereas lp28-3 and lp17 had higher overall AAI to B. garinii and B. afzelii (88.2% and 87.5%, respectively). Each of the proteins on these two plasmids was further interrogated to identify those showing higher AAI to genospecies other than B. burgdorferi. The B. mayonii lp28-3 plasmid encodes a single hypothetical protein, Bmayo_04835 that displays 82 to 92% AAI identity to hypothetical proteins present in other genospecies as compared to only 50% AAI to a hypothetical protein present in the B. burgdorferi database. On lp17, a single predicted hypothetical protein, Bmayo_05225, returned poor amino acid similarity when compared to B. burgdorferi, B. garinii, B. bissettii, and B. miyamotoi proteins (0–80% AAI, 0–40% query coverage), whereas strong identity (89% AAI, 100% query coverage) to a hypothetical protein in B. afzelii was noted. When Bmayo_05225 was compared against the NCBI non-redundant protein database significant identity (88–89% AAI) was also detected to hypothetical protein homologs present in both B. valaisiana and B. spielmanii.
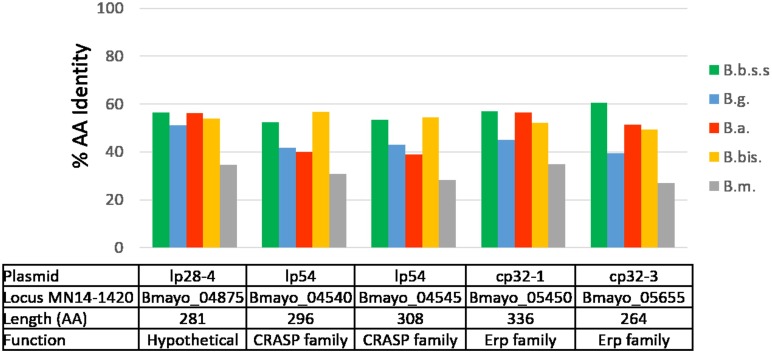
No proteins unique to B. mayonii were identified and only five plasmid encoded proteins were detected that were ≥ 100 amino acids in length and shared ≤ 60% identity to proteins from other Bbsl species (Fig 3). Four of these proteins, Bmayo_04540, Bmayo_04545, Bmayo_05450, Bmayo_05655 are part of the CRASP (Complement Regulator-Acquiring Surface Protein) family, which are outer bacterial surface proteins involved in complement resistance, tissue tropism, and host cell binding of Bbsl genospecies (Reviewed in [38]). The two CRASP-1-like proteins (Bmayo_04540 and Bmayo_04545) encoded within the PF54 gene array of B. mayonii lp54 share from 39 to 56% AAI with CRASP proteins from other Bbsl genospecies. The two CRASP-3/4/5-like, also known as Erp family proteins (OspEF-related protein), encoded on cp32-1 and cp32-3 (Bmayo_05450 and Bmayo_05655), share from 39 to 60% AAI with CRASP-3/4/5 proteins from other Bbsl genospecies. The remaining protein encoded on lp28-4 is predicted as a hypothetical protein in B. mayonii and other Bbsl genospecies.
Fig 3. B. mayonii proteins with greatest sequence diversity as compared to 4 other Bbsl genospecies and 1 RF Borrelia species.
Bar graph depicting B. mayonii proteins greater than 100 amino acids with less than 60% amino acid identity to other Bbsl proteins. Amino acid identity is shown with respect to homologs present in B. burgdorferi (B.b.s.s.; green), B. garinii (B.g.; blue), B. afzelii (B. a.; red), B. bissettii (B.b.; yellow) and B. miyamotoi (B.m.; grey).
B. mayonii cp26 and lp54
The conserved B. mayonii cp26 plasmid encodes a homolog of the OspC protein (Bmayo_06270). In B. burgdorferi, OspC is also encoded on cp26 and plays an important role in the invasion of tick salivary glands and mammalian infection [39, 40]. Between B. mayonii and B. burgdorferi B31, the OspC protein shares 76% AAI. The OspC proteins of B. mayonii MN14-1420 and MN14-1539 exhibit 100% AAI, which following the B. burgdorferi classification system, suggests both B. mayonii strains belong to the same allele group, termed here OspC type Bmayo1. Phylogenetic analysis of a ~6.5 kbp region of cp26, shown previously to be relatively free from recombination in Bbsl genospecies [22], is shown in Fig 4. The two B. mayonii strains form their own branch, distinct from B. burgdorferi and B. bissettii. In contrast to the B. burgdorferi strains, no genetic differences were detected across the 6.5 kbp region for the two B. mayonii strains. Within this ~6.5 kbp region, a 743 nucleotide stretch between genes Bmayo_06250 (hypothetical protein) and Bmayo_06255 (ABC transporter OppAIV) appears to be species specific for B. mayonii. BLAST analysis of the 743 nucleotide stretch against the NCBI non-redundant nucleotide database did not identify sequence similarity to any other Borrelia species.
Fig 4. Phylogenetic analysis of B. mayonii based on a ~6.5 kbp region of cp26.
The ~6.5 kbp region of cp26 encompasses open reading frames from BB_B14 through BB_B18 of B. burgdorferi B31. DNA sequences from 8 other Bbsl genospecies (22 strains) were retrieved from GenBank. Bootstrap values greater than 50% are shown. The scale bar corresponds to 0.02 substitutions per nucleotide position.
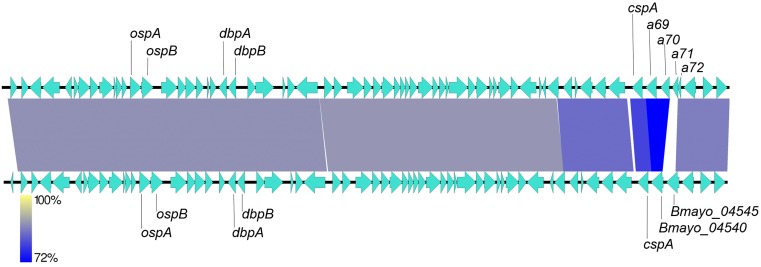
The lp54 plasmid is also present in B. mayonii and like other Bbsl lp54 plasmids, encodes the outer surface proteins, OspA and OspB (Bmayo_04320 and Bmayo_04325), which play an important role in colonization of tick vectors [41]. The OspA and OspB homologs in B. mayonii display 89% and 84% AAI to B. burgdorferi B31 OspA and OspB, respectively. Homologs to the lp54-borne decorin binding proteins, DbpA and DbpB (Bmayo_04355 and A7X69_04360), exhibit 67% and 73% AAI with B. burgdorferi B31, respectively. In addition to the aforementioned CRASP-1-like proteins (Bmayo_04540 and Bmayo_04545), a homolog of the CRASP CspA, (Bmayo_04535) is also encoded in the variable PF54 gene array and shares only 60% AAI to CspA from B. burgdorferi B31. The lower nucleotide identity between B. burgdorferi B31 and B. mayonii MN14-1420 across the PF54 gene array (3’ end of lp54) is highlighted in Fig 5. Nine proteins, BB_A05, 15, 25, 44, 52, 57, 65, 66, and 73, identified by Qiu et al [22] as highly variable among Bbsl genospecies and thus likely to be involved in host-pathogen interaction, were predicted as functional proteins in B. mayonii. Between B. burgdorferi B31 and B. mayonii, the AAI of these 9 proteins ranged from 67–90%.
Fig 5. Comparison of B. burgdorferi B31 and B. mayonii lp54.
Heat map of nucleotide identity between lp54 plasmids of B. burgdorferi B31 (top) and B. mayonii MN14-1420 (bottom). Nucleotide identity along the entire lp54 plasmids is displayed as colored blocks (Yellow = highest, Blue = lowest). Orientation of arrows indicates gene orientation on the forward or reverse DNA strand. The variable PF54 gene array is at the 3’ end of the plasmid. Specific gene loci discussed in text are indicated.
cp32 plasmids of B. mayonii
Multiple members of the cp32 plasmid family are present in all the analyzed Bbsl genospecies [42]. Despite the highly redundant sequence present among cp32 family members, maintenance in a single cell is possible due to unique plasmid partitioning genes present on each molecule that allow for discrete replication and division of the molecules amongst dividing cells [43, 44]. The B. mayoniii genome contains five different cp32s. The gene content and organization of the cp32-1, 3, 4, 6, and 13 plasmids in B. mayonii is highly conserved as compared to other Bbsl genospecies, with the exception of cp32-6, in which a truncation results in a 17 kbp circular plasmid and the loss of 21 ORFs as compared to other cp32’s. cp32-1, 3, 4, and 13 encode between 39 and 43 ORFs that share 74–100% AAI with proteins from the cp32 plasmids from other Bbsl species with the exception of the two Erp family proteins mentioned above. Each B. mayonii cp32 plasmid includes at least 1 Erp locus, while cp32-1 and cp32-4 each contain two 2 Erp loci.
B. mayonii lp25 and lp5
B. mayonii lp25 and lp5 are the only other linear plasmids that share overall gene organization with plasmids from B. burgdorferi. B. mayonii lp25 encodes a homolog of the B. burgdorferi BptA (BB_E16) protein (Bmayo_05095) which has been shown to be associated with persistence of B. burgdorferi in the tick and is present on lp25 in other Bbsl genospecies [45]. The B. mayonii BptA homolog shares 94% AAI with B. burgdorferi B31 BptA. Homologs for both the nicotamidase (PncA, BB_E22) and restriction modification enzyme (BB_E02) are also encoded on B. mayonii lp25 and share 100% and 88% AAI with B. burgdorferi B31 BB_E22 and BB_E02, respectively. The small linear plasmid, lp5, was only present in B. mayonii MN14-1420. No sequence reads corresponding to this plasmid were detected in the MN14-1539 dataset, suggesting its absence from this strain. Previous studies have shown that plasmid stability varies in B. burgdorferi, with lp5 being lost at the highest frequency during laboratory passage [46]. The B. mayonii lp5 plasmid encodes five highly conserved proteins, four hypothetical (Bmayo_05230, Bmayo_05235, Bmayo_05240, Bmayo_05255) and one Sua5/YciO/YrdC/YwlC family protein (Bmayo_05250).
vls containing lp28-10 in B. mayonii
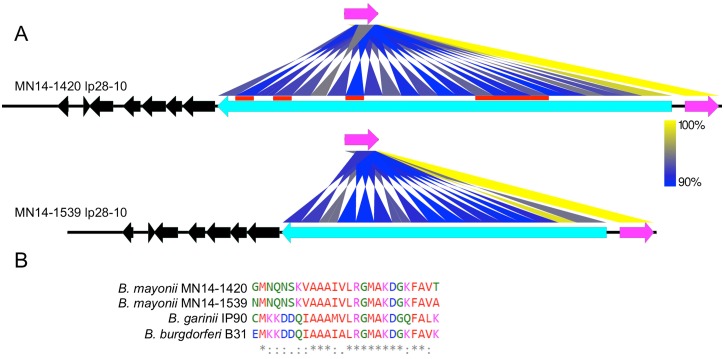
A common feature amongst the lp28 plasmids of Bbsl genospecies is the vls locus, which encodes an antigentic variation system comprised of the active vlsE expression site (3’ terminus of the linear plasmid on the template strand) as well as multiple silent cassettes (vls), arranged head to tail on the complementary strand (Fig 6A). To evade the host immune response, the silent cassettes undergo rearrangements with the central region of the active vlsE in vivo in order to alter the protein sequence (reviewed in [47]). Comparison of the vls locus of the two B. mayonii strains revealed 24 silent vls cassettes in MN14-1420 as compared to 17 silent vls cassettes in MN14-1539 (Fig 6A). This variation in the number of silent vls cassettes results in a ~4kb size difference in the lp28-10 plasmids, which was independently confirmed by pulsed-field gel electrophoresis (data not shown). Alignment of the silent vls cassettes in MN14-1420 and MN14-1539 indicates that MN14-1539 is missing vls2/4/8 and vls15-18 (numbered left-to-right) as compared to MN14-1420, suggesting the loss or expansion of silent vls cassettes is not the result of a single recombination event (see red bars in Fig 6A). Of the 17 silent vls cassettes shared by MN14-1420 and MN14-1539, nucleotide identity ranged from 93% to 100%. The amino acid sequence of the VlsE protein of B. mayonii MN14-1420 and MN14-1539 shows 92% identity and only ~50% AAI with VlsE of B. burgdorferi B31. The highest AAI to VlsE in other Bbsl genospecies is to B. finlandensis SV1, at 77% AAI. The 26 amino acid region of VlsE, known as the C6 peptide, is conserved in B. mayonii with 14 identical amino acids and 10 conservative amino acid substitutions as compared to the B. burgdorferi B31 and B. garinii IP90 C6 peptide (Fig 6B).
Fig 6. vls containing linear plasmid lp28-10 from B. mayonii MN14-1420 and MN14-1539.
A) Heat map displaying nucleotide identity (blue—lowest; yellow—highest) between the active vlsE nucleotide sequence (magenta arrow) and silent vls cassettes within the two B. mayonii strains. Position and copy number of the silent cassette array is indicated by the blue to yellow ribbons. Red bars indicate silent vls loci present in MN14-1420 and missing in MN14-1539. Genes flanking the vls locus are indicated by black arrows. Orientation of arrows indicates gene orientation on the forward or reverse DNA strand. B) Multi-alignment of the 26 amino acid C6 peptide within VlsE from B. mayonii, B. burgdorferi B31 and B. garinii IP90. Identical (*), conserved (:) and differentially charged (.) amino acid residues are indicated.
B. burgdorferi proteins lacking from B. mayonii
A notable feature of the B. mayonii genome is that it is comprised of fewer plasmids and is ~200 kbp smaller as compared to the B. burgdorferi B31 genome. Consistent with this, B. burgdorferi B31 plasmids encode 593 proteins, while B. mayonii plasmids encode only 329. The average plasmid encoded protein size is 222 amino acids for both B. burgdorferi B31 and B. mayonii. To investigate putative differences in the plasmid-borne protein composition of B. burgdorferi as compared to B. mayonii, sequences for the 593 B. burgdorferi B31 plasmid encoded proteins were compared to the predicted plasmid-borne proteome of B. mayonii. Sixty-eight plasmid encoded proteins in B. burgdorferi B31, which were present in at least one other B. burgdorferi strain, were also found to have an e-value greater than or equal to 10−2 when compared to B. mayonii plasmid encoded proteins (Table 4). In addition to e-value, percent identity, bit score, and query coverage were also evaluated (Table 4). Many of these proteins (52%) are located on plasmids in B. burgdorferi B31 for which there is no equivalent (e.g. lp28-2, lp38) in the B. mayonii genome. Fifty-five of the 68 proteins lacking from B. mayonii are predicted as hypothetical proteins (81%) with unknown function, 8 as lipoproteins (11%), and only 7 (10%) assigned a known function. Most interestingly, a homolog of the fibronectin binding protein (BB_K32) is not present on B. mayonii lp36 or elsewhere in the genome. Additionally, a homolog of the CRASP family protein, CspZ (BB_H06) is lacking in B. mayonii.
Table 4. Putative B. burgdorferi plasmid-borne proteins absent from the B. mayonii genome.
| Gene* | Protein | % ID | E-Value | Bit Score | Query Coverage (%) | Accession #* | Size |
|---|---|---|---|---|---|---|---|
| BB_A58 | hypothetical protein | 40 | 0.05 | 24.3 | 37 | NP_045731.1 | 67 |
| BB_D001 | lipoprotein | 50 | 0.062 | 23.9 | 28 | YP_004940415.1 | 64 |
| BB_D0031 | hypothetical protein | 36.36 | 0.4 | 20.8 | 60 | YP_004940417.1 | 53 |
| BB_D01 | hypothetical protein | 31.25 | 2.3 | 21.6 | 20 | NP_045385.1 | 157 |
| BB_D24 | hypothetical protein | 33.33 | 6.9 | 18.1 | 34 | NP_045406.1 | 76 |
| BB_F0039 | hypothetical protein | 41.67 | 3.4 | 18.5 | 23 | YP_004940412.1 | 52 |
| BB_F02 | hypothetical protein | 50 | 3.3 | 20.4 | 15 | NP_045438.1 | 118 |
| BB_F08 | hypothetical protein | 40.63 | 1.6 | 19.6 | 58 | NP_045444.1 | 55 |
| BB_F17 | putative transmembrane protein | 32.56 | 3.7 | 19.2 | 51 | YP_004940411.1 | 85 |
| BB_G0036 | hypothetical protein | 44.44 | 0.26 | 23.5 | 23 | YP_004940633.1 | 112 |
| BB_G02 | hypothetical protein | 41.94 | 0.05 | 27.7 | 11 | NP_045464.1 | 293 |
| BB_G12 | hypothetical protein | 28.24 | 0.13 | 25 | 51 | NP_045472.1 | 142 |
| BB_G13 | hypothetical protein | 31.43 | 0.53 | 24.6 | 13 | NP_045473.2 | 272 |
| BB_G14 | hypothetical protein | 50 | 1.4 | 21.9 | 14 | NP_045474.1 | 147 |
| BB_G15 | hypothetical protein | 30.91 | 0.52 | 23.1 | 42 | NP_045475.2 | 130 |
| BB_G16 | hypothetical protein | 29.55 | 0.011 | 27.3 | 39 | NP_045476.1 | 112 |
| BB_G17 | hypothetical protein | 22.12 | 4.7 | 21.6 | 38 | NP_045477.1 | 277 |
| BB_G18 | hypothetical protein | 36.84 | 2.5 | 21.2 | 11 | NP_045478.1 | 169 |
| BB_G19 | hypothetical protein | 23.38 | 0.21 | 25 | 42 | NP_045479.1 | 181 |
| BB_G20 | hypothetical protein | 23.58 | 0.37 | 25.4 | 29 | NP_045480.1 | 341 |
| BB_G21 | hypothetical protein | 40.74 | 2.5 | 23.1 | 7 | NP_045481.1 | 396 |
| BB_G22 | hypothetical protein | 38.3 | 1.2 | 23.5 | 17 | NP_045482.1 | 265 |
| BB_G23 | hypothetical protein | 40 | 2.3 | 22.3 | 12 | NP_045483.1 | 260 |
| BB_G24 | hypothetical protein | 23.64 | 5.8 | 23.1 | 6 | NP_045484.2 | 879 |
| BB_G26 | hypothetical protein | 25.68 | 0.87 | 22.3 | 52 | NP_045486.1 | 124 |
| BB_G28 | hypothetical protein | 36.59 | 0.36 | 23.5 | 32 | NP_045488.1 | 128 |
| BB_G30 | hypothetical protein | 28.38 | 2.1 | 21.2 | 54 | NP_045490.2 | 136 |
| BB_G32 | replicative DNA helicase | 40.91 | 0.36 | 24.6 | 8 | NP_045492.1 | 275 |
| BB_G34 | hypothetical protein | 24.32 | 1.2 | 23.1 | 29 | NP_045494.1 | 254 |
| BB_H02 | hypothetical protein | 23.13 | 1.8 | 21.9 | 73 | NP_045496.1 | 155 |
| BB_H06 | complement regulator-acquiring surface protein (CspZ) (lipoprotein) | 33.33 | 1.3 | 23.1 | 15 | NP_045500.1 | 236 |
| BB_H09a | hypothetical protein | 40.63 | 1.2 | 21.9 | 23 | YP_004940634.1 | 137 |
| BB_H17 | hypothetical protein | 35 | 5.4 | 17.7 | 52 | NP_045510.1 | 63 |
| BB_H25 | hypothetical protein | 33.33 | 4 | 19.2 | 46 | NP_045516.1 | 81 |
| BB_H40 | transposase-like protein | 31.37 | 1 | 22.3 | 33 | NP_045530.1 | 155 |
| BB_I18 | hypothetical protein | 27.27 | 1.3 | 19.6 | 44 | NP_045549.1 | 50 |
| BB_I41 | hypothetical protein | 32.69 | 1.8 | 20.4 | 56 | NP_045572.1 | 80 |
| BB_J0056 | hypothetical protein | 40.54 | 1 | 20 | 70 | YP_004940639.1 | 53 |
| BB_J0058 | hypothetical protein | 37.5 | 0.12 | 22.3 | 47 | YP_004940640.1 | 51 |
| BB_J11 | hypothetical protein | 31.67 | 0.31 | 21.6 | 95 | NP_045635.1 | 61 |
| BB_J13 | hypothetical protein | 28.57 | 0.52 | 21.9 | 53 | NP_045637.1 | 92 |
| BB_J23 | hypothetical protein | 28.3 | 0.23 | 25 | 19 | NP_045647.2 | 269 |
| BB_J25 | hypothetical protein | 28.89 | 0.54 | 24.6 | 13 | NP_045649.2 | 346 |
| BB_J27 | efflux ABC transporter permease | 32.31 | 0.79 | 24.6 | 15 | NP_045651.2 | 409 |
| BB_J28 | hypothetical protein | 24.69 | 0.083 | 26.6 | 61 | NP_045652.2 | 241 |
| BB_J29 | hypothetical protein | 50 | 0.11 | 25 | 7 | NP_045653.2 | 321 |
| BB_J31 | hypothetical protein | 17.74 | 0.67 | 23.9 | 26 | NP_045655.1 | 240 |
| BB_J36 | lipoprotein | 37.21 | 0.73 | 24.3 | 12 | NP_045660.1 | 352 |
| BB_J37 | hypothetical protein | 23.26 | 4.5 | 18.1 | 84 | NP_045661.2 | 51 |
| BB_J43 | hypothetical protein | 50 | 0.1 | 25 | 7 | NP_045667.1 | 306 |
| BB_J45 | lipoprotein | 17.74 | 0.55 | 24.3 | 26 | NP_045669.1 | 240 |
| BB_J46 | hypothetical protein | 29.63 | 0.7 | 21.9 | 49 | NP_045670.1 | 98 |
| BB_J48 | hypothetical protein | 27.17 | 0.12 | 25.8 | 86 | NP_045672.2 | 204 |
| BB_K0058 | hypothetical protein | 22 | 2.3 | 19.2 | 78 | YP_004940637.1 | 58 |
| BB_K0059 | hypothetical protein | 38.1 | 0.17 | 22.3 | 34 | YP_004940638.1 | 61 |
| BB_K07 | lipoprotein | 29.25 | 0.25 | 25.4 | 40 | NP_045581.1 | 250 |
| BB_K09 | hypothetical protein | 43.48 | 0.63 | 21.9 | 24 | NP_045583.1 | 94 |
| BB_K32 | fibronectin-binding protein (lipoprotein) | 43.48 | 0.07 | 27.7 | 6 | NP_045605.1 | 354 |
| BB_K34 | hypothetical protein | 53.33 | 2 | 20 | 21 | NP_045607.1 | 73 |
| BB_K35 | hypothetical protein | 40 | 5.5 | 17.7 | 26 | NP_045608.1 | 57 |
| BB_K41 | hypothetical protein | 27.78 | 1.1 | 22.7 | 35 | NP_045613.2 | 181 |
| BB_K42 | hypothetical protein | 58.82 | 0.15 | 22.7 | 24 | NP_045614.1 | 72 |
| BB_K47 | hypothetical protein | 30 | 0.069 | 27.7 | 18 | NP_045618.1 | 328 |
| BB_K48 | immunogenic protein P37 (lipoprotein) | 47.83 | 0.066 | 27.3 | 8 | NP_045619.1 | 288 |
| BB_M39 | hypothetical protein | 26.83 | 1.8 | 21.9 | 28 | NP_051330.1 | 222 |
| BB_Q62 | hypothetical protein | 27.5 | 0.15 | 24.3 | 32 | NP_051521.1 | 124 |
| BB_Q88 | hypothetical protein | 31.25 | 2.2 | 21.6 | 20 | NP_051536.1 | 157 |
| BB_Q89 | lipoprotein | 50 | 0.062 | 23.9 | 28 | NP_051537.1 | 64 |
*Gene name and accession numbers are derived from the B. burgdorferi B31 genome.
Discussion
The full genomes, including chromosome and plasmid sequences, for two B. mayonii strains (MN14-1420 and MN14-1539) cultured from the blood of two patients are presented in this report. The recently discovered cause of LB in the United States, B. mayonii has thus far only been detected in patients and ticks from the upper midwest [12, 13]. Overall, the genomes of the two strains are very similar, being comprised of a 904,387 bp linear chromosome with only 9 SNP differences. The MN14-1420 genome includes 407,158 bp of extrachromosomal plasmid (235,971 bp linear, 171,187 bp circular) DNA while the MN14-1539 genome contains 397,714 bp of extrachromosomal plasmid DNA (226,488 bp linear, 171,226 bp circular), with the primary difference due to a 4 kbp size difference in the vls containing plasmid, lp28-10, and the absence of the 5 kbp lp5 plasmid in MN14-1539. It’s likely the lp5 plasmid was lost from MN14-1539 during cultivation of the spirochete in vitro, as the corresponding plasmid in B. burgdorferi has been shown to be lost at the highest frequency during laboratory passage [46]. Notably, between the ~400,000 bp of plasmid sequence shared by MN14-1420 and MN14-1539 (excluding the vls locus), only 6 SNPs, 1 INDEL and 7 VNTRs were detected. As the two isolates were recovered from patients exposed in different geographic regions of the upper midwest, these findings suggest that B. mayonii strain diversity in this region may be limited [12, 13].
The genome sequence of B. mayonii confirms previous taxonomic findings from 8 housekeeping gene MLSA identifying it as a novel genospecies of Bbsl [12, 13]. Here, nucleotide comparison of the entire linear chromosome of B. mayonii with other Bbsl genospecies revealed an ANI ≤93.8%. An ANI value of 95% corresponds with a DNA-DNA hybridization value of 70%, the threshold historically used for bacterial species classification [35]. An additional, plasmid based phylogeny inferred from a 6.5 kbp segment of cp26 similarly demonstrates B. mayonii forms a clade distinct from other known Bbsl genospecies. Lastly, the B. mayonii linear plasmids lp28-3, lp28-4, lp28-10, lp36 and lp17 all possess unique gene order and gene content compared to similar plasmids in B. burgdorferi B31. For example, B. mayonii lp36 lacks 11 other proteins normally present on lp36. Likewise, the first 13 ORFs present on B. burgdorferi B31 lp17 are either missing from B. mayonii lp17 or present on alternative plasmids.
Features of the B. mayonii genome in common with other Bbsl genomes include a conserved, syntenic chromosome with high coding density (>95%), several linear and circular plasmids with significantly lower coding density (<72%), a high number of pseudogenes, and a large number of putative proteins of unknown function. Plasmids similar to lp54, lp25, lp5, cp26 and cp32 in other Bbsl genospecies, including largely conserved gene orders, were identified in B. mayonii. Of these plasmids, the lowest AAI to other Bbsl genospecies was for lp54. This is primarily due to the sequence diversity in the 3’ PF54 gene array which includes CspA (Bmayo_04535) and the two CRASP-1-like proteins, Bmayo_04540 and Bmayo_04545. Significant sequence divergence of the PF54 gene array between different Bbsl genospecies has been previously described and postulated to reflect adaptation to different host species [22].
During mammalian infection, the vlsE expression site of LB spirochetes undergoes extensive sequence recombination with the silent vls cassettes in order to evade host immune defenses [48, 49]. Despite overall amino acid diversity among VlsE proteins of various Bbsl genospecies, the C6 peptide is conserved and thus widely used as a serologic diagnostic marker for detection of antibodies produced by infected patients [50, 51]. Here, sequence analysis confirmed that the VlsE C6 peptide sequence in B. mayonii is also conserved. Consistent with this, serum samples collected ≥3 days of illness onset from B. mayonii infected patients were seropositive when tested using a C6 B. burgdorferi (Lyme) ELISA assay [11]. Across the entire vls locus, only limited comparisons with other Bbsl genospecies species was possible due to the small number of complete assemblies publicly available, two from B. burgdorferi, strains B31 and JD1, and one from B garinii, strain Far04 [20, 47, 52]. The two B. burgdorferi strains contain 15 and 14 silent vls cassettes while the B. garinii strain includes 19 silent vls cassettes. In comparison, the silent vls cassette of B. mayonii strains, MN14-1420 and MN14-1539, is comprised of 24 and 17 silent copies, respectively. Considering the small amount of nucleotide variation detected between the two B. mayonii strains, the observed differences in silent vls cassette copy number and nucleotide identity was notable. Moreover, the location of silent vls cassettes present in MN14-1420 as compared to MN14-1539 indicates that silent cassette expansion or reduction most likely occurs throughout the cassette rather than only at the 5’ or 3’ ends. The direct repeats bookending each of the silent vls cassettes typically make assembly of this region challenging, especially if sequencing platforms yielding short read lengths are utilized. Within the ~200X sequence read coverage of the B. mayonii lp28-10 plasmids, multiple overlapping reads greater than 10kbp spanned the entire vls locus as well as flanking regions which allowed for complete assembly. Thus, long read genome sequencing provides a new tool for studying the complete vls locus of Bbsl genospecies in greater detail.
Given the reported spirochetemia levels in B mayonii infected patients and detection of the organism in I. scapularis ticks, proteins encoded in the B. mayonii genome were analyzed for similarity to those present in the I. scapularis transmitted relapsing fever-like spirochete, B. miyamotoi. No proteins with significantly higher identity to proteins in B. miyamotoi as compared to other Bbsl genospecies were detected. The ability of relapsing fever Borrelia to grow to very high numbers in the blood of infected patients differs from LB Borrelia and has been attributed, in part, to an intact purine salvage pathway (hpt, purA and purB) and complete purine metabolism pathway (nrdIEF) [53]. As observed for other Bbsl genospecies, homologs of the proteins encoded in the nrdIEF/purAB operons of relapsing fever borreliae were also not present in the B. mayonii genome.
The genetic basis for differences among Bbsl genospecies with respect to tissue tropism and/or clinical manifestations is not well understood. Outer surface lipoproteins important for tissue colonization (DbpA, BBK_32) and evasion of complement-dependent killing (CRASPs) have been linked to tropism and/or pathogenic differences [38, 54–57]. In the B. mayonii genome, the proteins displaying the highest sequence variation as compared to other Bbsl genospecies include DpbA (Bmayo_04355) as well as five CRASP family proteins [Bmayo_04530 (CspA), Bmayo_04535, Bmayo_04540, Bmayo_04545, Bmayo_05450, Bmayo_05655], three of which are encoded within the PF54 gene array. Additionally, our analyses indicate that the B. mayonii genome lacks homologs to two proteins involved in tissue colonization and immune evasion by B. burgdorferi: the fibronectin binding protein, BB_K32 [57–59] and CspZ, a complement inhibitor with no known paralogs [60, 61]. Whether these differences might be linked to a distinct tissue tropism or pathogenicity of B. mayonii requires further characterization. Description of illness in additional patients infected with B. mayonii will also be important for increasing our understanding of the breadth of clinical manifestations.
Another notable feature of the B. mayonii genome is that at least 68 plasmid-borne proteins present in B. burgdorferi, including BBK_32 and CspZ, appear to be lacking. Interestingly, most of these proteins (52%) are located on plasmids in B. burgdorferi B31 for which there is no equivalent (e.g. lp28-2, lp38) in the B. mayonii genome. Of the 68 proteins, all are present in B. burgdorferi strains other than B31, indicating the identified proteins lacking from B. mayonii are not B. burgdorferi B31 strain specific. Given, the paralogous nature of Borrelia genomes, we cannot rule out the possibility that functional domains of these B. burgdorferi proteins may be present elsewhere in B. mayonii. Additionally, it’s possible other B. burgdorferi proteins may be lacking from the B. mayonii genome and were not detected here due to the stringency of the comparisons performed.
Currently, relatively few complete genome assemblies are available for Bbsl genospecies, despite being the causative agents of the most commonly reported tick-borne infection in the northern hemisphere. We have shown here the feasibility of using long read sequencing technology, specifically PacBio, coupled with high density sequence coverage, for non-hybrid, de novo assembly of complete Bbsl genomes. Future utilization of long read sequencing to generate complete genome assemblies should enable a more comprehensive picture of genetic diversity, gene content differences, and structural organization of Bbsl plasmid sequences. Sequencing of additional B. mayonii strains from both environmental and human samples will be important for understanding genetic diversity of this species as well as for confirming the detected differences between B. burgdorferi and B. mayonii. The genome sequences presented here should provide a valuable resource for future work to characterize the pathobiology and ecology of B. mayonii, and a foundation for diagnostic and vaccine work.
Supporting Information
ANI was calculated using 2-way reciprocal best hits between the chromosomes of B. mayonii, B. burgdorferi B31, B. garinii PBR, B. bavariensis PBi, B. afzelii PKo, B. bissettii DN127, B. finlandensis SV1, B. spielmanii A14S, B. valaisiana VS116.
(DOCX)
Acknowledgments
We would like to thank the Genomic Sequencing Lab (GSL) in Atlanta, GA for library preparation and PacBio sequencing. We would like to thank Martin Schriefer and Bob Gilmore for thoughtful discussions and Laurel Respicio-Kingry for performing pulsed field gel electrophoresis of both B. mayonii strains.
*The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Data Availability
Data have been deposited in the NCBI database under BioProject # PRJNA321302.
Funding Statement
This work was made possible through support from the Advanced Molecular Detection (AMD) initiative at the Centers for Disease Control and Prevention. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Adeolu M, Gupta RS. A phylogenomic and molecular marker based proposal for the division of the genus Borrelia into two genera: the emended genus Borrelia containing only the members of the relapsing fever Borrelia, and the genus Borreliella gen. nov. containing the members of the Lyme disease Borrelia (Borrelia burgdorferi sensu lato complex). Antonie Van Leeuwenhoek. 2014;105(6):1049–72. 10.1007/s10482-014-0164-x [DOI] [PubMed] [Google Scholar]
- 2.Cutler SJ, Ruzic-Sabljic E, Potkonjak A. Emerging borreliae—Expanding beyond Lyme borreliosis. Mol Cell Probes. 2016. [DOI] [PubMed] [Google Scholar]
- 3.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti JC, Assous M, et al. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42(3):378–83. 10.1099/00207713-42-3-378 [DOI] [PubMed] [Google Scholar]
- 4.Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease-a tick-borne spirochetosis? Science. 1982;216(4552):1317–9. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RC, Schmid GP, Hyde FW, Steigerwalt A, Brenner DJ. Borrelia burgdorferi sp. nov.: etiologic agent of Lyme disease. International Journal of Systematic and Evolutionary Microbiology. 1984;34(4):496–7. [Google Scholar]
- 6.Steere AC, Bartenhagen NH, Craft JE, Hutchinson GJ, Newman JH, Rahn DW, et al. The early clinical manifestations of Lyme disease. Ann Intern Med. 1983;99(1):76–82. [DOI] [PubMed] [Google Scholar]
- 7.Anderson JF. Mammalian and avian reservoirs for Borrelia burgdorferi. Ann N Y Acad Sci. 1988;539:180–91. [DOI] [PubMed] [Google Scholar]
- 8.Gern L, Estrada-Pena A, Frandsen F, Gray JS, Jaenson TG, Jongejan F, et al. European reservoir hosts of Borrelia burgdorferi sensu lato. Zentralbl Bakteriol. 1998;287(3):196–204. [DOI] [PubMed] [Google Scholar]
- 9.van Dam AP, Kuiper H, Vos K, Widjojokusumo A, de Jongh BM, Spanjaard L, et al. Different genospecies of Borrelia burgdorferi are associated with distinct clinical manifestations of Lyme borreliosis. Clin Infect Dis. 1993;17(4):708–17. [DOI] [PubMed] [Google Scholar]
- 10.Stanek G, Strle F. Lyme borreliosis. Lancet. 2003;362(9396):1639–47. 10.1016/S0140-6736(03)14798-8 [DOI] [PubMed] [Google Scholar]
- 11.Stanek G, Wormser GP, Gray J, Strle F. Lyme borreliosis. Lancet. 2012;379(9814):461–73. 10.1016/S0140-6736(11)60103-7 [DOI] [PubMed] [Google Scholar]
- 12.Pritt BS, Mead PS, Johnson DK, Neitzel DF, Respicio-Kingry LB, Davis JP, et al. Identification of a novel pathogenic Borrelia species causing Lyme borreliosis with unusually high spirochaetaemia: a descriptive study. Lancet Infect Dis. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritt BS R-K L.B.; Sloan L.M.; Schriefer M.E.; Replogle A.J.; Bjork J.; Gongping L.; Kingry L.C.; Mead P.S.; Neitzel D.F.; Schiffman E.; Johnson D.K.; Davis J.P.; Paskewitz S.M.; Boxrud D.; Deedon A.; Lee X.; Miller T.K.; Feist M.A.; Steward C.R.; Theel E.S.; Patel R.; Irish C.L.; Petersen J.M. Borrelia mayonii sp. nov., a member of the Borrelia burgdorferi sensu lato complex, detected in patients and ticks in the upper Midwestern United States. International Journal of Systematic and Evolutionary Microbiology 2016; Submitted July 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguero-Rosenfeld ME, Wang G, Schwartz I, Wormser GP. Diagnosis of lyme borreliosis. Clin Microbiol Rev. 2005;18(3):484–509. 10.1128/CMR.18.3.484-509.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liveris D, Schwartz I, McKenna D, Nowakowski J, Nadelman RB, DeMarco J, et al. Quantitation of cell-associated borrelial DNA in the blood of Lyme disease patients with erythema migrans. Eur J Clin Microbiol Infect Dis. 2012;31(5):791–5. 10.1007/s10096-011-1376-x [DOI] [PubMed] [Google Scholar]
- 16.Bil-Lula I, Matuszek P, Pfeiffer T, Wozniak M. Lyme Borreliosis—the Utility of Improved Real-Time PCR Assay in the Detection of Borrelia burgdorferi Infections. Adv Clin Exp Med. 2015;24(4):663–70. 10.17219/acem/28625 [DOI] [PubMed] [Google Scholar]
- 17.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390(6660):580–6. 10.1038/37551 [DOI] [PubMed] [Google Scholar]
- 18.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35(3):490–516. [DOI] [PubMed] [Google Scholar]
- 19.Casjens S, Eggers C., Schwartz I. Borrelia: Molecular Biology, Host Interaction and Pathogenesis. Samuels S, Radolf J., editor: Caister Academic Press; 2010. [Google Scholar]
- 20.Casjens SR, Mongodin EF, Qiu WG, Luft BJ, Schutzer SE, Gilcrease EB, et al. Genome stability of Lyme disease spirochetes: comparative genomics of Borrelia burgdorferi plasmids. PLoS One. 2012;7(3):e33280 10.1371/journal.pone.0033280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mongodin EF, Casjens SR, Bruno JF, Xu Y, Drabek EF, Riley DR, et al. Inter- and intra-specific pan-genomes of Borrelia burgdorferi sensu lato: genome stability and adaptive radiation. BMC Genomics. 2013;14:693 10.1186/1471-2164-14-693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu WG, Martin CL. Evolutionary genomics of Borrelia burgdorferi sensu lato: findings, hypotheses, and the rise of hybrids. Infect Genet Evol. 2014;27:576–93. 10.1016/j.meegid.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuler W, Bunikis I, Weber-Lehman J, Comstedt P, Kutschan-Bunikis S, Stanek G, et al. Complete genome sequence of Borrelia afzelii K78 and comparative genome analysis. PLoS One. 2015;10(3):e0120548 10.1371/journal.pone.0120548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–9. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- 25.Krumsiek J, Arnold R, Rattei T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23(8):1026–8. 10.1093/bioinformatics/btm039 [DOI] [PubMed] [Google Scholar]
- 26.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Setubal JC, Reis M, Matsunaga J, Haake DA. Lipoprotein computational prediction in spirochaetal genomes. Microbiology. 2006;152(Pt 1):113–21. 10.1099/mic.0.28317-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di L, Pagan PE, Packer D, Martin CL, Akther S, Ramrattan G, et al. BorreliaBase: a phylogeny-centered browser of Borrelia genomes. BMC Bioinformatics. 2014;15:233 10.1186/1471-2105-15-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cannon EK, Cannon SB. Chromosome visualization tool: a whole genome viewer. Int J Plant Genomics. 2011;2011:373875 10.1155/2011/373875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–7. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–10. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraiczy P, Stevenson B. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: Structure, function and regulation of gene expression. Ticks Tick Borne Dis. 2013;4(1–2):26–34. 10.1016/j.ttbdis.2012.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, et al. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun. 2006;74(6):3554–64. 10.1128/IAI.01950-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fingerle V, Rauser S, Hammer B, Kahl O, Heimerl C, Schulte-Spechtel U, et al. Dynamics of dissemination and outer surface protein expression of different European Borrelia burgdorferi sensu lato strains in artificially infected Ixodes ricinus nymphs. J Clin Microbiol. 2002;40(4):1456–63. 10.1128/JCM.40.4.1456-1463.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J Exp Med. 2004;199(5):641–8. 10.1084/jem.20031960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson B, Zuckert WR, Akins DR. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J Mol Microbiol Biotechnol. 2000;2(4):411–22. [PubMed] [Google Scholar]
- 43.Eggers CH, Caimano MJ, Clawson ML, Miller WG, Samuels DS, Radolf JD. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the lyme disease spirochaete. Mol Microbiol. 2002;43(2):281–95. [DOI] [PubMed] [Google Scholar]
- 44.Stewart PE, Thalken R, Bono JL, Rosa P. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol. 2001;39(3):714–21. [DOI] [PubMed] [Google Scholar]
- 45.Revel AT, Blevins JS, Almazan C, Neil L, Kocan KM, de la Fuente J, et al. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc Natl Acad Sci U S A. 2005;102(19):6972–7. 10.1073/pnas.0502565102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norris SJ, Howell JK, Odeh EA, Lin T, Gao L, Edmondson DG. High-throughput plasmid content analysis of Borrelia burgdorferi B31 by using Luminex multiplex technology. Appl Environ Microbiol. 2011;77(4):1483–92. 10.1128/AEM.01877-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Norris SJ. vls Antigenic Variation Systems of Lyme Disease Borrelia: Eluding Host Immunity through both Random, Segmental Gene Conversion and Framework Heterogeneity. Microbiol Spectr. 2014;2(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coutte L, Botkin DJ, Gao L, Norris SJ. Detailed analysis of sequence changes occurring during vlsE antigenic variation in the mouse model of Borrelia burgdorferi infection. PLoS Pathog. 2009;5(2):e1000293 10.1371/journal.ppat.1000293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang JR, Norris SJ. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1998;66(8):3698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang FT, Steere AC, Marques AR, Johnson BJ, Miller JN, Philipp MT. Sensitive and specific serodiagnosis of Lyme disease by enzyme-linked immunosorbent assay with a peptide based on an immunodominant conserved region of Borrelia burgdorferi vlsE. J Clin Microbiol. 1999;37(12):3990–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang FT, Alvarez AL, Gu Y, Nowling JM, Ramamoorthy R, Philipp MT. An immunodominant conserved region within the variable domain of VlsE, the variable surface antigen of Borrelia burgdorferi. J Immunol. 1999;163(10):5566–73. [PubMed] [Google Scholar]
- 52.Casjens SR, Mongodin EF, Qiu WG, Dunn JJ, Luft BJ, Fraser-Liggett CM, et al. Whole-genome sequences of two Borrelia afzelii and two Borrelia garinii Lyme disease agent isolates. J Bacteriol. 2011;193(24):6995–6. 10.1128/JB.05951-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersson J, Schrumpf ME, Raffel SJ, Porcella SF, Guyard C, Lawrence K, et al. Purine salvage pathways among Borrelia species. Infect Immun. 2007;75(8):3877–84. 10.1128/IAI.00199-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammerschmidt C, Klevenhaus Y, Koenigs A, Hallstrom T, Fingerle V, Skerka C, et al. BGA66 and BGA71 facilitate complement resistance of Borrelia bavariensis by inhibiting assembly of the membrane attack complex. Mol Microbiol. 2016;99(2):407–24. 10.1111/mmi.13239 [DOI] [PubMed] [Google Scholar]
- 55.Kraiczy P, Skerka C, Kirschfink M, Brade V, Zipfel PF. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and Factor H. Eur J Immunol. 2001;31(6):1674–84. [DOI] [PubMed] [Google Scholar]
- 56.Lin YP, Benoit V, Yang X, Martinez-Herranz R, Pal U, Leong JM. Strain-specific variation of the decorin-binding adhesin DbpA influences the tissue tropism of the lyme disease spirochete. PLoS Pathog. 2014;10(7):e1004238 10.1371/journal.ppat.1004238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin YP, Chen Q, Ritchie JA, Dufour NP, Fischer JR, Coburn J, et al. Glycosaminoglycan binding by Borrelia burgdorferi adhesin BBK32 specifically and uniquely promotes joint colonization. Cell Microbiol. 2015;17(6):860–75. 10.1111/cmi.12407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hyde JA, Weening EH, Chang M, Trzeciakowski JP, Hook M, Cirillo JD, et al. Bioluminescent imaging of Borrelia burgdorferi in vivo demonstrates that the fibronectin-binding protein BBK32 is required for optimal infectivity. Mol Microbiol. 2011;82(1):99–113. 10.1111/j.1365-2958.2011.07801.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moriarty TJ, Shi M, Lin YP, Ebady R, Zhou H, Odisho T, et al. Vascular binding of a pathogen under shear force through mechanistically distinct sequential interactions with host macromolecules. Mol Microbiol. 2012;86(5):1116–31. 10.1111/mmi.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hartmann K, Corvey C, Skerka C, Kirschfink M, Karas M, Brade V, et al. Functional characterization of BbCRASP-2, a distinct outer membrane protein of Borrelia burgdorferi that binds host complement regulators factor H and FHL-1. Mol Microbiol. 2006;61(5):1220–36. 10.1111/j.1365-2958.2006.05318.x [DOI] [PubMed] [Google Scholar]
- 61.Siegel C, Schreiber J, Haupt K, Skerka C, Brade V, Simon MM, et al. Deciphering the ligand-binding sites in the Borrelia burgdorferi complement regulator-acquiring surface protein 2 required for interactions with the human immune regulators factor H and factor H-like protein 1. J Biol Chem. 2008;283(50):34855–63. 10.1074/jbc.M805844200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ANI was calculated using 2-way reciprocal best hits between the chromosomes of B. mayonii, B. burgdorferi B31, B. garinii PBR, B. bavariensis PBi, B. afzelii PKo, B. bissettii DN127, B. finlandensis SV1, B. spielmanii A14S, B. valaisiana VS116.
(DOCX)
Data Availability Statement
Data have been deposited in the NCBI database under BioProject # PRJNA321302.