Abstract
Eukaryotic genomes are functionally organized into chromatin, a compact packaging of nucleoproteins with the basic repeating unit known as the nucleosome. A major focus for the chromatin field has been understanding what rules govern nucleosome positioning throughout the genome, and here we review recent findings using a novel, sequence-targeted remodeling enzyme. Nucleosomes are often packed into evenly spaced arrays that are reproducibly positioned, but how such organization is established and maintained through dramatic events such as DNA replication is poorly understood. We hypothesize that a major fraction of positioned nucleosomes arises from sequence-specific targeting of chromatin remodelers to generate “founding” nucleosomes, providing reproducible, predictable and condition-specific nucleation sites against which neighboring nucleosomes are packed into evenly spaced arrays.
Keywords: chromatin modification, chromatin remodeling, genome structure, Isw2, nucleosome positioning, transcription
Introduction
Compaction of eukaryotic genomes into chromatin facilitates global packaging of genetic material and regulates accessibility of the underlying DNA sequence. Nucleosomes, the fundamental repeating units of chromatin, consist of a histone core wrapped by ~147 bp of duplex DNA. Wrapping of DNA into nucleosomes weakens potential interactions with many sequence-specific factors, both by occluding potential DNA binding sites facing inwards toward the histone core and by widening the major and minor grooves of DNA facing away from the core, distorting DNA structure from canonical B-form [1]. Nucleosomes are therefore broadly repressive in nature, and appear to play an important role in regulating DNA-dependent processes. For example, the location and density of nucleosomes can prevent transcription initiation outside of promoters [2, 3], regulate the timing and efficiency of replication origin firing [4–6], and control access of DNA repair machinery to sites of DNA damage [7]. The importance of understanding the mechanisms of nucleosome positioning arises not only from the tight integration of chromatin architecture with basic cellular processes, but also from the finding that many factors involved in nucleosome positioning are often mutated in cancers and other human diseases [8–10].
Using genome sequencing technologies coupled with micrococcal nuclease (MNase) or exonuclease (MNase-seq or ChIP-exo), the locations of nucleosomes have been extensively mapped in human, fly, worm and yeast cells [11–15]. Within each organism, nucleosomes show a striking conservation in positioning and distribution, particularly with respect to transcription start sites (Fig. 1). The regularity and reproducibility of nucleosome positions has raised the perplexing question of how cells establish and maintain defined nucleosome positions over multiple cell divisions. Previous work showed that while nucleosome are favored at some locations due to DNA sequence preferences [16], additional cellular factors are required for recapitulating the genome-wide patterns observed in vivo [17–19].
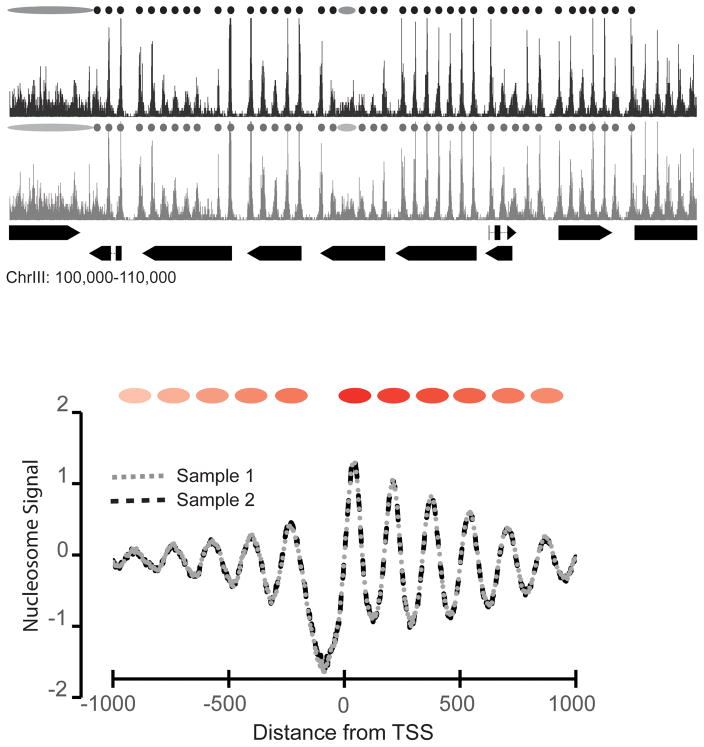
Figure 1.
Nucleosome positions are highly reproducible in S. cerevisiae cells. Top: Two representative genome browser images showing nucleosome dyads from MNase-seq experiments in independent S. cerevisiae isolates. Small circles denote well-positioned nucleosomes while large ovals represent poorly-positioned nucleosomes. Pointed rectangles denote annotated transcription units. Bottom: Overlay of nucleosome dyad signal at transcription start sites (TSS) for the two isolates in (from top). Red ovals represent positioned nucleosomes with respect to the TSS. Data was obtained from Gene Expression Omnibus (GEO) GSE72572 [27].
It is now well established that positioning nucleosomes into regularly spaced arrays relies on chromatin remodelers [17, 19]. Chromatin remodelers include several subfamilies of ATP-dependent enzymes that can alter chromatin structure using a helicase-like motor [20]. Each remodeler subfamily has a unique composition of domains and subunits that appear responsible for guiding the outcome of remodeling reactions, with distinct activities such as nucleosome assembly, nucleosome disassembly, histone exchange, and nucleosome repositioning [20]. How cells target particular remodeling activities to particular genomic loci, though, has been difficult to resolve. Many remodelers possess domains that recognize histone post-translational modifications (PTMs), such as bromodomains, chromodomains, and PHD fingers, that likely help localize remodeling activities [21]. However, PTMs are typically distributed over multiple neighboring nucleosomes, and it is difficult to envision how targeting via PTMs might be coupled to positioning nucleosomes over specific DNA sequences. Many remodelers also have DNA-binding domains, but these appear to be largely sequence-nonspecific in nature and therefore would not be sufficient for targeting [22–26].
Here we review our recent findings using a hybrid chromatin remodeler, where the native DNA-binding domain was replaced by a foreign, sequence-specific domain that targeted remodeling activity to defined loci throughout the yeast genome [27]. This work helped reveal a parallel endogenous mechanism for targeting a natural chromatin remodeler genome-wide via a transcription factor to produce precisely positioned nucleosomes. We also discovered that targeted nucleosomes were responsible for phasing local nucleosome arrays, and below we describe how sequence-targeting can help explain global nucleosome patterns.
Chimeric chromatin remodelers can specify nucleosome placement in S. cerevisiae
S. cerevisiae has proven to be an excellent model system for understanding the basis of nucleosome positioning, due to its relatively small genome size, ease of genetic manipulation, and extensive characterization of remodeling factors. We revisited nucleosome positioning at targets of the conserved Isw2 chromatin remodeler in S. cerevisiae to determine processes contributing to precise nucleosome placement across the genome. Early in vivo studies showed that the Isw2 remodeler is coupled to transcriptional repression of early meiotic genes, and identified targeted Isw2 remodeling at URS1 sites that required localization via the Ume6 transcription factor [28]. In contrast with Isw2, two remodelers with similar biochemical activities in vitro, Isw1 and Chd1, do not appear to have specific genomic targeting and instead are required for maintaining arrays of evenly spaced nucleosomes in coding regions [17]. Despite the relatively nonspecific functions in vitro and general lack of sequence preferences in vivo, Isw2 along with Isw1 and Chd1 are thought to be responsible for the organization of a large fraction of nucleosome positions in S. cerevisiae. In fact, for the isw1/chd1/isw2 triple mutant, the well-defined, reproducible positions of nucleosomes across the yeast genome are completely abrogated [17].
A major area of interest in the chromatin remodeling field has focused on how the direction of nucleosome sliding is determined. Studying yeast Chd1 in vitro, we previously found that that the sliding direction could be dictated by attaching a foreign, sequence-specific DNA-binding domain [25]. By fusing a foreign binding domain in place of the natural, sequence-nonspecific DNA-binding domain, we showed that chimeric Chd1 remodelers preferentially shifted nucleosomes on top of target sites on DNA. Interestingly, a chimeric remodeler containing monomeric streptavidin also showed targeted remodeling, but displayed distinct outcomes depending on the locations of biotinylation sites [29]. When biotinylation sites were restricted to the DNA flanking the nucleosome, repositioning by the Chd1-streptavidin remodeler shifted biotinylation sites onto the nucleosomes, similar to the behavior of other Chd1 chimeras made with sequence-specific DNA-binding domains. For both types of remodelers, directional sliding appeared to arise from burial of the binding site on the nucleosome, which reduced accessibility and thus remodeler binding. For the Chd1-streptavidin remodeler, biotinylation of the histone tails yielded uncharacteristic behaviors for Chd1, resulting in mononucleosomes shifting past the ends of DNA and nucleosomes colliding into their neighbors [29]. These unique behaviors were consistent with remodeling activity being largely regulated through binding: targeting via histone tails allowed for continued remodeling, regardless of nucleosome positioning, whereas targeting via DNA led to directional sliding, where movement of nucleosomes on top of the binding site significantly weakened remodeler binding, promoting accumulation of positions with buried sites.
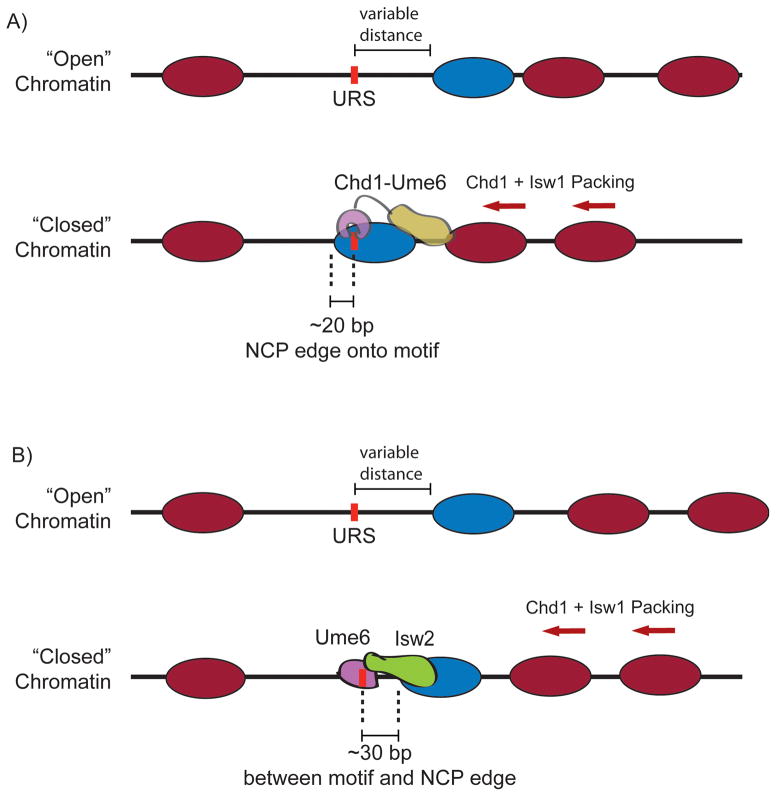
With their propensity for directional sliding, the chimeric Chd1 remodelers appeared well suited for challenging native nucleosome positioning systems in vivo, providing a unique tool for investigating how chromatin responds to site-specific perturbations. Given the known targeting of Isw2 remodeler via the Ume6 transcription factor, we generated a Chd1-Ume6 chimeric remodeler [27]. Consistent with in vitro results, in yeast cells the Chd1-Ume6 remodeler specifically repositioned nucleosomes adjacent to the URS1 motif, resulting in burial of these Ume6 recruitment sites within nucleosomes (Fig. 2A). The precision and specificity of these nucleosome movements were remarkable, because locations of recruitment motifs could be readily identified by simply finding the positions where Chd1-Ume6 shifted nucleosome dyads.
Figure 2.
Sequence-targeted chromatin remodeling in S. cerevisiae. A: Cartoon representation of sequence-targeted chromatin remodeling by a chimeric Chd1-Ume6 protein. Motif-proximal nucleosomes are mobilized toward the recruitment site until the recruitment motif is buried by ~20 base pairs of nucleosomal DNA. Distal nucleosomes are packed against this motif-proximal nucleosome to form a phased chromatin array. B: Cartoon representation of motif-proximal nucleosome positioning at Ume6 targets (URS sites) in S. cerevisiae. Through action of Ume6-recruited Isw2, motif proximal nucleosomes are moved toward the recruitment site to leave ~30 base pairs between the motif center and the nucleosome edge. Subsequent positioning of downstream nucleosomes is achieved through packing against the motif-proximal nucleosome barrier.
Endogenous sequence-targeted chromatin remodeling is predictable and precise
There has been significant debate about the relative contributions of cis-elements like poly-A tracts and other sequence motifs and trans- acting factors like ATP-dependent chromatin remodeling proteins to global nucleosome positioning under biological conditions [16–19, 30, 31]. We investigated whether specific recruitment of endogenous chromatin remodeling proteins, which can similarly be targeted through DNA sequence motifs, may explain the reproducible nucleosome positions observed in S. cerevisiae. Although previous observations indicated cooperation between Isw2 and Ume6 [28], genome-wide analysis revealed that the natural targeting of Isw2 via Ume6 yields strikingly precise and predictable nucleosome positioning at hundreds of sites throughout the genome [27] (Fig. 2B). In contrast to the hybrid fusion Chd1-Ume6 remodeler, which moves nucleosomes onto the recruitment site until the motif is occluded, the endogenous Isw2/Ume6 system leaves a considerable gap of 30 base pairs between the Ume6 binding site and the edge of the closest, repositioned nucleosome. This maintained exposure of target binding sites strongly suggests an inhibitory mechanism that attenuates Isw2 action. The unexpected precision in movement of a single motif-proximal nucleosome suggests that the sequence-specific recruitment of Isw2 at Ume6 binding sites is highly reproducible, and encoded in the underlying DNA sequence.
While we only demonstrated this precise, genome-wide targeting for the interaction between Isw2 and Ume6, recent high resolution ChIP-exo experiments have uncovered a highly specific interaction of Isw2 at Reb1 sites [32], and similar Isw2 recruitment has been observed or suggested at many other transcription factor sites [33, 34]. In humans, a large number of transcription factors have highly organized proximal nucleosome patterns, and many of these require the Isw2-related SNF2H and SNF2L remodelers [35, 36]. More recently, endogenous nucleosome positions were recapitulated on salt-dialyzed chromatin using purified Isw2 in combination with the sequence-specific general regulatory factors (GRFs) Reb1 or Abf1, specifically at Reb1/Abf1 binding sites [37]. While previous work has implicated GRFs in establishing nucleosome positioning [32, 38–40], this groundbreaking study from the Pugh and Korber labs demonstrated in a highly purified system that up to 1/3 of genomic +1 nucleosome positions can be explained by remodeler positioning at motif-encoded GRF binding locations [37]. Together, these results suggest that targeting a chromatin remodeler through interactions with a sequence-specific transcription factor is likely a pervasive system for precisely positioning specific nucleosomes throughout eukaryotic genomes.
Sequence-targeted remodeling sets the phasing of organized nucleosome arrays
Although the precise nucleosome positioning achieved through transcription factor targeting would help explain reproducible nucleosome peaks observed genome wide, it was not clear whether hundreds of TF sites could specify thousands of unique nucleosome positions. Unexpectedly, the chimeric Chd1-Ume6 remodeler helped disentangle direct from indirect effects. Both the chimeric Chd1-Ume6 remodeler and the natural Isw2/Ume6 system showed a limited range of influence, where nucleosomes were only shifted when within ~100 bp of the expected final locations. Despite this limited range, however, both remodelers catalyzed the repositioning of up to five nucleosomes neighboring the target site. This shifting of nucleosome arrays is consistent with the single targeted nucleosomes providing barriers against which neighboring nucleosomes are phased (Fig. 2). We expect that native, non-targeted remodelers such as Isw1 and Chd1 are likely responsible for these array shifts in S. cerevisiae, because these remodelers were previously shown to be required for array packing against transcriptional start sites in vivo and in purified systems [17, 37]. While barrier establishment and packing mechanisms have been proposed before [19, 41, 42], the underlying mechanisms were unresolved. The comparison of endogenous Isw2/Ume6 and synthetic Chd1-Ume6 remodeling at TF binding motifs clearly identify sequence-targeted nucleosomes as barriers themselves that can define the phasing for adjacent arrays. Through this array phasing from a remodeler-targeted barrier, precise positioning of nucleosomes covering roughly 1kb of genomic sequence can therefore be encoded in a single 6 base pair transcription factor binding motif.
Precise nucleosome positions may influence transcriptional effectors
Why might the cell require such a precise positioning mechanism? A straightforward explanation would be that reproducible nucleosome positions are required for faithful regulation of transcription. However, when Chd1-Ume6 was used to disrupt nucleosome positions in yeast, there was little discernible impact on mRNA transcription, although we found a modest role in regulation of cryptic ncRNAs [27]. Similarly, although deletion of Isw2 impacts the positions of nucleosomes in thousands of nucleosome depleted regions, there is minimal impact on steady-state mRNA transcription with modest induction of cryptic ncRNA [43–45]. These studies argue against a critical contribution of precise nucleosome positions to steady-state RNA levels. Perhaps exact nucleosome positions are more critical for directly regulating mRNA levels during large-scale changes in transcriptional program, such as those seen during diauxic shift [46] or quiescence [47]. An alternative explanation could be that nucleosome placement influences activity of other chromatin-regulated processes. Recently, in vitro studies have shown an activity dependence on inter-nucleosomal distances for chromatin modifying enzymes [48]. If histone modifying enzymes display a similar preference for specific nucleosome geometries in vivo, the precise positioning of nucleosomes imparted by a sequence-targeted chromatin remodeler might enhance or restrict local histone modification efficiencies (Fig. 3). Accordingly, there may be regulatory crosstalk between precisely positioned nucleosomes and histone modifying enzymes. Interestingly, in addition to localizing the Isw2 chromatin remodeling factor, Ume6 also recruits the histone deacetylase Rpd3 [49], although it remains to be tested whether Rpd3 activity is influenced by nucleosome positioning or spacing.
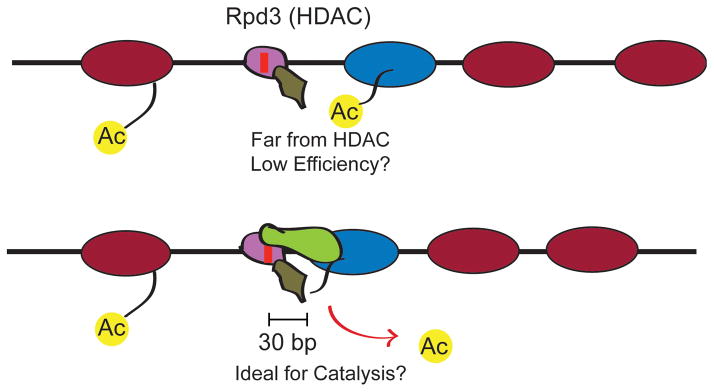
Figure 3.
Precisely-positioned motif-proximal nucleosomes may affect histone modification catalysis. Hypothetical situation where the action of a histone deacetylase (HDAC) such as Rpd3 is dependent on the distance of nucleosome substrate with respect to a recruitment site. (top) Low Rpd3 activity is achieved when nucleosome positions are not properly established. (bottom) Optimal histone deacetylation by Rpd3 occurs when the motif-proximal nucleosome is specifically positioned by sequence-targeted chromatin remodeling (eg the Isw2/Ume6 system).
A “spring-loaded” mechanism, where chromatin remodelers initially position nucleosomes onto unfavorable sequences to allow for rapid relaxation to thermodynamically-preferred locations, has been observed during Kaposi’s sarcoma-associated herpesvirus reactivation in human cells [50]. In a case such as this, precise nucleosome positioning may help govern the activity of transcriptional activators. In one scenario, nucleosome placement may occlude binding sites and thus directly compete with binding of transcriptional activators. It has been suggested that movement of nucleosomes on top of transcription factor sites leads to eviction of bound activators [51–55]. By controlling positions of nucleosome arrays, targeted chromatin remodelers likely regulate activator-mediated transcriptional programs. A second example of modulating activator function could occur when the proximity of a nucleosome to a DNA-associated activator can physically promote or restrict histone eviction. If sufficiently far from bound activators that recruit histone evicting remodelers like SWI/SNF [56–58] or Rsc [59], distal nucleosome positioning may prevent or limit histone removal, whereas closer placement could favor eviction. Regulation of histone eviction, likely commonly coupled to transcriptional activation, is therefore the product of a competition among one or more pairs of TF-remodelers and the thermodynamically preferred, “spring-loaded” nucleosome positions. We believe that the precision that is intrinsic to sequence-targeted chromatin remodeling is well-suited to crosstalk with transcriptional effectors, and future research efforts should help improve our understanding of the scope and impact of these interactions.
Sequence-directed nucleosome positioning supports fidelity and plasticity
Chromatin remodelers are known to shift nucleosomes from their thermodynamically preferred positions, providing a means for cells to switch between two defined chromatin states [50]. In addition to protecting nucleosome positions from thermodynamic fluctuations, the TF targeting we describe is well suited for quickly re-establishing nucleosome positions in the wake of disruptive processes like DNA replication. Such a mechanism offers a simple explanation for how precise nucleosome positions can be persist in a population of dividing cells (Fig. 4). Recent reports find that packing against GRFs including Abf1, Reb1, and Rap1 occurs immediately after passage of the replication fork [60], and that transcription factors are in direct competition with nucleosomes after replication [61]. Since transcription factors are thought to bind throughout mitosis [62] and nucleosomes can be organized immediately after replication [60, 63], TFs and GRFs can use transcriptional history, DNA-encoded sequence motifs, and sequence-targeted chromatin remodeling to quickly and faithfully reestablish proper nucleosome positioning after each cell division. The remarkably predictable nucleosome positioning at target loci in budding yeast reinforces the idea that precise, sequence-directed nucleosome sliding can be encoded in the genome, and an important question for future investigations is determining unique preferences for different remodelers and remodeler-TF combinations with regard to recruitment sites and nucleosome positioning.
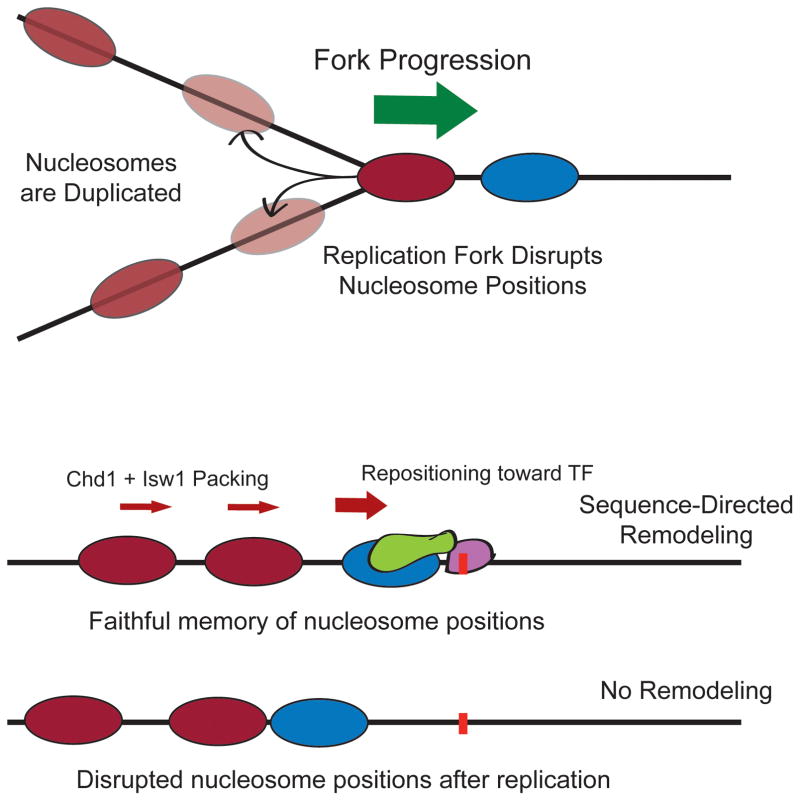
Figure 4.
Sequence-targeted chromatin remodeling can explain memory of nucleosome positions after replication. Top: As the replication fork moves through a DNA sequence, nucleosome positions are disrupted and need to be established on both DNA strands after replication. Bottom: Theoretical nucleosome positions after fork passage are shown in the presence or absence of targeted chromatin remodeling. Sequence-targeted recruitment of a chromatin remodeling factor can establish a precise motif-proximal nucleosome position on nascent DNA strands. Packing of distal nucleosomes against the motif-proximal boundary can reproduce the same nucleosome positions on newly-replicated DNA.
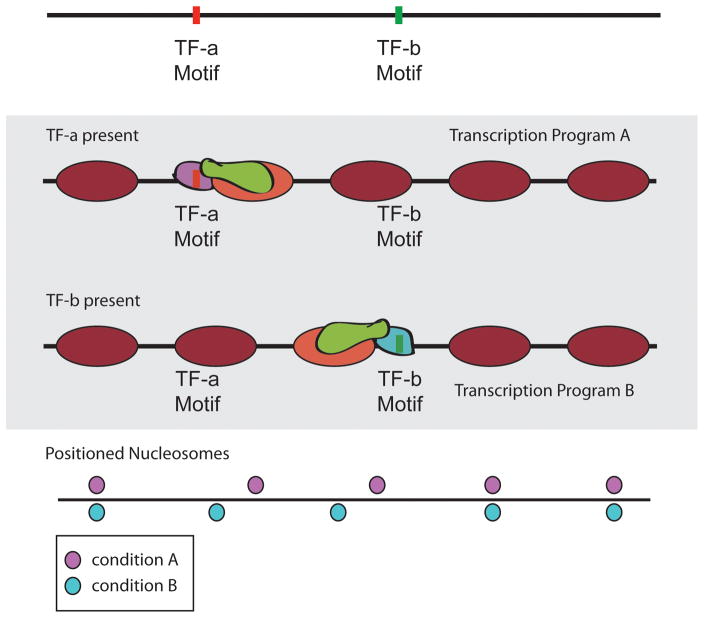
An emergent property of global nucleosome positioning based on transcription factors is that it naturally allows for plasticity. While partnered TF/remodelers are well suited to faithfully re-establish nucleosome positions after dramatic events that erase the chromatin landscape such as replication, widespread transcriptional reprogramming that requires distinct, condition-specific nucleosome positions can be easily accomplished by toggling the availability of transcription factors that direct chromatin remodelers (Fig. 5). As previously described, the removal of specific remodelers can allow nucleosomes to shift to more thermodynamically preferred positions [50]. However, by varying the TF availability, either through transcriptional induction of condition-specific TFs, or transport different TFs into/out of the nucleus, the pattern of nucleosomes could easily be altered in a locus-specific manner.
Figure 5.
Sequence-targeted chromatin remodeling allows for nucleosome positioning plasticity in different conditions. Top: Schematic of two hypothetical transcription factor (TF) binding sites on a DNA strand. Middle: Hypothetical nucleosome positions if a chromatin remodeling factor is targeted through TF-a or TF-b in a condition where the competing TF is not present. Bottom: Comparison of nucleosome positions under two distinct environmental conditions established through sequence-targeted chromatin remodeling.
The dynamics of transcription factor binding is likely a critical parameter in sequence-targeted chromatin remodeling. For example, a distinct set of nucleosome positions has been demonstrated for budding yeast in a quiescent state [47], and we expect that changes in TF targeting of remodelers is likely responsible. When S. cerevisiae enters quiescence, the Xbp1 repressor is transcriptionally induced while Stb3 is translocated from the cytoplasm to the nucleus [64, 65]. If these sequence-specific transcription factors similarly interact with Isw2, they could reposition nucleosomes near Stb3 and/or Xbp1 binding sites genome-wide, thus imparting a genomic nucleosome repositioning response that is preprogrammed in the underlying DNA sequence. In agreement with this notion, nucleosome positions around Xbp1, Stb3, and other transcription factor binding motifs change reproducibly during yeast entry into quiescence, although the dependence on Isw2 has not yet been confirmed [47]. Similarly, for differentiated cells in multicellular organisms, the cell type-specific transcription factor repertoire may instruct unique yet programmed nucleosome positioning patterns through targeted chromatin remodeling. Notably, in humans, the IKAROS transcription factor anchors the NuRD chromatin remodeling complex at DNA targets [66], which may similarly lead to cell lineage-specific motif-proximal nucleosome positioning.
Other aspects of transcription factor dynamics can shape nucleosome positioning through associated chromatin remodeling. In response to specific stimuli, dynamic interactions of pioneer factors with ATP-dependent chromatin remodeling proteins can make “closed chromatin” regions more amenable to binding of secondary transcription factors [53, 67]. Conversely, steroid receptor binding can alter dynamics of pioneer factor associations [68, 69]. Pioneer factor activity therefore helps determine which chromatin regions are accessible for local nucleosome rearrangements thus fine-tuning the scope of targeted nucleosome positioning. On a larger scale, the immediate accessibility of nascently-replicated DNA adjacent to origins of replication may dictate the order of nucleosome domain establishment, where early-replicating DNA nucleates primary chromatin arrays, which may preclude or favor binding of transcription factors in later-replicating regions. The idea of self-organizing “ground states” for remodeler-driven nucleosome positioning was recently postulated for genome-utilizing processes like replication and transcription, and is an elegant potential mechanism for mediating genome-wide organization of chromatin through TF-remodeler interactions at specific DNA motifs [37]. A system that uses DNA-encoded motifs to bridge a sequence-specific binding factor to chromatin remodeling machinery is thus ideal for providing fidelity and precision in nucleosome positioning while simultaneously allowing for rapid and tunable response to changing conditions.
Conclusions and outlook
In S. cerevisiae, targeted remodeling occurs through TF-mediated remodeler recruitment at specific DNA motifs [27, 28, 37]. Targeting can also be achieved synthetically by creation of hybrid, sequence-specific chromatin remodeling proteins like Chd1-Ume6 [25, 27]. Is precise nucleosome positioning by TF-remodeler pairs fundamentally conserved in eukaryotes? An important future goal will be determining the extent of TF cooperation with chromatin remodeling factors throughout eukaryotic genomes. The existence of precise nucleosome positioning at TF sites suggests a regulatory mechanism may exist for dictating the final position of remodeled nucleosomes, so it will be enlightening to uncover the mechanistic basis and evolutionary conservation of this precision. In some eukaryotic systems, histone modifications may significantly influence recruitment of chromatin remodeling proteins and/or transcription factors, so future work should consider interdependence of histone modifications, chromatin remodeling proteins, transcription factors, and nucleosome positioning. Additionally, since nucleosome positioning on genomic DNA arises from different remodeler classes working together [37], an important future undertaking will be deciphering the relative contributions of each remodeler type in vivo.
Sequence-targeting of chromatin remodelers both ensures high fidelity and enables plasticity of nucleosome positions required to support dynamic cellular processes. Given the striking changes in cellular programming and phenotype that can accompany widespread changes in chromatin organization, we expect that transcription factor recruitment of chromatin remodelers likely underlies global reprogramming observed in cell differentiation. Likewise, aberrant targeting or regulation of chromatin remodeling factors can explain some instances of global shifts in transcriptional programs correlated with cancer. An intriguing area for future research includes investigating the specific local effects on nucleosome placement by chromatin remodelers targeted through noncoding RNAs or three-dimensional folding [33, 70]. We expect that bridging chromatin remodeling to sequence-specific factors is a widespread mechanism for precise nucleosome placement contributing to the creation, maintenance, and dynamics of genomic nucleosome positions.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.DeGennaro CM, Alver BH, Marguerat S, Stepanova E, et al. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol Cell Biol. 2013;33:4779–92. doi: 10.1128/MCB.01068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rando OJ, Winston F. Chromatin and transcription in yeast. Genetics. 2012;190:351–87. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deniz O, Flores O, Aldea M, Soler-Lopez M, et al. Nucleosome architecture throughout the cell cycle. Sci Rep. 2016;6:19729. doi: 10.1038/srep19729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton ML, Galani K, Kang S, Bell SP, et al. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–53. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez J, Tsukiyama T. ATR-like kinase Mec1 facilitates both chromatin accessibility at DNA replication forks and replication fork progression during replication stress. Genes Dev. 2013;27:74–86. doi: 10.1101/gad.202978.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dion V, Gasser SM. Chromatin movement in the maintenance of genome stability. Cell. 2013;152:1355–64. doi: 10.1016/j.cell.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Langst G, Manelyte L. Chromatin remodelers: From function to dysfunction. Genes (Basel) 2015;6:299–324. doi: 10.3390/genes6020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 10.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee W, Tillo D, Bray N, Morse RH, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–44. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 12.Mavrich TN, Jiang C, Ioshikhes IP, Li X, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–62. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee HS, Bataille AR, Zhang L, Pugh BF. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell. 2014;159:1377–88. doi: 10.1016/j.cell.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valouev A, Ichikawa J, Tonthat T, Stuart J, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–63. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valouev A, Johnson SM, Boyd SD, Smith CL, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–20. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gkikopoulos T, Schofield P, Singh V, Pinskaya M, et al. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–60. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, et al. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16:847–52. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Wippo CJ, Wal M, Ward E, et al. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332:977–80. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou CY, Johnson SL, Gamarra NI, Narlikar GJ. Mechanisms of ATP-Dependent Chromatin Remodeling Motors. Annu Rev Biophys. 2016;45:153–81. doi: 10.1146/annurev-biophys-051013-022819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–27. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem. 2006;281:16279–88. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan DP, Sundaramoorthy R, Martin D, Singh V, et al. The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J. 2011;30:2596–609. doi: 10.1038/emboj.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada K, Frouws TD, Angst B, Fitzgerald DJ, et al. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472:448–53. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- 25.McKnight JN, Jenkins KR, Nodelman IM, Escobar T, et al. Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol Cell Biol. 2011;31:4746–59. doi: 10.1128/MCB.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma A, Jenkins KR, Heroux A, Bowman GD. Crystal structure of the chromodomain helicase DNA-binding protein 1 (Chd1) DNA-binding domain in complex with DNA. J Biol Chem. 2011;286:42099–104. doi: 10.1074/jbc.C111.294462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKnight JN, Tsukiyama T, Bowman GD. Sequence-targeted nucleosome sliding in vivo by a hybrid Chd1 chromatin remodeler. Genome Res. 2016;26:693–704. doi: 10.1101/gr.199919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldmark JP, Fazzio TG, Estep PW, Church GM, et al. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–33. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 29.Patel A, Chakravarthy S, Morrone S, Nodelman IM, et al. Decoupling nucleosome recognition from DNA binding dramatically alters the properties of the Chd1 chromatin remodeler. Nucleic Acids Res. 2013;41:1637–48. doi: 10.1093/nar/gks1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorch Y, Maier-Davis B, Kornberg RD. Role of DNA sequence in chromatin remodeling and the formation of nucleosome-free regions. Genes Dev. 2014;28:2492–7. doi: 10.1101/gad.250704.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan GC, Liu YJ, Dion MF, Slack MD, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–30. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 32.Yen K, Vinayachandran V, Batta K, Koerber RT, et al. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–73. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yadon AN, Singh BN, Hampsey M, Tsukiyama T. DNA looping facilitates targeting of a chromatin remodeling enzyme. Mol Cell. 2013;50:93–103. doi: 10.1016/j.molcel.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zentner GE, Tsukiyama T, Henikoff S. ISWI and CHD chromatin remodelers bind promoters but act in gene bodies. PLoS Genet. 2013;9:e1003317. doi: 10.1371/journal.pgen.1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Zhuang J, Iyer S, Lin X, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiechens N, Singh V, Gkikopoulos T, Schofield P, et al. The Chromatin Remodelling Enzymes SNF2H and SNF2L Position Nucleosomes adjacent to CTCF and Other Transcription Factors. PLoS Genet. 2016;12:e1005940. doi: 10.1371/journal.pgen.1005940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krietenstein N, Wal M, Watanabe S, Park B, et al. Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell. 2016;167:709–21. e12. doi: 10.1016/j.cell.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–58. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–48. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Bakel H, Tsui K, Gebbia M, Mnaimneh S, et al. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet. 2013;9:e1003479. doi: 10.1371/journal.pgen.1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kornberg RD, Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16:6677–90. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–83. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–5. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 44.Yadon AN, Van de Mark D, Basom R, Delrow J, et al. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol Cell Biol. 2010;30:5110–22. doi: 10.1128/MCB.00602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fazzio TG, Kooperberg C, Goldmark JP, Neal C, et al. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol. 2001;21:6450–60. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–6. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 47.McKnight JN, Boerma JW, Breeden LL, Tsukiyama T. Global promoter targeting of a conserved lysine deacetylase for transcriptional shutoff during quiescence entry. Mol Cell. 2015;59:732–43. doi: 10.1016/j.molcel.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee CH, Wu J, Li B. Chromatin remodelers fine-tune H3K36me-directed deacetylation of neighbor nucleosomes by Rpd3S. Mol Cell. 2013;52:255–63. doi: 10.1016/j.molcel.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–71. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 50.Sexton BS, Avey D, Druliner BR, Fincher JA, et al. The spring-loaded genome: nucleosome redistributions are widespread, transient, and DNA-directed. Genome Res. 2014;24:251–9. doi: 10.1101/gr.160150.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M, Hada A, Sen P, Olufemi L, et al. Dynamic regulation of transcription factors by nucleosome remodeling. Elife. 2015;4:e06249. doi: 10.7554/eLife.06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miranda TB, Morris SA, Hager GL. Complex genomic interactions in the dynamic regulation of transcription by the glucocorticoid receptor. Mol Cell Endocrinol. 2013;380:16–24. doi: 10.1016/j.mce.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voss TC, Schiltz RL, Sung MH, Yen PM, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–54. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo Y, North JA, Rose SD, Poirier MG. Nucleosomes accelerate transcription factor dissociation. Nucleic Acids Res. 2014;42:3017–27. doi: 10.1093/nar/gkt1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–74. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Owen-Hughes T, Utley RT, Cote J, Peterson CL, et al. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–6. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 58.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–92. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 59.Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–19. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yadav T, Whitehouse I. Replication-coupled nucleosome assembly and positioning by ATP-dependent chromatin-remodeling enzymes. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.03.059. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramachandran S, Henikoff S. Transcriptional regulators compete with nucleosomes post-replication. Cell. 2016;165:580–92. doi: 10.1016/j.cell.2016.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teves SS, An L, Hansen AS, Xie L, et al. A Dynamic Mode of Mitotic Bookmarking by Transcription Factors. bio Rxiv. 2016 doi: 10.7554/eLife.22280. doi: http://dx.doi.org/10.1101/066464. [DOI] [PMC free article] [PubMed]
- 63.Fennessy RT, Owen-Hughes T. Establishment of a promoter-based chromatin architecture on recently replicated DNA can accommodate variable inter-nucleosome spacing. Nucleic Acids Res. 2016;44:7189–203. doi: 10.1093/nar/gkw331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huber A, French SL, Tekotte H, Yerlikaya S, et al. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011;30:3052–64. doi: 10.1038/emboj.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miles S, Li L, Davison J, Breeden LL. Xbp1 directs global repression of budding yeast transcription during the transition to quiescence and is important for the longevity and reversibility of the quiescent state. PLoS Genet. 2013;9:e1003854. doi: 10.1371/journal.pgen.1003854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bottardi S, Mavoungou L, Pak H, Daou S, et al. The IKAROS interaction with a complex including chromatin remodeling and transcription elongation activities is required for hematopoiesis. PLoS Genet. 2014;10:e1004827. doi: 10.1371/journal.pgen.1004827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–41. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swinstead EE, Miranda TB, Paakinaho V, Baek S, et al. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell. 2016;165:593–605. doi: 10.1016/j.cell.2016.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swinstead EE, Paakinaho V, Presman DM, Hager GL. Pioneer factors and ATP-dependent chromatin remodeling factors interact dynamically: A new perspective. Bio Essays. 2016;38:1150–7. doi: 10.1002/bies.201600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bohmdorfer G, Wierzbicki AT. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015;25:623–32. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]