Abstract
Human patient-derived induced pluripotent stem cells (hiPSCs) provide unique opportunities for disease modeling and drug development. However, adapting hiPSCs or their differentiated progenies to high throughput assays for phenotyping or drug screening has been challenging. Fragile X syndrome (FXS) is the most common inherited cause of intellectual disability and a major genetic cause of autism. FXS is caused by mutational trinucleotide expansion in the FMR1 gene leading to hypermethylation and gene silencing. One potential therapeutic strategy is to reactivate the silenced FMR1 gene, which has been attempted using both candidate chemicals and cell-based screening. However, molecules that effectively reactivate the silenced FMR1 gene are yet to be identified; therefore, a high throughput unbiased screen is needed. Here we demonstrate the creation of a robust FMR1-Nluc reporter hiPSC line by knocking in a Nano luciferase (Nluc) gene into the endogenous human FMR1 gene using the CRISPR/Cas9 genome editing method. We confirmed that luciferase activities faithfully report FMR1 gene expression levels and showed that neural progenitor cells derived from this line could be optimized for high throughput screening. The FMR1-Nluc reporter line is a good resource for drug screening as well as for testing potential genetic reactivation strategies. In addition, our data provide valuable information for the generation of knock-in human iPSC reporter lines for disease modeling, drug screening, and mechanistic studies.
Keywords: Fragile X syndrome, FMR1, FMRP, drug discovery, induced pluripotent stem cells, luciferase, high throughput
Graphical abstract
We have created robust FMR1-luciferase reporter cell lines using genome editing. The reporter line faithfully reports endogenous FMR1 gene activation therefore will provide a critical resource for drug screening
Introduction
Fragile X syndrome (FXS), with a prevalence of 1 in 4000 in boys and 1 in 7000 in girls, is the most common inherited intellectual disability and the largest known single gene contributor to autism [1]. FXS patients suffer from severe learning deficits, hyperactivity, attention deficit disorder, and autistic-like behavior [2]. About one third of FXS individuals meet the diagnostic criteria for autism [3-9]. FXS is caused by mutational trinucleotide expansion in the 5’ region of the FMR1 gene leading to DNA hypermethylation and gene silencing. The mechanism of FMR1 gene silencing only occurs in human cells. Mice engineered to mimic the human mutation in the Fmr1 gene do not show hypermethylation and silencing of the gene [10]. Therefore, FMR1 gene silencing mechanism is unique to humans, which precludes the possibility of studying FMR1 silencing in mouse models of FXS.
Because the protein coding sequence of the silenced FMR1 gene is normal in the majority of FXS patients, a potential therapeutic strategy is to reactivate FMR1 and restore the expression of Fragile X Mental Retardation Protein (FMRP). Indeed, rare individuals who have FMR1 full-length CGG expansion but lack DNA hypermethylation show mild or no intellectual disability [11-17]. In addition, expression of exogenous FMRP in FMRP-deficient mice rescues certain FXS phenotypes [18-22]. We have discovered that genetically restoring FMR1 expression in adult-born new neurons in FMRP-deficient background restores adult neurogenesis-dependent learning and memory in mice [23]. Therefore, developmental deficits in FMRP-deficient neurons might be reversible.
Reactivation of FMR1 has been attempted in vitro through using epigenetic modulators, such as inhibiting DNA methylation and changing chromatin structure. For example, after treatment of human FXS lymphoblastoid cell lines with a DNA methyltransferase (DNMT) inhibitor 5-azacytidine (5-aza-C) or 5-azadeoxycytidine (5-aza-dC), the promoter of FMR1 gene becomes passively unmethylated through cell division, resulting in partial reactivation of the FMR1 gene and FMRP expression [24, 25]. Importantly, the increases in FMR1 mRNA production are associated with increased active chromatin markings and decreased repressive chromatin markings to the FMR1 promoter [17, 24, 26, 27]. However, these approaches suffer from numerous pitfalls including a lack of robust reactivation, dependence on cell division, and toxicity [28]. The limited success of using known epigenetic reagents to reactivate FMR1 has prompted studies to identify novel chemical reagents and molecules.
We and others have generated human iPSCs from somatic cells of FXS individuals [29-36]. Studying these FXS iPSCs has enabled us to assess the functional consequences of the lack of FMRP in human neural development and explore potential mechanisms of epigenetic reactivation[36]. Without exception, the methylated, silenced FMR1 mutation in the patient fibroblasts is retained through the reprogramming process [29-36], therefore FXS patient-derived iPSCs might be valuable to discover molecules that can reactivate FMR1 in human neural cells. Adaptation of human iPSCs or their differentiated progeny to high throughput screening (HTS) has undergone significant progress in the past few years with an increased ability to scale iPSC-derived neural cells to high throughput platforms [37]. Yet, many iPSC assays emphasize phenotyping for disease modeling and the success of drug screen relies on mRNA, protein, or morphological analysis as readouts. These methods suffer from low reproducibility, high variability and are time consuming. Two recent reports to identify compounds that reactivate FMR1 in hiPSC-derived neural progenitors rely on antibodies to detect FMRP, which are limited in signal-to-background ratio, time consuming and expensive [34, 38]. Therefore, a more robust, simpler, and more economical assay that is adaptable to high throughput screening is needed.
Here we report the creation of a reporter for FMR1 gene activation in human FXS neural cells. Using newly developed CRISPR/Cas9 gene editing methods [39], we inserted the Nano luciferase gene (Nluc) into the endogenous FMR1 gene locus in FXS-patient-derived iPSCs to create FXS-FMR1-Nluc reporter iPSC lines (FX-iPSC-Nluc). We confirmed that Nluc activity faithfully reports FMR1 gene expression and that the cells retain their ability to differentiate into neural progenitor cells (NPCs) and neurons. We screened a small molecule library of known epigenetic chemical probes using NPCs derived from our iPSC reporter line in a 384-well format. We also adapted our screening to 1536-well format and screened an FDA-approved drug library. Our results show that our high-throughput screening platform with reporter NPCs has high signal to background (S/B) ratio and a good Z’ factor, and can reproducibly identify positive hits. Therefore, this FXS-FMR1-Nluc reporter iPSC line offers a valuable resource to identify novel compounds for FMR1 gene reactivation studies.
Materials and Methods
(See also Supplemental Methods)
Generation of reporter cell lines
For expression of sgRNA targeting FMR1, a DNA fragment, 5’-CTCGTGAATGGAGTACCCTA-3’ (sequences of primers and DNA oligos used in this study are listed in Supplemental methods), was inserted into a plasmid downstream of U6 promoter (Addgene # 49535; http://www.addgene.org/49535/) that also expresses SpCas9 protein. To construct a donor plasmid, 5’ and 3’ homology arms (1072 bp and 1115 bp, respectively) were amplified from genomic DNA of H1 hESCs using primers HA-L-F plus HA-L-R and HA-R-F plus HA-R-R, respectively. To minimize the possibility of cleavage by Cas9 after integration of the donor plasmid, four point mutations were introduced at the end of FMR1 coding sequence without changing the amino acid sequence of FMRP (see Fig S1A). To generate a DNA fragment coding for P2A-Nluc, PCR was performed using template plasmid pNL1.1 (Promega) and primers Nluc-F plus Nluc-R. The PCR product was used as template for a second PCR with primers P2A-F plus Nluc-R. The P2A-Nluc DNA fragment and homology arms were inserted into vector (Addgene# 31938) digested by enzymes BamH I and Not I (New England Biolabs).
To generate reporter cells, FXS iPSCs or H1 hESCs were dissociated to single cells with TrypLE Express and washed. 2-4 × 106 cells were electroporated (Gene Pulser Xcell, Bio-Rad; 250 V, 500 μF, 4 mm cuvette, infinite resistance) using 10 μg of Cas9-sgRNA plasmid plus 15 μg of donor plasmid. As the Cas9-sgRNA plasmid carries a puromycin-resistant gene, cells were transiently selected with puromycin (Thermo Fisher Scientific; 0.5 μg/ml 48-72 hours after electroporation and 0.25 μg/ml 72-96 hours afterwards). About two weeks later, colonies were picked for expansion and PCR screening.
To screen colonies, a pair of primers was designed to span the Nluc region to downstream of the 3’ homology arm to ensure proper site-specific targeting by donor vector (red arrows in Fig. 1A; gel in Fig. 1C; ~1.3 kb). To ensure the positive colonies do not have addition random insertion of donor plasmid, two more pairs of primers were designed to span donor plasmid backbone upstream of 5’ homology arm to 5’ homology arm and 3’ homology arm to donor plasmid backbone downstream of 3’ homology arm, respectively (grey arrows, 1072 bp, and black arrows, 1105 bp, respectively, in Fig. 1A; gels in Fig. 1D, 1E, respectively). Correct targeting was confirmed by PCR-amplification with a pair of primers spanning this region (green arrows in Fig. 1A; gel in Fig. 1B; ~0.6kb) followed by sequencing.
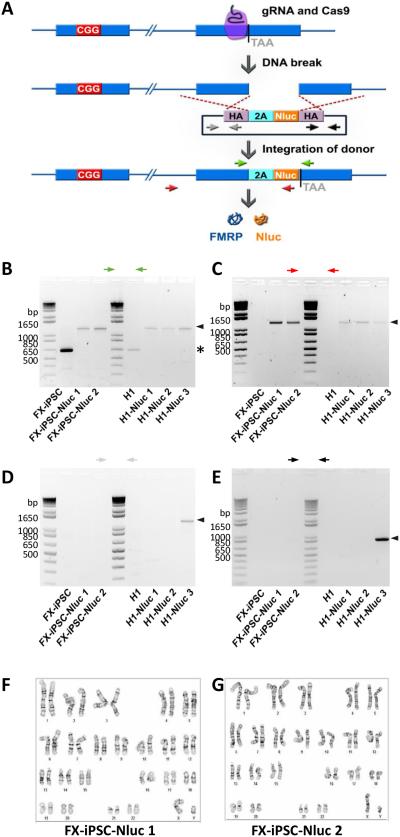
Figure 1. Generation of FMR1-Nluc reporter cell lines from H1 ESCs and FXS-iPSCs.
(A) Generation of reporter lines by using CRISPR/Cas9-mediated genome editing. Cleavage of FMR1 DNA by a guide RNA (gRNA)-Cas9 complex was followed by homologous recombination with the homology arms (HA) in a donor plasmid containing Nano luciferase gene (Nluc) and the 3’ coding sequence of FMR1 gene. (B, C) PCR validation of expected donor integration through homologous recombination in reporter lines. One pair of primers (green arrows) amplified the region of FMR1 gene with Nluc insertion yielding difference sizes from parental or targeted alleles. Another pair of primers (red) detected only targeted allele, but not parental allele. Black Arrowheads indicate expected bands of reporter lines and asterisk indicates bands of parental lines. (D, E) PCR validation of the absence of random integration of donor plasmid in reporter lines. FX-iPSC-Nluc 1 and 2 and H1-Nluc 1 and 2 were negative while H1-Nluc 3 was positive for random inserted donor plasmid. Black Arrowheads indicate expected bands from random, non-homology directed integration of the donor plasmid. (F, G) Karyotypes of FX-iPSC-Nluc 1 (F) and FX-iPSC-Nluc 2 (G). Normal 46XY karyotypes were observed.
Cell culture and neural differentiation
Human embryonic cell (hESC) line H1 (WA01) was obtained from WiCell. FXS iPSCs were previously described (Doers et al., 2014). Pluripotent stem cells were cultured on MEF feeder layers (WiCell) with a daily change of hESC medium of DMEM/F12 (Thermo Fisher Scientific), 20% knockout serum replacement (KSR, Thermo Fisher Scientific), 0.1 mM 2-mercaptoethanol (Sigma), 200 mM L-Glutamine (Thermo Fisher Scientific), 4 ng/ml FGF-2 (Waisman Biomanufacturing). Cells were passaged using 6 U/ml of dispase (Thermo Fisher Scientific) in hESC medium, washed and replated at a dilution of 1:5 to 1:10.
Neural induction was performed using a dual SMAD method [40]with modifications. In brief, hPSCs were dissociated using TrypLE Express (Thermo Fisher Scientific), washed, and plated on Matrigel (Corning)-coated plates at a density of 500,000-1,000,000 cells/cm2 in MEF-conditioned medium (CM) supplemented with 10 ng/ml FGF-2 and 10 μM ROCK inhibitor (Y-27632 dihydrochloride, Tocris). The cells were nearly confluent the next day. Neural differentiation was induced with a chemically defined medium, CDM (DMEM/F12: neural basal medium (Thermo Fisher Scientific) 1:1, 200 mM L-Glutamine, 1% N2 (Thermo Fisher Scientific), 0.5 × B27 without vitamin A (Thermo Fisher Scientific), 10 μM SB432542 (Selleck), 100 nM LDN193189 (Selleck)). Cells were cultured in CDM for 10 days with a daily medium change. Cells were then passaged with TrypLE and cultured on Matrigel-coated plates in NPC medium (neural basal medium, 1x GlutaMAX (Thermo Fisher Scientific), 1% N2, 0.5% B27, 10 ng/ml FGF-2). For the first few passages, 10 μM ROCK inhibitor was added when plating and was washed out the day after passaging cells. Most cells at this point expressed markers of neural progenitor cells (NPCs), NESTIN and PAX6 (Fig. 3B). For neuronal differentiation, NPCs were plated on Matrigel-coated coverslips at density of 5 × 104 cells/ cm2 in NDM medium (neural basal medium, 1% GlutaMAX, N2, B27, 0.2 μM ascorbic acid (Sigma), 1 μM cAMP (Sigma), 10 ng/ml BDNF (Peprotech), 10 ng/ml GDNF (Peprotech) supplemented with ROCK inhibitor and 0.1 μM Compound E (Calbiochem). Half NDM medium was changed every 3 days. Cells were fixed for immunostaining at 2 weeks after plating.
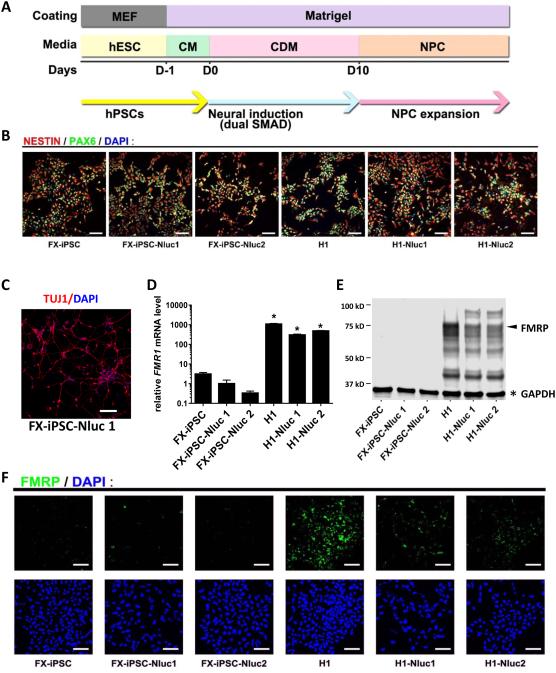
Figure 3. Neural differentiation and characterization of NPCs derived from reporter and parental lines.
(A) Schematic diagram of neural differentiation. (B) Differentiated NPCs are positive for neural progenitor markers, NESTIN and PAX6, as assessed by immunofluorescence (scale bar = 100 μm). (C) NPCs derived from the FXS reporter line were expanded for 23 passages and then differentiated into neurons (Tuj1+, red; scale bar = 50 μm). (D) Relative FMR1 mRNA levels assessed by quantitative RT-PCR. The mean expression of FX-iPSC-Nluc1 cells was set as 1 (Y axis in log scale; n=3; mean ± SEM; * indicates significant p value < 0.05). (E) A representative western blot result showing FMRP (arrow) and GAPDH (asterisk). (F) FMRP in reporter and parental NPCs detected using immunofluorescence and confocal imaging (scale bar = 100 μm).
Small molecule treatment and luciferase assay
For treatment with 5-aza-dC (Sigma, Cat # A3656) or 5-aza-C (Sigma, Cat #A2385), NPCs were plated on Matrigel-coated plates at density of approximately 2 × 104 cells/well (or otherwise specified) into wells of 96-well plates for luciferase assay experiments or 2 × 105 cells/well of 12-well plates for qPCR analysis. Two days after plating, 5-aza-dC or 5-aza-C or DMSO control were added to cells. Treated cells were either assayed for luciferase activity or collected in TRIzol for qPCR. For luciferase assay, medium was aspirated and cells were lysed with 50 μl of 1 × passive lysis buffer (Promega) for 10 min at room temperature. 20 μl of the lysate was mixed with 50 μl of Nano-Glo reagent (Promega) for Nluc activity and another 20 μl of the lysate was mixed with 50 μl of CellTiter-Glo 2.0 reagent (Promega) for an estimate of number of live cells. Both Nluc activity and Cell Titer signal were measured with GloMax-Multi+ Detection System (Promega). Luciferase assay and Cell Titer assay were performed three days after cell treatment or otherwise specified. Statistical significance was analyzed by one-way or two-way ANOVA with post-hoc tests.
Small molecule library screening
Library screenings were performed in 384-well plates coated with Matrigel (Corning, Cat #: 3707). NPCs derived from FX-iPSC-Nluc 1 line were dissociated with TrypLE express, washed, and resuspended in NPC medium supplemented with 10 μM ROCK inhibitor. NPCs were mixed with ice-cold NPC medium containing 0.13 mg/ml Matrigel at a cell density of 1 × 106 cells/ml and 40 μl of the cell suspension per well was immediately plated with a MicroFlo Select dispenser (BioTek). The cells were grown for 24 hours in a 37°C, 5% CO2 incubator. An Epigenetics Compound Library (Selleck, Cat #: L1900) was reformatted to 384-well plates with Biomek FX (Beckman) and added to cells using Echo 550 (Labcyte). Each plate included wells of positive controls treated with final concentration of 0.03 μM of 5-aza-dC and wells of negative controls treated with DMSO. Three days after adding small molecule libraries, 20 μl of Nano-Glo reagent per well was dispensed. The plates were incubated at room temperature for 5 min followed by detection of luciferase activity using EnSpire Multimode Reader (PerkinElmer). Z’-factors were calculated from data of positive control and negative control wells using equation [41].
σP, σN: standard deviation of positive controls and negative controls, respectively
μP, μN: mean of positive controls and negative controls, respectively
Experiments with 1536-well plates were performed as 384-well plates with modifications as following: 1000 cells in 6 μl of ice-cold NPC medium containing 0.05 mg/ml Matrigel were plated in each well of 1536-well plates (Greiner Bio-one, Cat #: 782095) and plates were sealed with sealing films (Axygen, Cat #: UC-500). On day 4, 4 μl of Nano-Glo reagents were added to each well and Nluc luciferase assay was performed with PHERAstar FS (BMG LABTECK).
Results
Generation of FMR1-Nluc reporter cell lines
To create a FMR1 reporter cell line for high throughput screening, we introduced Nano-luciferase (Nluc) into the endogenous FMR1 gene in both FXS-iPSCs and H1 human ESCs (WA01) using CRISPR/Cas9 gene editing. As shown in Fig.1A, a DNA cassette coding for Nluc following a “self-cleaving” peptide, P2A [42], was inserted immediately upstream of the stop codon of FMR1. Different isoforms of FMRP have been reported [43]. In our design, Nluc is co-expressed with the major isoforms of FMRP and cleaved at P2A, yielding separate FMRP and Nluc proteins. Colonies with correct insertion of P2A-Nluc were screened by PCR (Fig. 1B and C) and validated by sequencing (Fig. S1). Random insertion of donor vectors was evaluated using primers specific for donor vectors (Fig. 1D and E). Only targeted clones without random insertions were selected for further analysis. Two positive colonies derived from each parental FXS-iPSCs (FX-iPSC-Nluc1 and FX-iPSC-Nluc2) and H1 hESCs (H1-Nluc 1 and H1-Nluc 2) were expanded and used for further experiments. To assess the integrity of the reporter lines, we used hPSC markers SOX2, Oct4 and TRA-1-81 and immunocytochemical analysis. All the reporter lines exhibited similar marker expression as their parental hPSC lines (Fig. S2). In addition, normal karyotypes were confirmed for both FX-iPSC-Nluc1 and FX-iPSC-Nluc2 lines (Fig. 1F and 1G), indicating that gene editing did not generate genomic instability. Further, the CGG repeat length in the FMR1 gene was not altered during genetic manipulation, as validated by PCR amplification of the repeats (Fig. S2). Therefore, the FX-iPSC-Nluc reporter lines retain the pluripotent signature and genomic integrity of the parental lines.
We next determined whether the FMR1-Nluc reporter cell lines faithfully report FMR1 gene expression. Quantitative RT-PCR showed that mRNA level of FMR1 in the reporter lines resembled their parental lines (Fig. 2A), indicating that the insertion of Nluc had no effect on transcriptional silencing of FMR1 in the FX-iPSCs nor active transcription of FMR1 in the H1 hESCs. In addition, FMRP protein was detected in both the H1 hESCs parental line and H1-hESC-Nluc lines but not in any of the FXS-iPSC lines, as assessed by both western blot and immunofluorescence for FMRP (Fig. 2B, 2C). Importantly, the insertion of Nluc had no significant effect on the levels of FMRP protein expression in H1 hESC lines. To validate whether Nluc is co-expressed with FMRP as expected, Nluc activity was measured in both reporter cell lines and parental lines. As expected, high levels of Nluc activities were detected in the H1-Nluc lines whereas FXS-iPSC-Nluc lines had very low Nluc activity (Fig. 2D). Therefore, insertion of Nluc does not affect FMR1 activation status and Nluc activity can be used to monitor expression levels of FMRP in both FX-iPSCs and H1 hESC cells.
Figure 2. FMR1-Nluc reporter hPSC lines retain FMR1 expression status of their parental lines.
(A) Relative FMR1 mRNA levels assessed by qRT-PCR. The mean expression of FX-iPSC-Nluc1 cells was set as 1 (Y axis in log scale; n=3; mean ± SEM); * indicates significant p value < 0.05). (B) A representative western blot result showing FMRP (arrow) and GAPDH (asterisk, loading control) protein expression. (C) FMRP in reporter and parental lines detected using immunocytochemistry and confocal imaging (scale bar = 200 μm). (D) Relative Nluc activities of reporter and parental lines. The mean activity of FX-iPSC-Nluc 1 cells is set as 1 (Y axis in log scale; n=3; mean ± SEM; * indicates significant p value < 0.05).
Differentiation of FMR1-Nluc reporter cell lines to NPCs
HTS requires a large number of cells that can be maintained and expanded stably. FXS is a neurological disorder therefore drug screening needs to be carried out with cells that are relevant to the disorder. NPCs are neural-lineage cells that can be easily expanded and stably maintained for an extended period of time. NPCs can also be frozen and thawed for long-term storage and use. These characteristics make NPCs suitable for high throughput screening for neurological diseases. In fact, NPCs derived from FXS patient iPSCs have been used for small molecule screening[34, 38]. Neural induction of our reporter lines was carried out using an established protocol [40, 44] (Fig. 3A). Both FX-iPSC and H1 FMR1-Nluc reporter cell lines efficiently differentiated into NPCs expressing NESTIN and PAX6 (Fig. 3B). Importantly, the FXS-iPSC-Nluc1 and Nluc2 lines could differentiate into >95% NESTIN+ or PAX6+ cells with >91% of the cells expressing both (Fig 3B). These NPCs can be maintained for many passages without obvious changes in their neuronal differentiation potency. For example, NPCs derived from the FX-iPSC-Nluc1 line cultured for over 20 passages differentiated into neurons efficiently (Fig 3C).
We assessed the expression of FMR1 and Nluc expression in these NPCs. Similar to undifferentiated hPSC reporter cell lines (Fig. 2), NPCs differentiated from the H1 reporter lines exhibited high levels of FMR1 mRNA expression (Fig 3D) and FMRP protein expression (Fig 3E-F), at similar levels as NPCs differentiated from the H1 hESC parental line. However, NPCs differentiated from FX-iPSC or FX-iPSC reporter lines had very low or undetectable FMR1 mRNA (Fig. 3D) or FMRP protein levels (Fig. 3E-F). Further, the CGG repeat length in the FMR1 gene was not altered during differentiation, as validated by PCR amplification of the repeats (Fig. S4). Therefore, NPCs differentiated from FMR1-Nluc reporter cell lines have NPC characteristics and retain FMR1 gene silencing status, which make them suitable for studying reactivation of FMR1 gene expression.
Characterization of FMR1-Nluc reporter NPCs for HTS
To ensure that luciferase can serve as a specific reporter for endogenous FMR1 gene expression, we first performed acute knockdown of FMR1 in H1-reporter lines using lentivirus expressing small inhibitory RNAs (shRNAs). We found that in both H1 reporter lines (H1-Nluc1 and H1-Nluc2), FMR1 shRNA treatment led to reduction in both luciferase activities and FMR1 mRNA levels to similar extent (Fig. S5). To define parameters for successful HTS for FMR1 reactivation using NPCs generated from our FMR1-Nuc reporter cell lines, we used 5-aza-dC, a DNMT inhibitor known to partially activate FMR1 in FXS cells [28] as a positive control (Fig. 4A). NPCs differentiated from both FXS-iPSC-Nluc1 and FXS-iPSC-Nluc2 lines showed similar dosage-dependent induction of Nluc activity with a single treatment of 5-aza-dC with minimal cell death up to 10 μM concentration (Fig. 4C-E). We chose the FX-iPSC-Nluc1 cell line for subsequent experiments. To validate that the increase in Nluc activity in response to 5-aza-dC reflects the activation of FMR1, we measured FMR1 mRNA levels, Nluc mRNA levels, and Nluc activity in parallel FX-iPSC-Nluc1 NPC samples treated with different concentrations of 5-aza-dC. We found that the induction of Nluc mRNA expression (Fig. 4F red line) and Nluc activity (Fig. 4G) by 5-aza-dC coincided with the changes in expression levels of FMR1 mRNA (Fig 4F blue line), demonstrating that Nluc activities in the FXS-iPSC-Nluc cells faithfully report the expression levels of FMR1 mRNA. Since 5-aza-dC is known to partially reactivate the human FMR1 promoterthrough demethylation in other cell types, we assessed the methylation status of 5' regions of FMR1gene. We found that 5-aza-dC treatment indeed led to decreased methylation of the FMR1 promoter (Fig S6).
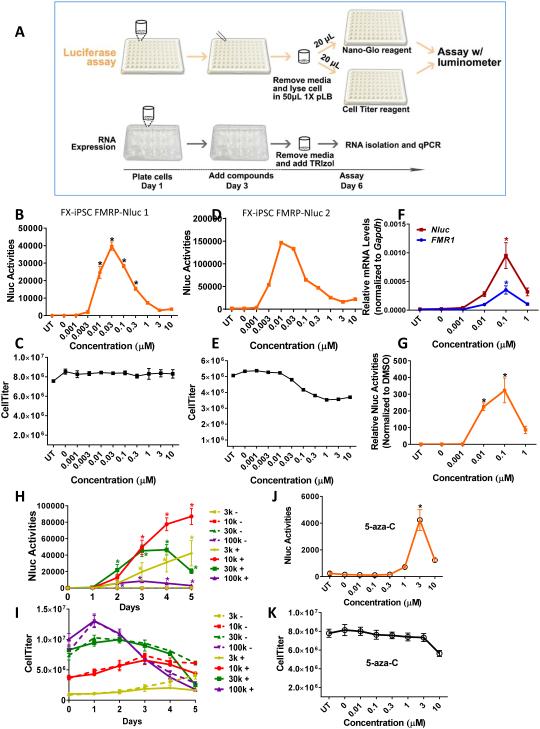
Figure 4. Responses of FX-iPSC-Nluc NPCs to 5-aza-dC and 5-aza-C.
(A) Schematic diagram of experimental timeline showing treatment of cells with compounds followed by luciferase assay or quantitative RT-PCR. (B, C) Dose-response curve of Nluc activities (B) and Cell Titer (cell viability, C) of FX-iPSC-Nluc1 NPCs treated with 5-aza-dC (UT, untreated; n=3; mean ± SEM; * indicates significant p value < 0.05). (D, E) Dose-response curve of Nluc activity (D) and Cell Titer (E) of FX-iPSC-Nluc2 NPCs treated with 5-aza-dC (n=1). (F, G) Dose-response curves of FMR1 mRNA (F, blue), Nluc mRNA (F. red), and normalized Nluc activity (G, normalized to control DMSO) of NPCs treated with 5-aza-dC (n=4; mean ± SEM). (H, I) Time-course of Nluc activities (H) and Cell Titer (I) of NPCs treated with 5-aza-dC (solid lines, n=4; mean ± SEM) compared to cells treated with vehicle (DMSO, doted lines). Four cell densities (3,000 or 3k, 10,000 or 10k, 30,000 or 30k, and 100,000 or 100k cells per well) were tested in 96-well plates. (J, K) Dose-response curve of Nluc activity (J) and Cell Titer (K) of NPCs treated with 5-aza-C (n=4; mean ± SEM). * indicates significant p value < 0.05.
We next assessed how cell density might affect the ability of Nluc to report FMR1 expression in FX-iPSC-Nluc1 NPCs. A single treatment of 5-aza-dC of cells plated in a 96-well plate induced significant increase in Nluc activity at all cell densities over days (Fig. 4H). Even 100,000 per well (100k) cell density exhibited >10-fold induction. Since both 10,000 cells per well (10k) and 30k cell densities reached a similar Nluc activity at 3 days post-treatment without significantly affecting cell viability (Fig. 4I), we decided to use 20,000 cell per well (20k) as the optimal cell density for our HTS screen. Moreover, since 5-aza-C has also been shown to reactivate FMR1 in FXS patient-derived iPSCs [35], we assessed its ability in reactivating FMR1 gene in our assay system. A single treatment of 5-aza-C also induced Nluc activity in FMR1-Nluc reporter NPCs (Fig. 4J); however, Nluc activity was lower and higher concentration of compound was needed when compared to 5-aza-dC (Fig. 4K). These data indicate that our FMR1-Nluc reporter line is a robust platform for small molecule screening
Library screening with FMR1-Nluc reporter cells
Encouraged by the robust responses of FMR1-Nluc reporter NPCs to 5-aza-dC and 5-aza-C, we set out to screen a library consisting of 128 small molecules of epigenetic modulators, including 5-aza-dC and 5-aza-C (Table S1). Based on our optimization data (Fig. 4), we adapted our screen to 384-well plate format for a 3-day drug treatment scheme (Fig. 5A). We plated 4000-5000 NPCs into each well of a 384-well plate, which was scaled down from 20,000 NPCs per well (of a 96-well plate). One 384-well plate contains the library at one concentration in duplicate wells and wells treated with either positive controls (5-aza-dC) or negative controls (DMSO) and we screened this library at three different concentrations (Fig. 5B). The Z’-factors calculated from positive and negative controls for all three plates were greater than 0.5, indicating that our platform is sufficient for library screening. Although no novel hit was identified from this library, 5-aza-dC and 5-aza-C were detected as hits (Fig. 5B) and their responsive dosages were consistent with our dose-response results (Fig. 4).
Figure 5. Screening small molecule library in 384-well plates.
(A) Schematic diagram of HTS screening of small molecule library. (B) Screening results of an epigenetics library (128 compounds; see Table S1 for a description of compounds in each well). Each heat map represents one 384-well plate treated with the epigenetics library in duplicate wells at specified concentration (0.1, 1 and 10 μM). Wells G11 and K11 were duplicate wells of 5-aza-C. Wells K7 and L7 were duplicate wells of 5-aza-dC.
To achieve higher throughput, we assessed the feasibility of HTS screening with 1536-well plates. We tested a number of parameters including cell plating densities, types of plates used for assay, and methods for reducing evaporation. We finally achieved an optimized condition (see Methods for further details). We plated 1000 NPCs into each well of a 1536-well plate which is also scaled down from 20,000 cells per well of a 96-well plate. To evaluate HTS platform, we divided the 1536-well plates into quadrants and treated each quadrant with either DMSO or 5-aza-dC (Fig. 6A). We employed the same experimental timeline as for 384-well pate (Fig. 5A). Sample images from randomly selected wells showed that FX-iPSC-Nluc NPCs grew uniformly in wells of the 1536-well plates on both day 2 (the day after plating) and day 5 (the day of luciferase assay) (Fig. 6B, Fig S7). Our results showed that NPCs plated in 1536-well plates responded robustly and consistently to 5-aza-dC with 240 X fold induction (5-aza-dC/DMSO) and a good Z’ factor (Z’= 0.57). Moreover, good Z’ factors (~0.5) were also obtained consistently from most rows and columns of the plates (Fig. 6C), demonstrating a robust platform for HTS library screening with 1536-well plates.
Figure 6. Platform setup and library screening in 1536-well plates.
(A) Nluc activity from a 1536-well plate treated with DMSO in quadrant 2 and 4 and 5-aza-dC in quadrant 1 and 3. The Z’=0.57 for the screen. (B) Cells in a corner well (A1) and a center well (P24) in 1536-well plates were imaged in bright field on day 2 (the day after plating) and day 5 (the day of cell collection and luciferase assay), showing good cell survival and growth in 1536-well plates (scale bar = 400 μm). Larger images are shown in Figure S3. (C) Z’-factors were calculated for each column and each row of the 1536-well plate. (D) Screening of an FDA-approved Compound Library (1134 compounds, 3 μM) in 1536-well plates. Columns 23 and 47 were treated with negative control DMSO. Columns 24 and 48 were treated with positive control 5-aza-dC. Well A25 was 5-aza-C and well I11 was 5-aza-dC.
We then used this 1536-well plate method to screen an FDA-approved Drug Library that contains 1134 FDA-approved drugs including both 5-aza-dC and 5-aza-C (Table S2). As expected, both 5-aza-dC and 5-aza-C in the library were positive hits (Fig. 6D). Although we did not identify any new hits, the background and false positive rate were low. These results demonstrate that we have created a robust platform for HTS of small molecules for FMR1 reactivation in FXS patient-derived reporter NPCs. The specific and sensitive novel human PSC reporter line for reporting FMR1 gene activation will enable discovery at high throughput.
Discussion
There is an urgent need for high throughput screens for FMR1 gene reactivation. Human patient-derived induced pluripotent stem cells (iPSCs) provide unique opportunities for disease modeling and drug development. A few FXS iPSC-based screens have been recently published using NPCs differentiated from FXS iPSCs [34, 38]. These strategies rely on commercial FMRP antibodies to detect FMRP in high content imaging or time-resolved fluorescence resonance energy transfer (TR-FRET). Library screening that included 50,000 compounds covering epigenetic targets and known FMRP regulated pathways revealed several compounds (identity not revealed) that induced weak FMR1 reactivation [38]. Screening of ~5000 compounds including a FDA-approved drug library identified six hits that modestly enhanced FMR1 gene transcription, although no significant amount of FMRP protein was detected. Interestingly, one of the identified compounds is SB216763 that we have previously found to rescue learning deficits in FMR1-null mice through enhancing Wnt signaling [45]. Yet, none of the compounds identified so far can reactivate FMR1 expression to near normal levels, necessitating new and better strategies to discover new compounds. These negative results suggest that we must explore large collections of compounds using unbiased approaches. Neither of the above strategies is amenable for HTS due to limited sensitivities and high cost. Our FMR1-Nluc reporter cell line provides a clean assay for such a purpose. In addition to its utility for small molecule screens to identify FMR1 reactivating compounds, the FMR1-Nluc reporter line is potentially valuable for arrayed genetic screens using shRNAs or novel guide RNAs to restore FMR1 gene expression.
A number of studies have shown that treatment of human FXS lymphoblastoid cell lines with 5-aza-C or 5-aza-dC results in partial reactivation of the FMR1 gene and FMRP expression and the promoter of FMR1 gene become passively unmethylated after treatment through cell division [24, 25]. The increases in FMR1 mRNA production are associated with increased active chromatin markings and decreased repressive chromatin markings to the FMR1 promoter [17, 24, 26, 27]. The inhibition of DNA methylation seems to be a key aspect of 5-aza-dC action because methotrexate, a folate antagonist that does not reduce DNA methylation in the FMR1 promoter, cannot reactivate the FMR1 gene [27]. Chemicals affecting histone modification have also been explored for FMR1 reactivation. Most studies have so far focused on Class I, II, and IV HDAC inhibitors include butyrate, and trichostatin A. Using human FXS lymphoblastoid cell lines, Chiurazzi et al., showed that treatment with histone deacetylatase inhibitors, phenylbutyrate, sodium butyrate and TSA, leads to moderate increased FMR1 gene transcription, which is significantly smaller compared to the effect of 5azaC or 5-azadC [46]. However other studies show no FMR1 transcription after TSA treatment [47, 48] or VPA treatment [17]. The libraries that we screened have several other DNA (cytosine-5)-methyltransferase (Dnmt) inhibitors; however none but 5-aza-dC and 5-aza-C showed positive response. It will be important to determine the mechanistic differences between 5-aza-dC and other Dnmt inhibitors and design novel drugs for gene reactivation.
Supplementary Material
Acknowledgements
We thank Yina Xing, Erich Berndt, and Song Guo for technical assistance, the Bhattacharyya and Zhao lab members for discussion, and members of the Su-Chun Zhang and Qiang Chang labs for helpful advice on gene editing. This work was supported by grants from the John Merck Fund (to XZ, AB, KS), NIH (R01MH080434, R01MH078972 to XZ; R21HD085288 to AB), NSF (CBET-1350178 to KS) and a Center grant from the NIH to the Waisman Center (P30HD03352).
Footnotes
Author contributions
Meng Li: Conception and design, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
Huashan Zhao: Collection and/or assembly of data, Data analysis and interpretation.
Gene Ananiev: Conception and design, Collection and/or assembly of data, Final approval of manuscript
Michael Musser: Collection and/or assembly of data.
Kathryn Ness: Collection and/or assembly of data.
Dianne Maglaque: Collection and/or assembly of data.
Krishanu Saha: Conception and design, Financial support, Data analysis and interpretation, Final approval of manuscript
Anita Bhattacharyya: Conception and design, Financial support, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
Xinyu Zhao: Conception and design, Financial support, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
References
- 1.Coffee B, Keith K, Albizua I, et al. Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA. American journal of human genetics. 2009;85:503–514. doi: 10.1016/j.ajhg.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagerman RJ, Hagerman PJ. Johns Hopkins University Press; Baltimore MD: 2002. Fragile X syndrome. [Google Scholar]
- 3.Cohen IL, Sudhalter V, Pfadt A, et al. Why are autism and the fragile-X syndrome associated? Conceptual and methodological issues. AmJHumGenet. 1991;48:195–202. [PMC free article] [PubMed] [Google Scholar]
- 4.Fisch GS, Cohen IL, Wolf EG, et al. Autism and the fragile X syndrome. AmJPsychiatry. 1986;143:71–73. doi: 10.1176/ajp.143.1.71. [DOI] [PubMed] [Google Scholar]
- 5.Hagerman RJ, Ono MY, Hagerman PJ. Recent advances in fragile X: a model for autism and neurodegeneration. Curr Opin Psychiatry. 2005;18:490–496. doi: 10.1097/01.yco.0000179485.39520.b0. [DOI] [PubMed] [Google Scholar]
- 6.Kaufmann WE, Cortell R, Kau AS, et al. Autism spectrum disorder in fragile X syndrome: communication, social interaction, and specific behaviors. AmJMedGenetA. 2004;129:225–234. doi: 10.1002/ajmg.a.30229. [DOI] [PubMed] [Google Scholar]
- 7.Reiss AL, Feinstein C, Rosenbaum KN. Autism and genetic disorders. SchizophrBull. 1986;12:724–738. doi: 10.1093/schbul/12.4.724. [DOI] [PubMed] [Google Scholar]
- 8.Hatton DD, Sideris J, Skinner M, et al. Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. Am J Med Genet A. 2006;140A:1804–1813. doi: 10.1002/ajmg.a.31286. [DOI] [PubMed] [Google Scholar]
- 9.Lathe R. Fragile X and autism. Autism : the international journal of research and practice. 2009;13:194–197. doi: 10.1177/13623613090130020402. [DOI] [PubMed] [Google Scholar]
- 10.Brouwer JR, Mientjes EJ, Bakker CE, et al. Elevated Fmr1 mRNA levels and reduced protein expression in a mouse model with an unmethylated Fragile X full mutation. Experimental cell research. 2007;313:244–253. doi: 10.1016/j.yexcr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loesch DZ, Huggins R, Hay DA, et al. Genotype-phenotype relationships in fragile X syndrome: a family study. American journal of human genetics. 1993;53:1064–1073. [PMC free article] [PubMed] [Google Scholar]
- 12.Loesch DZ, Huggins RM, Hagerman RJ. Phenotypic variation and FMRP levels in fragile X. Mental retardation and developmental disabilities research reviews. 2004;10:31–41. doi: 10.1002/mrdd.20006. [DOI] [PubMed] [Google Scholar]
- 13.Hagerman RJ, Hull CE, Safanda JF, et al. High functioning fragile X males: demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. American journal of medical genetics. 1994;51:298–308. doi: 10.1002/ajmg.1320510404. [DOI] [PubMed] [Google Scholar]
- 14.Loesch D, Hagerman R. Unstable mutations in the FMR1 gene and the phenotypes. Advances in experimental medicine and biology. 2012;769:78–114. doi: 10.1007/978-1-4614-5434-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smeets HJ, Smits AP, Verheij CE, et al. Normal phenotype in two brothers with a full FMR1 mutation. Hum Mol Genet. 1995;4:2103–2108. doi: 10.1093/hmg/4.11.2103. [DOI] [PubMed] [Google Scholar]
- 16.Pietrobono R, Tabolacci E, Zalfa F, et al. Molecular dissection of the events leading to inactivation of the FMR1 gene. Human Molecular Genetics. 2005;14:267–277. doi: 10.1093/hmg/ddi024. [DOI] [PubMed] [Google Scholar]
- 17.Tabolacci E, De Pascalis I, Accadia M, et al. Modest reactivation of the mutant FMR1 gene by valproic acid is accompanied by histone modifications but not DNA demethylation. Pharmacogenetics and genomics. 2008;18:738–741. doi: 10.1097/FPC.0b013e32830500a1. [DOI] [PubMed] [Google Scholar]
- 18.Gholizadeh S, Arsenault J, Xuan ICY, et al. Reduced Phenotypic Severity Following Adeno-Associated Virus-Mediated Fmr1 Gene Delivery in Fragile X Mice. Neuropsychopharmacology. 2014;39:3100–3111. doi: 10.1038/npp.2014.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musumeci SA, Calabrese G, CM Bonaccorso, et al. Audiogenic seizure susceptibility is reduced in fragile X knockout mice after introduction of FMR1 transgenes. Experimental neurology. 2007;203:233–240. doi: 10.1016/j.expneurol.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Paylor R, Yuva-Paylor LA, Nelson DL, et al. Reversal of sensorimotor gating abnormalities in Fmr1 knockout mice carrying a human Fmr1 transgene. Behavioral neuroscience. 2008;122:1371–1377. doi: 10.1037/a0013047. [DOI] [PubMed] [Google Scholar]
- 21.Peier AM, McIlwain KL, Kenneson A, et al. (Over)correction of FMR1 deficiency with YAC transgenics: behavioral and physical features. Hum Mol Genet. 2000;9:1145–1159. doi: 10.1093/hmg/9.8.1145. [DOI] [PubMed] [Google Scholar]
- 22.Zeier Z, Kumar A, Bodhinathan K, et al. Fragile X mental retardation protein replacement restores hippocampal synaptic function in a mouse model of fragile X syndrome. Gene Ther. 2009;16:1122–1129. doi: 10.1038/gt.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo W, Allan AM, Zong R, et al. Ablation of Fmrp in adult neural stem cells disrupts hippocampus-dependent learning. Nature medicine. 2011;17:559–565. doi: 10.1038/nm.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietrobono R, Pomponi MG, Tabolacci E, et al. Quantitative analysis of DNA demethylation and transcriptional reactivation of the FMR1 gene in fragile X cells treated with 5-azadeoxycytidine. Nucleic Acids Res. 2002;30:3278–3285. doi: 10.1093/nar/gkf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiurazzi P, Pomponi MG, Willemsen R, et al. In vitro reactivation of the FMR1 gene involved in fragile X syndrome. Hum Mol Genet. 1998;7:109–113. doi: 10.1093/hmg/7.1.109. [DOI] [PubMed] [Google Scholar]
- 26.Kumari D, Bhattacharya A, Nadel J, et al. Identification of Fragile X Syndrome-Specific Molecular Markers in Human Fibroblasts: A Useful Model to Test the Efficacy of Therapeutic Drugs. Human mutation. 2014 doi: 10.1002/humu.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brendel C, Mielke B, Hillebrand M, et al. Methotrexate treatment of FraX fibroblasts results in FMR1 transcription but not in detectable FMR1 protein levels. Journal of neurodevelopmental disorders. 2013;5:23. doi: 10.1186/1866-1955-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doers ME, et al. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells and Development. 2014;23:1777–1787. doi: 10.1089/scd.2014.0030. Musser Mt Fau - Nichol R, Nichol R Fau - Berndt ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Esch Celine EF, Ghazvini M, Loos F, et al. Epigenetic Characterization of the FMR1 Promoter in Induced Pluripotent Stem Cells from Human Fibroblasts Carrying an Unmethylated Full Mutation. Stem Cell Reports. 2014 doi: 10.1016/j.stemcr.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halevy T, Czech C, Benvenisty N. Molecular Mechanisms Regulating the Defects in Fragile X Syndrome Neurons Derived from Human Pluripotent Stem Cells. Stem Cell Reports. 2015;4:37–46. doi: 10.1016/j.stemcr.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brick DJ, Nethercott HE, Montesano S, et al. The Autism Spectrum Disorders Stem Cell Resource at Children's Hospital of Orange County: Implications for Disease Modeling and Drug Discovery. Stem Cells Transl Med. 2014;3:1275–1286. doi: 10.5966/sctm.2014-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doers ME, Musser MT, Nichol R, et al. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem cells and development. 2014;23:1777–1787. doi: 10.1089/scd.2014.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheridan SD, Theriault KM, Reis SA, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaufmann M, Schuffenhauer A, Fruh I, et al. High-Throughput Screening Using iPSC-Derived Neuronal Progenitors to Identify Compounds Counteracting Epigenetic Gene Silencing in Fragile X Syndrome. Journal of Biomolecular Screening. 2015 doi: 10.1177/1087057115588287. [DOI] [PubMed] [Google Scholar]
- 35.Bar-Nur O, Caspi I, Benvenisty N. Molecular analysis of FMR1 reactivation in fragile-X induced pluripotent stem cells and their neuronal derivatives. Journal of Molecular Cell Biology. 2012;4:180–183. doi: 10.1093/jmcb/mjs007. [DOI] [PubMed] [Google Scholar]
- 36.Urbach A, Bar-Nur O, Daley GQ, et al. Differential Modeling of Fragile X Syndrome by Human Embryonic Stem Cells and Induced Pluripotent Stem Cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schadt EE, Buchanan S, Brennand KJ, et al. Evolving toward a human-cell based and multiscale approach to drug discovery for CNS disorders. Front Pharmacol. 2014;5:252. doi: 10.3389/fphar.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumari D, Swaroop M, Southall N, et al. High-Throughput Screening to Identify Compounds That Increase Fragile X Mental Retardation Protein Expression in Neural Stem Cells Differentiated From Fragile X Syndrome Patient-Derived Induced Pluripotent Stem Cells. Stem Cells Transl Med. 2015 doi: 10.5966/sctm.2014-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Gaetano CM, Williams KR, et al. FMRP interacts with G-quadruplex structures in the 3’-UTR of its dendritic target Shank1 mRNA. RNA Biology. 2014;11:1364–1374. doi: 10.1080/15476286.2014.996464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers SM, Fasano CA, Papapetrou EP, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature biotechnology. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of Biomolecular Screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 42.Kim JH, Lee SR, Li LH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pretto DI, Eid JS, Yrigollen CM, et al. Differential increases of specific FMR1 mRNA isoforms in premutation carriers. Journal of Medical Genetics. 2014 doi: 10.1136/jmedgenet-2014-102593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barkess G, West AG. Chromatin insulator elements: establishing barriers to set heterochromatin boundaries. Epigenomics. 2012;4:67–80. doi: 10.2217/epi.11.112. [DOI] [PubMed] [Google Scholar]
- 45.Guo W, Murthy AC, Zhang L, et al. Inhibition of GSK3beta improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21:681–691. doi: 10.1093/hmg/ddr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiurazzi P, Pomponi MG, Pietrobono R, et al. Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum Mol Genet. 1999;8:2317–2323. doi: 10.1093/hmg/8.12.2317. [DOI] [PubMed] [Google Scholar]
- 47.Coffee B, Zhang F, Warren ST, et al. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nature genetics. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 48.Coffee B, Zhang F, Ceman S, et al. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. American journal of human genetics. 2002;71:923–932. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.