Abstract
Worldwide, approximately one billion people smoke cigarettes. Cigarette smoking persists in part because long-term smoking cessation rates are modest on existing treatments. Smoking cessation outcomes are influenced by genetic factors, including genetic variation in enzymes that metabolize nicotine and smoking cessation medications, as well as in receptor targets for nicotine and treatment medications. For example, smokers with genetically slow nicotine metabolism have higher cessation success on behavioural counseling and nicotine patch compared to smokers with genetically fast nicotine metabolism. This review highlights new progress in our understanding of how genetic variation in the pharmacological targets of nicotine and smoking cessation medications could be used to tailor smoking cessation therapy, increase quit rates, and reduce tobacco-related harm.
Keywords: smoking cessation, tobacco, pharmacogenetics, treatment, nicotine, metabolism
Applying pharmacogenetics to reduce cigarette smoking
Tobacco smoking persists despite decades of research demonstrating its harms to health and the availability of three types of pharmacotherapy for treating tobacco dependence: nicotine replacement therapy (NRT), bupropion, and varenicline, which bind selected subtypes of nicotinic acetylcholine receptors (nAChRs) to exert their varying effects [1–3]. Bupropion can additionally bind to the dopamine transporter which is thought to also contribute to its pharmacological action [4]. A large proportion of the ability to quit smoking is heritable (~50–60%) [5]. In the late-1990s, genetic variation in the major pathway of nicotine metabolism was shown to alter the quantity of cigarettes smoked [6]. In more recent years, genetic variation in nicotine metabolism and receptor genes, and in dopaminergic pathway genes, has been implicated in the ability to quit smoking both in the absence and presence of pharmacotherapy. Genetic factors with more general effects on cessation are likely to influence both shorter (e.g., 8-weeks) and longer (e.g., 6-months) term abstinence, while those that influence cessation on pharmacotherapy are likely to be more apparent during the period of active treatment but can have lasting effects. Recently, several meta-analyses have indicated the robustness of some of these specific genetic findings [7, 8]. Single gene and polygenic influences on smoking cessation, within and between treatment conditions, have been studied and are reviewed here; the identification of gene variants associated with higher quit rates on one treatment relative to another will likely be especially valuable in clinical settings for personalizing treatment. Genomics-based personalization of tobacco dependence treatment will, in turn, increase smoking cessation rates and reduce the burden of tobacco-related disease.
Optimizing cessation using genetics of drug metabolism enzymes
Nicotine’s metabolic pathways
Nicotine is the major psychoactive compound in cigarette smoke [9]. The rate at which nicotine is metabolically inactivated, or cleared, from the body has important implications for nicotine dependence and the ability to quit smoking. Up to 80% of a nicotine dose is metabolically inactivated to cotinine, principally (~90%) by the hepatic enzyme cytochrome P450 (CYP) 2A6 [10] (Figure 1), with minor contributions from CYP2B6 (10%) [11]. Nicotine is also metabolized to more minor metabolites by additional enzymes, including FMO3 and UGT2B10 [12]. The majority of cotinine undergoes further metabolism to 3′hydroxycotinine in a reaction exclusively mediated by CYP2A6 [13] (Figure 1). The ratio of 3′hydroxycotinine to cotinine, known as the nicotine metabolite ratio (NMR), is an established and validated phenotypic indicator of CYP2A6 activity in daily smokers; faster CYP2A6 activity is reflected by a higher NMR [14–18].
Figure 1.
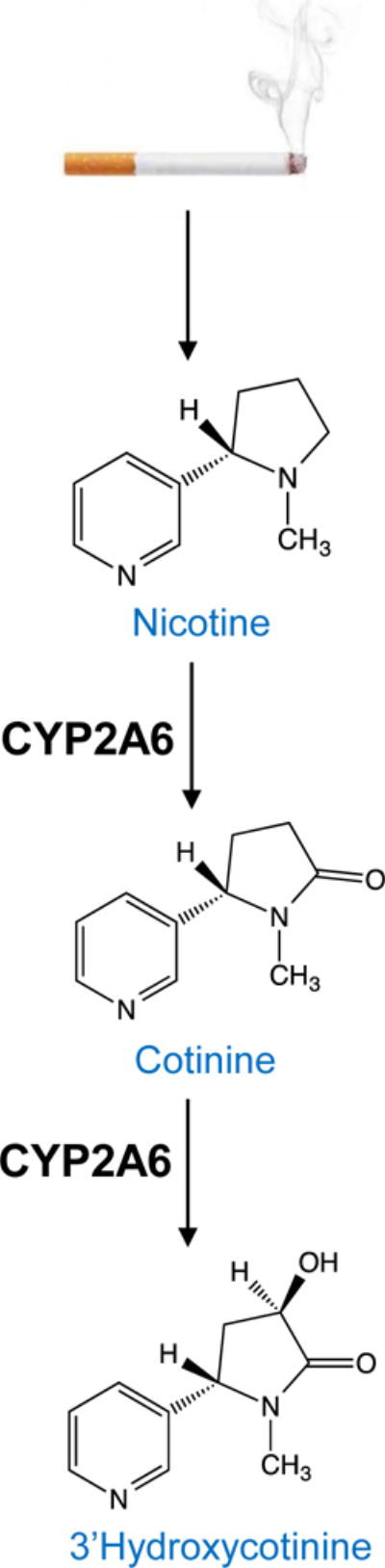
Role of CYP2A6 in the metabolic pathway of nicotine.
Up to 80% of a nicotine dose inhaled from cigarette smoke undergoes metabolic inactivation to cotinine. CYP2A6 is responsible for approximately 90% of nicotine’s inactivation to cotinine. The majority of cotinine is then further metabolized to 3′hydroxycotinine, in a reaction mediated wholly by CYP2A6. The ratio of these two metabolites, 3′hydroxycotinine/cotinine, known as the nicotine metabolite ratio or NMR, serves as a biomarker of CYP2A6 activity. Faster CYP2A6 activity is reflected by a higher NMR.
CYP2A6 genetics and smoking cessation outcomes
The CYP2A6 gene is highly polymorphic, with many variants altering CYP2A6 function; individuals can be genotyped for these variants and grouped into CYP2A6 activity groups (e.g., faster and slower metabolizers) based on the predicted metabolic impact of their CYP2A6 genotype on nicotine clearance [19]. In addition to capturing the influence of any existing CYP2A6 polymorphisms, the NMR also reflects environmental sources of variation in CYP2A6 activity (e.g., use of estrogen-containing hormonal therapy [20]). Similar to genotype-based activity groupings, smokers can be dichotomized as faster and slower metabolizers based on NMR. There is currently no single optimal NMR cut-point used to distinguish slower from faster metabolizers for cessation optimization or other smoking phenotypes (see Outstanding Questions). Different investigations have selected NMR cut-points based on sensitivity and specificity analyses of smoking cessation outcomes and/or pragmatic reasons (e.g., recruitment goals, statistical power); across different studies slow metabolizers generally represent the lowest 25–40% of the NMR distribution [21, 22]. Slower nicotine metabolizers (determined by CYP2A6 activity group or NMR) have lower cigarette consumption [23], dependence [24, 25], nAChR availability [26], and brain response to smoking (versus control) cues [27, 28], compared to faster nicotine metabolizers. Slower nicotine metabolizers also display higher smoking cessation rates in the absence of pharmacotherapy [29–31].
Outstanding Questions Box.
Would the refinement of genetic risk scores, accounting for variation in multiple genes, improve the personalization of tobacco dependence treatment?
How does pharmacogenetic variation in smoking cessation medication target genes influence treatment side effect profiles?
Will the pharmacogenetic findings from smoking cessation clinical trials replicate in community-based smokers, and in those with additional/psychiatric comorbidities?
As the vast majority of pharmacogenetic investigations have been conducted in clinical trials in heavy smokers (10+ cigarettes/day), will these findings replicate in light smokers?
As many of the pharmacogenetic outcomes have been determined in smokers of Caucasian and African American descent, will these pharmacogenetic findings replicate in smokers from other ethnic and cultural backgrounds and smoking phenotypes?
What is the optimal nicotine metabolite ratio cut-point to distinguish slower from faster nicotine metabolizers when attempting to optimize smoking cessation outcomes?
Could a more complete understanding of CYP2A6 genomics lead to the development of a CYP2A6 genetics algorithm to optimally treat nicotine dependence?
How feasible will it be to implement the nicotine metabolite ratio and/or CYP2A6 genetics in clinical settings to tailor treatment choice and/or adjust treatment dose?
What mechanism(s) underpin the increased ability of CYP2A6 slow nicotine metabolizers to quit smoking?
Could CYP2B6 genetic information be used to optimize bupropion dosing or the selection of an alternative smoking cessation treatment?
In addition to NMR, an alternative CYP2A6 genotype-based measure of nicotine metabolism rate exists that uses the ratio of deuterated (D2) cotinine/(D2cotinine + D2nicotine) determined 30 minutes after the oral administration of D2 nicotine [32]. In Caucasian treatment-seeking smokers randomized to placebo, those within the predicted lowest quartile of nicotine metabolism rate had lower relapse risk versus faster metabolizers (Hazard Ratio=0.40; P=0.013). Similarly, on placebo, Caucasian smokers with slower rates of CYP2A6 activity (lower NMR) were less likely to relapse than those with faster rates [31]. Variation in NMR is also associated with smoking cessation outcomes on active pharmacotherapy, as discussed below.
The nicotine metabolite ratio and smoking cessation outcomes
In smokers randomized to nicotine patch therapy, slower nicotine metabolizers (lowest NMR quartile) displayed higher quit rates compared to faster metabolizers [33, 34]. In a separate trial, smokers within the lowest NMR quartile had higher quit rates on extended (6-month) versus standard (8-week) nicotine patch therapy; in contrast, those with faster rates of nicotine metabolism did not benefit from extended therapy [35]. In another small trial of faster nicotine metabolizers (NMR≥0.18), there was a trend toward higher end-of-treatment quit rates on 42 mg patch versus 21 mg patch (46% versus 30%; P=0.08) [21]. Thus, the nicotine patch, at higher doses, may be effective in those with higher NMR. Follow-up studies are required to determine whether these findings replicate beyond clinical trial settings. In community-based smokers receiving nicotine patch therapy, faster metabolizers (NMR≥0.47) displayed lower end-of-treatment quit rates compared to slower metabolizers (24% versus 33%; P=0.03) [36]. To our knowledge, this is the first study demonstrating that relationships between NMR and cessation outcomes replicate in community-based smokers.
In smokers prospectively randomized to treatment based on NMR, varenicline was more efficacious than nicotine patch in faster metabolizers (NMR≥0.31) as hypothesized; in slower metabolizers, quitting did not differ by treatment [22] (Figure 2a, Key Figure). In faster metabolizers, the number needed to treat (NNT) for patch (versus placebo) and varenicline (versus placebo) was 26 and 5, respectively; for slower metabolizers, the NNT values were 10 and 8, respectively [22] (Figure 2b). Moreover, varenicline (versus placebo) was associated with greater side effect severity in slower (versus faster) metabolizers. In contrast, the nicotine patch (versus placebo) was well-tolerated in both groups. These findings suggest that varenicline would be more suitable for faster metabolizers, whereas the patch would be more suitable for slower metabolizers [22]. Follow-up studies in community-based smokers evaluating NMR and comparing cessation outcomes on varenicline and patch will help inform the implementation of NMR as a clinical tool to personalize smoking cessation therapy.
Figure 2.
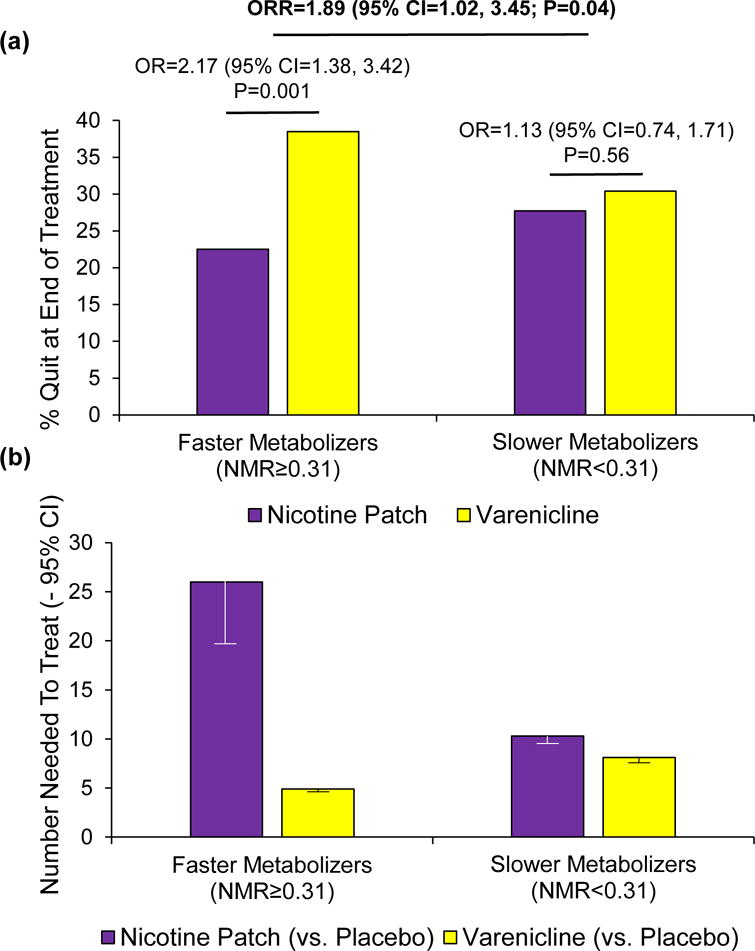
Key Figure. The nicotine metabolite ratio influences smoking cessation outcomes on varenicline and the nicotine patch.
In smokers prospectively randomized to varenicline, nicotine patch, or placebo based on the nicotine metabolite ratio or NMR, end-of-treatment quit rates and odds ratios (OR) with 95% confidence intervals (CI) comparing the efficacy of varenicline versus the nicotine patch as a function of NMR group (faster vs. slower nicotine metabolizers) are shown in (a). There was a significant NMR-by-treatment interaction on end-of-treatment quit rates: ratio of odds ratios (ORR) = 1.89 (95% CI = 1.02, 3.45; P=0.04) (a). In a longitudinal model including end-of-treatment as well as 6-month and 12-month quit rates, the NMR-by-treatment interaction was also significant: ORR = 1.96 (95% CI = 1.11, 3.46; P=0.02). Number needed to treat analyses comparing the effectiveness of varenicline versus placebo, and nicotine patch versus placebo, as a function of NMR group (faster vs. slower nicotine metabolizers) are shown in (b). For example, 26 faster metabolizers would need to be treated with the nicotine patch to have one smoker quit. The full results from this clinical trial (NCT01314001) are found in a publication by Lerman et al., 2015 [22]. Abbreviation: CI, confidence interval
CYP2B6 genetics and bupropion treatment outcomes
Like CYP2A6, CYP2B6 is also highly polymorphic. CYP2B6 protein is expressed in the liver and in extra-hepatic tissues including the brain, and metabolizes bupropion to its pharmacologically active metabolite hydroxybupropion [37]. Thus, variation in CYP2B6 activity that alters hydroxybupropion levels could influence bupropion-assisted cessation. Variability in the activity of CYP2A6, which does not metabolize bupropion, was not associated with bupropion-assisted cessation [31, 32].
The common CYP2B6*6 haplotype (e.g., ~25% in Caucasians [38]) is comprised of the CYP2B6*4 (rs2279343) and CYP2B6*9 (rs3745274) non-synonymous variants, and is associated with lower hepatic CYP2B6 protein expression [11] and reduced metabolism of bupropion [39]. In Caucasian smokers, end-of-treatment quit rates on bupropion did not differ between individuals with no copies of CYP2B6*6 versus those with 1–2 copies of CYP2B6*6 (31% vs 33%, respectively; P=0.84). However, bupropion and hydroxybupropion levels were not assessed in this study, thus it is not possible to determine treatment adherence [40]. In verified treatment-adherent African American light smokers (‹10 cigarettes/day) randomized to bupropion, slow CYP2B6 metabolizers (including those with the CYP2B6*6 haplotype) had lower plasma hydroxybupropion levels versus normal metabolizers; lower hydroxybupropion levels, in turn, were associated with poorer bupropion-assisted cessation [41]. In this study, CYP2B6 genotype was not associated with bupropion side effects. Together with the efficacy data, these findings suggest that genetically slow CYP2B6 metabolizers may benefit from a higher bupropion dose [41].
In a separate study of bupropion-treated Caucasian smokers, several additional CYP2B6 variants were associated with cessation [42]. The G allele of rs8109525, present in a non-coding region ~5kb upstream of CYP2B6, was associated with a higher likelihood of continuous abstinence at weeks 9–12 (OR=1.8; P=0.0008). This SNP is not in linkage disequilibrium (i.e., not inherited together) with key CYP2A6 functional polymorphisms and has been linked to lower CYP2B6 mRNA expression in human liver samples [43]. Thus, the CYP2B6 rs8109525 G allele may be associated with lower CYP2B6 activity and bupropion metabolism to hydroxybupropion; the mechanism behind its association with higher bupropion-assisted cessation remains to be determined.
In addition to its role in bupropion metabolism, CYP2B6 is hypothesized to play a role in the central (brain) metabolism of nicotine. In rats, the selective inhibition of brain CYP2B, which is thought to mimic genetically slow CYP2B6 metabolism in humans, was associated with higher brain (but not plasma) nicotine levels and a greater number of sessions required to extinguish nicotine self-administration behaviour [44]. In placebo-treated Caucasian heavy smokers, those with one or two copies of CYP2B6*6 had lower end-of-treatment quit rates versus those with no copies of CYP2B6*6 (14% versus 32%, respectively; P=0.01) [40]. Thus, it appears that slow CYP2B6 activity may be associated with increased relapse risk on placebo [40] and possibly bupropion [41].
Genetic variation in CYP2C19, which can also metabolize bupropion [45], has also been investigated for possible involvement in bupropion-assisted cessation [46]. CYP2C19*2 (rs4244285) was associated with higher bupropion area under the plasma concentration-time curve (AUC), but not hydroxybupropion AUC, in 42 healthy volunteers. Consistent with its lack of association with hydroxybupropion AUC, there was no association between CYP2C19*2 and cessation outcomes in bupropion-treated African American smokers [46]. Thus, CYP2C19 variation may not substantially influence bupropion-assisted cessation.
OCT2 genetics and varenicline treatment outcomes
Varenicline is the most effective smoking cessation drug, with odds ratios of ~3 versus placebo and ~2 versus bupropion or NRT for ≥6-month abstinence [47]. Varenicline undergoes virtually no metabolism [48] and is therefore unlikely to be influenced by polymorphisms in drug-metabolizing enzymes. Variation in the transport of varenicline throughout the body, however, may alter varenicline pharmacokinetics and cessation outcomes. Varenicline is transported by organic cation transporter 2 (OCT2) [49], which is expressed in renal proximal tubule cells and endothelial cells of the blood-brain barrier [50]. The rs316006 T allele in the OCT2 gene (SLC22A2) was associated with greater smoking abstinence at end-of-treatment and 6-months in varenicline-treated Caucasian smokers [51]. Whether OCT2 variation represents a useful target for the optimization of smoking cessation treatment remains to be determined.
Optimizing cessation using genetics of central nervous system targets
The reinforcing effects of nicotine are thought to result from its actions in the brain’s mesolimbic dopaminergic system. Nicotine binds nAChRs in the ventral tegmental area, which contains the cell bodies of the ventral mesolimbic pathway, evoking dopamine release in the nucleus accumbens [52]. Genetic variation in nicotine’s targets within the central nervous system, including nAChRs and components of the dopaminergic system (e.g., dopamine receptors and transporters), is associated with differences in smoking cessation. Nicotine replacement therapies, including patch, gum, and nasal spray which deliver nicotine more slowly than via smoking, also activate nAChRs and evoke dopamine release [1, 53]. Like nicotine, bupropion also binds nAChRs; however, it has antagonistic activity at multiple nAChR subtypes [54]. Bupropion can also bind the dopamine transporter [4]. While nicotine has full agonist activity at α4β2-containing nAChRs, varenicline is a partial agonist of this receptor subtype but has higher relative binding affinity [55].
nAChRs
Variation in nAChR genes, particularly the CHRNA5-CHRNA3-CHRNB4 cluster located on chromosome 15q25, has been examined for associations with smoking cessation success in the absence of treatment and with active pharmacotherapy. While the CHRNA5-CHRNA3-CHRNB4 cluster is robustly associated with small differences in cigarette consumption and nicotine dependence [56], the associations between this cluster and cessation outcomes have often differed between studies. Of the SNPs investigated within this cluster, rs16969968, located in CHRNA5, has been studied most frequently.
nAChR gene variation and smoking cessation: non-treatment-seeking smokers
In a meta-analysis of 24 studies in non-treatment-seeking Caucasian smokers, those with the CHRNA5 rs16969968 AA risk genotype quit a median of four years later compared to GG smokers [8]. In a separate analysis of community-based Caucasian smokers, a high-risk haplotype defined by rs16969968 (A allele) and rs680244 (C allele) delayed smoking cessation (self-reported) by a median of two years compared to lower-risk groups [57]. The CHRNA5 rs16969968 A allele, which has also been associated with higher cigarette consumption and cotinine levels, a biomarker of tobacco exposure [58], is thought to lead to a lower maximal nAChR response to nicotine [59]. In mice, CHRNA5 gene knock-out led to greater levels of nicotine intake [60]. Together these data suggest that reduced CHRNA5 activity may increase heaviness of smoking and decrease cessation.
nAChR gene variation and smoking cessation: treatment-seeking smokers
In addition to delaying cessation in non-treatment-seeking smokers, the high-risk haplotype defined by rs16969968 (A allele) and rs680244 (C allele) was associated with relapse on placebo, but not active treatment, in an NRT and bupropion clinical trial in Caucasian smokers [57]. Similarly, in a separate study, the risk allele for rs1051730 (in near perfect linkage disequilibrium with rs16969968) increased relapse on placebo [61]. Together these data suggest that pharmacotherapy could blunt relapse risk in smokers with the high-risk haplotype [57]. However, the association between rs16969968 variation and relapse on placebo has not been replicated in all trials [62, 63].
In contrast to unaided quitting or cessation on placebo [8, 57, 61], variation in rs16969968 does not appear to be associated with quitting on active pharmacotherapy. A recent meta-analysis in smokers (>95% Caucasian) receiving NRT showed no associations between rs16969968 or rs1051730 and end-of-treatment or 6-month quit rates [7] (Figure 3). This suggests that the influence of high-risk rs16969968/rs1051730 genotypes on increasing relapse [8, 57, 61] may be mitigated by treatment with NRT.
Figure 3.
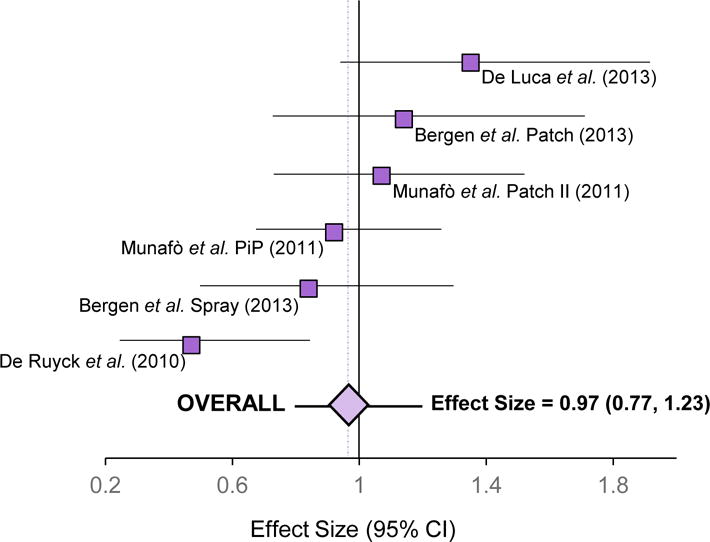
The rs1051730–rs16969968 genetic variant in the nicotinic receptor does not influence smoking cessation outcomes on NRT.
Effect sizes (odds ratios) from four clinical studies [61, 79–81] comprising NRT treatment arms show the likelihood of successful smoking cessation at end-of-treatment for smokers with the rs1051730-rs16969968 risk allele (rs1051730 T allele or rs16969968 A allele) relative to smokers with the rs1051730-rs16969968 reference allele (rs1051730 C or rs16969968 G allele). This figure is adapted from a meta-analysis performed by Leung et al, 2015 [7]. The overall effect size from the meta-analyzed data is also depicted. Results were generated from additive genetic models adjusting for age and sex. No significant heterogeneity was present in the meta-analysis (I2=50.6%; P=0.07). The dashed vertical line represents the overall effect size, which was 0.97. Abbreviations: PiP, Patch in Practice study; CI, confidence interval
In smokers receiving varenicline, CHRNA5 rs16969968 was not associated with cessation [62], suggesting, as for NRT, that varenicline may reverse the elevated relapse risk associated with the rs16969968 AA genotype. In Caucasian smokers prospectively randomized to nicotine patch or varenicline based on NMR [22], no associations for rs16969968 with end-of-treatment quit rates on nicotine patch or varenicline were observed [63]. Two other SNPs tagging loci within the CHRNA5-CHRNA3-CHRNB4 cluster (rs588765 and rs578776) that were previously robustly associated with minor influences on cigarette consumption and dependence in Caucasians were also not associated with cessation outcomes in any arm [63]. Three separate placebo-controlled trials also found no association for rs16969968 with abstinence on varenicline during the last four weeks of treatment [42]. In Caucasian smokers receiving both nicotine patch and bupropion, there was also no association between rs16969968 and cessation; however, an alternative CHRNA5 SNP, rs680244, was associated with abstinence at 52 weeks in those that received bupropion and nicotine patch in combination [64]. Together with the findings from untreated and placebo-treated smokers [8, 57, 61], these data suggest that pre-treatment determination of rs16969968 genotype may be useful for identifying subsets of smokers who are more likely to benefit from active pharmacotherapy versus behavioural counseling or no cessation interventions. The effects of treatment across rs16969968 genotype groups could be explicitly tested prospectively in a rs16969968-stratified clinical trial, similar to the approach used in the NMR-stratified trial [22], which may improve clinical recommendations for treating tobacco dependence.
In African American light smokers, neither rs16969968 nor rs578776 was associated with smoking abstinence on active treatment (nicotine gum or bupropion) or placebo [65]. In the placebo-controlled nicotine gum trial, smokers with the rs588765 T allele had higher quit rates on nicotine gum; however, in those receiving placebo gum, the rs588765 T allele was associated with lower abstinence. Similar magnitudes and directions of effect for active versus placebo arms were observed in the bupropion trial. The A allele of another SNP, rs2036527, located ~6.2kb 5′ of CHRNA5, was associated with lower quit rates on nicotine gum and bupropion during and at end-of-treatment (unadjusted ORs=0.31–0.59). In the placebo arms of both trials, however, there was no association between rs2036527 and abstinence [65].
Beyond the CHRNA5-CHRNA3-CHRNB4 cluster, genetic variation in other nAChR subtypes has been associated with smoking cessation in Caucasians. The CHRNB2 rs2072661 A allele was associated with lower quitting in both bupropion and placebo-treated smokers [66]. Analyses in three separate placebo-controlled clinical trials identified additional CHRNB2 SNPs (rs3811450 and rs4292956), as well as SNPs in other nAChR subunits including CHRNA4 (rs3787138 and rs2236196) and CHRNA7 (rs6494212), which influenced abstinence on varenicline [42]. Whether these findings will replicate in other varenicline-treated smokers or extend to other treatments remains to be determined.
Together, the lack of replicated findings for nAChR gene variants and smoking cessation outcomes reduces the likelihood that this genomic region will be useful in the personalization of smoking cessation therapy. Comparing cessation outcomes on active pharmacotherapy with a single, well-selected placebo group may facilitate the interpretation of findings from such studies [62].
Dopaminergic pathway gene variation and smoking cessation
Genetic variation in components of the dopaminergic system has also been investigated as a potential source of variability in smoking cessation rates. In general, functional polymorphisms that lead to lower dopaminergic tone (i.e., reduced dopaminergic activity) are thought to contribute to lower smoking cessation success [67].
Variation in the dopamine D4 receptor gene (DRD4) has been examined in placebo-controlled clinical trials. The exon-III VNTR polymorphism in DRD4 was not associated with overall abstinence in Caucasian smokers receiving placebo or bupropion [68]. However, in smokers with ≥1 copy of the long-allele (≥7-repeats), which may lead to lower DRD4 mRNA expression [69], bupropion increased cessation (versus placebo); smokers with two copies of the short-allele (<7-repeats) did not benefit from bupropion [68]. In a separate sample of Caucasian smokers, Bergen and colleagues similarly observed a larger, albeit non-significant, benefit of bupropion (versus placebo) in those with the long-allele (versus those homozygous for the short-allele) [70]. Together these data suggest that bupropion may be a more suitable treatment for smokers possessing the DRD4 exon-III VNTR long-allele. In a placebo-controlled nicotine patch trial in Caucasian smokers, there was an overall association for the DRD4 exon-III VNTR long-allele with reduced cessation at end-of-treatment (12 weeks); however, the effect of nicotine patch (versus placebo) on cessation did not differ as a function of DRD4 genotype, suggesting nicotine patch therapy worked equally well in both groups of smokers [67]. These latter findings do not support the use of this DRD4 polymorphism in tailoring cessation treatment with nicotine patch.
Polymorphisms associated with the dopamine transporter (SLC6A3) and dopamine D2 receptor (DRD2) genes have also been examined as potential modulators of smoking cessation outcomes. At end-of-treatment, neither the 3′ VNTR polymorphism in SLC6A3 nor the Taq1A2 restriction fragment length polymorphism located ~10kb 3′ of DRD2 were associated with overall abstinence in Caucasian smokers randomized to placebo or bupropion [71]. However, the Taq1A2 polymorphism was associated with bupropion-assisted quitting at 6-months follow-up [71]. In those with the DRD2-Taq1A2/A2 genotype, quit rates were higher on bupropion (versus placebo); in contrast, in Taq1A1 individuals, who are thought to have lower DRD2 receptor density [72], bupropion was not associated with greater cessation (versus placebo) [71]. Of note, the higher bupropion-assisted quit rates in those with the DRD2-Taq1A2/A2 genotype were restricted to those with CYP2B6 rs3211371 TT or CT genotypes [71]. These findings highlight the potential importance of assessing multiple genes and gene-gene interactions, as opposed to single gene analyses, to identify sub-groups of smokers who are more likely to benefit from a certain treatment, discussed further below.
Optimizing smoking cessation through multiple genetic predictors
Smoking cessation is influenced by a multitude of genetic and environmental factors (e.g., gender, socioeconomic status, and ethnicity) [73–75]. Treatment approaches that consider multiple predictors of cessation, versus single predictors, may show additional improvements in smoking cessation rates.
Chen and colleagues derived a four-level genetic risk score based on CYP2A6 and CHRNA5 in Caucasian smokers [32]. On NRT, but not bupropion, cessation outcomes differed according to risk score. In the highest-risk group (CYP2A6 fast metabolism plus CHRNA5 high-risk rs16969968-rs680244 diplotype), NRT had the largest treatment effect (versus placebo). Individuals with intermediate levels of risk showed an intermediate level of benefit from NRT, while individuals in the lowest-risk group (CYP2A6 slow metabolism plus CHRNA5 low-risk rs16969968-rs680244 diplotype) did not benefit from NRT [32].
An additive genetic efficacy score (AGES) has also been used to assess functional polymorphisms within the dopaminergic system and relapse risk in Caucasian smokers [76]. The AGES, calculated based on the number of alleles hypothesized to promote cessation on bupropion (versus placebo), comprised four variants selected based on their associations with smoking cessation [77]: DRD4 exon-III VNTR, SLC6A3 3′ VNTR, and DRD2 Taq1A (discussed previously), and COMT V158M (rs4680). In bupropion-treated smokers, AGES was not associated with time to first lapse [76]; however, bupropion mitigated relapse risk (versus placebo) in those with higher AGES scores, suggesting that these individuals should be treated with bupropion. The refinement of AGES and the development of other genetic scoring tools informed by biological pathways involved in smoking cessation and treatment pharmacology may show clinical utility in personalizing tobacco dependence treatment. The addition of environmental influences on smoking cessation to such approaches may further improve cessation.
Concluding Remarks
In summary, a growing body of literature has demonstrated the importance of genes involved in the metabolism and central nervous system effects of both nicotine and smoking cessation medications in altering smoking cessation. Smokers with faster rates of nicotine metabolism, as determined by the NMR, have higher quit rates on varenicline versus the nicotine patch; in contrast, varenicline is not superior to the patch in slower metabolizers [22] (Figure 2). NMR genome-wide association studies have identified novel sources of genetic variation in CYP2A6 activity which may improve the optimization of treatment using CYP2A6 genomics and phenotype (NMR) in the future [78]. Future studies should aim to determine a) the optimal NMR cut-point for distinguishing between faster and slower metabolizers (see Outstanding Questions), and b) whether the utility of NMR, as opposed to using a purely genetics-based approach, may be limited by transient environmental influences. Cessation studies in community-based smokers as well as cost-benefit analyses will enhance the implementation of NMR and other tools into clinical decision-making. Smokers with slower rates of bupropion metabolic activation to hydroxybupropion, as determined by CYP2B6 genetics, may benefit from a higher dose of bupropion or alternative treatments [41]. Genetic variation in nAChRs and components of the dopaminergic system is also associated with smoking cessation; however, the findings from these studies have been less consistent, requiring further investigation to establish their potential for personalizing treatment. The development of treatment approaches that consider multiple genetic and environmental factors in tandem may provide additional improvements in smoking cessation rates and reductions in tobacco-related harm.
Trends Box.
Slower (vs. faster) nicotine metabolizers, by CYP2A6 genotype or CYP2A6 phenotype (the nicotine metabolite ratio; NMR), have higher smoking abstinence rates on behavioural counseling, nicotine patch, and in the absence of treatment (“cold turkey”)
In smokers assigned to treatment based on their NMR, slower metabolizers had similar quit rates on varenicline and nicotine patch, while faster metabolizers quit better on varenicline (vs. nicotine patch)
In smokers with genetically slow (vs. normal) CYP2B6 activity which metabolically activates bupropion, smoking cessation rates are lower on bupropion, and placebo
Genetic variation in nicotine’s central nervous system targets (e.g., nicotinic receptors and dopaminergic pathways) has been associated with smoking cessation, albeit inconsistently
Acknowledgments
We acknowledge our sources of funding: Endowed Chair in Addictions (R. F. Tyndale), National Institutes of Health (NIH) PGRN grant DA020830, Canadian Institutes of Health Research (CIHR) grants MOP86471 and TMH-109787, the Campbell Family Mental Health Research Institute of the Centre for Addiction and Mental Health (CAMH), the CAMH Foundation, the Canada Foundation for Innovation (#20289 and #16014), and the Ontario Ministry of Research and Innovation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
R. F. Tyndale has consulted for Apotex. Funds were not received from Apotex for this work, nor was the manuscript reviewed by individuals affiliated with Apotex. M. J. Chenoweth declares no conflicts of interest.
References
- 1.Henningfield JE. Nicotine medications for smoking cessation. N Engl J Med. 1995;333:1196–1203. doi: 10.1056/NEJM199511023331807. [DOI] [PubMed] [Google Scholar]
- 2.Hurt RD, et al. A comparison of sustained-release bupropion and placebo for smoking cessation. N Engl J Med. 1997;337:1195–1202. doi: 10.1056/NEJM199710233371703. [DOI] [PubMed] [Google Scholar]
- 3.Faessel HM, et al. A review of the clinical pharmacokinetics and pharmacodynamics of varenicline for smoking cessation. Clin Pharmacokinet. 2010;49:799–816. doi: 10.2165/11537850-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 4.Learned-Coughlin SM, et al. In vivo activity of bupropion at the human dopamine transporter as measured by positron emission tomography. Biol Psychiat. 2003;54:800–805. doi: 10.1016/s0006-3223(02)01834-6. [DOI] [PubMed] [Google Scholar]
- 5.Broms U, et al. Genetic architecture of smoking behavior: a study of Finnish adult twins. Twin Res Hum Genet. 2006;9:64–72. doi: 10.1375/183242706776403046. [DOI] [PubMed] [Google Scholar]
- 6.Pianezza ML, et al. Nicotine metabolism defect reduces smoking. Nature. 1998;393:750. doi: 10.1038/31623. [DOI] [PubMed] [Google Scholar]
- 7.Leung T, et al. Effect of the rs1051730–rs16969968 variant and smoking cessation treatment: a meta-analysis. Pharmacogenomics. 2015;16:713–720. doi: 10.2217/pgs.15.34. [DOI] [PubMed] [Google Scholar]
- 8.Chen LS, et al. CHRNA5 risk variant predicts delayed smoking cessation and earlier lung cancer diagnosis–a meta-analysis. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benowitz NL. Nicotine addiction. N Engl J Med. 2010;362:2295–2303. doi: 10.1056/NEJMra0809890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakajima M, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 1996;24:1212–1217. [PubMed] [Google Scholar]
- 11.Al Koudsi N, Tyndale RF. Hepatic CYP2B6 is altered by genetic, physiologic, and environmental factors but plays little role in nicotine metabolism. Xenobiotica. 2010;40:381–392. doi: 10.3109/00498251003713958. [DOI] [PubMed] [Google Scholar]
- 12.Benowitz NL, et al. Nicotine chemistry, metabolism, kinetics and biomarkers. Handb Exp Pharmacol. 2009:29–60. doi: 10.1007/978-3-540-69248-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakajima M, et al. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996;277:1010–1015. [PubMed] [Google Scholar]
- 14.Dempsey D, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76:64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Tanner JA, et al. Nicotine metabolite ratio (3-hydroxycotinine/cotinine) in plasma and urine by different analytical methods and laboratories: implications for clinical implementation. Cancer Epidemiol Biomarkers Prev. 2015;24:1239–1246. doi: 10.1158/1055-9965.EPI-14-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lea RA, et al. Within-subject variation of the salivary 3HC/COT ratio in regular daily smokers: prospects for estimating CYP2A6 enzyme activity in large-scale surveys of nicotine metabolic rate. J Anal Toxicol. 2006;30:386–389. doi: 10.1093/jat/30.6.386. [DOI] [PubMed] [Google Scholar]
- 17.Mooney ME, et al. Stability of the nicotine metabolite ratio in ad libitum and reducing smokers. Cancer Epidemiol Biomarkers Prev. 2008;17:1396–1400. [Google Scholar]
- 18.St Helen G, et al. Stability of the nicotine metabolite ratio in smokers of progressively reduced nicotine content cigarettes. Nicotine Tob Res. 2013;15:1939–1942. doi: 10.1093/ntr/ntt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benowitz NL, et al. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin Pharmacol Ther. 2006;80:457–467. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Chenoweth MJ, et al. Known and novel sources of variability in the nicotine metabolite ratio in a large sample of treatment-seeking smokers. Cancer Epidemiol Biomarkers Prev. 2014;23:1773–1782. doi: 10.1158/1055-9965.EPI-14-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnoll RA, et al. High dose transdermal nicotine for fast metabolizers of nicotine: a proof of concept placebo-controlled trial. Nicotine Tob Res. 2013;15:348–354. doi: 10.1093/ntr/nts129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerman C, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. Lancet Resp Med. 2015;3:131–138. doi: 10.1016/S2213-2600(14)70294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoedel KA, et al. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14:615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Wassenaar CA, et al. Relationship Between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 Variation and Smoking Behaviors and Lung Cancer Risk. J Natl Cancer I. 2011;103:1342–1346. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sofuoglu M, et al. Rapid Nicotine Clearance is Associated with Greater Reward and Heart Rate Increases from Intravenous Nicotine. Neuropsychopharmacology. 2012;37:1509–1516. doi: 10.1038/npp.2011.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubroff JG, et al. Decreased Nicotinic Receptor Availability in Smokers with Slow Rates of Nicotine Metabolism. J Nucl Med. 2015;56:1724–1729. doi: 10.2967/jnumed.115.155002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang DW, et al. Genetic variation in CYP2A6 predicts neural reactivity to smoking cues as measured using fMRI. Neuroimage. 2012;60:2136–2143. doi: 10.1016/j.neuroimage.2012.01.119. [DOI] [PubMed] [Google Scholar]
- 28.Falcone M, et al. Brain Responses to Smoking Cues Differ Based on Nicotine Metabolism Rate. Biol Psychiatry. 2015;80:190–197. doi: 10.1016/j.biopsych.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu DF, et al. The use of long PCR to confirm three common alleles at the CYP2A6 locus and the relationship between genotype and smoking habit. Ann Hum Genet. 2000;64:383–390. doi: 10.1046/j.1469-1809.2000.6450383.x. [DOI] [PubMed] [Google Scholar]
- 30.Chenoweth MJ, et al. CYP2A6 slow nicotine metabolism is associated with increased quitting by adolescent smokers. Pharmacogenet Genom. 2013;23:232–235. doi: 10.1097/FPC.0b013e32835f834d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patterson F, et al. Toward personalized therapy for smoking cessation: A randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther. 2008;84:320–325. doi: 10.1038/clpt.2008.57. [DOI] [PubMed] [Google Scholar]
- 32.Chen LS, et al. Pharmacotherapy effects on smoking cessation vary with nicotine metabolism gene (CYP2A6) Addiction. 2014;109:128–137. doi: 10.1111/add.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lerman C, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther. 2006;79:600–608. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Schnoll RA, et al. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: a validation study. Pharmacol Biochem Behav. 2009;92:6–11. doi: 10.1016/j.pbb.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lerman C, et al. Genetic variation in nicotine metabolism predicts the efficacy of extended-duration transdermal nicotine therapy. Clin Pharmacol Ther. 2010;87:553–557. doi: 10.1038/clpt.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufmann A, et al. Rate of nicotine metabolism and smoking cessation outcomes in a community-based sample of treatment-seeking smokers. Addict Behav. 2015;51:93–99. doi: 10.1016/j.addbeh.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kharasch ED, et al. Stereoselective bupropion hydroxylation as an in vivo phenotypic probe for cytochrome P4502B6 (CYP2B6) activity. J Clin Pharmacol. 2008;48:464–474. doi: 10.1177/0091270008314254. [DOI] [PubMed] [Google Scholar]
- 38.Rotger M, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther. 2007;81:557–566. doi: 10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 39.Thorn CF, et al. PharmGKB summary: very important pharmacogene information for CYP2B6. Pharmacogenet Genom. 2010;20:520–523. doi: 10.1097/fpc.0b013e32833947c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee AM, et al. CYP2B6 genotype alters abstinence rates in a bupropion smoking cessation trial. Biol Psychiatry. 2007;62:635–641. doi: 10.1016/j.biopsych.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Zhu AZ, et al. CYP2B6 and bupropion’s smoking-cessation pharmacology: the role of hydroxybupropion. Clin Pharmacol Ther. 2012;92:771–777. doi: 10.1038/clpt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King DP, et al. Smoking cessation pharmacogenetics: analysis of varenicline and bupropion in placebo-controlled clinical trials. Neuropsychopharmacology. 2012;37:641–650. doi: 10.1038/npp.2011.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bloom AJ, et al. CYP2B6 Non-Coding Variation Associated with Smoking Cessation Is Also Associated with Differences in Allelic Expression, Splicing, and Nicotine Metabolism Independent of Common Amino-Acid Changes. Plos One. 2013;8 doi: 10.1371/journal.pone.0079700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia KLP, et al. Effect of Brain CYP2B Inhibition on Brain Nicotine Levels and Nicotine Self-Administration. Neuropsychopharmacology. 2015;40:1910–1918. doi: 10.1038/npp.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, et al. The in vitro metabolism of bupropion revisited: concentration dependent involvement of cytochrome P450 2C19. Xenobiotica. 2010;40:536–546. doi: 10.3109/00498254.2010.492880. [DOI] [PubMed] [Google Scholar]
- 46.Zhu AZ, et al. Gene variants in CYP2C19 are associated with altered in vivo bupropion pharmacokinetics but not bupropion-assisted smoking cessation outcomes. Drug Metab Dispos. 2014;42:1971–1977. doi: 10.1124/dmd.114.060285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cahill K, et al. Pharmacological interventions for smoking cessation: an overview and network meta-analysis. Cochrane Database Syst Rev. 2013:CD009329. doi: 10.1002/14651858.CD009329.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obach RS, et al. Metabolism and disposition of varenicline, a selective alpha4beta2 acetylcholine receptor partial agonist, in vivo and in vitro. Drug Metab Dispos. 2006;34:121–130. doi: 10.1124/dmd.105.006767. [DOI] [PubMed] [Google Scholar]
- 49.Feng B, et al. Effect of human renal cationic transporter inhibition on the pharmacokinetics of varenicline, a new therapy for smoking cessation: an in vitro-in vivo study. Clin Pharmacol Ther. 2008;83:567–576. doi: 10.1038/sj.clpt.6100405. [DOI] [PubMed] [Google Scholar]
- 50.Koepsell H. The SLC22 family with transporters of organic cations, anions and zwitterions. Mol Aspects Med. 2013;34:413–435. doi: 10.1016/j.mam.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 51.Bergen AW, et al. Organic cation transporter variation and response to smoking cessation therapies. Nicotine Tob Res. 2014;16:1638–1646. doi: 10.1093/ntr/ntu161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laviolette SR, van der Kooy D. The neurobiology of nicotine addiction: bridging the gap from molecules to behaviour. Nat Rev Neurosci. 2004;5:55–65. doi: 10.1038/nrn1298. [DOI] [PubMed] [Google Scholar]
- 53.Stead LF, et al. Nicotine replacement therapy for smoking cessation. Cochrane Database Syst Rev. 2012;11:CD000146. doi: 10.1002/14651858.CD000146.pub4. [DOI] [PubMed] [Google Scholar]
- 54.Slemmer JE, et al. Bupropion is a nicotinic antagonist. Journal of Pharmacology and Experimental Therapeutics. 2000;295:321–327. [PubMed] [Google Scholar]
- 55.Coe JW, et al. Varenicline: an alpha4beta2 nicotinic receptor partial agonist for smoking cessation. J Med Chem. 2005;48:3474–3477. doi: 10.1021/jm050069n. [DOI] [PubMed] [Google Scholar]
- 56.Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen LS, et al. Interplay of genetic risk factors (CHRNA5-CHRNA3-CHRNB4) and cessation treatments in smoking cessation success. Am J Psychiatry. 2012;169:735–742. doi: 10.1176/appi.ajp.2012.11101545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Munafo MR, et al. Association Between Genetic Variants on Chromosome 15q25 Locus and Objective Measures of Tobacco Exposure. J Natl Cancer I. 2012;104:740–748. doi: 10.1093/jnci/djs191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bierut LJ, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fowler CD, et al. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471:597–601. doi: 10.1038/nature09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bergen AW, et al. Nicotinic acetylcholine receptor variation and response to smoking cessation therapies. Pharmacogenet Genomics. 2013;23:94–103. doi: 10.1097/FPC.0b013e32835cdabd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen LS, et al. Genetic variation (CHRNA5), medication (combination nicotine replacement therapy vs. varenicline), and smoking cessation. Drug Alcohol Depend. 2015;154:278–282. doi: 10.1016/j.drugalcdep.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tyndale RF, et al. Lack of Associations of CHRNA5-A3-B4 Genetic Variants with Smoking Cessation Treatment Outcomes in Caucasian Smokers despite Associations with Baseline Smoking. Plos One. 2015;10:e0128109. doi: 10.1371/journal.pone.0128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarginson JE, et al. Markers in the 15q24 nicotinic receptor subunit gene cluster (CHRNA5-A3-B4) predict severity of nicotine addiction and response to smoking cessation therapy. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:275–284. doi: 10.1002/ajmg.b.31155. [DOI] [PubMed] [Google Scholar]
- 65.Zhu AZ, et al. Association of CHRNA5-A3-B4 SNP rs2036527 with smoking cessation therapy response in African-American smokers. Clin Pharmacol Ther. 2014;96:256–265. doi: 10.1038/clpt.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conti DV, et al. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17:2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.David SP, et al. Genetic variation in the dopamine D4 receptor (DRD4) gene and smoking cessation: follow-up of a randomised clinical trial of transdermal nicotine patch. Pharmacogenomics J. 2008;8:122–128. doi: 10.1038/sj.tpj.6500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leventhal AM, et al. Dopamine D4 receptor gene variation moderates the efficacy of bupropion for smoking cessation. Pharmacogenomics J. 2012;12:86–92. doi: 10.1038/tpj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simpson J, et al. The DRD4 receptor Exon 3 VNTR and 5′ SNP variants and mRNA expression in human post-mortem brain tissue. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1228–1233. doi: 10.1002/ajmg.b.31084. [DOI] [PubMed] [Google Scholar]
- 70.Bergen AW, et al. The DRD4 exon III VNTR, bupropion, and associations with prospective abstinence. Nicotine Tob Res. 2013;15:1190–1200. doi: 10.1093/ntr/nts245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.David SP, et al. Pharmacogenetic clinical trial of sustained-release bupropion for smoking cessation. Nicotine Tob Res. 2007;9:821–833. doi: 10.1080/14622200701382033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jonsson EG, et al. Polymorphisms in the dopamine D2 receptor gene and their relationships to striatal dopamine receptor density of healthy volunteers. Mol Psychiatr. 1999;4:290–296. doi: 10.1038/sj.mp.4000532. [DOI] [PubMed] [Google Scholar]
- 73.Bjornson W, et al. Gender differences in smoking cessation after 3 years in the Lung Health Study. Am J Public Health. 1995;85:223–230. doi: 10.2105/ajph.85.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Broms U, et al. Smoking cessation by socioeconomic status and marital status: the contribution of smoking behavior and family background. Nicotine Tob Res. 2004;6:447–455. doi: 10.1080/14622200410001696637. [DOI] [PubMed] [Google Scholar]
- 75.Trinidad DR, et al. A nationwide analysis of US racial/ethnic disparities in smoking behaviors, smoking cessation, and cessation-related factors. Am J Public Health. 2011;101:699–706. doi: 10.2105/AJPH.2010.191668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.David SP, et al. Influence of a dopamine pathway additive genetic efficacy score on smoking cessation: results from two randomized clinical trials of bupropion. Addiction. 2013;108:2202–2211. doi: 10.1111/add.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, Li MD. Common and unique biological pathways associated with smoking initiation/progression, nicotine dependence, and smoking cessation. Neuropsychopharmacology. 2010;35:702–719. doi: 10.1038/npp.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loukola A, et al. A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism. PLoS Genet. 2015;11:e1005498. doi: 10.1371/journal.pgen.1005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Munafo MR, et al. CHRNA3 rs1051730 genotype and short-term smoking cessation. Nicotine Tob Res. 2011;13:982–988. doi: 10.1093/ntr/ntr106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Ruyck K, et al. Genetic variation in three candidate genes and nicotine dependence, withdrawal and smoking cessation in hospitalized patients. Pharmacogenomics. 2010;11:1053–1063. doi: 10.2217/pgs.10.75. [DOI] [PubMed] [Google Scholar]
- 81.De Luca V, et al. Analysis of Nicotinic Receptor Genes in Nicotine Replacement Treatment. Eur Psychiat. 2013;28 [Google Scholar]


