Abstract
The naïve state of pluripotency is actively being explored by a number of labs. There is some controversy in the field as to the true identity of naïve human pluripotent cells as they are not exact mirrors of the mouse. The various reports published, though in basic agreement, present discrepancies in the characterization of the various lines, which likely reflect the etiology of these lines. The primary lesson learned from these contributions is that a human naïve state reflecting the pre-implantation human is likely to exist. The essential factors that will universally maintain the naïve state in human cells in vitro are not yet fully understood. These first need to be identified in order to describe the definitive characteristics of this state. Comparisons of naïve and primed human pluripotent cells have also highlighted consistencies between states and broadened our understanding of embryonic metabolism, epigenetic change required for development, embryonic DNA repair strategies and embryonic expression dynamics.
Keywords: naïve, primed, pluripotent, pre-implantation, hESC
Introduction
In vitro pluripotency encompasses more than one early developmental state from which the cells can form all body tissues. The naïve state for mammalian pluripotent stem cells approximates cells from the pre-implantation inner cell mass (ICM), while the primed state approximates the early post-implantation epiblast, a developmental state previously inaccessible in humans. Mouse embryonic stem cells (mESC) are readily isolated (1, 2) and maintained in the naïve state (3). As with mouse, human ESC (hESC) are isolated from pre-implantation ICM (4), but several characteristics vary from mESC. A mouse equivalent to hESC can be isolated and maintained as EpiSC from post-implantation epiblast using hESC culture conditions that include FGF and Activin A (5, 6). For this reason, hESC are considered to be in a “primed” post-implantation epiblast state.
Pluripotent cells in both the naïve and primed states have benefits for research and clinical use. More specifically, having both states of pluripotency in a self-renewing state in vitro allows a complementary comparison between the states of pluripotency to gain a deeper understanding of a state that must both protect genomic integrity between generations as a precursor of the germ lineage and maintain integrity of the individual that will develop. Thus, these cells enhance our understanding of human development that was previously inaccessible.
The first growth factors for naïve cell growth were defined in mESC culture. The mouse naïve state requires the presence of leukemia inhibitory factor (LIF), which signals through both the JAK-STAT and MAP kinase (MAPK) pathways (7). The JAK-STAT pathway maintains naïve pluripotency in mESC (8-10), while the MAPK pathway maintains primed pluripotency (11,12). Thus, inhibition of the MAPK pathway using PD0325901, a MEK/ERK inhibitor along with the presence of LIF maintains the naïve state, but will cause primed ESC to differentiate (11). LIF plus a MEK/ERK inhibitor combined with GSK3α,β inhibition (2iL) pull naïve mESC back into a more homogeneous naïve state referred to as the “ground state”, when serum supplementation is absent (13). The exact function of GSK3 inhibition in the context of human pluripotency has not been fully clarified. In mESC, GSK3 inhibition centers upon elevating Esrrb (14). GSK3 inhibition, induces canonical Wnt signaling which stimulates β-catenin (15-17) and subsequent inhibiton of Tcf3 (Tcf7l1; 14). It was noted in rat ESC that over inhibition of GSK3 leads to differentiation (18) influenced by the balance between Tcf3 and Lef1 (19). Thus, caution as to the level of GSK3 inhibition should be practiced in the context of naïve ESC until the effect is clarified for each individual species
Naïve mESC can be passaged as single-cells, which has clear advantages for ease of culture and for genetic manipulation by transfection. Also developmental capacity of naïve mESC when replaced into a pre-implantation embryo (8-cell to blastocyst) is robust, while primed EpiSC contribute poorly, at least in part due to mismatch in developmental timing between the host and the primed cells, which are better attuned to a post-implantation epiblast (20). Thus, from the mouse model we know that the naïve state can maintain appropriate epigenetic cues to allow full development of the complete individual, including a functional germline for generational continuity (21). Consequently, obtaining a stable human naïve culture has become a valid goal in the stem cell field. It must be noted that humans are quite genetiically heterogeneous relative to the laboratory mouse, so that individual variation can make drawing conclusions from a single line difficult.
Stabilizing hESC in a naïve state
Can cells from the human ICM stabilize as naïve in response to 2i and LIF as do cells from the mouse ICM? This question was first explored by reversing development of primed hESC through addition of histone deacetylase inhibitors (HDACi; 22). The subtle reversal of the pluripotent state in response to HDACi is then assisted into the naïve state through switching the medium to contain 2i plus FGF (2iF; 23). Generating new hESC lines in low O2 also allowed the primed cells to be subtly earlier in the primed state (24), which, like HDACi, allowed for reactivation of the inactive X in female lines. Alternatively, primed hESC established in low oxygen conditions can be converted to a naïve state by use of transgenes (25). Studies that use transgenes to achieve stable naïve cultures are summarized in Table 1 (25, 26-29). Once the naïve state is achieved, the transgenes are no longer required, as with induced pluripotency from somatic cells. Interestingly, several reports that utilize transgenes to guide primed cells into the naïve state indicate that the culture medium can be simplified to one more reflective of mESC, i.e. 2iL, following transgene exposure and silencing. Valamehr et al. (30) took the use of reprogramming genes one step further by generating naïve human iPSC cells from somatic cells using episomal reprogramming. Their final medium composition to maintain the naïve state in the absence of episomal influence includes 2iL with ROCKi and FGF2.
Table 1. Summary of preferred conditions to establish naïve hESC from primed using transgenes.
| Reference by first author | Inhibitors used | growth factors | Transgenes |
|---|---|---|---|
| Hanna (25) | 2i* | hLIF | OCT4, SOX2 & KLF4 |
| Theunissen (26) | 2i*, BRAFi, SRCi, ROCKi | hLIF, FGF2, Activin A | NANOG & KLF2 |
| Takashima (27) | 2i*, PKCi | hLIF | NANOG & KLF4 |
| Chen (28) | 2i* | hLIF | STAT3-ER |
| Qin (29) | 2i* | hLIF, forskolin | YAP |
Inhibitors used:
2i = PD0325901 + CHIR99021 (1 μM each)
MEKi = PD0325901
GSK3i = CHIR99021
BRAFi = SB590885
ROCKi =Y27632
SRCi = WH-4-023
PKCi = Gö6983
JNKi = SP600125
P38i = SB202190; BIRB796
Overall, there is a glaring lack of consensus with regard to the appropriate medium for naïve human pluripotent cell growth. This can stem from differences in medium requirements whether the naïve cells were established directly from human embryos or established by reversing development of pre-established primed cells. Culture of primed cells varies considerably, often dependent upon whether the culture media contain a defined protein source or if undefined by the inclusion of fetal bovine serum (FBS; 24-26, 31). FBS is thought to contain variable amounts of components that can drive differentiation (32) and so is never used in naïve cell culture. When FGF2 is not added to primed culture medium, FBS is sufficient to push the cells out of pluripotency and down all three embryonic lineages of differentiation. Thus, culture of primed cells in FBS could provide a heterogeneous starting population from those cultured in defined protein sources, such as Knockout Serum Replacer (KOSR) or mTeSR1 with possible alternate requirements for transition to naïve culture.
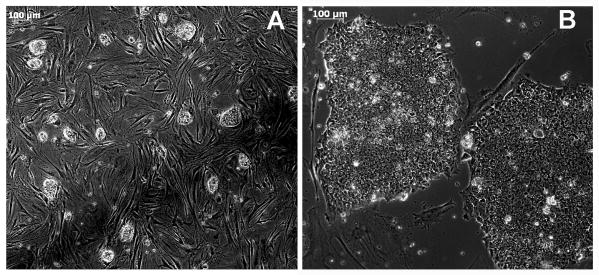
The function of FGF in naïve hESC culture is controversial. FGF is most often touted in the literature as a primed cell factor through the effect on mitogen-activated protein kinase (MAPK) signaling (33). Other than a proliferation effect through MAPK signaling, FGF also has a cell survival effect through phosphoinositide 3 kinase (PI3K) signaling and a cell motility effect through calcium activation in the phospholipase C gamma (PLCγ) pathway (34). It has been shown that MAPK inhibition through MEK inhibitors causes differentiation of primed mESC (11), but supports naïve mESC (35). Thus, FGF may have a positive effect on cell survival in the naïve state, although it is not a mandatory media component. Although FGF addition causes the cells to flatten slightly they appear to maintain naïve properties, including resilience in the face of single cell passage (23). Supplementation of the medium with IGF1 counters the colony flattening effect of FGF. 2i LIF + IGF1 + FGF holds Elf1 in what appears to be a homogenous early state by morphology (Figure 1). Possibly FGF responsiveness may be a feature of a late naïve state, preventing the cells from crossing into a primed state through FGF induced survival that would otherwise lead to differentiation in the presence of 2i (11). In support of this, primed cells pre-conditioned in HDACi followed by a switch to 2i plus FGF, with or without added LIF, are able to survive as naïve in the presence of FGF (23). Thus, these cells may not be in the earliest naïve state, but are naïve by other measures, as described later. Transgenic approaches to reverse development appear to convert the primed cells closer to a “ground state” and have been invaluable for definition of culture conditions to support naïve hESC.
Figure 1.
Elf1 hESC. A. Naïve B. Primed. Size bar within panels indicates 100 μM.
Reversion of primed cells into the naïve state using inhibitor cocktails
Several cocktails that allow human cells to cross the primed to naïve barrier without the use of transgenes have been described (Table 2;23, 26, 36-39). These resulting cells may be judged as naïve by many of the criteria that define the naïve state, though each cocktail engenders a distinct RNA expression pattern that makes it difficult to identify a unified pattern crucial to the naïve state. This may be due to the use of small molecule inhibitor cocktails that often have off-target effects. Unlike the transient use of transgenes, the small molecule forcing of naïve induction likely causes the cells to become reliant upon the particular cocktail used in derivation, such that the apparent naïve state is lost upon passage to media containing 2iL or 2iF. Our experience suggests that use of cocktails with 4-6 inhibitors to hold cells in the naïve state may delay responsiveness to differentiation signals relative to the behavior of cells grown in 2i. Thus, our experience found that Elf1 visually convert to primed in one passage,while it took cells grown as naïve in 4i (37) or 6i (26) three passages until they were primed by morphological criteria.
Table 2. Summary of preferred conditions to establish naïve hESC from established primed lines, without transgenes.
| Reference by first author | Inhibitors used | growth factors |
|---|---|---|
| Chan (36) | 2i, dorsomorphin | hLIF |
| Gafni (37) | 2i, JNKi, p38MAPKi | hLIF, FGF2, TGFβ1 |
| Ware (23) | 2i | FGF2 (HDACi pretreatment*) |
| Theunissen (26) | 2i*, BRAFi, SRCi, ROCKi | hLIF, FGF2, Activin A |
| Theunissen (31) | 2i (or without GSK3i) BRAFi, SRCi, ROCKi | hLIF, Activin A |
| Duggal (38) | 2i | ascorbic acid, forskolin, hLIF, FGF2 |
| Carter (39) | (ROCKi) | NME7AB, |
HDACi – sodium butyrate + vorinostat/SAHA
hESC lines established directly as naïve
De novo derivation of naïve hESC from pre-implantation embryos in culture conditions that support the naïve state are summarized in Table 3 (23, 26, 31, 37, 40). Interestingly, most media to support derivation of naïve hESC without transgenes appear to require more than 2iL to stablilize these cells directly from a pre-implantation embryo. The means of deriving new naïve hESC prior to achieving the primed state have a common message that supports our lessons in mice, that signaling through STAT3 following LIF exposure, while inhibiting the LIF-driven signaling through MAP/MEK, is a primary requirement to support the naïve state (3,12). Other tweaks are beneficial in capturing the cells directly from the ICM, most notably by inhibition of protein kinase C (PKCi, Gö6983; 40, 41). PKCζ appears to be the primary isoform responsible for maintaining mESC pluripotency (41). Curiously, PKCζ inhibition can antagonize GSK3 activity in non-pluripotent cells (42), though the PKC inhibitor, Gö6983, used most often in conjunction with naïve hESC is not as potent against this isoform as against other PKC isoforms. PKC is effective in insulin stimulated glucose transport (43) and many isoforms serve as MAPK agonists.(44). Inhibition of these PKC effects could be beneficial for naïve culture. Overall, effect of PKC inhibition on naïve hESC requires further study and possibly refinement.
Table 3. Summary of preferred conditions to establish naïve hESC de novo from embryos.
It should be noted that the Elf1 line (23), though isolated from a cryopreserved 8-cell embryo cultured to blastocyst and the naïve cells established from the ICM in 2i plus FGF, can be maintained in 2iL, without FGF. The currrent preferred medium is 2iL plus FGF2 and IGF1. Neither day 5 nor 6 blastocysts have successfully yielded hESC directly from ICM in our hands without the use of a further inhibitor. We assume we could isolate Elf1 using 2i and FGF in part due to the robust survival to blastocyst following cryopreservation of the 8-cell embryo. Could this be due to the lingering influence of maternal factors influencing survival at this embryonic developmental time-point since zygotic genome activation occurs at the 8-cell stage in humans? A role for the maternal histone 1 linker, H1foo, has been identified as normalizing the epigenome of cells during the process of pluripotency induction (45). H1foo is still present in the 8-cell embryo while it is absent shortly after morula formation (46). This is one possible reason for a greater resilience of human 8-cell embryos, relative to blastocyst, to cryopreservation and further in vitro development.
Epigenetic differences in naïve vs. primed hESC
Epigenetic factors are likely to impact the ultimate quality of any pluripotent cell line as they do for induced pluripotent cells. Overall histone and CpG methylation patterning is discussed below with regard to analyzing the stage of hESC development. Deeper inquiry into, epigenetic patterns are the focus of current studies.
Naïve hESC have notably reduced H3K27me3 histone marks than in the primed equivalents. H3K27me3 marks in primed hESC indicate transcriptional silencing, while the absence in naïve is an indicator of open chromatin. The relative level of this histone mark has become an accepted criterion of the naïve vs. primed hESC state (23-27, 36-38, 40). Further definition of the histone marks in the naïve and primed states is currently under study.
During pre-implantation mammalian development, DNA CpG methylation patterns are actively erased and reestablished (47, 48). Though mouse early embryonic methylation dynamics are similar to those in human, maternally contributed methylation to specific CpG island promoters differs between species, while paternal imprints are generally demethylated prior to implantation (49). DNA methylation in somatic cells is thought to be one of the primary means to prevent transposable element (TE) expression. In the early embryo, almost complete global DNA demethylation occurs in preparation for repatterning to set the stage for a new generation of development (47, 48). Alternate methods of TE suppression are required in the pre-implantation embryo, and by extension, naïve ESC (50). These early embryo strategies alternatively regulate transposons and the retrotransposons of long terminal repeats (LTRs), long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs) and micro RNAs (miRNA) in ways that are not yet fully defined (50). In particular, pluripotent hESC are associated with elevated expression levels of human retrovirus type H (HERV-H gamma retrovirus) that in turn provide binding sites for transcription factors that establish the pluripotency of hESC (51). Subsequently, it was found that HERV-H elements are expressed more highly in primed cells (31). HERV-K levels are specifically elevated in naïve as opposed to primed hESC (31, 52). It was found that members of the SINE-VNTR-Alu (SVA) family of TEs were transcribed almost exclusively in the naïve state (31).
Parentally inherited DNA methylation patterns in somatic cells are expected to reflect those parental imprinting patterns in the primed state. However, because imprinting is still an active process in the pre-implantation embryo, there is a concern that induction of the in vitro naïve state, whether from primed or somatic cells, may not perfectly reflect the active and appropriate pre-implantation parental imprint process. Also, a recent report (53) questions if naïve hESC grown in 5iLAF (26) when converted to primed can establish appropriate DNA epigenetic patterning, including appropriate parental monoallelic imprint patterns. Currently, this would imply that primed cells may be more epigenetically appropriate than naïve induced from primed or from somatic cells. It is unknown if this may be influenced by the inhibitors used in the induction of both naïve from primed or from somatic cells to primed, though it could be caused by many factors. Because mouse ESC and induced pluripotent stem cells (iPSC) are capable of generating germline chimeras via tetraploid complementation (21, 54-56), it would appear that epigenetic patterning can be fully restored to the naïve state in mouse and that some of the epigenetic discrepancies seen in hESC may be due to non-ideal culture conditions.
Differences between naïve and primed hESC for TE control, among other dangers inherent in cell growth, raise the question of whether the naïve or the primed state is the more karyotypically stable. Essentially, the answer may rest in turn on whether the mechanisms for DNA integrity and repair that operate during the naïve state are as effective as those that operate in the primed. This is directly relevant to protection of the germ lineage chromatin to survive intact into the next generation. Our data with naïve Elf1 indicate trisomy drift in the late passage 20's, while the primed cultures appear to be far more karyotypically stable, as determined by G-banding. In this fashion, genomic integrity of naïve hESC reflects mESC, in that the conventional wisdom when working with mESC is to begin gene editing experiments prior to passage 20. However, primed mouse EpiSC are fragile in our hands and though we have actively cultured three separate EpiSC lines that were of normal karyotype upon shipment, they all developed varying degrees of abnormality shortly upon thaw. The understanding of in vitro DNA surveillance and repair in these pluripotent states deserves further study.
hESC quality
There are several considerations impacting the utility of hESC, whether naïve or primed. The primary criterion is that the line should differentiate effectively to a broad range of tissues, as assessed by both in vitro and in vivo and protocols. If differentiation from the pluripotent stage halts prior to full development and retains primitive elements, there is a risk that the line can lead to cancer development. This can currently be assessed by close scrutiny of teratomas to detect aberrant development by limited differentiation ability and through in vitro differentiation efficiency. Because karyotype anomalies are linked to the incomplete differentiation seen in cancer, it is important to monitor cells for presence of aneuploidy.
Other desirable characteristics of a pluripotent line destined for clinical use are robust and reasonably uniform growth, ability to survive cryopreservation, and ability to generate the target tissue required efficiently. Do naïve hESC need to transition through the primed state prior to heading down lineage pathways? The Elf1 line, when transitioned to primed conditions does not achieve as primed a state as cells that were directly established as primed when analyzed by RNA expression patterns (57). It can differentiate efficiently down a mesodermal lineage from primed Elf1 cultures, but not naïve, indicating that it is either the pre-established differentiation protocol that demands primed cells or the need to transition to primed reflects in vivo biology.
Assays to determine whether cells are in the naïve state
There is general agreement in some aspects regarding the characteristics of naïve hESC. Metabolic switch from the ability to generate energy via oxidative phosphorylation by naïve cells to purely glycolytic metabolism in primed cells (27, 40, 57) is conserved between human and mouse (58). CpG methylation and H3K27me3 levels are reduced in naïve relative to primed (23-27, 36-38, 40). Also, there is agreement that growth rate slows and cloning efficiency decreases as cells transition from naïve to primed (23, 25, 37, 38).
Controversy arises with regard to RNA expression, as mentioned above. The standard genes defining the pluripotent state, such as POU5F1 (OCT4), SOX2, NANOG and KLF4 are expressed in both the naïve and primed states. Most have seen that SSEA-4 is expressed in both states (23, 26, 28, 29, 36, 37), but one report stresses that SSEA-4 is lower in naïve 5i cells and that this defines a human ground state (53). This observation requires validation by other investigators. Recently, in vitro culture of human embryos showed that the human ICM, unlike mouse, does not segregate the epiblast (OCT4-expressing) from the hypoblast (GATA6-expressing) within the pre-implantation ICM, but waits to segregate the hypoblast shortly after implantation/culture adherence (59). This may explain why GATA6 expression is found to be present in naïve human stem cells, if hypoblast precursors are part of a naïve human culture. This follows the pattern detected in human embryos by single-cell RNA-Seq, wherein expression of GATA-6 is detected in 8-cell, morula and pre-implantation epiblast, but not in primed hESC (46). Because the definition of naïve culture conditions is still being refined, it is useful to cross compare in vivo single cell RNA-Seq data from human embryos and the naïve line to determine appropriate expression patterns (46, 60). For example, embryo data (46) support the expectation that naïve hESC are likely to have robust DNMT3L expression, as does the human pre-implantation epiblast, but not primed hESC, while primed hESC have robust DNMT3B expression, but not the pre-implantation epiblast. There is the caveat that naïve and primed ESC are not actively differentiating, so should vary from pre- and post-implantation embryos by a cohort of expression variables involved in developmental arrest. Both pluripotent states may reflect aspects of diapause linked to reduction of MYC expression (61).
Also contested is whether both X's should be active in the naïve state. Theunissen et al. (26) indicated the presence of an inactive X, contrary to other reports that find two active X's. This has since been clarified and this group confirms that both X;s should be active in the naïve state (31). This report also indicated the inability of the 5i cells to contribute to xeno-chimerism in the mouse embryo, while other naïve human cells could contribute to the developing mouse embryo (36). The suggestion that 5i cells are karyotypically fragile (53) could impact the ability to contribute to the mouse embryo, could confound X-inactivation and possibly could impact SSEA-4 expression.
Finally, the accepted preferential use of the distal OCT4 enhancer by naïve cells was counter-indicated by Duggal et al. (38) in that the DNA of their naïve cells was relatively hypomethylated in both the proximal and distal enhancers. This might be explained by features of their assay rather than by true enhancer usage. Overall, naïve human pluripotent cells appear to preferentially use the distal OCT4 enhancer.
Currently, criteria to establish a line as naïve requires first that it is deemed pluripotent, as shown by expression of pluripotent markers; such as, OCT4, NANOG, SSEA-4, Tra-1-60 and/or Tra-1-81. Beyond this, distinguishing naïve from primed largely relies on cell and colony morphology, although culture conditions heavily influence this judgment. Reduced DNA CpG methylation and H3K27me3 marks (relative to primed), ability to utilize mitochondria for oxidative phosphorylation (while primed cells rely on glycolysis), the use of the distal enhancer of OCT4 (as opposed to the primed usage of the proximal enhancer) are all generally agreed upon features of the naïve state. Finally, Shakiba et al. (62) have shown that CD24 expression can distinguish primed hESC (high) from naïve (low). Table 4 summarizes current assays that are generally accepted as distinguishing the naïve from primed states.
Table 4. Differing characteristics between the naïve and primed states from combined literature.
| Naïve | Primed | |
|---|---|---|
| Morphology | mounded | flat |
| Single-cell passage (w/o ROCKi) | yes | no |
| Relative gene expression: | ||
| TBX3 | high | low |
| DNMT3L | high | low |
| IL6ST (GP130) | high | low |
| DNMT3B | low | high |
| CD24 | low | high |
| Transposable elements | high | low |
| HERVK | high | low |
| OCT4 enhancer use | distal | proximal |
| Relative H3K27me3 marks | low | high |
| X-activation | XaXa | XaXi |
| Metabolism | Ox-Phos | Glycolytic |
Conclusion
In summary, true human naïve pluripotent cells are possible to maintain in culture. However, we need to better define culture conditions to be confident that a line retains full developmental competence. Once culture conditions are perfected, which requires tighter definition of small molecule inhibitors and optimal growth factor use, naïve lines that may best correlate to the in vivo ICM will likely be derived de novo from pre-implantation human embryos, similar to mESC. Also, improvement in understanding medium requirements may allow naïve iPSC lines to be developed with broadened clinical utility. This window into the stage-specific mechanics of the peri-implantation stage in humans enriches the toolbox to answer research questions while clinical utility will span a deeper understanding of human infertility to tissue repair via regenerative medicine.
Significance Statement.
Although the existence of the human naïve pluripotent state is coming into acceptance, discrepancies in the literature defining the naïve human pluripotent state have served to undermine confidence in these cells as a reflection of normal development. Naïve mouse cells are the accepted paradigm of the naïve state. Because early human development does not exactly mirror mouse, work remains to be done to understand the differences between the naïve human lines and to understand the meaning of “normal” in this context. This will serve to broaden the understanding of human pluripotency, to define the factors that will best preserve the biology of these early embryonic cells, and to bring them into acceptance for further research into human development with clinical consequences for regenerative medicine.
Acknowledgments
I would like to thank Breck Byers for helpful discussions and for critical reading of the manuscript. We are greatly indebted to The Ellison Foundation for non-federal gift funds to the Institute for Stem Cell and Regenerative Medicine in support of this study and to the NIH/NIGMS P01 GM081619-06, for funding to study human ESC lines on the NIH Stem Cell Registry.
Footnotes
Human Subjects Approval: Review and approval by the University of Washington Human Subjects Division to receive donated human embryos following informed consent was obtained. Elf1, the line that resulted from this application is listed on the NIH Registry, indicating that the required oversight and consent requirements were adhered to. In addition, all embryonic stem cell research is carried out under the oversight of the University of Washington Embryonic Stem Cell Research Oversight Committee.
Bibliography
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 5.Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 6.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 7.Kishimoto T, Taga T, Akira S. Cytokine signal transduction. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 8.Niwa H, Burdon T, Chambers I, et al. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuda T, Nakamura T, Nakao K, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raz R, Lee CK, Cannizzaro LA, d'Eustachio P, et al. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 1999;96:2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo G, Yang J, Nichols J, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martello G, Sugimoto T, Diamanti E, et al. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato N, Meijer L, Skaltsounis L, et al. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nature Med. 2003;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 16.Lyanshenko N, Winter M, Migliiorini D, et al. Differential requirement for the dual functions of β-catenin in embryonic stem cell self-renewal and germ layer formation. Nature Cell Biol. 2011;13:753–761. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wray J, Kalkan T, Gomez-Lopez S, et al. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nature Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meek S, Wei J, Sutherland L, et al. Tuning of β-catenin activity is required to stabilize self-renewal of rat embryonic stem cells. Stem Cells. 2013;331:2104–2115. doi: 10.1002/stem.1466. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Blair K, Smith A. Robust self-renewal of rat embryonic stem cells requires fine-tuning of glycogen synthase kinase-3 inhibition. Stem Cell Reports. 2013;1:209–217. doi: 10.1016/j.stemcr.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Osorno R, Tsakiridis A, et al. In vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep. 2012;2:1571–1578. doi: 10.1016/j.celrep.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Nagy A, Rossant J, Nagy R, et al. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ware CB, Wang LL, Mecham BH, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4:359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ware CB, Nelson AM, Mecham B, et al. Derivation of naïve human embryonic stem cells. Proc Nat Acad Sci USA. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lengner CJ, Gimelbrant AA, Erwin JA, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–83. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Hanna J, Cheng AW, Saha K, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Nat Acad Sci USA. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theunissen TW, Powell BE, Wang H, et al. Systematic identification of defined conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takashima Y, Guo G, Loos R, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Aksoy I, Gonnot F, et al. Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency. Nature Comm. 2015;6:7095. doi: 10.1038/ncomms8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin H, Hejna M, Liu Y, et al. YAP induces human naive pluripotency. Cell Reports. 2016;14:2301–2312. doi: 10.1016/j.celrep.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valamehr B, Robinson M, Abujarour R, et al. Platform for induction and maintenance of transgene-free hiPSCs resembling ground state pluripotent stem cells. Stem Cell Reports. 2014;2:366–381. doi: 10.1016/j.stemcr.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theunissen T, Friedli, He Y, et al. Molecular Criteria for defining the naïve human pluripotent state. Cell Stem Cell. 2016;19:1–14. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovisto H, Hyvärinen M, Strömberg, et al. Cultures of human embryonic stem cells: serum replacement medium or serum-containing media and the effect of basic fibroblast growth factor. Reprod Biomed Online. 2014;9:330–337. doi: 10.1016/s1472-6483(10)62150-5. [DOI] [PubMed] [Google Scholar]
- 33.van Oosten AL, Costa Y, Smith A, Silva JC. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naïve pluripotency. Nat Commun. 2012;3:817. doi: 10.1038/ncomms1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunath T, Saba-El-Leil MK, Almousaileakh M, et al. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 36.Chan Y-S, Göke J, Ng J-H, et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Gafni O, Weinberger L, Mansour AA, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 38.Duggal G, Warrier S, Ghimire S, et al. Alternative routes to induce naïve pluripotency in human embryonic stem cells. Stem Cells. 2015;33:2686–2698. doi: 10.1002/stem.2071. [DOI] [PubMed] [Google Scholar]
- 39.Carter MG, Smagghe BJ, Stewart AK, et al. A primitive growth factor, NME7AB, is sufficient to induce stable naïve state human pluripotency; reprogramming in this novel growth factor confers superior differentiation. Stem Cells. 2016;34:847–859. doi: 10.1002/stem.2261. [DOI] [PubMed] [Google Scholar]
- 40.Guo G, von Meyenn F, Santos F, et al. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports. 2016;6:437–446. doi: 10.1016/j.stemcr.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dutta D, Ray S, Home P, et al. Self renewal vs. lineage commitment of embryonic stem cells: protein kinase C signaling shifts the balance. Stem Cells. 2011;29:618–628. doi: 10.1002/stem.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tejeda-Muñoz N, González-Aguilar H, Santoyo-Ramos P, et al. Glycogen synthase kinase 3β is positively regulated by protein kinase Cζ-mediated phosphorylation induced by Wnt Agonists. Mol Cell Biol. 2013;36:731–741. doi: 10.1128/MCB.00828-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu LZ, He AB, Liu XJ, et al. Protein kinase Czeta and glucose uptake. Biochemistry (Mosc) 2006;71:701–706. doi: 10.1134/s0006297906070017. [DOI] [PubMed] [Google Scholar]
- 44.Iankov I, Praskova M, Kalenderova S, et al. The effect of chemical blockade of PKC with Gö6976 and Gö6983 on proliferation and MAPK activity in IL-6-dependent plasmacytoma cells. Lek Res. 2002;26:363–368. doi: 10.1016/s0145-2126(01)00132-1. [DOI] [PubMed] [Google Scholar]
- 45.Kunitomi A, Yuasa S, Sugiyama F, et al. H1foo has a pivotal role in qualifying induced pluripotent stem cells. Stem Cell Reports. 2016;6:825–833. doi: 10.1016/j.stemcr.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan L, Yang M, Guo H, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 47.Okae H, Chiba H, Hiura H, et al. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 2014;10:e1004868. doi: 10.1371/journal.pgen.1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith ZD, Chan MM, Humm KC, et al. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 50.Hutchins AP, Pei D. Transposable elements at the center of the crossroads between embryogenesis, embryonic stem cells, reprogramming, and long non-coding RNAs. Sci Bull (Beijing) 2015;60:1722–1733. doi: 10.1007/s11434-015-0905-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Xie G, Singh M, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 52.Grow EJ, Flynn RA, Chavez SL, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pastor WA, Chen D, Liu W, et al. Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell. 2016;18:323–329. doi: 10.1016/j.stem.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carey BW, Markoulaki S, Hanna JH, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–98. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Boland MJ, Hazen JL, Nazor KL, et al. Generation of mice derived from induced pluripotent stem cells. J Vis Exp. 2012;69:4003. doi: 10.3791/4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buganim Y, Markoulaki S, van Wietmarschen N, et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15:295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sperber H, Mathieu J, Wang Y, et al. The metabolome regulates the epigenetic landscape during naïve-to-primed human embryonic stem cell transition. Nature Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou W, Choi M, Margineantu D, et al. HIF1a induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:1203–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shahbazi MN, Jedrusik A, Vuoristo S, et al. Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol. 2016;18:700–708. doi: 10.1038/ncb3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blakeley P, Fogarty NM, del Valle I, et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142:3151–3165. doi: 10.1242/dev.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scognamiglio R, Cabezas-Wallscheid N, Thier MC, et al. Myc depletion induces a pluripotent dormant state mimicking diapause. Cell. 2016;164:668–680. doi: 10.1016/j.cell.2015.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shakiba N, White CA, Lipsitz YY, et al. CD24 tracks divergent pluripotent states in mouse and human cells. Nat Commun. 2015;6:7329. doi: 10.1038/ncomms8329. [DOI] [PMC free article] [PubMed] [Google Scholar]