Abstract
For nanobiotechnology to achieve its potential, complex organic-inorganic systems must grow to utilize the sequential functions of multiple biological components. Critical challenges exist: immobilizing enzymes can block substrate-binding sites or prohibit conformational changes, substrate composition can interfere with activity, and multistep reactions risk diffusion of intermediates. As a result, the most complex tethered reaction reported involves only 3 enzymes. Inspired by the oriented immobilization of glycolytic enzymes on the fibrous sheath of mammalian sperm, here we show a complex reaction of 10 enzymes tethered to nanoparticles. Although individual enzyme efficiency was higher in solution, the efficacy of the 10-step pathway measured by conversion of glucose to lactate was significantly higher when tethered. To our knowledge, this is the most complex organic-inorganic system described, and it shows that tethered, multi-step biological pathways can be reconstituted in hybrid systems to carry out functions such as energy production or delivery of molecular cargo.
Keywords: Biomimicry, Enzymes, Glycolysis, Nanoparticles, Tethered Enzymes
Multiple strategies have been used to interface biological components with inorganic surfaces. In engineering studies and industrial applications, carboxyl-amine binding chemistry is the most common attachment strategy[1]. However, this chemistry is not site-specific and does not inherently impart proper molecular orientation—two factors that can lead to significant reduction in function of the tethered enzyme[2]. Use of self-assembly templates can immobilize and orient enzymes; for instance sequential reactions of glucose oxidase and horseradish peroxidase have been demonstrated using a DNA scaffold to control enzyme spacing[3]. Recently, the sequential reaction of three enzymes conjugated to quantum dots was demonstrated[4]. Despite the different approaches available, the current state of the art remains limited to two- or three-step, coupled tethered reactions[5]. This is in stark contrast to multi-step reactions in solution, in which complex reactions have been demonstrated ranging up to a 13-step production of ethanol from glucose[6].
Inspired by the organization of glycolytic enzymes on the fibrous sheath of mammalian sperm[7], we previously showed that replacement of germ cell-specific targeting domains with a bioaffinity tag provided oriented immobilization of glycolytic enzymes. This translated into significant advantages in specific activity of both individual enzymes and for sequential reactions when compared against the same enzymes tethered via random adsorption without a histidine tag or carboxyl-amine chemistry[8] These prior studies suggested that biomimetic oriented immobilization increased the activity of individual tethered enzymes for several reasons.
First, oriented immobilization facilitates substrate access and required conformational changes. Indeed, contrary to earlier reports using non-oriented attachment strategies, the advantages of oriented enzyme immobilization were so great that for 3 enzymes representing 3 different enzyme classes, we found no trends in changes in kM, kcat or kcat/kM when enzymes were tethered in monolayers to nanoparticles (NPs) of different sizes.[9] It is also possible that oriented immobilization might promote formation of catalytically active structures such as homo-dimers or –tetramers, and/or reduce possible interference between neighboring multimers. This possibility was suggested by advantages for oriented immobilization shown by those same 3 enzymes when tethered in multilayers,[9] although further studies are needed.
Here, we investigated whether the model of glycolytic enzymes from mammalian sperm would enable us to create tethered systems with the same degree of complexity as biological pathways. Eleven histidine-tagged glycolytic enzymes were engineered from mouse testis cDNA (Table 1S), expressed in mammalian cells, purified, and examined with Coomassie gel staining and immunoblotting using enzyme-specific antibodies and antibodies against the histidine tag (Figure 1 and Figure 1S). Activity of the recombinant enzymes in solution was detected using coupled reactions involving exogenous enzymes and kinetic measurements were obtained (Table 2S and 3S).
Figure 1.
Glycolysis and recombinant histidine-tagged enzymes. a. Schematic model of 10 tethered glycolytic enzymes on Ni-NTA functionalized silica NPs. b. Representative Coomassie staining of recombinant histidine-tagged enzymes (left panel) and purity analysis by densitometry (right panel; performed with ImageJ).
The composition and dimensions of the scaffold can affect the function of tethered enzymes. In early studies, we found the activity of several tethered enzymes on planar surfaces was <10% compared to when in solution (Figure 2S). Testing the activity of enzymes when tethered to various scaffolds, we found that magnetic NPs significantly reduced the activ ity of certain enzymes. When comparing NPs of different composition, we found that enzymes tethered to 500 nm diameter Ni-NTA functionalized silica nanoparticles (NPs) showed higher specific activity than when on magnetic NPs or plain silica NPs (Figure 2S). Although magnetic nanoparticles had advantages in separation of the NPs from the reactions, they tended to form aggregates, which might have contributed to the lower specific activities (data not shown, our observation).
In preliminary studies, we divided the sequential reactions of 10 enzymes into three subsets (Figure 2) and tested their coupled, sequential activities by mixing enzymes in various ratios based on their individual specific activities in solution (Table 3S). The first reaction (reaction A) included four enzymes, hexokinase, glucose-6-phosphate isomerase, phosphofructokinase, and aldolase (HK, GPI, PFK and Aldo) tethered to the same NP, and converted glucose to glyceraldehyde phosphate (GAP) and dihydroxyacetone phosphate (DHAP). Activity was measured by a change in absorbance through the use of the exogenous enzyme, glycerol phosphate dehydrogenase (GPDH), which converted NADH to NAD+ (Figure 2A and Table 4S). Activity of the coupled 4-step reaction was observed only in the presence of the initial substrate glucose (Figure 2A).
Figure 2. Enzymes tethered on Ni-NTA-modified silica NPs performed sequential reactions.
Representative results are shown for triplicate reactions from single protein preparations, with identical results obtained from at least 3 preparations. a. Schematic of reaction A and sequential enzyme activity. Absorbance values were normalized from time 0 (in all panels, mean and standard deviation are shown). Results are shown for the 4 enzymes in reaction A tethered to NPs with glucose (A-NP +) and without added glucose (A-NP −), and for a control with no enzymes (NP only). b. Schematic of reaction B and corresponding sequential enzyme activity (center: change in absorbance, right: ATP production). Results are shown for NPs with GAPDHS alone (G), PGK alone (P), GAPDHS and PGK (GP), with (+) and without (−) added GAP. c. Schematic of reaction C and sequential enzyme activity (center: ATP production, right: change in absorbance). Results are shown for NPs with PGAM alone (M), PGAM and ENO (ME), PGAM-ENO-PK (MEP), and PGAM-ENO-PK-LDH (MEPL), with (+) and without (−) added 3PGA.
The second reaction (reaction B) was performed with two tethered enzymes, glyceraldehyde-3-phosphate dehydrogenase-S and phosphoglycerate kinase (GAPDHS and PGK). GAPDHS catalyzes GAP to 1,3 bisphosphoglycerate and converts NAD+ to NADH. PGK then catalyzes 1,3 bisphosphoglycerate to 3-phosphoglyceric acid (3PGA), thereby producing ATP detected through luminescence emitted from exogenous luciferase and luciferin (Figure 2B and Table 4S). Both GAPDHS-NPs and GAPDHS-PGK-NPs showed changes in absorbance (left panel) reflecting GAPDHS activity. ATP production was only observed when both enzymes were present and acting sequentially (right panel).
The last reaction (Figure 2C) was performed with tethered phosphoglycerate mutase, enolase, pyruvate kinase and lactate dehydrogenase (PGAM, ENO, PK and LDH). In the first three steps, the product from PGK, 3PGA, is catalyzed to pyruvate and ATP is produced by PK. Pyruvate is then converted to lactate by LDH coupled with the conversion of NADH into NAD+. Note that this extra-glycolytic step of including LDH was included because NAD+ would need to be replenished to keep Reaction B active. Reaction C was monitored by both change in absorbance and ATP production as in reaction B (Figure 2C and Table 4S). PGAM-ENO-PK-NPs and PGAM-ENO-PK-LDH-NPs produced ATP, reflecting PK activity (left panel), but only PGAM-ENO-PK-LDH-NPs showed change of absorbance deriving from the LDH activity.
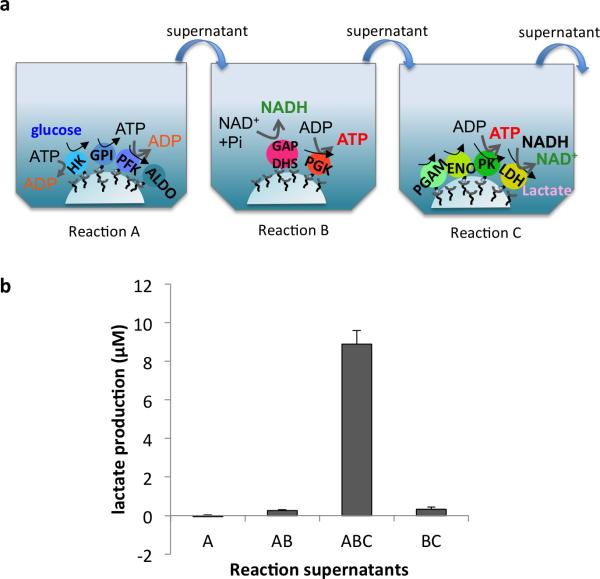
After analyzing the three reactions (A, B and C) separately, we next examined whether transferring the supernatant from one tethered reaction to the next would allow the entire 10-step pathway to function in series. This design also enabled us to investigate where any potential blockages might occur. The assay buffer for Reaction A containing glucose, ATP and MgCl2 was added to the well containing HK-GPI-PFK-Aldo-NPs. After a 30 min incubation, this supernatant (A) was transferred to a well containing GAPDHS-PGK-NPs and the assay buffer for Reaction B, minus GAP. This supernatant (AB) was then transferred to the well containing PGAM-ENO-PK-LDH-NPs and reagents for Reaction C, minus 3PGA. We quantified lactate production from each supernatant (A, AB, and ABC). As a control to ensure that the initial substrate of glucose was in fact being acted upon by the enzymes in Reaction A, and not downstream enzymes or intermediates, we performed the same steps as above, but with no enzymes attached to the NPs in Reaction A. That is, we added the reaction mix for Reaction A to Reaction B, and then the supernatant of that reaction to Reaction C. The resulting supernatant, BC, did not show lactate production (Figure 3). These results demonstrate that glucose was sequentially metabolized by all ten tethered enzymes to yield a final product of lactate. To the best of our knowledge, this is the first demonstration of reconstitution of a physiological metabolic pathway in vitro on scaffolds through the use of genetically engineered, tethered enzymes.
Figure 3. Lactate production from 10 tethered enzymes.
a. Schematic of experimental design. b. A lactate quantification assay was performed on various supernatants. A: Reaction A; AB: reactions A and B; ABC: reactions A, B and C; BC: control in which reaction buffer and reagents from Reaction A (no tethered enzymes), were added to Reactions B and C, showing that lactate production required metabolism of the glucose in Reaction A. Columns represent means and standard dev iations (n=3).
Enzyme reaction kinetics are often studied and discussed as simplified models assuming thorough mixing in solution. In both cells and in hybrid organic-inorganic devices, this assumption is not always valid. Enzymes and the pathways in which they function are often compartmentalized to specific sub-cellular regions or functional domains in a device. Positive effects, such as substrate channeling, can be provided from this organization. However, negative effects, such as diffusion of intermediates away from subsequent steps of the reaction, can also occur.
To investigate the impact of tethering on the coupled enzyme reactions, we compared the overall reaction efficacy of tethered enzymes vs. enzymes free in solution. We made these comparisons between two single microplate wells, one containing all ten enzymes tethered versus one with all ten enzymes in solution. We quantified glucose consumption and lactate production at three different time points using enzyme-based colorimetric assays. Values for tethered vs. free enzymes were calculated in nanomol (nmol)/micrograms of protein. Note that for each protein, the ratios of protein in-solution vs when tethered were similar according to densitometry analysis (Figure 3S), with the exception of relatively increased binding for PFK and ALDO in reaction A, which had no deleterious effect on that coupled reaction. To confirm and quantify lactate production by another method, we performed LC-MRM (Liquid Chromatography-multiple reaction monitoring) analysis of lactate, using 13C-lactate as an internal standard. Comparison of results obtained with the two methods showed high correlation, with an R2 value > 0.99 (Figure 4e). As expected given the enhanced access to substrate from all sides and freedom to undergo needed conformational changes, enzymes in solution showed significantly higher glucose consumption (Figure 4b) and lactate production (Figure 4c) at these time points.
Figure 4. Conversion of glucose to lactate using 10 sequential enzyme reactions on NPs.
a. Schematic of experimental design. In panels b, c, and d, open bars show results for enzymes in solution; black bars show results for tethered enzymes. b. Glucose consumption over time, calculated as nmol glucose that were enzymatically reacted upon/mg of enzyme. c. Lactate production over time, calculated as nmol lactate produced/mg of enzyme. d Lactate production efficiency, calculated as the percentage of the amount of lactate produced over the amount of glucose consumed (nmol/mg of lactate)/ (nmol/mg of glucose). N=12, mean + SEM, *p=0.0038, **p=0.0013, ***p=0.0012. e. Lactate production as measured by LC-MRM. Black columns show lactate concentrations measured by lactate oxidase enzymatic assay, and white columns show values obtained by LC-MRM. The inset graph shows the correlation between the enzyme based assay and LC-MRM. R2=0.991.
However, of great interest, the tethered enzymes showed a significantly higher efficiency of lactate produced from each nmol of glucose consumed (Figure 4d). This demonstrates that flux through the entire system was more efficient for the tethered enzymes. In biological systems, several mechanis ms of substrate channeling have been described[10], with demonstrations that intermediates can move within the hydration shells around adjacent proteins, and that other intervening proteins can provide a hydration bridge[3a-c, 11]. Such models are highly compatible with our data, in which multiple enzymes were tethered to single NPs.
In nature, it is well established that the fifth enzyme of glycolysis, TPI, is essential for conversion of DHAP to GAP, leading to downstream doubling of ATP production. However, we found that TPI, without exception, had enhanced catalysis of GAP to DHAP (In solution: Table 3S, Tethered: Figure 4S), a phenomenon which has been well documented with both cell extracts and free enzymes [12]. Although production of DHAP might be useful for other biosynthetic pathways, here it prevented net production of ATP by not providing a second molecule of GAP to proceed through glycolysis. Further studies of regulation of TPI in highly glycolytic cells are needed to understand how cells regulate production of ATP vs DHAP.
The elegant high-throughput glycolytic system in mammalian sperm provided inspiration for overcoming the challenges of retaining enzyme function when tethered, enabling a three-fold jump in complexity of sequential tethered reactions over prior reports. Our new findings create a foundation demonstrating that complex biological systems can be integrated into hybrid organic-inorganic devices, enabling other advantages of tethering such as spatiotemporal control of the reactions, and improved stability[3b, 3c]. Future applications for tethered enzymes range from in vivo (e.g. provision of metabolic functions, drug delivery), to in vitro (e.g. enzymatic detection of disease biomarkers), while enabling investigation of fundamental principles of regulation of enzyme biochemistry.
Experimental Section
Supplementary Material
Acknowledgements
This work was supported by a U.S. National Institutes of Health Pioneer Award (8DP1-EB016541), NYSTAR Center for Life Science Enterprise grant (AJT), and by JSPS Grants-in-Aid for Scientific Research (Wakate B: 21770105) and JSPS Institutional Program for Young Researcher Overseas Visits R10 (CM). We thank Drs. Takashi Ijiri and George Gerton of the University of Pennsylvania for providing information regarding the germ cell-specific TPI isoform.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- 1.Rodrigues RC, Berenguer-Murcia A, Fernandez-Laf uente R. Adv Synth Catal. 2011;353:2216–2238. [Google Scholar]
- 2.a Jia F, Narasimhan B, Mallapragada S. Biotechnol Bioeng. 2014;111:209–222. doi: 10.1002/bit.25136. [DOI] [PubMed] [Google Scholar]; b Garcia-Galan C, Berenguer-Murcia A, Fernandez-Lafuente R, Rodrigues RC. Adv Synth Catal. 2011;353:2885–2904. [Google Scholar]
- 3.a Wilner OI, Weizmann Y, Gill R, Lioubashevski O, Freeman R, Willner I. Nat Nanotechnol. 2009;4:249–254. doi: 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]; b Fu J, Liu M, Liu Y, Woodbury NW, Yan H. Journal of the American Chemical Society. 2012;134:5516–5519. doi: 10.1021/ja300897h. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Liu Y, Du J, Yan M, Lau MY, Hu J, Han H, Yang OO, Liang S, Wei W, Wang H, Li J, Zhu X, Shi L, Chen W, Ji C, Lu Y. Nat Nanotechnol. 2013;8:187–192. doi: 10.1038/nnano.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Kuchler A, Yoshimoto M, Luginbuhl S, Mavelli F, Walde P. Nat Nanotechnol. 2016;11:409–420. doi: 10.1038/nnano.2016.54. [DOI] [PubMed] [Google Scholar]
- 4.Kang W, Liu J, Wang J, Nie Y, Guo Z, Xia J. Bioconjugate chemistry. 2014;25:1387–1394. doi: 10.1021/bc5002399. [DOI] [PubMed] [Google Scholar]
- 5.a Fessner WD, Walter C. Angew Chem Int Edit. 1992;31:614–616. [Google Scholar]; b Keighron JD, Keating CD. Langmuir. 2010;26:18992–19000. doi: 10.1021/la1040882. [DOI] [PMC free article] [PubMed] [Google Scholar]; c You C, Zhang YH. ACS synthetic biology. 2013;2:102–110. doi: 10.1021/sb300068g. [DOI] [PubMed] [Google Scholar]; d You C, Zhang YH. ACS synthetic biology. 2014;3:380–386. doi: 10.1021/sb4000993. [DOI] [PubMed] [Google Scholar]; e You C, Myung S, Zhang YH. Angew Chem Int Ed Engl. 2012;51:8787–8790. doi: 10.1002/anie.201202441. [DOI] [PubMed] [Google Scholar]
- 6.a Fessner WD, Eyrisch O. Angew Chem Int Edit. 1992;31:56–58. [Google Scholar]; b Welch P, Scopes RK. J Biotechnol. 1985;2:257–273. [Google Scholar]
- 7.a Mukai C, Travis AJ. Reprod Domest Anim. 2012;47:164–169. doi: 10.1111/j.1439-0531.2012.02071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Cao W, Gerton GL, Moss SB. Mol Cell Proteomics. 2006 doi: 10.1074/mcp.M500322-MCP200. [DOI] [PubMed] [Google Scholar]; c Krisfalusi M, Miki K, Magyar PL, O'Brien DA. Biol Reprod. 2006;75:270–278. doi: 10.1095/biolreprod.105.049684. [DOI] [PubMed] [Google Scholar]; d Miki K, Qu W, Goulding EH, Willis WD, Bunch DO, Strader LF, Perreault SD, Eddy EM, O'Brien DA. Proc Natl Acad Sci U S A. 2004;101:16501–16506. doi: 10.1073/pnas.0407708101. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Mukai C, Okuno M. Biol Reprod. 2004;71:540–547. doi: 10.1095/biolreprod.103.026054. [DOI] [PubMed] [Google Scholar]; f Travis AJ, Jorgez CJ, Merdiushev T, Jones BH, Dess DM, Diaz-Cueto L, Storey BT, Kopf GS, Moss SB. J Biol Chem. 2001;276:7630–7636. doi: 10.1074/jbc.M006217200. [DOI] [PubMed] [Google Scholar]
- 8.a Mukai C, Bergkvist M, Nelson JL, Travis AJ. Chem Biol. 2009;16:1013–1020. doi: 10.1016/j.chembiol.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mukai C, Gao L, Bergkvist M, Nelson JL, Hinchman MM, Travis AJ. PloS one. 2013;8:e61434. doi: 10.1371/journal.pone.0061434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lata JP, Gao L, Mukai C, Cohen R, Nelson JL, Anguish L, Coonrod S, Travis AJ. Bioconjugate chemistry. 2015;26:1931–1938. doi: 10.1021/acs.bioconjchem.5b00354. [DOI] [PubMed] [Google Scholar]
- 10.Wheeldon I, Minteer SD, Banta S, Barton SC, Atanassov P, Sigman M. Nature chemistry. 2016;8:299–309. doi: 10.1038/nchem.2459. [DOI] [PubMed] [Google Scholar]
- 11.Zhang YHP. Biotechnol Adv. 2011;29:715–725. doi: 10.1016/j.biotechadv.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 12.a Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Bujara M, Schumperli M, Pellaux R, Heinemann M, Panke S. Nat Chem Biol. 2011;7:271–277. doi: 10.1038/nchembio.541. [DOI] [PubMed] [Google Scholar]; c Harris TK, Cole RN, Comer FI, Mildvan AS. Biochemistry. 1998;37:16828–16838. doi: 10.1021/bi982089f. [DOI] [PubMed] [Google Scholar]; d Rozovsky S, McDermott AE. Proc Natl Acad Sci U S A. 2007;104:2080–2085. doi: 10.1073/pnas.0608876104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.