Abstract
Background
Recurrent abdominal pain is a common and costly health‐care problem attributed, in part, to visceral hypersensitivity. Increasing evidence suggests that gut bacteria contribute to abdominal pain perception by modulating the microbiome‐gut‐brain axis. However, specific microbial signals remain poorly defined. γ‐aminobutyric acid (GABA) is a principal inhibitory neurotransmitter and a key regulator of abdominal and central pain perception from peripheral afferent neurons. Although gut bacteria are reported to produce GABA, it is not known whether the microbial‐derived neurotransmitter modulates abdominal pain.
Methods
To investigate the potential analgesic effects of microbial GABA, we performed daily oral administration of a specific Bifidobacterium strain (B. dentium ATCC 27678) in a rat fecal retention model of visceral hypersensitivity, and subsequently evaluated pain responses.
Key Results
We demonstrate that commensal Bifidobacterium dentium produces GABA via enzymatic decarboxylation of glutamate by GadB. Daily oral administration of this specific Bifidobacterium (but not a gadB deficient) strain modulated sensory neuron activity in a rat fecal retention model of visceral hypersensitivity.
Conclusions & Inferences
The functional significance of microbial‐derived GABA was demonstrated by gadB‐dependent desensitization of colonic afferents in a murine model of visceral hypersensitivity. Visceral pain modulation represents another potential health benefit attributed to bifidobacteria and other GABA‐producing species of the intestinal microbiome. Targeting GABAergic signals along this microbiome‐gut‐brain axis represents a new approach for the treatment of abdominal pain.
Keywords: Bifidobacterium, brain gut axis, GABA, microbiome, neuromodulation
1.
Key Points
Recurrent abdominal pain is a common and costly health‐care problem attributed, in part, to visceral hypersensitivity.
Intestinal bacteria capable of producing the primary inhibitory CNS neurotransmitter, γ‐aminobutyric acid (GABA), modulate primary sensory neurons of the enteric nervous system. The production of GABA by the intestinal microbiome can be increased by supplementing bacteria encoding the glutamate decarboxylase gene gadB.
GABA produced by the intestinal microbiome may form the basis of future nutritional interventions or microbiome‐based therapeutics that ameliorate abdominal pain.
1. Introduction
Bidirectional communication between the gastrointestinal (GI) tract and the central nervous system (CNS) appears to be functionally linked to the intestinal microbiome, collectively termed the microbiome‐gut‐brain axis.1, 2, 3, 4, 5, 6 The microbiome‐gut‐brain axis may contribute to pathophysiology in human diseases such as autism spectrum disorder and Parkinson's disease, whereby GI comorbidities have been associated with disturbances of the intestinal microbiome, and may precede CNS symptoms.7, 8 Whether gut microbes play a role in such disease etiologies is not clearly established, although recent reports in animal models implicate microbiome‐gut‐brain signaling.9, 10, 11, 12, 13, 14, 15
Functional GI disorders, for example, irritable bowel syndrome (IBS) and constipation, represent a spectrum of diseases with active bidirectional gut‐brain communication.9 A subset of patients with IBS (30–40%) have been reported to experience enhanced sensitivity to colonic distension, accompanied by a reduced threshold for pain and increased intensity of sensation.16 Visceral hypersensitivity has therefore been used as a clinical marker in this subset of patients with IBS. The cause of visceral hypersensitivity is unknown; however, the recurrent abdominal pain that is attributed, in part, to visceral hypersensitivity is a common and costly health‐care problem. This amplification of visceral signals to the brain may occur at various levels, including at the level of the intestinal mucosa. Responses of these visceral afferents are affected by local chemical and mechanical stimuli, but may also be influenced by interactions with the intestinal microbiota. Through these interactions, the intestinal microbiota is proposed to play a regulatory role. Support for regulatory roles of intestinal microbes in human disease is provided by studies of probiotics. Numerous studies have shown that Lactobacillus and Bifidobacterium species10, 17, 18, 19, 20, 21, 22, 23 ameliorate abdominal symptoms in functional GI disorders, such as IBS and constipation. Probiotic induction of μ‐opioid and cannabinoid receptor expression on intestinal epithelial cells may promote analgesic effects, as was observed in studies with Lactobacillus acidophilus NCFM supplementation.10 These conceptual findings support a major regulatory role of the intestinal microbiome in enteric neurotransmission and visceral sensitivity. However, interoceptive mechanisms by which gut microbes signal to the enteric and central nervous system, ultimately influencing pain perception and GI function, are poorly understood.
Metagenomic studies revealed that the human microbiome has the potential to generate numerous bioactive molecules, including but not limited to histamine, epinephrine, and γ‐aminobutyric acid (GABA).3, 4, 24, 25 Although Lactobacillus 26 and Bifidobacterium 27, 28 species are reported as GABA‐producing bacteria, the links between microbial production and neuromodulatory activity in vivo have not been established. γ‐aminobutyric acid is a primary inhibitory CNS neurotransmitter in mammals,29 exerting its major functional effects via two GABA receptor subclasses—GABAA and GABAB.30 However, GABA bioactivity is evolutionally conserved throughout the animal kingdom. In mammals, GABA has diverse physiologic effects, such as modulation of blood pressure31 and immune function,32 and it is also known to promote stress tolerance and ATP production in microbial communities.33, 34, 35, 36 A major source of bioactive GABA is the enzymatic conversion of l‐glutamate by glutamate decarboxylase by both microbes and host.37, 38 The potential health benefits of GABA have fueled a growing interest in GABA‐enriched dietary supplements and naturally fermented food products.27 We hypothesized that in vivo production of GABA by commensal bacterial species would modulate the excitability of colonic sensory afferents. We aimed to demonstrate the neuromodulatory properties of microbial‐derived GABA in an animal model of constipation and visceral pain and to show that GABAergic signaling via the microbiome‐gut‐brain axis is mechanistically linked to glutamate decarboxylation by the common human commensal bacterium, Bifidobacterium dentium.
2. Methods
2.1. Bacterial strains, cell lines, media, and culture conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Bifidobacteria were cultured in Reinforced Clostridium Medium (Oxoid, Mampshire, UK) or modified de Man, Rogosa and Sharpe (mMRS) medium,39 supplemented with 0.05% (w/v) l‐cysteine‐HCl (Sigma‐Aldrich, Steinhein, Germany) and 1% (w/v) of sodium glutamate. Strains were grown under anaerobic conditions in a Modular Atmosphere Controlled System (Davidson & Hardy Ltd., Dublin, Ireland) at 37°C. Lactobacillus sp. strains were grown at 37°C in mMRS medium or MRS medium supplemented with 0.05% (w/v) l‐cysteine‐HCl (Sigma‐Aldrich). Alistipes sp. strains were cultured in MTGE medium (Anaerobe Systems, Morgan Hill, CA, USA) or MTGE medium supplemented with 1% l‐glutamate (mMTGE). Escherichia coli strains were grown aerobically at 37°C in Luria Bertani (LB) on a rotary shaker (150 rpm). Where appropriate, media were supplemented with 5 μg/mL erythromycin (Erm) or 50 μg/mL to maintain plasmids in B. breve or E. coli, respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Relevant features | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli strains | ||
| E. coli XL1‐Blue | (supE44 hsdR17 recA1 gyrA96 thi relA1 lac F′ [proAB + lacl q lacZΔM15 Tn10(Tetr)]) | Stratagene |
| E. coli XL1‐Blue‐pQE‐GadB | XL1‐Blue harboring pQE60 derivative containing gadB | This study |
| E. coli XL1‐Blue‐pQE‐GadB‐T | XL1‐Blue harboring pQE60 derivative containing gadB, which has mutation of Threonine225 to alanine | This study |
| E. coli XL1‐Blue‐pQE‐GadB‐D | XL1‐Blue harboring pQE60 derivative containing gadB, which has mutation of Aspartate256 to alanine | This study |
| E. coli XL1‐Blue‐pQE‐GadB‐K | XL1‐Blue harboring pQE60 derivative containing gadB, which has mutation of Lysine289 to alanine | This study |
| Bifidobacterium sp. strains | ||
| B. dentium ATCC27678 | ATCC | |
| B. angulatum ATCC27535 | ||
| B. bifidum MIMBb13CS | ||
| B. bifidum MIMBb23SG | ||
| B. breve NCIMB8807(also known as UCC2003) | Isolate from nursling stool | NCIMB |
| B. breve NCIMB8807+pESH‐GadB | Recombinant B. breve NCIMB8807 harboring pESH40 derivative containing gadB | This study |
| Lactobacillus sp. strains | ||
| L. acidophilus | ||
| L. farciminis DSM | DSMZ | |
| L. gasseri ATCC33323 | ATCC | |
| L. plantarum NCIMB8826 | NCIMB | |
| L. plantarum ATCC14917 | ATCC | |
| L. reuteri ATCC23272 | ATCC | |
| L. reuteri DSM17932 | DSMZ | |
| L. reuteri ATCC PTA 6475 | ATCC | |
| L. reuteri 100‐23 | Su et al.34 | |
| L. rhamnosus ATCC53103 (GG) | ATCC | |
| Alistipes sp. strains | ||
| A. indistinctus DSM22520 | DSMZ | |
| A. finegoldii DSM17242 | DSMZ | |
| A. putredinis DSM17216 | DSMZ | |
| Plasmids | ||
| pQE60 | Ampr, IPTG‐inducible expression vector | De Ruyter et al.78 Qiagen |
| pQE‐gadB | Ampr, pQE60 derivative containing translational fusion of gadB‐encoding DNA fragment to IPTG‐inducible promoter | This study |
| pQE‐gadB‐A | Ampr, pQE60 derivative containing translational fusion of gadB‐encoding DNA fragment to IPTG‐inducible promoter | This study |
| pQE‐gadB‐B | Ampr, pQE60 derivative containing translational fusion of gadB‐encoding DNA fragment to IPTG‐inducible promoter | This study |
| pQE‐gadB‐C | Ampr, pQE60 derivative containing translational fusion of gadB‐encoding DNA fragment to IPTG‐inducible promote | This study |
| pESH46 | Emr, Ampr, repA−, ori+, cloning vector | Shkoporov et al.47 |
| pESHGadB | Emr, Ampr, pESH46 derivative containing translational fusion of gadB‐encoding DNA fragment | This study |
ATCC, American Type Culture Collection; DSMZ, German Collection of Microorganisms and Cell Cultures; JCM, Japan Collection of Microorganisms; NCIMB, National Collection of Industrial and Marine Bacteria; MIM, collection from University of Milan.
2.2. In silico analysis of human microbiome and metagenomic data
To determine the distribution and relative abundances (RAs) of the glutamate decarboxylase‐encoding gad genes at various sites across the human body, Human Microbiome Project (HMP)24 metabolic reconstruction profiles, as generated by the HUMAnN software package,40 were obtained from the HMP Data Analysis and Coordination Center (http://www.hmpdacc.org/HMMRC/). From these profiles, the RA of the KEGG ortholog for gadB (K01580), across subjects and body sites, was obtained.
To identify potential genomes within the stool microbiome containing glutamate decarboxylases, the Integrated Microbial Genomes (IMG)/HMP database (http://img.jgi.doe.gov)41 was used to search the annotations of the stool‐associated reference genomes from the HMP (most recent annotation updated 20 April 2016). These genome annotations were queried using the IMG pathway term for glutamate decarboxylation (PathwayID 390: Glutamate decarboxylation to 4‐aminobutyrate). This search returned 163 genes from 142 unique genomes. The genomes identified in this analysis were summarized at the genus level to characterize the distribution of putative glutamate decarboxylases among gut‐associated bacteria. The data were parsed to the species level for Bifidobacterium.
The RA of B. dentium in the intestinal tract of healthy children (n=22), young adults (n=22), and older adults (n=85) was evaluated by generating taxonomic profiles of metagenomic sequence libraries from the HMP,24 Metahit cohort,41 and a school‐aged pediatric cohort42. Raw sequence libraries were obtained from each project's respective SRA repository or project ftp site (HMP: ftp://public-ftp.hmpdacc.org/Illumina/, Metahit: ftp://public.genomics.org.cn/BGI/gutmeta/Raw_Reads/, Pediatric: SRA project accession PRJNA74951). Paired‐end libraries were concatenated into a single input file per subject, and each library was profiled using the Metaphlan software package (version 1.7.0)43 with the Bowtie244 search algorithm and “sensitive‐local” settings. The RA of B. dentium was compared across the three cohorts using a Kruskal‐Wallis H‐test and Tukey‐Kramer post hoc test, as employed by the STAMP software package.45
2.3. Screening for bacterial GABA production
Nine Lactobacillus sp., five Bifidobacterium sp., and four Alistipes sp. strains were screened for their ability to consume l‐glutamate and secrete GABA (Table 1). Lactobacilli and Bifidobacterium sp. strains were cultured in MRS or mMRS, while Alistipes sp. strains were cultured in MTGE and mMTGE. Growth medium was inoculated with 1% inoculum of overnight cultures (12 hours), followed by incubation anaerobically at 37°C for 48 hours. Cells were removed by centrifugation and cell‐free supernatants were subjected to filtration through 3 kDa MWCO filters (Amicon, Bedford, MA, USA), following LC‐MS analysis (Data S1) for the detection of secreted GABA and l‐glutamate.
2.4. In silico analysis of GadB structure
Sequence data were obtained from the non‐redundant sequence database accessible at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). The genome sequence of B. dentium ATCC 27678 (accession number: ABIX02000002.1) in combination with Artemis Informatics Software (Wellcome Trust Sanger Institute) was used for genome resource, display, and analysis. BLAST searches were performed using the National Center for Biotechnology Information website. Sequence analysis was performed using the VectorNTI software package. GadB 3D structure modeling was achieved by means of Phyre2 software (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). Visualization file was generated with Molmol.46
2.5. DNA manipulation
Chromosomal DNA was isolated from B. dentium ATCC 27678 using Qiagen All Prep kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. An initial lysis step involving cell resuspension in lysis buffer with 30 mg/mL lysozyme (Sigma, St Louis, MO, USA) and incubation at 37°C for 30 minutes was used. Plasmid DNA minipreps were obtained from E. coli using the QIAprep Spin Plasmid Miniprep kit (Qiagen) according to the manufacturer's instructions. Microbial genomic DNA was isolated from mouse and rat fecal samples using ZR Fecal DNA Miniprep Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer's instructions. Single‐stranded oligonucleotide primers used in this study were synthesized by Lone Star Labs. Standard PCR reactions were performed using Takara polymerase Takara Bio USA, Inc. (Clontech), Mountain View, CA, USA. PCR fragments were purified using Qiagen PCR purification kit (Qiagen). Electroporation of plasmid into B. breve NCIMB8807 was performed as described previously.47
2.6. Construction of plasmids pQEgadB and overexpression of recombinant GadB proteins
The entire coding region of gadB (gene locus BIFDEN01403) was amplified by PCR with genomic DNA from B. dentium ATCC 27678 serving as a template and primer pair GadBFw and GadBRv (Table 2). These primers allowed the incorporation of a His15‐encoding sequence into the 3′ end of gadB gene (designated here as recombinant gadB [rgadB]). The rgadB PCR product was amplified with Takara Ex Taq DNA polymerase (Takara Bio USA). Overlap extension PCR was used to create substitutions in three conserved amino acid residues in gadB from B. dentium to alanine (A). Therefore, six individual forward and reverse DNA fragments were designed with amino acid changes in the Threonine225, Aspartate256, and Lysine289 codon positions of the gadB gene with mutagenic primers (Table 2). The amplified DNA fragments were annealed to change amino acid residues to GCC, GCT, and GCG alanine variants and generate a full‐length gadB gene product to express in E. coli M15pREP4 cells. Wild‐type and mutated rgadB amplicons were cloned in the pSMARTGC HK cloning vector (Lucigen), transformed into E. coli 10G ELITE cells (Lucigen, Middleton, WI, USA), digested with NcoI and HindIII endonucleases (New England Biolabs, Ipswich, MA, USA), and ligated (T4 DNA ligase; New England Biolabs) with the expression vector pQE‐60 (Qiagen) which had been treated with the same enzymes and transformed into E. coli M15pREP4 electrocompetent cells. The selection of gadB transformants was accomplished by restriction digestion with NcoI and HindIII endonucleases and by sequencing of positive clones. The resulting plasmid with the wild‐type gadB was designated as pQEgadB. Recombinant GadB proteins (rGadB) were overexpressed and analyzed by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis as previously described.48
Table 2.
Oligonucleotide primers used in this study
| Primer name | Sequence code (5′→3′) |
|---|---|
| GadBFw | AGCTCTCCATGGCGATAATGCATTCCAACATCAa |
| GadBRv | AGCTCTAAGCTTTCAGTGATGGTGATGGTGATGGTGATGGTGATGGTGATGGTGATGGAAGCCGGAGACCCb |
| GadB‐T‐Fw | CATGGGCGTGGCCTACACGc |
| GadB‐D‐Fw | CCACGTGGCTGCCGCTTCGd |
| GadB‐K‐Fw | CGGACACGCGTACGGTCTGe |
| GadB‐T‐Rv | CGTGTAGGCCACGCCCATGf |
| GadB‐D‐Rv | CGAAGCGGCAGCCACGTGGg |
| GadB‐K‐Rv | CAGACCGTACGCGTGTCCGh |
| GadB‐XhoI‐Fw | CAACTCGAGGGAAACAGAAGGTTTGACAGi |
| GadB‐BamHI‐Fw | CTGGGATCCTCAGTGATGGAAGCCGj |
| Bd_gadB_pL_4_L | GCCAAGGCCTTCTCTAAGTTC |
| Bd_gadB_pL_4_R | CCACCTGATAGGCGGTCTC |
| bglC_pL_6_L | TGGAAGGACGAGGTGACTGk |
| bglC_pL_6_R | CTACGCGCTTCGGATCATk |
| h‐gabra1_pL_2_L | GGACAAACAGTAGACTCTGGAATTG |
| h‐gabra1_pL_2_R | TCAAGTGGAAATGAGTGGTCA |
| h‐gabrb2_pL_27_L | ATGGCACCGTCCTTTATGG |
| h‐gabrb2_pL_27_R | TGGGTACCTCCTTAGGTCCA |
| h‐gabbr1_pL_87_L | CATGGACGCTTATCGAGCA |
| h‐gabbr1_pL_87_R | GATCATCCTTGGTGCTGTCA |
| h‐gabbr2_pL_1_L | GACACCATCAGGTTCCAAGG |
| h‐gabbr2_pL_1_R | GATGCTGTAGAGAGGTAGGGAGA |
| h‐gabdh_pL_45_L | GAGTCCACTGGCGTCTTCAC |
| h‐gabdh_pL_45_R | TTCACACCCATGACGAACAT |
The underlined sequence corresponds to an NcoI site.
The underlined sequence corresponds to an HindIII site.
The underlined sequence corresponds to the introduced mutation site for Threonine.
The underlined sequence corresponds to the introduced mutation site for Aspartate.
The underlined sequence corresponds to the introduced mutation site for Lysine.
The underlined sequence corresponds to the introduced mutation site for Threonine.
The underlined sequence corresponds to the introduced mutation site for Aspartate.
The underlined sequence corresponds to the introduced mutation site for Lysine.
The underlined sequence corresponds to an XhoI site.
The underlined sequence corresponds to an BamHI site.
B. breve NCIMB8807 strain‐specific primers have been described previously by Pokusaeva et al.79
2.7. Enzyme assay and determination of glutamate decarboxylase activity by rGadB
In vitro enzymatic reactions with rGadB and its derivatives were performed as previously described by Hiraga et al.49 Cell lysates were prepared by sonicating bacterial pellets six cycles for 15 seconds and collected by centrifugation at 3,300 x g for 20 minutes at 4°C. After centrifugation, soluble and insoluble fractions were collected and separated on a 10% SDS polyacrylamide gel at 200 V for 30 minutes; 100 μL of soluble and insoluble fractions was suspended in equal volumes of 4 M ammonium sulfate (Sigma), and the resulting mixture was then incubated for 3 minutes. Subsequently, 1.3 mL of substrate solution, consisting of 20 mM sodium glutamate (Sigma), pyridoxal phosphate (PLP) cofactor (Sigma), and Pyridine‐HCl (Sigma), was added to the mixture, followed by an incubation at 37°C for 20 minutes. The mixture was then heat inactivated by boiling for 5 minutes to stop the decarboxylation reaction. Reaction mixtures were subsequently analyzed for the presence of GABA and glutamate using LC‐MS analysis (details of LC‐MS analysis are provided in Data S1).
2.8. Construction of recombinant B. breve NCIMB8807
Construction of B. dentium ATCC 27678 gadB plasmid was performed as follows. B. dentium ATCC 27678 genomic DNA served as a template to amplify gadB by PCR using gadB‐specific oligonucleotide primers (Table S2). The gadB PCR product was amplified with Takara Ex Taq DNA polymerase (Takara Bio USA), cloned in the TOPO 2.1‐TA cloning vector (Invitrogen, Carlsbad, CA, USA), transformed into E. coli Top10 cells (Invitrogen), digested with XhoI and BamHI endonucleases (New England Biolabs), and ligated (T4 DNA ligase; New England Biolabs) with the expression vector pESH46,47 which had been treated with the same enzymes. B. breve NCIMB8807 cells were transformed, as previously described50 with the ligated mixture, and several transformants were isolated. The selection of gadB transformants was accomplished by restriction digestion with NdeI and BamHI endonucleases and by sequencing of positive clones. Sequencing of all constructs was performed by Lonestar sequencing labs Inc. The resulting plasmid was designated as pESHgadB, and the strain was designated as B. breve NCIMB8807 pESHgadB.
The recombinant B. breve NCIMB8807 harboring pESHgadB plasmid was screened for its ability to secrete GABA as follows. The recombinant strain, wild‐type B. breve NCIMB8807 (negative control), and B. dentium ATCC 27678 (positive control) were grown in MRS medium supplemented with 0.05% (w/v) l‐cysteine‐HCl for 48 hours, and the cell free supernatants were analyzed using LC‐MS analysis (Data S1).
2.9. In vivo GABA measurement
2.9.1. Animals
Six‐week‐old, male, Swiss Webster mice (n=6–8 per group, 30 total) were purchased from Taconic Biosciences and were maintained under specific‐pathogen‐free (SPF) conditions. Mice were kept under filter‐top cages (3–4 mice per cage) and had free access to distilled water and LabDiet 5V5R rodent chow, containing 5.09% glutamate. All mouse experiments were performed in the SPF animal facility according to an Institutional Animal Care and Use Committee–approved mouse protocol at Baylor College of Medicine, Houston, TX.
2.9.2. Bifidobacterial strain administration
Bifidobacterium strains (B. dentium ATCC 27678, B. breve NCIMB 8807, and B. breve NCIMB8807 pESHgadB) were grown in culture conditions described above. Bacteria were harvested in early‐stationary phase (12 hours growth in mMRS medium or mMRS supplemented with 5 μg/mL Erm to maintain the plasmid in B. breve pESHgadB). OD600 of the culture was used to approximate cell viability based on previous growth curve data (Fig. S1). The culture volume required for approximately 8 × 108 CFU/mL was centrifuged at 5,000 x g for 5 minutes to pellet the bacterial cells. The cells were then washed in pre‐reduced PBS (PBS + 0.05% l‐cysteine) and re‐pelleted. The wash was performed to minimize the amount of GABA administered to the mice in the gavage mixture. The cells were suspended to a final concentration of 8 × 108 CFU/mL in PBS with 0.05% l‐cysteine and 1% sodium glutamate. Cell viability in the gavage mixture was also verified by dilution plating on MRS agar immediately before gavage procedure. Mice were administered 4 × 108 CFU/mL (in 0.5 mL) of B. dentium, B. breve, or B. breve pESH suspended in PBS with 0.05% l‐cysteine and 1% glutamate. Control mice received only sterile PBS with 0.05% l‐cysteine and 1% glutamate. Oral gavages were administered once per day for 5 days. Weight was monitored over the course of treatment, and mice were euthanized 24 hours after the last gavage. The GI tract was carefully removed. Luminal contents in the cecum were collected, flash frozen immediately in liquid nitrogen, and stored at −80°C until use.
2.9.3. Fecal water preparation
Samples of cecal content (50–80 mg) were manually homogenized in 1 mL of DEPC‐treated water by vigorous vortexing until slurry was produced. The mixture was centrifuged at 5000 g for 5 minutes. The supernatant (fecal water) was carefully decanted into 1.5 mL Eppendorf tubes and stored frozen (−20°C) until analysis; 100 μL of this preparation was used in subsequent GABA ELISA.
2.9.4. GABA measurement via ELISA
γ‐aminobutyric acid concentrations in cecum content were measured via GABA ELISA (LDN Immunoassays, Nordhorn, Germany) according to manufacturer instructions. Briefly, after extraction and derivatization, GABA was quantitatively determined by competitive ELISA using an anti‐rabbit IgG‐peroxidase conjugate with 3,3′,5,5′‐tetramethylbenzidine as a substrate. All samples and standards were run in duplicate, and the reaction was monitored at 450 nm. The resulting ng/mL values were background adjusted using 650 nm as a reference wavelength, and results were normalized to initial weight of sample.
2.10. Rat fecal retention model
2.10.1. Animals
Male Sprague‐Dawley rats (n=4 per group) of 225–250 g were used for this study. The Institutional Animal Care and Use Committee of the University of Texas Medical Branch at Galveston, TX, approved the study using Sprague‐Dawley rats. Rats were anesthetized with 2% isoflurane with an E‐Z Anesthesia System. A 20‐mm‐circumference glass rod was inserted into the rat anus. A purse‐string suture was placed in the external sphincter to keep the lumen of the anus around the rod, and then the rod was removed. The outlet occlusion was kept for 3 days. This treatment significantly reduced fecal output and led to lumen distention in the distal colon, which significantly increased colon‐specific sensory neuron excitability. Age‐matched control rats were treated similarly, except that the suture was removed immediately after being placed. B. dentium ATCC 27678 (4 × 108 CFU in 0.5 mL mMRS) was administered to control and FR rats via oral gavage daily for 5 days (2 days before and 3 days during fecal retention [FR]).
2.10.2. Retrograde fluorescence label injections
Labeling of colon‐specific dorsal root ganglia (DRG) neurons was performed as previously described.51 Under general 2% isoflurane anesthesia, the lipid soluble fluorescence dye, 1,1′‐dioleyl‐3,3,3′,3′‐tetramethylindocarbocyanine methanesulfonate (DiI, Invitrogen) (50 mg/mL), was injected into muscularis externae of the distal colon in 8–10 sites (2 μL each site). To prevent leakage, the needle was kept in place for 1 minute following each injection.
2.10.3. Dissociation of DRG neurons
Control and FR rats were euthanized by cervical dislocation, followed by decapitation. Lumbosacral (L6–S2) DRGs were collected in ice‐cold and oxygenated dissecting solution, containing (in mM) 130 NaCl, 5 KCl, 2 KH2PO4, 1.5 CaCl2, 6 MgSO4, 10 glucose, and 10 HEPES, pH 7.2 (305 mOsm), as described previously.51 After removal of the connective tissue, the ganglia were transferred to a 5 mL dissecting solution containing collagenase D (1.8 mg/mL; Roche, Indianapolis, IN, USA) and trypsin (1.0 mg/mL; Sigma) and incubated for 1.5 hours at 34.5°C. Dorsal root ganglia were then taken from the enzyme solution, washed, and put in 0.5 mL of the dissecting solution containing DNase (0.5 mg/mL; Sigma). Cells were subsequently dissociated by gentle trituration for 10–15 times with fire‐polished glass pipettes and placed on acid‐cleaned glass coverslips. The dissociated DRG neurons were kept in the dissecting solution for at least 1 hour in room temperature before recording.
2.10.4. Whole‐cell patch clamp recordings from dissociated DRG neurons
Before each experiment, the glass coverslip with DRG neurons was transferred to recording chamber perfused (1.5 mL/min) with external solution containing (in mM) 130 NaCl, 5 KCl, 2 KH2PO4, 2.5 CaCl2, 1 MgCl2, 10 HEPES, and 10 glucose, pH adjusted to 7.4 with NaOH (300 mOsm) at room temperature. Recording pipettes, pulled from borosilicate glass tubing, with resistance of 1–5 MΩ, were filled with solution containing (in mM): 100 KmeSO3, 40 KCl, and 10 HEPES, pH 7.25 adjusted with KOH (290 mOsm). DiI‐labeled neurons were identified under fluorescent microscope. Whole‐cell currents and voltage were recorded from DiI‐labeled neurons using Dagan 3911 patch clamp amplifier. Data were acquired and analyzed by pCLAMP 9.2 (Molecular Devices, Sunnyvale, CA, USA). The currents were filtered at 2–5 kHz and sampled at 50 or 100 μs per point. While still under voltage clamp, the Clampex Membrane Test program (Molecular Devices) was used to determine membrane capacity, Cm and membrane resistance, Rm, during a 10 ms, 5 mV depolarizing pulse form a holding potential of −60 mV. The configuration was then switched to current clamp (0 pA) for determining other electrophysiological properties. After stabilizing for 2–3 minutes, resting membrane potential (RMP) was measured. The minimum acceptable RMP was −40 mV. Between 20 and 24 neurons from four rats in each group were used for analysis (methods employed for Liquid Chromatography coupled with Mass Spectrometry are provided in Data S1).
3. Results
3.1. Microbial glutamate decarboxylase (gadB) is enriched in human intestine
GadB is the primary bacterial enzyme responsible for generating bioactive, microbe‐derived GABA. The availability of metagenomic data from the HMP allowed us to determine the relative abundance (RA) of gadB in the microbial genomes found at different body sites in 96 healthy adult individuals. Our analysis demonstrates that although bacterial gadB is present at multiple body sites, it is most abundant in adult human stool specimens (Fig. 1A). Our results indicate that the gadB gene signature is present at nearly three‐fold greater percentage in the stool microbiome vs the microbiomes at other body sites. This result is not surprising, as the glutamate decarboxylase system is known for its role in acid‐resistance, and contributes to bacterial survival during transit through the GI tract.
Figure 1.
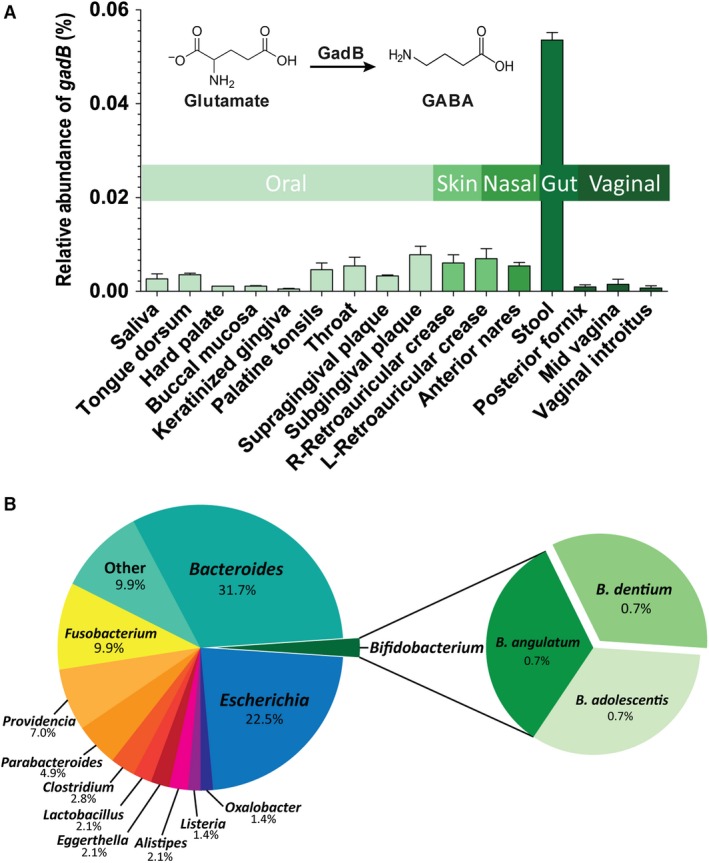
Microbial glutamate decarboxylase gene (gadB) in the human microbiome. (A) The relative abundance of the gadB gene among different body sites in 96 healthy adult individuals is depicted as a bar graph. The vertical bars indicate the mean relative abundances (and body site) of gadB (±SEM). The chemical structure of glutamate and GABA and conversion of glutamate to GABA by GadB are shown within the graph. The colored horizontal bars indicate body sites. (B) Bacterial genera/species of the human gut microbiome harboring putative glutamate decarboxylases are depicted as a pie chart. The prevalence of glutamate decarboxylases among the members of the healthy human gut microbiome was estimated from data deposited at the Integrated Microbial Genomes/Human Microbiome Project (IMG/HMP) database (http://img.jgi.doe.gov). Percentages displayed represent genus‐level distribution among these genomes, with species‐level distribution shown for the Bifidobacterium species
The RA of gadB in stool prompted us to investigate which microbes harbor glutamate decarboxylases (Fig. 1B). The prevalence of glutamate decarboxylases among the members of the healthy human gut microbiome was estimated from data deposited at the IMG/HMP database (http://img.jgi.doe.gov). From this database, genomes associated with the human GI tract were selected for analysis. The genomes of 26 unique genera were found to possess glutamate decarboxylase orthologs, including intestinal bacteria such as Bacteroides spp. (RA, 31.7%), Escherichia spp. (RA, 22.5%), and Fusobacterium spp. (RA, 9.9%). Interestingly, the commensal bacteria B. dentium, which contains a gadB gene, was found to be present at a RA of 0.7% (Fig. 1B). Thus, microbial GadB is a potentially rich enzymatic source of bioactive GABA in the gut lumen, and this species represents a model organism for GABA‐producing microbes.
3.2. GadB‐derived GABA secretion by B. dentium
In order to quantify GABA production by specific intestinal bacterial strains, 16 different commensal microorganisms were screened for GABA production capacity by LC‐MS. Based on the metagenomics‐based screening, several human‐derived isolates of Alistipes spp. and Bifidobacterium spp. (including B. dentium) were selected, along with various commensal and probiotic species (Fig. 2A). From this microbial screening library, only four strains actively secreted increased quantities of GABA into MRS medium supplemented with glutamate (Fig. 2A), including L. plantarum NCIMB8826, L. plantarum ATCC14917, B. angulatum ATCC27535, and B. dentium ATCC 27678. This B. dentium strain was the only significant GABA secretor (P<0.0001) with the greatest amount of GABA produced among this group, and more than 1 mg/mL of GABA detected in vitro. Our efforts, therefore, focused on characterizing the mechanisms of GABA production in this human‐derived commensal strain.
Figure 2.
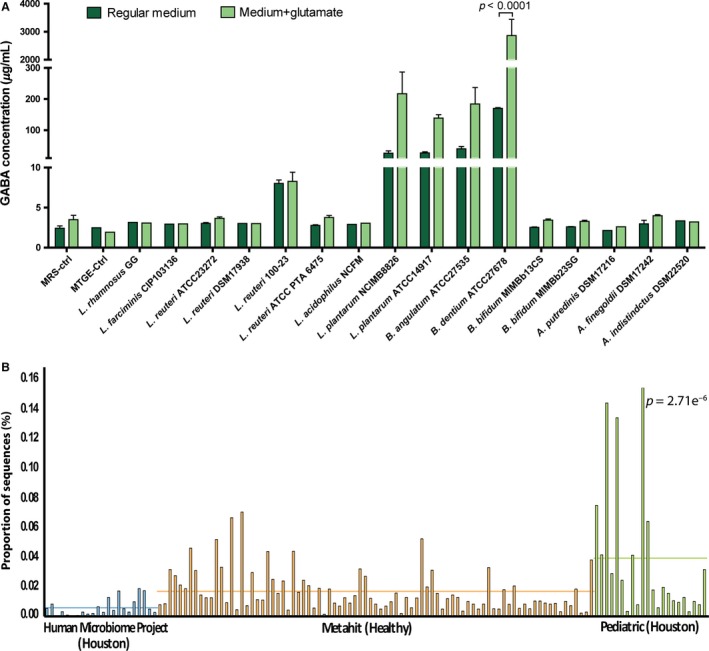
GABA production by commensal intestinal strains and Bifidobacterium dentium in the human gut microbiome. (A) Screening of 16 different intestinal commensal and/or probiotic isolates identified B. dentium ATCC 27678 as a major GABA producer. GABA concentrations were measured using LC‐MS after 48 hours of anaerobic growth at 37°C in either regular MRS (dark green) or MRS medium supplemented with 1% w/v glutamate (light green). Error bars represent standard error of 3 independently performed experiments. B. dentium was the only species that produced significantly more GABA (P<.0001; two‐way ANOVA with Bonferroni correction for multiple comparisons). (B) B. dentium in the healthy human gut microbiome, as detected by Metaphlan profiling of shotgun metagenomic sequence libraries. Different colors represent subject cohorts, while bars show proportion of sequences with hits to B. dentium found in each individual. The horizontal bars represent the average relative abundance (as determined by the proportion of sequences with hits to B. dentium) in each cohort
Taxonomic profiling of metagenomic sequencing data suggests that B. dentium may account for up to 0.18% (median value across all subjects evaluated) of the healthy human gut microbiome (Fig. 2B). The observed enrichment in pediatric samples (P=2.71e−6) may be partially explained by the increased overall abundance of Bifidobacterium species typically detected in children relative to adult samples.42 To characterize the dynamics of GABA production by B. dentium ATCC 27678, its growth in MRS medium, glutamate consumption, and GABA secretion were monitored over 72 hours (Fig. S1). Unlike B. bifidum MIMBb13CS, which lacks the gadB gene, B. dentium secreted GABA by consuming glutamate in a stationary phase environment associated with low pH. These results confirmed and extended prior reports that B. dentium is a human‐associated GABA‐producing microbe.27, 28
To confirm that GadB is the enzyme responsible for GABA production in B. dentium, we generated recombinant His‐tagged wild‐type and mutant GadB proteins to characterize the catalytic mechanism of glutamate decarboxylation. Homology modeling of GadB in B. dentium identified three conserved active site amino acids52: lysine (K289), threonine (T225), and aspartic acid (D256) (Fig. 3A). Each of these amino acids was separately mutated to alanine, and relative enzyme activities were measured in the presence of glutamate and PLP cofactor (Fig. 3B). Using this quantitative approach, wild‐type GadB converted approximately 80% of the substrate, glutamate, into GABA, whereas mutant proteins were deficient in enzymatic activity. Furthermore, to test whether GadB is sufficient for GABA production by bifidobacteria, gadB was cloned into the Bifidobacterium‐E. coli shuttle vector pESH46,47 transformed into B. breve NCIMB8807, and analyzed for GABA production. Since it is currently not possible to genetically delete gadB in B. dentium, we chose this highly transformable B. breve strain because it lacks gadB and does not produce GABA.53 Constitutively expressed recombinant GadB in B. breve harboring pESHgadB produced GABA in comparable amounts to B. dentium in vitro (Fig. 3C), demonstrating that GadB functions as a glutamate decarboxylase in B. dentium with conserved catalytic amino acid residues involved in PLP cofactor binding and glutamate decarboxylase activity.
Figure 3.
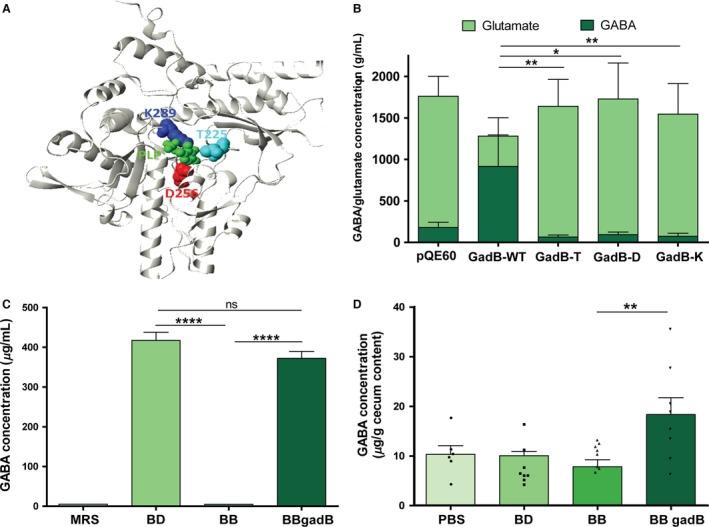
In vitro and in vivo activity of glutamate decarboxylase (GadB) from Bifidobacterium dentium. (A) GadB 3D structure with its proposed active site highlighted. Dark blue color depicts catalytic lysine at position 289 (K289), while threonine (T225) and aspartate (D256) are colored in light blue and red, respectively. Position of co‐factor pyridoxal phosphate, PLP, is shown in green. (B) Site directed mutagenesis effect on recombinant GadB activity. pQE60—negative control of crude extract from Escherichia coli strain harboring empty pQE60 vector. GadB‐WT—crude extract with recombinant wild‐type GadB overexpressed in E. coli. GadB‐T, GadB‐D, and GadB‐K are crude extracts of recombinant GadB with mutated amino acids from T225, D256, and K289 to alanine, respectively. Bars demonstrate GABA (dark green) or l‐glutamate (light green) concentration. Error bars represent standard error of 3 independently performed experiments (*P<.01, **P<.05; one‐way ANOVA of log transformed data, Bonferroni correction for multiple comparisons). (C) Expression of GadB from B. dentium in B. breve by complementation. gadB from B. dentium ATCC 27678 (BD) was cloned into the pESH46 (pESHgadB) expression vector and transformed into B. breve NCIMB8807 (BB) allowing for constitutive expression (BBgadB). GABA was measured via LC‐MS method (Data S1). Error bars represent standard error of 3 independently performed experiments (****P<.0001, ns=not significant; one‐way ANOVA of log transformed data, Bonferroni correction for multiple comparisons). (D) Six‐week‐old male Swiss Webster mice (n=6–8 per group) were orally administered 1% glutamate plus B. dentium ATCC 27678 (BD), B. breve NCIMB8807 (BB), B. breve NCIMB8807 pESHgadB (BBgadB), or saline (PBS) for 5 days. Mice administered B. breve pESHgadB had significantly more GABA in their cecal content as measured by ELISA (**P<.01; one‐way ANOVA with Bonferroni correction)
3.3. Microbial GABA production in vivo is mediated by GadB
To confirm that GadB‐mediated GABA production occurs in an in vivo setting as well as in vitro, we used a murine model to examine how host GABA concentrations are affected by short‐term colonization with these Bifidobacterium strains (Fig. 3D). Six‐week‐old male Swiss Webster mice (n=6–8 per group) were orally administered 1% glutamate plus B. dentium, B. breve, B. breve pESHgadB, or saline (PBS) for 5 days. This oral gavage mimicked the natural route of gut microbial colonization. Inter‐subject variation was observed in each group; however, mice administered B. breve pESHgadB had significantly more GABA in their cecal content than mice that received non‐modified B. breve (P<0.01; 18.4 ± 3.4 vs 7.8 ± 1.4 μg/g cecal content). A trend toward increased GABA production was also observed in the mice that received B. dentium relative to those that received B. breve (10.1 ± 2.5 vs 7.8 ± 1.4 μg/g cecal content); however, GABA concentrations in the PBS‐treated mice were also within a similar range (10.3 ± 1.7 μg/g cecal content).
Increased GABA production in cecal content of B. dentium colonized mice has also been demonstrated in previous pilot studies using mice pretreated with antibiotics (Fig. S2). A trend toward increased GABA production was observed in mice receiving gadB‐positive B. dentium compared with the gadB‐negative B. breve (P>0.05; 14.2 ± 2.5 vs 5.4 ± 0.9 μg/g cecal content; n=6 per group). The increased GABA accumulation was not due to altered survival or transit of the bacteria since similar bacterial counts were measured in extracted cecal content (P>0.05; 1.7 × 107 and 6.7 × 107 copies of B. dentium and B. breve, respectively; n=6 per group) (Fig. S2). Taken altogether, these data demonstrate that the gadB gene from B. dentium ATCC 27678 is sufficient to facilitate GABA production in vivo when cloned into other bacterial species, and that expression of the microbial gadB gene in vivo can result in significant increases in host intestinal GABA concentrations.
3.4. B. dentium ATCC 27678 desensitizes sensory neuron activity in a rat model of visceral hypersensitivity
To determine if GABA‐producing bifidobacterial strains have beneficial effects on abdominal pain in FR‐associated constipation, we induced FR in rats by partial occlusion of the external anal sphincter as previously described.54 This procedure results in abdominal distention, constipation, and visceral pain, as confirmed by behavioral and electrophysiological studies.54, 55 After daily oral gavage of 4 × 108 B. dentium or B. breve for 5 days, electrophysiologic patch clamp recordings of retro‐labeled colon‐specific afferent neurons in the dorsal root ganglion (DRG) were used as a measure of afferent neuron sensitivity56 (Fig. 4A–L). This technique is well established as a surrogate indicator of visceral sensitization and abdominal pain in animal models.54, 57 Colon‐specific DRG neurons in FR rats treated with gadB‐negative B. breve (n=4 rats, n=24 neurons) yielded significant depolarization of RMP (P<0.001; −48.33 ± 0.48 vs −56.19 ± 0.46), decreased rheobase (P<0.001; 0.16 ± 0.01 vs 0.25 ± 0.01), and an increased number of action potentials evoked at 2× rheobase (P<0.01; 1.38 ± 0.12 vs 1.00 ± 0.0) and 3× rheobase (P<0.001; 2.25 ± 0.20 vs 1.33 ± 0.11), compared with neurons from the sham B. breve‐treated rats (n=4 rats, n=21 neurons). Importantly, these differences were largely absent in neurons from rats with FR receiving GABA‐producing B. dentium. There was no significant difference in rheobase (0.25 ± 0.006 vs 0.23 ± 0.007), or the number of action potentials at 2× rheobase (1.05 ± 0.05 vs 1.17 ± 0.08) or 3× rheobase (1.25 ± 0.1 vs 1.5 ± 0.12) in DRG neurons from sham B. dentium‐treated rats (n=4 rats, 20 neurons) when compared with FR B. dentium‐treated rats (n=4 rats, n=24 neurons). Fecal retention and treatment with Bifidobacterium species by itself did not alter other electrophysiologic characteristics of DRG neurons, including cell diameter, capacitance, input resistance, action potential amplitude, duration, threshold, or overshoot (P>0.05 between sham and FR rats treated with both B. dentium and B. breve; n=4 rats and 20–24 neurons recorded per group; Fig. 4E–L). Furthermore, similar numbers of both Bifidobacterium strains were detected in stool specimens from treatment and control animals (Fig. S3) and demonstrated a corresponding trend toward increased GABA in B. dentium‐treated animals only. These data indicate that colonic‐specific sensory neurons are hypersensitized following luminal distention associated with FR, and this hyperexcitability phenotype is inhibited by GABA‐producing, gadB‐positive B. dentium and not by a genetically similar non‐GABA producer.
Figure 4.
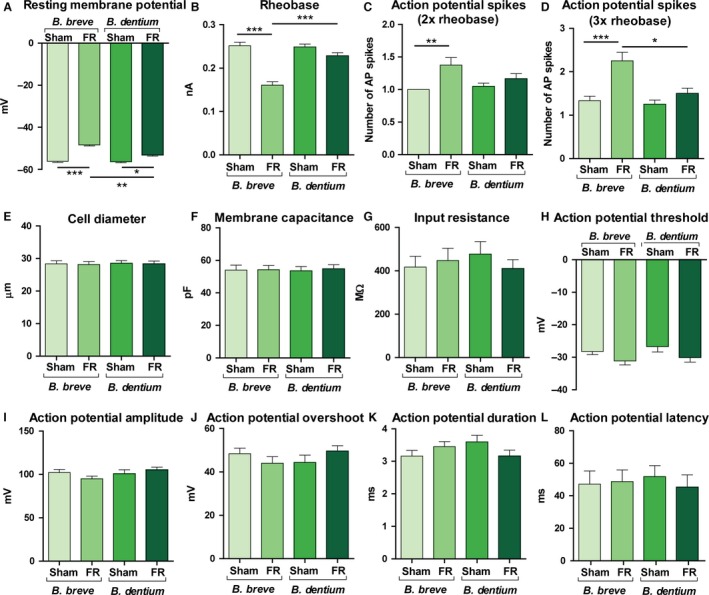
Neuromodulatory effects of GABA‐producing Bifidobacterium dentium ATCC 27678 administration on colonic sensory neuron activity. Control (Sham) and fecal retention (FR) rats were gavaged daily with GABA‐producing, gadB‐positive B. dentium or gadB‐negative B. breve strains (n=4 rats and 20–24 neurons per treatment group). Colon‐specific DRG neurons were isolated and used for the measurements of cell excitability by patch clamp recordings. The following parameters are displayed: (A) Resting membrane potential (RMP), (B) rheobase, (C) action potential spikes at 2× rheobase, (D) action potential spikes at 3× rheobase, (E) cell diameter, (F) membrane capacitance, (G) input resistance, (H) action potential threshold, (I) action potential amplitude, (J) action potential overshoot, (K) action potential duration, (L) action potential latency. Bars represent mean values with standard error (*P<.05, **P<.01, ***P<.001; Kruskal‐Wallis with Dunn correction for multiple comparison)
4. Discussion
We have shown that visceral hypersensitivity may be modulated by select GABA‐producing members of the gut microbiota, implicating neurotransmitter production by the microbiota as a means of microbiome‐gut‐brain communication. Many amino acids can be metabolized by microbes to form biologically active amine compounds via decarboxylation. These reactions consume intracellular protons, resulting in increased pH within bacterial cells. In this way, the decarboxylation of amino acids (including glutamate) plays a central role in acid stress resistance among enteric58 and lactic acid bacteria.33, 34, 35 Protection against the acidity of the mammalian stomach and acidic microenvironments within the intestinal lumen helps these bacteria survive and even colonize their host.
Amino acid decarboxylation can also generate strong proton motive forces in bacterial cells, which can increase ATP production, and metabolic activity by these cells. Glutamate decarboxylase (GAD or GadB, EC 4.1.1.15) catalyzes the irreversible α‐decarboxylation of glutamate (glutamic acid) to produce GABA. The gadB gene is present in numerous bacterial species residing within the GI tract, and we have demonstrated that microbial GadB is a potentially rich enzymatic source of bioactive GABA from the gut lumen. A small subset of bacteria that are especially adept at this particular enzymatic reaction may indeed be sufficient to elicit large changes in the host metabolome.
Our metagenomic analysis confirmed the presence of B. dentium in the gut microbiome of healthy children and adults. Bacterial screening revealed B. dentium ATCC 27678 as the major GABA‐producer among those strains encoding the gadB gene. Bifidobacteria have been recently hypothesized and demonstrated to produce GABA27, 28; however, this is the first in‐depth study that investigates GABA secretion and its possible effect on the host. B. dentium is a representative taxon of the healthy human gut microbiome,24 and strain ATCC 27678 was isolated from healthy human feces. Analysis of metagenomic datasets demonstrated that B. dentium is common in healthy pediatric and adult stool specimens, with a relative enrichment in children suggesting that microbe‐derived GABA signaling may be developmentally important during childhood.
With our finding that microbial gadB is highly enriched in human stool, relative to other body sites, we explored the possibility that local GABAergic signals originating from the GI tract regulate gut‐brain function. We have shown that the addition of the microbial gadB gene in vivo can significantly increase luminal GABA concentrations in the intestine. Since GABA is a well‐characterized inhibitory neurotransmitter involved in central and visceral pain perception, we chose to examine the physiologic effects of B. dentium‐associated GABA in a rat FR model of visceral sensitivity.54, 55 Here, we provide the first experimental demonstration of a direct link between a GABA‐producing microbe and neuromodulation of colonic sensory afferents in an animal model of constipation and visceral pain. We have shown that sensory afferents are less sensitive to the colonic distension in the presence of B. dentium, as is demonstrated by the increased rheobase, and fewer number of action potential spikes. This would indicate that these neurons are less inclined to reach the depolarization threshold, suggesting that nociception is diminished. With the findings in this study, we suggest that GABA‐producing bacterial strains may be potential probiotics, benefitting visceral pain in these conditions.
Neuromodulatory signals that emanate from luminal microorganisms and influence gut‐brain function have been studied extensively during the past decade. Proposed mechanisms include activation of Toll‐like59, 60 and histamine receptors,61 as well as short‐chain fatty acids,62 but these signal transduction pathways likely do not target neuronal synapses exclusively. In vitro studies support the notion that intestinal commensals have the capacity to secrete bioactive compounds and neurotransmitters, such as histamine,61 nitric oxide,63 hydrogen sulfide,64 and GABA,27 at concentrations that can directly regulate neuronal activity. Changes in the gut microbiome also result in fluctuations of amino acid precursors to serotonin, an important neurotransmitter in the enteric and CNSs.65, 66 Serotonin is released by the population of enterochromaffin cells with consequent effects on enteric neurotransmission and gut motility.67 By responding to host‐derived catecholamines, pathogenic E. coli elegantly provided an additional example of communication between the enteric nervous system and the gut microbiome.68 However, production of these neuroactive compounds in vivo is currently understudied, and the functional significance of their effects on the host system is not well understood.
We provide in vivo evidence that visceral hypersensitivity in response to FR is preferentially inhibited by microbes that produce GABA, possibly by direct modulation of GABAergic signaling via colonic afferent neurons. Previous reports localized GABAA and GABAB receptors on murine and human enteric neurons, and these cells are optimally positioned to receive microbial‐derived GABA signals.69, 70 As reported by other groups,30 this receptor distribution may coordinate GABAergic regulation of intestinal motility,71 gastric emptying,72 gastric acid secretion,69 and transient lower esophageal sphincter relaxation.73 Interestingly, a recent in vitro study showed that the GABA agonist baclofen decreased DRG neuron activity by acting on GABAB receptors and inhibiting both low‐voltage activated and high‐voltage activated Ca2+ currents.74 γ‐aminobutyric acid has also recently been shown to act via the GABAB1 receptor on peripheral nociceptive terminals to modulate nociceptor sensitization and counteract inflammatory pain via interaction with the pain receptor, TRPV1.75 In our current study, we demonstrate that a GABA‐producing species, but not a closely related non‐GABA‐producing species of commensal gut bacteria, can modulate the hyperexcitability of peripheral somatosensory neurons in response to FR. Taken altogether, these findings provide additional support for the role of GABA in visceral nociception and reinforce the notion that GABA receptors on peripheral somatosensory neurons may be targeted for regulation of pain signaling. In future studies, we will examine GABA receptor localization on sensory colonic neurons and the mechanistic role of this signaling pathway in modulation of visceral hypersensitivity by B. dentium. Future directions also include using isotope‐labeled glutamate to track microbial conversion of glutamate to GABA and assessing host utilization of GABA. Studies are needed to carefully delineate whether other microbial GABA targets colonic afferents directly, or whether indirect neuromodulatory signals activate receptors on other cell types, such as enteroendocrine cells.
In conclusion, metagenomic data analysis of diverse human microbial communities identified genes involved in GABA synthesis as being particularly enriched in the intestine. Qualitative metabolomic studies supported this observation by reporting GABA in human and mouse luminal contents,76, 77 but a direct demonstration of microbial production in the gut was missing. Here, we identify the common commensal microbe, B. dentium, as an active GABA producer within the human gut microbiome. The presence of the glutamate decarboxylase encoding gene, gadB, in this species provides the genetic basis for glutamate to GABA conversion. By modulating the hyperexcitability of peripheral somatosensory neurons, candidate probiotic B. dentium and other GABA‐producing gut microbes may represent future therapeutics for recurrent abdominal pain and functional bowel disorders.
Figures S1, S2, and S3, as well as method employed for liquid chromatography coupled with mass spectrometry, are provided in Data S1.
Funding
This work was supported by grants from the National Institute of Diabetes, Digestive, and Kidney Diseases (UH3 DK083990 and R01 DK065075 to JV, R21DK096323 to TS, and R01 DK082563 to XZS), the National Center for Complementary and Alternative Medicine (RO1 AT004326 to JV), the National Cancer Institute (U01 CA170930 to JV), and R01DK56338 funded from the National Institute of Allergy and Infectious Diseases (TS).
Disclosure
J. Versalovic received unrestricted research support from Biogaia AB (Stockholm, Sweden). T. Savidge received research support from Merck.
Author Contribution
KP, CJ, BL, GU, YF, NO, RM, YMA, EH, SD, XZS, and DE substantially contributed to the design and acquisition of experiments, as well as analysis and interpretation of the data. They also contributed to drafting the work and revising it critically for important intellectual content; ML and AM substantially contributed to the acquisition of some experiments as well as analysis of the data; TS and JV contributed to study concept and design, analysis and interpretation of the data, and drafting of the manuscript, while JV supervised the conduct of these studies.
Supporting information
Acknowledgements
We thank Dr. Shkoporov for generously providing us with the plasmid pESH46. We thank Anne Hall for critical review of the manuscript and technical assistance in the data analysis.
References
- 1. Bercik P, Collins SM, Verdu EF. Microbes and the gut‐brain axis. Neurogastroenterol Motil. 2012;24:405–413. [DOI] [PubMed] [Google Scholar]
- 2. Forsythe P, Kunze WA. Voices from within: gut microbes and the CNS. Cell Mol Life Sci. 2013;70:55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reid G. Neuroactive probiotics. BioEssays. 2011;33:562. [DOI] [PubMed] [Google Scholar]
- 4. Lyte M. Probiotics function mechanistically as delivery vehicles for neuroactive compounds: microbial endocrinology in the design and use of probiotics. BioEssays. 2011;33:574–581. [DOI] [PubMed] [Google Scholar]
- 5. Cryan JF, Dinan TG. Mind‐altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. [DOI] [PubMed] [Google Scholar]
- 6. Cryan JF, O'Mahony SM. The microbiome‐gut‐brain axis: from bowel to behavior. Neurogastroenterol Motil. 2011;23:187–192. [DOI] [PubMed] [Google Scholar]
- 7. van De Sande MM, van Buul VJ, Brouns FJ. Autism and nutrition: the role of the gut‐brain axis. Nutr Res Rev. 2014;27:199–214. [DOI] [PubMed] [Google Scholar]
- 8. Unger MM, Ekman R, Bjorklund AK, Karlsson G, Andersson C, Mankel K, et al. Unimpaired postprandial pancreatic polypeptide secretion in Parkinson's disease and REM sleep behavior disorder. Mov Disord. 2013;28:529–533. [DOI] [PubMed] [Google Scholar]
- 9. Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108:16050–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rousseaux C, Thuru X, Gelot A, Barnich N, Neut C, Dubuquoy L, et al. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat Med. 2007;13:35–37. [DOI] [PubMed] [Google Scholar]
- 11. Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety‐like behavior and central neurochemical change in germ‐free mice. Neurogastroenterol Motil. 2011;23:255–264, e119. [DOI] [PubMed] [Google Scholar]
- 12. Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108:3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishino R, Mikami K, Takahashi H, Tomonaga S, Furuse M, Hiramoto T, et al. Commensal microbiota modulate murine behaviors in a strictly contamination‐free environment confirmed by culture‐based methods. Neurogastroenterol Motil. 2013;25:521–528. [DOI] [PubMed] [Google Scholar]
- 14. Dupont HL. Review article: evidence for the role of gut microbiota in irritable bowel syndrome and its potential influence on therapeutic targets. Aliment Pharmacol Ther. 2014;39:1033–1042. [DOI] [PubMed] [Google Scholar]
- 15. Collins SM. A role for the gut microbiota in IBS. Nat Rev Gastroenterol Hepatol. 2014;11:497–505. [DOI] [PubMed] [Google Scholar]
- 16. Zhou Q, Verne GN. New insights into visceral hypersensitivity–clinical implications in IBS. Nat Rev Gastroenterol Hepatol. 2011;8:349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whorwell PJ, Altringer L, Morel J, Bond Y, Charbonneau D, O'Mahony L, et al. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am J Gastroenterol. 2006;101:1581–1590. [DOI] [PubMed] [Google Scholar]
- 18. McKernan DP, Fitzgerald P, Dinan TG, Cryan JF. The probiotic Bifidobacterium infantis 35624 displays visceral antinociceptive effects in the rat. Neurogastroenterol Motil. 2010;22:1029–1035, e268. [DOI] [PubMed] [Google Scholar]
- 19. Francavilla R, Miniello V, Magista AM, De Canio A, Bucci N, Gagliardi F, et al. A randomized controlled trial of Lactobacillus GG in children with functional abdominal pain. Pediatrics. 2010;126:e1445–e1452. [DOI] [PubMed] [Google Scholar]
- 20. Nobaek S, Johansson ML, Molin G, Ahrne S, Jeppsson B. Alteration of intestinal microflora is associated with reduction in abdominal bloating and pain in patients with irritable bowel syndrome. Am J Gastroenterol. 2000;95:1231–1238. [DOI] [PubMed] [Google Scholar]
- 21. Kamiya T, Wang L, Forsythe P, Goettsche G, Mao Y, Wang Y, et al. Inhibitory effects of Lactobacillus reuteri on visceral pain induced by colorectal distension in Sprague‐Dawley rats. Gut. 2006;55:191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ait‐Belgnaoui A, Han W, Lamine F, Eutamene H, Fioramonti J, Bueno L, et al. Lactobacillus farciminis treatment suppresses stress induced visceral hypersensitivity: a possible action through interaction with epithelial cell cytoskeleton contraction. Gut. 2006;55:1090–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Verdu EF, Bercik P, Verma‐Gandhu M, Huang XX, Blennerhassett P, Jackson W, et al. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut. 2006;55:182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Li W, Liu X, Cao Y. gadA gene locus in Lactobacillus brevis NCL912 and its expression during fed‐batch fermentation. FEMS Microbiol Lett. 2013;349:108–116. [DOI] [PubMed] [Google Scholar]
- 27. Barrett E, Ross RP, O'Toole PW, Fitzgerald GF, Stanton C. gamma‐Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol. 2012;113:411–417. [DOI] [PubMed] [Google Scholar]
- 28. Liu S, Zhao L, Ren F, Sun E, Zhang M, Guo H. Complete genome sequence of Bifidobacterium adolesentis BBMN23, a probiotic strain from healthy centenarian. J Biotechnol. 2015;198:44–45. [DOI] [PubMed] [Google Scholar]
- 29. Wong CG, Bottiglieri T, Snead OC 3rd. GABA, gamma‐hydroxybutyric acid, and neurological disease. Ann Neurol. 2003;54(Suppl 6):S3–S12. [DOI] [PubMed] [Google Scholar]
- 30. Hyland NP, Cryan JF. A gut feeling about GABA: focus on GABA(B) receptors. Front Pharmacol. 2010;1:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen CY, Bonham AC. Postexercise hypotension: central mechanisms. Exerc Sport Sci Rev. 2010;38:122–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jin Z, Mendu SK, Birnir B. GABA is an effective immunomodulatory molecule. Amino Acids. 2013;45:87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Small PL, Waterman SR. Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli . Trends Microbiol. 1998;6:214–216. [DOI] [PubMed] [Google Scholar]
- 34. Su MS, Schlicht S, Ganzle MG. Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb Cell Fact. 2011;10(Suppl 1):S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sanders JW, Leenhouts K, Burghoorn J, Brands JR, Venema G, Kok J. A chloride‐inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol. 1998;27:299–310. [DOI] [PubMed] [Google Scholar]
- 36. Cotter PD, Gahan CG, Hill C. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol Microbiol. 2001;40:465–475. [DOI] [PubMed] [Google Scholar]
- 37. Feehily C, Karatzas KA. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J Appl Microbiol. 2013;114:11–24. [DOI] [PubMed] [Google Scholar]
- 38. Sze PY. L‐Glutamate decarboxylase. Adv Exp Med Biol. 1979;123:59–78. [DOI] [PubMed] [Google Scholar]
- 39. De Man JC, Rogosa A, Sharpe ME. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 40. Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Pillay M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42(Database issue):D560–D567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hollister EB, Riehle K, Luna RA, Weidler EM, Rubio‐Gonzales M, Mistretta TA, et al. Structure and function of the healthy pre‐adolescent pediatric gut microbiome. Microbiome. 2015;3:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade‐specific marker genes. Nat Methods. 2012;9:811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Langmead B, Salzberg SL. Fast gapped‐read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. [DOI] [PubMed] [Google Scholar]
- 46. Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55, 29‐32. [DOI] [PubMed] [Google Scholar]
- 47. Shkoporov AN, Efimov BA, Khokhlova EV, Kafarskaia LI, Smeianov VV. Production of human basic fibroblast growth factor (FGF‐2) in Bifidobacterium breve using a series of novel expression/secretion vectors. Biotechnol Lett. 2008;30:1983–1988. [DOI] [PubMed] [Google Scholar]
- 48. Pokusaeva K, O'Connell‐Motherway M, Zomer A, Fitzgerald GF, Van Sinderen D. Characterization of two novel alpha‐glucosidases from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2009;75:1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hiraga K, Ueno Y, Oda K. Glutamate decarboxylase from Lactobacillus brevis: activation by ammonium sulfate. Biosci Biotechnol Biochem. 2008;72:1299–1306. [DOI] [PubMed] [Google Scholar]
- 50. Maze A, O'Connell‐Motherway M, Fitzgerald GF, Deutscher J, Van Sinderen D. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2007;73:545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winston JH, Xu GY, Sarna SK. Adrenergic stimulation mediates visceral hypersensitivity to colorectal distension following heterotypic chronic stress. Gastroenterology. 2010;138:294–304 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim S‐H, Shin B‐H, Kim Y‐H, Nam S‐W, Jeon S‐J. Cloning and expression of a full‐length glutamate decarboxylase gene from Lactobacillus brevis BH2. Biotechnol Bioprocess Eng. 2007;12:707–712. [Google Scholar]
- 53. O'Connell Motherway M, O'Driscoll J, Fitzgerald GF, Van Sinderen D. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol. 2009;2:321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heredia DJ, Grainger N, McCann CJ, Smith TK. Insights from a novel model of slow‐transit constipation generated by partial outlet obstruction in the murine large intestine. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1004–G1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shi XZ, Lin YM, Powell DW, Sarna SK. Pathophysiology of motility dysfunction in bowel obstruction: role of stretch‐induced COX‐2. Am J Physiol Gastrointest Liver Physiol. 2011;300:G99–G108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu GY, Shenoy M, Winston JH, Mittal S, Pasricha PJ. P2X receptor‐mediated visceral hyperalgesia in a rat model of chronic visceral hypersensitivity. Gut. 2008;57:1230–1237. [DOI] [PubMed] [Google Scholar]
- 57. Arbuckle RA, Carson RT, Abetz‐Webb L, Hyams J, Di Lorenzo C, Lewis BE, et al. Measuring the symptoms of pediatric constipation and irritable bowel syndrome with constipation: expert commentary and literature review. Patient. 2014;7:343–364. [DOI] [PubMed] [Google Scholar]
- 58. Tsai MF, McCarthy P, Miller C. Substrate selectivity in glutamate‐dependent acid resistance in enteric bacteria. Proc Natl Acad Sci U S A. 2013;110:5898–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rhee SH, Keates AC, Moyer MP, Pothoulakis C. MEK is a key modulator for TLR5‐induced interleukin‐8 and MIP3alpha gene expression in non‐transformed human colonic epithelial cells. J Biol Chem. 2004;279:25179–25188. [DOI] [PubMed] [Google Scholar]
- 60. Rhee SH, Kim H, Moyer MP, Pothoulakis C. Role of MyD88 in phosphatidylinositol 3‐kinase activation by flagellin/toll‐like receptor 5 engagement in colonic epithelial cells. J Biol Chem. 2006;281:18560–18568. [DOI] [PubMed] [Google Scholar]
- 61. Thomas CM, Hong T, van Pijkeren JP, Hemarajata P, Trinh DV, Hu W, et al. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE. 2012;7:e31951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. De Vadder F, Kovatcheva‐Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota‐generated metabolites promote metabolic benefits via gut‐brain neural circuits. Cell. 2014;156:84–96. [DOI] [PubMed] [Google Scholar]
- 63. Sobko T, Huang L, Midtvedt T, Norin E, Gustafsson LE, Norman M, et al. Generation of NO by probiotic bacteria in the gastrointestinal tract. Free Radic Biol Med. 2006;41:985–991. [DOI] [PubMed] [Google Scholar]
- 64. Rey FE, Gonzalez MD, Cheng J, Wu M, Ahern PP, Gordon JI. Metabolic niche of a prominent sulfate‐reducing human gut bacterium. Proc Natl Acad Sci U S A. 2013;110:13582–13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG. Minireview: gut microbiota: the neglected endocrine organ. Mol Endocrinol. 2014;28:1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Raghupathi R, Duffield MD, Zelkas L, Meedeniya A, Brookes SJ, Sia TC, et al. Identification of unique release kinetics of serotonin from guinea‐pig and human enterochromaffin cells. J Physiol. 2013;591(Pt 23):5959–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Freestone PP, Haigh RD, Lyte M. Specificity of catecholamine‐induced growth in Escherichia coli O157:H7, Salmonella enterica and Yersinia enterocolitica . FEMS Microbiol Lett. 2007;269:221–228. [DOI] [PubMed] [Google Scholar]
- 69. Li Y, Xiang YY, Lu WY, Liu C, Li J. A novel role of intestine epithelial GABAergic signaling in regulating intestinal fluid secretion. Am J Physiol Gastrointest Liver Physiol. 2012;303:G453–G460. [DOI] [PubMed] [Google Scholar]
- 70. Watanabe M, Maemura K, Kanbara K, Tamayama T, Hayasaki H. GABA and GABA receptors in the central nervous system and other organs. Int Rev Cytol. 2002;213:1–47. [DOI] [PubMed] [Google Scholar]
- 71. Auteri M, Zizzo MG, Mastropaolo M, Serio R. Opposite role played by GABAA and GABAB receptors in the modulation of peristaltic activity in mouse distal colon. Eur J Pharmacol. 2014;731:93–99. [DOI] [PubMed] [Google Scholar]
- 72. Collares EF, Vinagre AM. Effect of baclofen on liquid and solid gastric emptying in rats. Arq Gastroenterol. 2010;47:290–296. [DOI] [PubMed] [Google Scholar]
- 73. Beaumont H, Smout A, Aanen M, Rydholm H, Lei A, Lehmann A, et al. The GABA(B) receptor agonist AZD9343 inhibits transient lower oesophageal sphincter relaxations and acid reflux in healthy volunteers: a phase I study. Aliment Pharmacol Ther. 2009;30:937–946. [DOI] [PubMed] [Google Scholar]
- 74. Huang D, Huang S, Peers C, Du X, Zhang H, Gamper N. GABAB receptors inhibit low‐voltage activated and high‐voltage activated Ca(2+) channels in sensory neurons via distinct mechanisms. Biochem Biophys Res Commun. 2015;465:188–193. [DOI] [PubMed] [Google Scholar]
- 75. Hanack C, Moroni M, Lima WC, Wende H, Kirchner M, Adelfinger L, et al. GABA blocks pathological but not acute TRPV1 pain signals. Cell. 2015;160:759–770. [DOI] [PubMed] [Google Scholar]
- 76. Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, et al. Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep. 2012;2:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol. 2011;60(Pt 6):817–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. de Ruyter PG, Kuipers OP, de Vos WM. Controlled gene expression systems for Lactococcus lactis with the food‐grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pokusaeva K, Neves AR, Zomer A, O'Connell Motherway M, MacSharry J, Curley P, et al. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb Biotechnol. 2009;3:311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


