Summary
Inflammation triggers the differentiation of Ly6Chi monocytes into microbicidal macrophages or monocyte-derived dendritic cells (moDCs). Yet, it is unclear whether environmental inflammatory cues control the polarization of monocytes toward each of these fates or whether specialized monocyte progenitor subsets exist before inflammation. Here, we have shown that naive monocytes are phenotypically heterogeneous and contain an NR4A1- and Flt3L-independent, CCR2-dependent, Flt3+CD11c−MHCII+PU.1hi subset. This subset acted as a precursor for FcγRIII+PD-L2+CD209a+, GM-CSF-dependent moDCs but was distal from the DC lineage, as shown by fate-mapping experiments using Zbtb46. By contrast, Flt3−CD11c−MHCII−PU.1lo monocytes differentiated into FcγRIII+PD-L2−CD209a−iNOS+ macrophages upon microbial stimulation. Importantly, Sfpi1 haploinsufficiency genetically distinguished the precursor activities of monocytes toward moDCs or microbicidal macrophages. Indeed, Sfpi1+/− mice had reduced Flt3+CD11c−MHCII+ monocytes and GM-CSF-dependent FcγRIII+PD-L2+CD209a+ moDCs but generated iNOS+ macrophages more efficiently. Therefore, intercellular disparities of PU.1 expression within naive monocytes segregate progenitor activity for inflammatory iNOS+ macrophages or moDCs.
Keywords: monocytes, monocyte-derived dendritic cells, GM-CSF, macrophages, PU.1 transcription factor
Graphical Abstract
Highlights
-
•
Murine Ly6ChiCD115+ monocytes are heterogeneous
-
•
DC-related genes (Cd209a and MHCII) are expressed in a subset of FcγRIII+ monocytes
-
•
GM-CSF-dependent CD209a+ moDCs are generated by FcγRIII+CD209a+MHCII+ monocytes
-
•
iNOS+ macrophages are generated by FcγRIII+CD209a−MHCII− monocytes
Monocytes can differentiate into multiple progenies during inflammation. Here, Menezes et al. show that monocytes from naive mice are heterogeneous and contain distinct precursor subsets giving rise to iNOS+ inflammatory macrophages or GM-CSF-induced CD209a+ monocyte-derived dendritic cells.
Introduction
Haematopoietic stem cells continually give rise to mononuclear phagocytes, including monocytes and conventional dendritic cells (DCs) (Steinman and Cohn, 1973). Both monocytes and DCs arise from common early bone marrow (BM) myeloid progenitors called MDPs (Fogg et al., 2006, Lee et al., 2015). MDPs further differentiate into (1) monocyte-committed progenitors (cMoPs) (Hettinger et al., 2013), giving rise to Ly6C+ monocytes unable to differentiate into DCs, and (2) common DC progenitors (CDPs) (Lee et al., 2015, Naik et al., 2007, Onai et al., 2007), which do not give rise to monocytes but generate circulating precursors for DCs (pre-DCs) (Breton et al., 2015, Liu et al., 2009). More recently, MDPs have been shown to generate granulocytes as well (Sathe et al., 2014).
Initially defined by their ability to drive the priming of naive T cells after activation (Nussenzweig et al., 1980), DCs are now regarded as a specific hematopoietic lineage defined by their dependency on growth factor Flt3L (McKenna et al., 2000), which engages the Flt3 receptor tyrosine kinase (CD135) (Waskow et al., 2008), and the expression of the transcription factor (TF) ZBTB46 (Meredith et al., 2012, Satpathy et al., 2012). Fate-mapping (Schraml et al., 2013) and barcoding (Naik et al., 2013) studies have firmly established that DCs are distinct from other lineages.
Monocytes are BM-derived mononuclear phagocytes that circulate in the blood stream. In mice, circulating monocytes are classically defined by expression of CD115 (CSF1R), a receptor for the macrophage growth factor CSF1 (M-CSF). Two categories of monocytes have been identified on the basis of the expression of Ly6C and CX3CR1 according to GFP intensity in Cx3cr1GFP/+ mice: Ly6C+CX3CR1int and Ly6C−CX3CR1hi monocytes (Geissmann et al., 2003). Various studies support the notion that Ly6C+ monocytes can convert to blood Ly6C− monocytes (Hettinger et al., 2013, Sunderkötter et al., 2004, Varol et al., 2007, Yona et al., 2013). However, selective impairment of Ly6C+ monocytes in Irf8−/− mutant mice suggests an independent developmental pathway for Ly6C− monocytes (Kurotaki et al., 2013). The egress of BM Ly6C+ monocytes at steady state requires the engagement of the chemokine receptor CCR2 (Serbina and Pamer, 2006). By contrast, most Ly6C− monocytes gain access to the bloodstream independently of CCR2 and rely on the TF NR4A1 (Hanna et al., 2012). They exhibit a “patrolling” behavior (Auffray et al., 2007) and scavenge damaged endothelia during inflammation (Carlin et al., 2013). A subset of Ly6C− monocytes expressing extracellular major histocompatibility complex II (MHCII) has also been described (Jakubzick et al., 2013).
Inflammatory monocytes have multiple fates. Pamer and colleagues have elegantly shown that the sensing of Listeria monocytogenes (L.m.) infection activates the release of CCR2 ligands to mediate the recruitment of Ly6C+ monocytes, which differentiate into TNF-α+iNOS+ microbicidal phagocytes (Serbina et al., 2003). iNOS+Ly6C+ phagocytes are distinct from the DC lineage (Meredith et al., 2012, Satpathy et al., 2012) and are essential for the control of Listeria infection, as demonstrated by infection of Nos2−/− (MacMicking et al., 1995), Ccr2−/− (Serbina et al., 2003), and monocyte-depleted (Schreiber et al., 2013) mice.
In addition to differentiating into iNOS+ phagocytes, Ly6C+ monocytes can differentiate into CCR2-dependent monocyte-derived DCs (moDCs) (Bain et al., 2013, Zigmond et al., 2012). Accordingly, moDCs can be generated upon adoptive transfer of Ly6C+ monocytes that progressively lose Ly6C and acquire MHCII when differentiating in inflamed tissues (Bain et al., 2013, Zigmond et al., 2012). FcγRI (CD64), FcεRI, and CD206 have emerged as markers of inflammatory phagocytes distinct from the DC lineage (Cheong et al., 2010, Langlet et al., 2012, Plantinga et al., 2013).
The processes regulating the polarization of Ly6C+ monocytes toward iNOS+ macrophages or moDCs remain unclear. Local inflammatory cues might control the nature of monocyte progeny. Alternatively, monocyte subpopulations might be endowed with a selective potential to generate iNOS+ phagocytes or moDCs. Here, we report the description and functional characterization of monocyte subsets endowed with the selective ability to generate iNOS+ phagocytes or moDCs. We show that the amount of PU.1 arbitrates the commitment of monocytes toward either cell fate.
Results
Ly6C+ Monocytes Are Heterogeneous
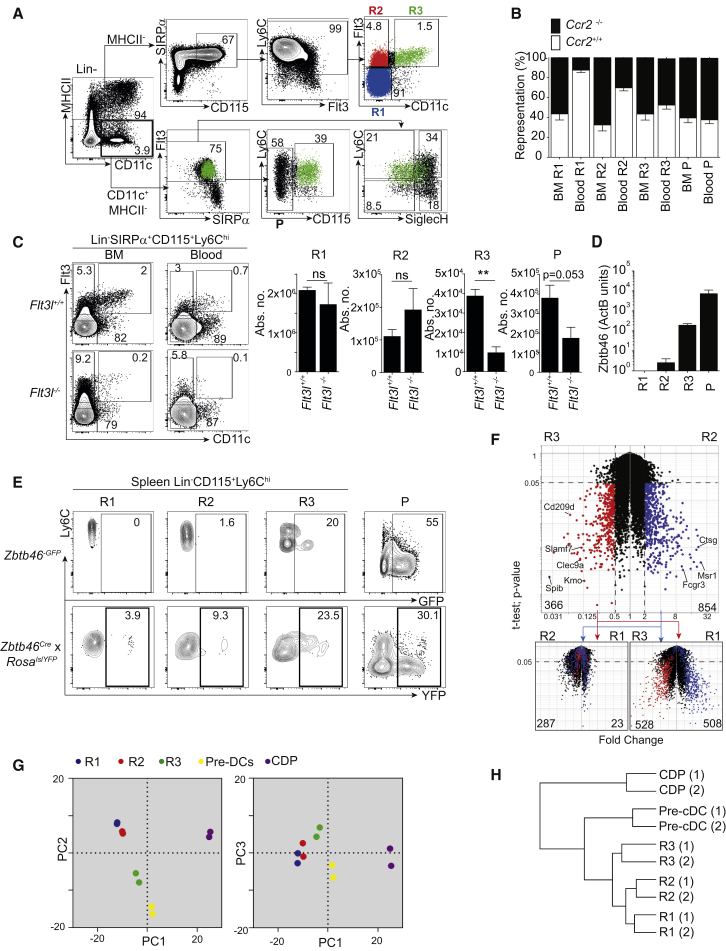
As an initial approach to addressing the heterogeneity of BM mononuclear phagocyte precursors, we analyzed the expression of CD135 (Flt3) and CD115 (CSF1R) in Lin− BM cells (Figure 1A; isotype controls in Figure S1A). MHCII+ cells were not considered because they correspond to F4/80hi BM macrophages (Figure S1B). We noticed that MHCII−CD172a (SIRPα)+CD115+Ly6C+ cells contained three sub-populations: a major CD11c−Flt3− (R1) and two minor CD11c−Flt3+ (R2) and CD11c+Flt3+ (R3) populations (Figures 1A and S1C). R3 corresponded phenotypically to a subset of pre-DCs (CD11c+Flt3+SIRPαint; Liu et al., 2009; Figure 1A) and was distinct from cKit+CD115+Flt3+ CDPs (Naik et al., 2007, Onai et al., 2007; Figure S1D). Within pre-DCs, R3 coexisted with CD115− pre-DCs (P) and aligned with both the Ly6C+SiglecH− and Ly6C+SiglecH+ pre-DC subsets (Figure 1A; Schlitzer et al., 2015). R1 and R2 monocytes expressed heterogeneous amounts of CD11b and CX3CR1 (Figure S1E), had horse-shoe-shaped nuclei (Figure S1F), and were distinct from Nr4a1-dependent Ly6Clo monocytes (Figure S1G).
Figure 1.
Identification of BM Ly6C+CD115+ Subsets
(A) BM Lin−MHCII−Ly6C+CD115+ monocytes consist of three subsets. Shown is a representative flow cytometry analysis of WT BM at steady state. (Top) Lineage− (CD19−, CD3ε−, Ly6G−, Ter119−, CD45RA−, NK1.1−, cKit−) MHCII− cells analyzed by a conventional monocyte gating strategy. CD115+Ly6Chi cells can be subdivided into three sub-populations: R1(Flt3−CD11c−), R2(Flt3+CD11c−), and R3(Flt3+CD11c+). (Bottom) Pre-DC gating of Lin−MHCII−CD11c+Flt3+SIRPαint cells. Pre-DCs can be subdivided into CD115− (P) and CD115+(R3) subsets. Overlay of R3 is shown in green in the lower panels.
(B) Graphical summary of WT and Ccr2−/− mixed BM chimera. Shown are steady-state percentages of WT (CD45.1, white bars) and Ccr2−/− (CD45.2, black bars) cells within R1–R3 and P in the BM and blood of WT (CD45.1) reconstituted recipients. Data represent five chimeric mice over two experiments.
(C) Flow cytometry analysis of R1–R3 and P in Flt3l−/− and Flt3l+/+ BM and blood. Quantification shows the absolute number of each cell population within the blood (n = 4 mice per group).
(D) qPCR for Zbtb46 in WT BM R1–R3 and P in β-actin units.
(E) Zbtb46 labeling and reporter expression. Steady-state GFP reporter expression and YFP expression in splenic R1–R3 and P cells in Zbtb46GFP/+ and Zbtb46Cre x RosalslYFP mice, respectively.
(F) Genes differentially expressed among R1–R3. Volcano plots of R2 versus R3 (main plot) show genes with a fold change ≥ 2 and a p value of p < 0.05 in R2 (blue) and R3 (red). These genes are overlaid on volcano plots of R1 versus R2 (left) and R1 versus R3 (right) with the same axes of fold change and p value. Numbers indicate differentially expressed genes in each comparison (gene list available in Table S1).
(G) Clustering of R1–R3 with pre-DCs and CDPs. Principal-component analysis compares microarray data of R1 (blue), R2 (red), and R3 (green) with previously published data of pre-DCs (yellow) and CDPs (violet) on PC1 (72% variance), PC2 (24% variance), and PC3 (2% variance).
(H) Hierarchical clustering analysis (1 − Pearson correlation) of monocytes (R1 and R2), pre-DCs (R3), total pre-DCs, and CDPs.
Data represent the mean ± SEM (∗p < 0.05;∗∗p < 0.005; ns, not significant; Student’s t test). Please also refer to Figure S1.
Blood Lin−CD115+Ly6C+ cells, like their Ly6C+ BM counterparts, also contained sub-populations R1–R3 (Figures 1B and 1C). R1 expressed higher amounts of CCR2 than R2 and R3 (Figure S1E), and Ccr2 inactivation drastically restricted the size of circulating R1 and R2 monocytes, but not pre-DCs (Figure S1H). Mixed BM chimeras of wild-type (WT) and Ccr2−/− cells showed that CCR2 controls the egress of R1 and R2 monocytes by a cell-intrinsic effect (Figures 1B and S1I).
Unlike pre-DCs and DCs (McKenna et al., 2000), BM and blood R1 and R2 cells were largely independent of growth factor Flt3L (Figure 1C). The DC-specific TF-encoding gene Zbtb46 (BTBD4) was highly expressed only in R3 and P pre-DCs (Figure 1D). Accordingly, reporter expression in Zbtb46GFP/+ mice and fate mapping in the Zbtb46Cre x RosalslYFP model (Loschko et al., 2016; Figures 1E and S1J) showed that splenic R3 and P pre-DCs, but not R1 or R2 monocytes, belonged to the DC lineage.
Genes with higher expression in R3 pre-DCs than in R2 monocytes largely overlapped the genes with higher expression in R3 pre-DCs than in R1 monocytes (e.g., Clec9a and Slamf7; Figure 1F, red dots; Table S1) and, to a lesser extent, overlapped genes with higher expression in R2 monocytes than in R1 monocytes (Ctsg and Flt3; Figure 1F, red dots in lower left plot). R3 pre-DCs expressed genes belonging to the DC signature (Miller et al., 2012) (e.g., Clec9a and Slamf7; Figures 1F and S1K) and clustered with total pre-DCs and CDPs (Figures 1G and 1H).
Genes with higher expression in R2 monocytes than in R3 pre-DCs (Figure 1F, blue dots in the main panel) largely overlapped the genes with higher expression in R1 monocytes than in R3 pre-DCs (e.g., Msr1 and Fcgr3; Figure 1F, lower right plot; Table S1). However, most of these genes were not differentially expressed between R2 and R1 monocytes (Figure 1F, blue dots in lower left panel). Furthermore, R1 and R2 expressed a macrophage signature (e.g., Fcgr3 and Csf3r; Figure S1K; Gautier et al., 2012) while clustering close to each other (Figures 1G and 1H) apart from the DC-committed precursors.
Overall, R1 and R2 were more similar to each other than to R3 (Figures 1F–1H; Table S1). We conclude that R1 and R2 qualify as bona fide monocytes given that both are largely CCR2 dependent for BM egress, do not rely on Flt3L, and do not express Zbtb46. R3 met all the criteria for bona fide pre-DCs because it was largely CCR2 independent and Flt3L dependent and expressed the DC-specific Zbtb46 (Meredith et al., 2012, Satpathy et al., 2012).
R2 Monocytes Bear a Mixed Transcriptional Profile
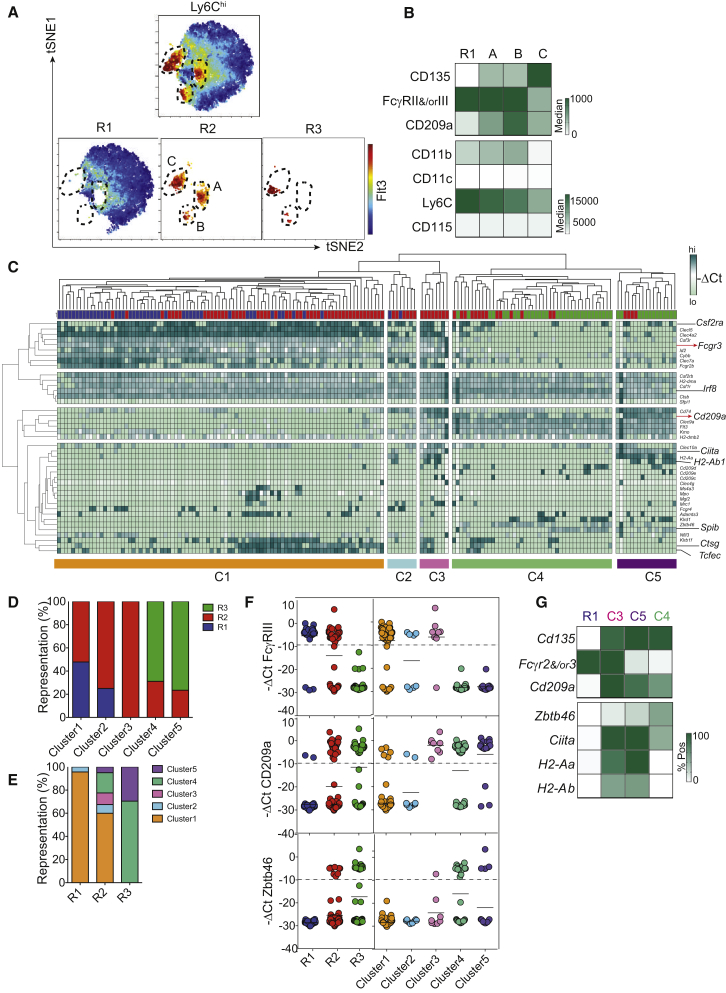
We next aimed to assess the diversity of Ly6C+ monocytes by using unsupervised analyses. To this end, we used multi-dimensional reduction analysis of multi-parametric flow cytometry. BM Ly6C+CD115+ cells were divisible into one major Flt3− and three minor Flt3+ subsets with distinct t-distributed stochastic neighbor embedding (t-SNE) coordinates (populations A–C) (Figure 2A). Population A was CD11c− and found only within R2. By contrast, populations B and C overlapped R2 and R3. Unlike population C, A and B phenotypically shared high expression of FcγRII and/or FcγRIII with R1 monocytes. However, like population C but unlike R1 monocytes, A and B expressed CD209a (Figure 2B). All together, these data were corroborated by microarray analysis of BM R1–R3 populations (Figure S2A) and flow cytometry analysis of blood Ly6C+ cells (Figure S2B). In addition, population C had lower CD11b expression than A and B (Figure 2B).
Figure 2.
Transcriptional Profiling of Monocyte Subsets R1 and R2
(A) Nonlinear dimensionality reduction analysis of Ly6ChiCD115+cells. t-SNE maps of total Lin−MHCII−CD115+SIRPα+ cells, R1 and R2 monocytes, and R3 pre-DCs are based on the parameters CD115, SIRPα, Ly6C, Flt3, CD11c, CD209a, and FcγRII and/or FcγRIII. Color scale indicates Flt3 expression.
(B) Expression analysis of t-SNE-generated sub-populations of R2 and expression of Flt3, FcγRII and/or FcγRIII, CD209a, CD11b, CD11c, Ly6C, and CD115 of t-SNE-generated Flt3+ populations A–C with R1 monocytes. CD209a expression is shown as the difference between MFI and fluorescence minus one (FMO) control for all four populations.
(C) Heterogeneity in population R2. The hierarchical clustering dendogram (top and left margins) is based on −ΔCt values from single-cell multiplex qPCR analysis of 44 R1 (blue), 81 R2 (red), and 44 R3 (green) single cells for 42 genes.
(D) Representation of populations R1–R3 within the five clusters defined in (C).
(E) Representation of the five clusters within populations R1–R3.
(F) Single-cell expression of Fcgr3, CD209a, and Zbtb46 in populations R1–R3 or clusters C1–C5. Each dot represents the −ΔCt value of a single cell.
(G) Analysis of the defining genes of population R1 with clusters C3–C5. The heatmap compares mRNA expression of MHCII-related genes Cd135, Zbtb46, Cd209a, and Fcgr2 and/or Fcgr3 on single cells between clusters C3–C5 and monocyte population R1.
Please also refer to Figure S2.
As a parallel unsupervised approach, we used gene-expression profiling at the single-cell level to assess the diversity of Ly6C+CD115+ cells. We performed single-cell qPCR by using a set of 42 genes and 3 house-keeping controls. Unsupervised clustering of gene expression at the single-cell level revealed the existence of five clusters within Ly6C+CD115+ cells (Figure 2C). Clusters 1 and 2 aligned mostly with R1 and also partially with R2. Cluster 3 was exclusively represented within R2, whereas clusters 4 and 5 were enriched in R3 but also present in R2 (Figures 2C–2E). Cluster 3 was unique in its mixed expression pattern of monocyte (Fcgr3, Fcgr2b, and Csf3r) and DC (Kmo, Cd209a, and Flt3) genes (Figures 2C and 2F and S2A and S2B). However, cluster 3 (in addition to clusters 1 and 2) showed low transcription of Zbtb46, which was found in clusters 4 and 5 (Figure 2F).
Of interest, we noticed that the expression of Ciita and MHCII-related genes was found mostly in clusters 3 and 5 (Figure S2C). This is consistent with MHCII expression in BM and blood R2 and R3, as assessed by flow cytometry and microarray analysis (Figures S2D and S2E). R2 and R3 thus aligned with previously described Ly6C+CD115+MHCII+ cells in the blood (Carlin et al., 2013, Jakubzick et al., 2013; Figure S2F). Using mice deficient of various CIITA promoters (pI−/−, pIV−/−, or pIII+pIV−/−), we showed that R2, like DCs, expressed MHCII after pI-dependent induction of Ciita (LeibundGut-Landmann et al., 2004; Figures S2G and S2H).
All together, both unsupervised flow cytometry and gene-expression analyses revealed that R2 monocytes contained unique populations of Zbtb46− cells that were distinct from the cDC lineage and expressed transcriptional profiles with mixed features of monocytes and cDCs (Zbtb46−Flt3+FcγRII and/or FcγRIII+CD209a+CD11c−). Using MHCII as a surrogate marker for these cells, we showed that they are independent of Flt3L and rely on CCR2 for their mobilization from the BM to the blood (Figures S2I and S2J).
PU.1 Controls the Formation of Flt3+MHCII+ R2 Monocytes
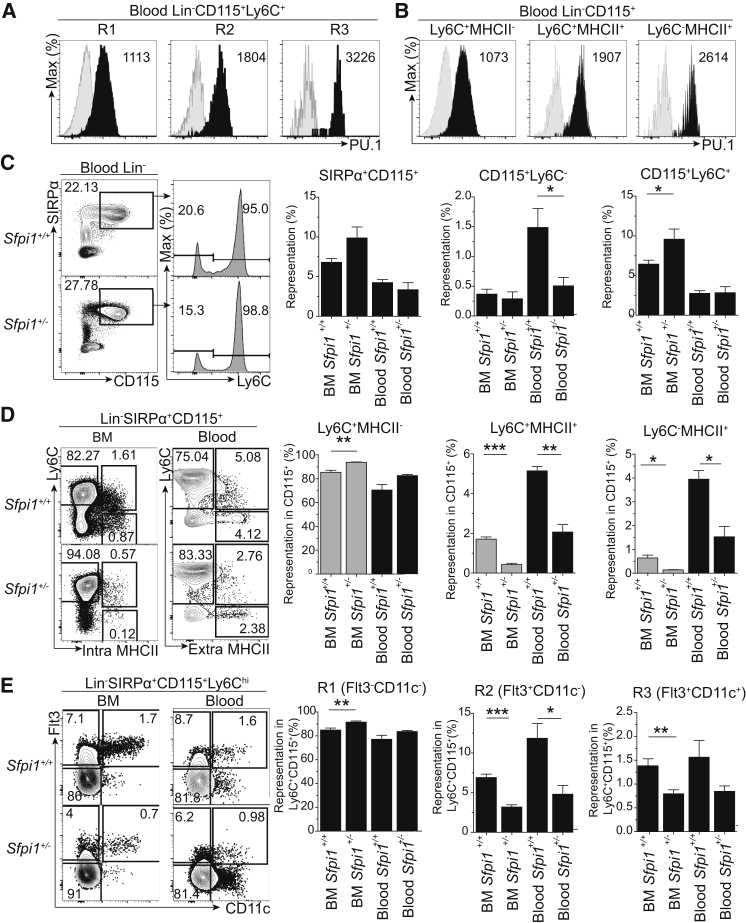
We next sought to identify TFs regulating the formation of R2 Flt3+MHCII+ monocytes. Sfpi1 (PU.1) is an attractive candidate because it promotes Flt3 expression (Carotta et al., 2010) and MHCII through the induction of Ciita (Bakri et al., 2005). Intracellular flow cytometry staining for PU.1 indicated that R3 pre-DCs and R2 monocytes expressed higher amounts of PU.1 than R1 monocytes (Figure 3A). Accordingly, PU.1 was expressed more in MHCII+ than in MHCII− blood CD115+ cells (Figure 3B).
Figure 3.
PU.1 Controls the Development of PU.1hiFlt3+MHCII+ R2 Monocytes at Steady State
(A and B) Expression of PU.1 (black shading) and isotype control (gray shading) within Ly6ChiCD115+ monocytes (R1 and R2) and pre-DCs (R3) (A) and in Ly6ChiMHCII−, Ly6C+MHCII+, and Ly6C−MHCII+CD115+ cells (B) in the blood as seen by intra-nuclear staining of PU1 by flow cytometry. Numbers within plots indicate the MFI of PU.1.
(C–E) Representative flow cytometric analysis of the blood of Sfpi1+/+ and Sfpi1+/− mice at steady state. (C) Comparison of total (SIRPα+CD115+), Ly6C+, and Ly6Clo blood monocytes by flow cytometry and quantification as the percentage of total live cells. (D and E) Comparison of Ly6C+MHCII−, Ly6C+MHCII+, and Ly6C−MHCII+ cells within SIRPα+CD115+ monocytes (D) and of R1–R3 within Ly6Chi MHCII+/− monocytes (E) in the BM and blood. Gray bars indicate intracellular staining for MHCII.
Data represent the mean ± SEM of three mice per group from three identical experiments (∗p < 0.05,∗∗p < 0.005, ∗∗∗p < 0.0005; Student’s t test). Please also refer to Figure S3.
The effects of PU.1 were tightly dependent on its expression; therefore, we analyzed Sfpi1+/− mice that had reached adulthood without any obvious phenotype. Sfpi1+/− mice had reduced numbers of blood CD115+Ly6Clo cells, whereas blood Ly6C+ were not affected despite an increase in BM Ly6C+ cells (Figure 3C). Sfpi1+/− mice had reduced MHCII+ (Ly6C+ and Ly6C−) CD115+ populations in the BM and blood (Figure 3D). Finally, Sfpi1+/− mice had smaller populations of R2 monocytes (including the MHCII+ fraction of R2) and R3 pre-DCs than WT mice in BM and blood (Figures 3E and S3A–S3C). In agreement with R3 pre-DC reduction, Sfpi1+/− mice displayed a selective deficiency in spleen CD11b+ DCs, mostly in the ESAM1loFlt3lo compartment, whereas plasmacytoid DCs remained unchanged (Figures S3D–S3F). In contrast to R2 monocytes and R3 pre-DCs, R1 monocytes were slightly increased in the BM of Sfpi1+/− mice (Figure 3E). However, this increase did not reach significance in the blood (Figures 3E, S3A, and S3B). Sfpi1 hemizygosity reduced the numbers of MDPs and CDPs, but not cMoPs (Figure S3F). We conclude that high expression of PU.1 is selectively required for the development of Flt3+MHCII+ R2 monocytes and R3 pre-DCs at steady state.
PU.1loFlt3−MHCII− R1 Monocytes Generate iNOS+ Macrophages upon Microbial Stimulation
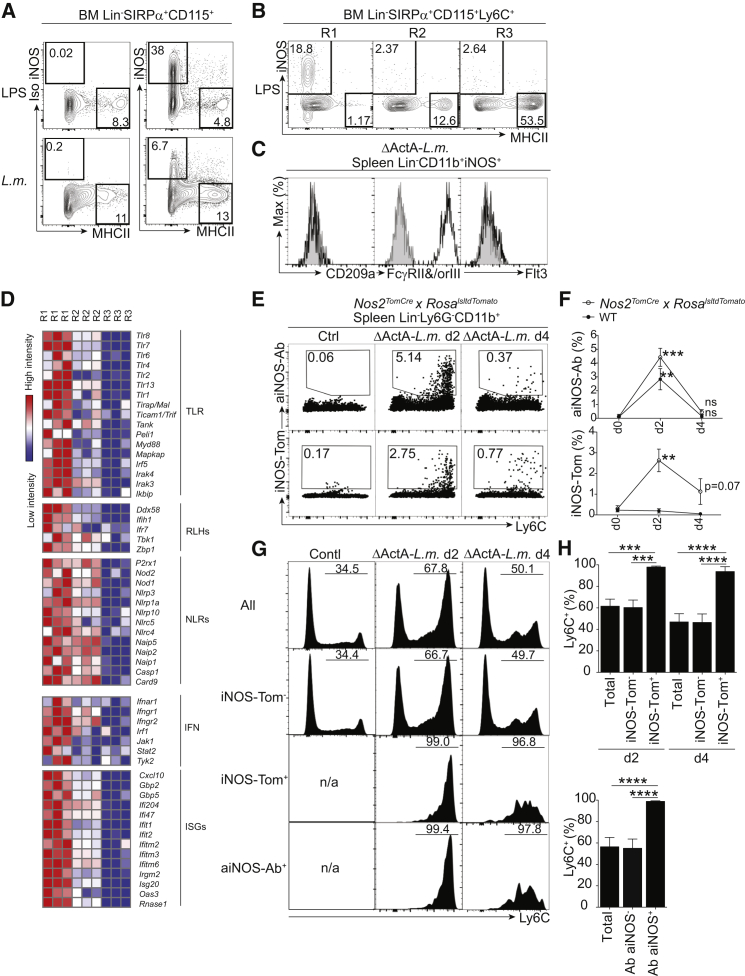
In the next set of experiments, we aimed to determine which Ly6C+ monocytes are precursors for iNOS+ macrophages. We chose to use Listeria infection because it efficiently induces the recruitment and differentiation of iNOS+ phagocytes from Ly6C+ monocytes (Serbina et al., 2003). In agreement with their independence of Flt3L (Meredith et al., 2012), L.m.-induced iNOS+ macrophages were not identified in Zbtb46crexROSAlslYFP mice (Loschko et al., 2016), thus confirming their monocytic origin (Figure S4A). In vitro exposure to lipopolysaccharide (LPS) or infection with L.m. resulted in iNOS expression in a subset of responding BM CD115+Ly6C+ cells (up to 38%) that remained mostly MHCIIlo (Figure 4A), unless these cells were treated with interferon-γ (IFN-γ), which increased MHCII expression in iNOS+ cells (Figure S4B). Overnight culture of sorted R1 or R2 monocytes or R3 pre-DCs with L.m. or LPS (Figures 4B and S4C) revealed the selective ability of R1 to produce iNOS+MHCII− macrophages, whereas R2 and R3 generated iNOS−MHCII+ cells (Figures 4B and S4C). We conclude that the formation of iNOS+ macrophages upon microbial exposure is a selective feature of R1 monocytes or possibly a subset of them. Like R1 monocytes (Figures 2F and 2G), in-vivo-generated Ly6C+CD11b+iNOS+ macrophages (Serbina et al., 2003) expressed FcγRII and/or FcγRIII, but not CD209a or Flt3 (Figure 4C).
Figure 4.
Pu.1loFlt3−MHCII− R1 Monocytes Differentiate into iNOS+ Phagocytes upon Microbial Stimulation
(A) iNOS production by CD115+ cells in vitro. Shown is surface MHCII and intracellular iNOS or isotype control (Iso iNOS) staining after overnight culture of BM Lin−CD115+ cells in the presence of LPS or L.m. (MOI = 0.1).
(B) In vitro microbial stimulation of R1–R3. Shown is the analysis of surface MHCII and intracellular iNOS on sorted R1–R3 cells cultured overnight in the presence of LPS (1 μg/ml).
(C) Cell-surface phenotype of Lin−CD11b+iNOS+ cells during L.m. infection (day 2). Shown is flow cytometry analysis of CD209a, FcγRII and/or FcγRIII (CD16/32), and Flt3 (black lines) against respective isotype controls (gray shading).
(D) Pathway analysis of differentially expressed genes in flow-cytometry-sorted steady-state BM R1–R3. Abbreviations are as follows: TLR, toll-like receptor; RLH, RIG-like helicase; NLR, NOD-like receptor; IFN, interferon; and ISG, interferon-stimulated gene.
(E–H) Fate mapping of L.m.-induced iNOS-expressing splenocytes. (E) Nos2TomatoCre x RosalsltdTomato mice infected with the ΔActA mutant of L.m. were analyzed for intracellular anti-iNOS staining (top) and tomato labeling (bottom) in Lin−CD11b+ splenocytes in control or L.m.-infected mice (days 2 and 4). (F) Mean ± SEM of the percentage of iNOS-Ab+ (top) and Tomato+ (bottom) cells in Bl/6 (WT) or Nos2TomatoCre x RosalsltdTomato mice in untreated (d0) or L.m.-infected mice on days 2 and 4 (n = 3). (G) Histograms show Ly6C expression in cells either stained or unstained with anti-iNOS antibody or iNOS-Tomato. (H) Percentages of Ly6C+ cells within each indicated population (n = 4 mice per group; ∗∗p < 0.005,∗∗∗p < 0.0005, ∗∗∗∗p < 0.00005; Student’s t test).
Please also refer to Figure S4.
Transcriptomic analysis of BM R1–R3 demonstrated an upregulation of toll-like receptor (TLR) signaling, RIG-like helicase, NOD-like receptor genes, and IFN-γ signaling and its target genes in R1 monocytes (Figure 4D). We conclude that R1 cells are efficiently equipped for innate sensing prior to microbial exposure.
We next wanted to determine whether iNOS+ macrophages lose iNOS and enter a differentiation pathway of monocytes characterized by the loss of Ly6C in inflamed tissues (Bain et al., 2013, Zigmond et al., 2012). We used a fate-mapping approach to irreversibly label iNOS-expressing cells in Nos2tomato-crexROSAlsltdTomato mice. L.m. infection triggered the labeling (Tomato+) of a subset of Ly6C+ cells 2 days after infection; these cells were also stained with anti-iNOS antibody (Figures 4E, 4F, and S4D). We found that by day 4, the iNOS+ cells (both Tomato+ and anti-iNOS-Ab+ cells) had reduced drastically (Figure 4F). Like iNOS-Ab+ cells, tomato+ cells remained Ly6C+ and did not become Ly6C− (Figures 4G and 4H). Therefore, we conclude that Ly6C+iNOS+ macrophages do not convert efficiently to Ly6C−. This result is compatible with the existence of a subset of Ly6C+ monocytes specialized to generate iNOS+ macrophages.
High Amounts of PU.1 Inhibit the Production of iNOS+ Macrophages
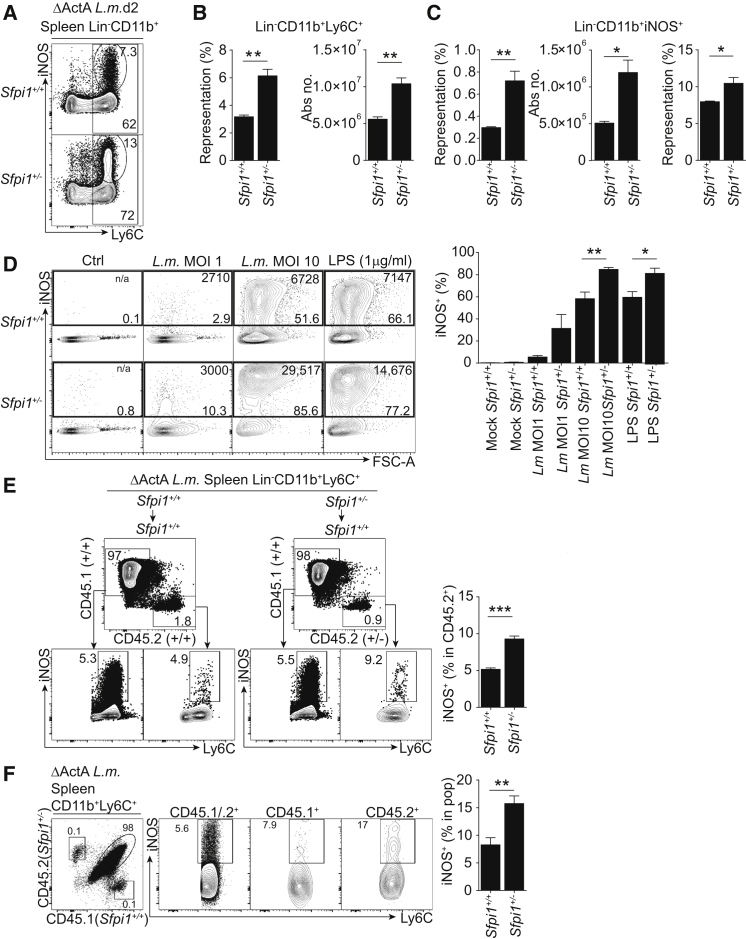
From the experiments performed so far, we noticed that PU.1 concentration was inversely correlated with the ability of monocyte subsets to generate iNOS+ macrophages (Figures 3A and 4B). We therefore hypothesized that PU.1 acts as a regulator of iNOS expression. To test this hypothesis, we infected WT and Sfpi1+/− mice with L.m. and analyzed the generation of CD11b+Ly6C+iNOS+ macrophages in the spleens of these animals 2 days later. Using the avirulent ΔActA mutant of L.m., we were able to assess innate sensing of the bacteria independently of infectious load (Schreiber et al., 2013, Serbina et al., 2003). Sfpi1+/− mice accumulated higher numbers of total CD11b+Ly6C+ and CD11b+Ly6C+iNOS+ macrophages than did their WT counterparts (Figures 5A–5C). Importantly, the percentage of iNOS+ cells in Ly6C+CD11b+ macrophages was increased in Sfpi1+/− mice, whereas MHCII expression remained unchanged (Figures 5C and S5A).
Figure 5.
PU.1 Inhibits the Generation of iNOS+ Macrophages upon Microbial Stimulation
(A–C) Generation of Ly6C+iNOS+ cells upon microbial stimulation of Sfpi1+/+ or Sfpi1+/− mice. (A) Intracellular flow cytometry staining of spleen Lin− (CD19−, CD3ε−, Ly6G−, Ter119−, CD45RA−, NK1.1−, cKit−) CD11b+ cells in Sfpi1+/+ and Sfpi1+/− mice infected with ΔActA L.m. (B and C) Quantification of the percentage and absolute number of Lin−CD11b+Ly6C+ cells (B) and Lin−CD11b+iNOS+ cells (C).
(D) Microbial stimulation of BMDMs generated from Sfpi1+/+ or Sfpi1+/− mice and flow cytometry analysis of iNOS+ BMDMs from Sfpi1+/+ and Sfpi1+/− mice. BMDMs were cultured with MCSF alone, MCSF and L.m. at a MOI of 1 or 10, or MCSF and 1 μg/ml LPS. Numbers at the top of each gate indicate MFI. The percentage of iNOS+ cells obtained within each culture is quantified.
(E) PU.1 reduction in the BM compartment results in an increase in iNOS+ macrophages in vivo. CD45.2+Sfpi1+/+ or Sfpi1+/− whole BM was adoptively transferred into WT ΔActA L.m.-infected CD45.1+ recipients. Representative flow cytometry analysis of iNOS expression in Lin−CD11b+Ly6C+ splenocytes from recipients (CD45.1+) or donors (CD45.2+) is shown.
(F) PU.1 in monocytes regulates the production of iNOS+ macrophages in vivo. Shown is flow cytometry analysis of iNOS expression in Lin−CD11b+Ly6C+ cells of CD45.1+Sfpi1+/+ and CD45.2+Sfpi1+/− sorted BM monocytes adoptively transferred intravenously into WT ΔActA L.m.-infected CD45.1/.2+ recipients (n = 4 mice per group).
Data represent the mean ± SEM (∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005; ns, not significant; Student’s t test). Please also refer to Figure S5.
In order to test whether the regulatory role of PU.1 is cell intrinsic, we infected normalized numbers of BM macrophages (BMDMs) from WT or Sfpi1+/− mice infected with L.m. (or LPS) overnight. We found that Sfpi1+/− macrophages expressed higher amounts of iNOS than did WT cells both in percentage and staining intensity (Figures 5D and S5B). We conclude that Sfpi1 acts as a negative regulator of iNOS acquisition in monocytes responding to microbial stimulation.
In order to address the cell-intrinsic role of PU.1 in vivo, we performed adoptive transfers of CD45.2+Sfpi1+/+or CD45.2+Sfpi1+/− BM cells into L.m.-infected CD45.1+ recipients. Sfpi1+/− donor cells expressed more iNOS than their WT counterparts (Figure 5E). Finally, to address the role of Sfpi1 in the monocyte compartment, we engrafted a mixture of Sfpi1+/− (CD45.2) and Sfpi1+/+ (CD45.1) BM monocytes into L.m.-infected CD45.1 and CD45.2 recipient mice. We found that the percentage of iNOS+ macrophages within Ly6C+CD11b+ cells was higher in the progeny of Sfpi1+/− monocytes than in that of Sfpi1+/+ monocytes (Figure 5F).
We conclude that PU.1 acts as a cell-intrinsic negative regulator of the differentiation of monocytes into iNOS+ macrophages upon microbial exposure.
PU.1hiFlt3+MHCII+ R2 Monocytes Differentiate into PDL2+CD209a+ moDCs upon GM-CSF Exposure
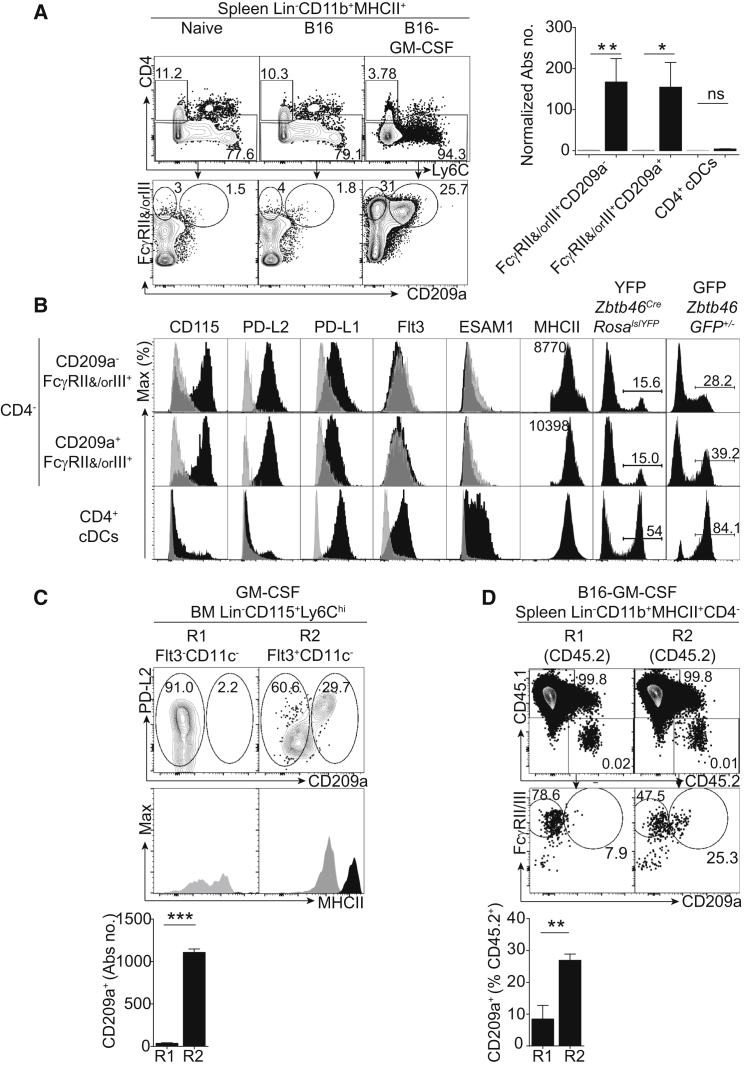
Having established that R1 monocytes are poised to generate iNOS+ macrophages, we next assessed the ability of monocyte subsets to give rise to moDCs. Elevation of granulocyte-macrophage colony-stimulating factor (GM-CSF) concentrations induces the accumulation of CD11b+MHCII+ spleen cells that resemble CD11b+ DCs (Daro et al., 2000, Mach et al., 2000). Engraftment of live GM-CSF-producing B16 melanoma (B16-GM-CSF) (Dranoff et al., 1993) triggered the expansion of (1) circulating MHCII+ monocytes with varying expression levels of Ly6C (Figure S6A) and (2) Lin−Ly6G−CD11b+Ly6Chi–loMHCII+ spleen cells largely overlapping the CD11b+ DC phenotype (Figure S6B). GM-CSF-induced (or LPS-induced; Figure S6C) Ly6Chi–loMHCII+ cells can be subdivided according to their expression of FcγRII and/or FcγRIII and CD209a (Figures 6A and 6B). Both CD209a− and CD209a+ FcγRII+ and/or FcγRIII+ populations rely on CCR2 (Figure S6D). Compared with CD4+ DCs, CD209a− and CD209a+ FcγRII+ and/or FcγRIII+ cells expressed greater CD115 and lower Flt3 and ESAM1 (Figures 6B and S6E). In stark contrast with CD4+CD11b+ DCs (Figures 6B and S6E), both expressed PD-L2 and low amounts of PD-L1 (Figures 6B and S6C). Additionally, both CD209a− and CD209a+ FcγRII+ and/or FcγRIII+ cells had lower expression of YFP than CD4+ DCs from naive or B16-GM-CSF-bearing Zbtb46Cre x RosalslYFP mice (Loschko et al., 2016; Figures 6B and S6F). A similar hierarchy of labeling was obtained in the Zbtb46GFP/+ reporter mice (Satpathy et al., 2012; Figure 6B). Together, these results support a monocytic origin of CD209a− and CD209a+ FcγRII+ and/or FcγRIII+ cells.
Figure 6.
PU.1hiFlt3+MHCII+ R2 Monocytes Differentiate into PD-L2+CD209a+ moDCs upon Exposure to GM-CSF
(A) Phenotype of spleen cells from control, B16, or B16-GM-CSF-engrafted mice. FACS analysis for CD4, Ly6C, FcγRII and/or FcγRIII, and CD209a expression within Lin− (Ly6G−, CD3ε−, NK1.1−, Ter119−, CD45RA−, cKit−) CD11b+MHCII+ spleen cells in naive, B16, or B16-GM-CSF-bearing mice. Absolute numbers of CD4−FcγRII+ and/or FcγIII+CD209a−, CD4−FcγRII+ and/or FcγIII+CD209a+, and CD4+ cells in Lin−CD11b+MHCII+ cells in spleens from B16-GM-CSF-engrafted mice. Data are normalized to the naive control of each population.
(B) Phenotype of CD4−FcγRII+ and/or FcγIII+CD209a−, CD4−FcγRII+ and/or FcγRIII+CD209a+, and CD4+ cells in Lin−Ly6G−CD11b+MHCII+ splenocytes from B16-GM-CSF-bearing mice. Extracellular flow cytometry analysis of each population is shown for CD115, PDL2 (CD273), PDL1 (CD274), Flt3, ESAM1, and MHCII (black) against isotype controls (gray). YFP labeling of the indicated populations in Zbtb46Cre x RosalslYFP mice and GFP labeling in Zbtb46GFP/+ mice is also shown. Numbers above the histograms indicate MFI. Data represent five mice over two experiments.
(C) GM-CSF culture of flow-cytometry-sorted R1 and R2 BM monocytes. Shown is PDL2 and CD209a expression on DAPI−CD45+ cells after 2 days of culture and MHCII expression of CD209a− (gray) and CD209a+ (black). Data represent three independent cultures.
(D) Cell fate of flow-cytometry-sorted R1 or R2 BM monocytes adoptively transferred into B16-GM-CSF-engrafted mice. Shown is FcγRII and/or FcγRIII and CD209a expression on recipient CD45.1+ (Figure S6G) and donor CD45.2+ cells in the spleens of CD45.1+ B16-GM-CSF-bearing recipients, as well as quantification of CD209a+ cells within Lin−CD11b+MHCII+CD4−CD45.2+ cells.
Data represent the mean ± SEM (∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005; ns, not significant; Student’s t test). Please also refer to Figure S1.
Finally, we addressed whether R1 and/or R2 BM monocytes could differentiate into PDL2+CD209a+ cells upon short-term culture in GM-CSF. We found that, reminiscent of macrophages produced during long-term GM-CSF culture (Barthélémy et al., 2015), R1 cells produced exclusively CD209a−MHCIIloPDL2+ cells. Unlike R1, R2 monocytes produced CD209a+MHCIIhiPDL2+ cells (Figure 6C). In vivo, adoptive transfer of R2, but not R1, monocytes in B16-GM-CSF-engrafted mice generated CD209a+ moDCs, as was seen in the CD45.1+ recipients (Figures 6D and S6G). We conclude that R2, but not R1, monocytes exhibit a precursor ability for CD209a+PDL2+ moDCs in vitro and in vivo.
An Increase in PU.1 Promotes the Generation of Monocyte-Derived PDL2+CD209a+ moDCs
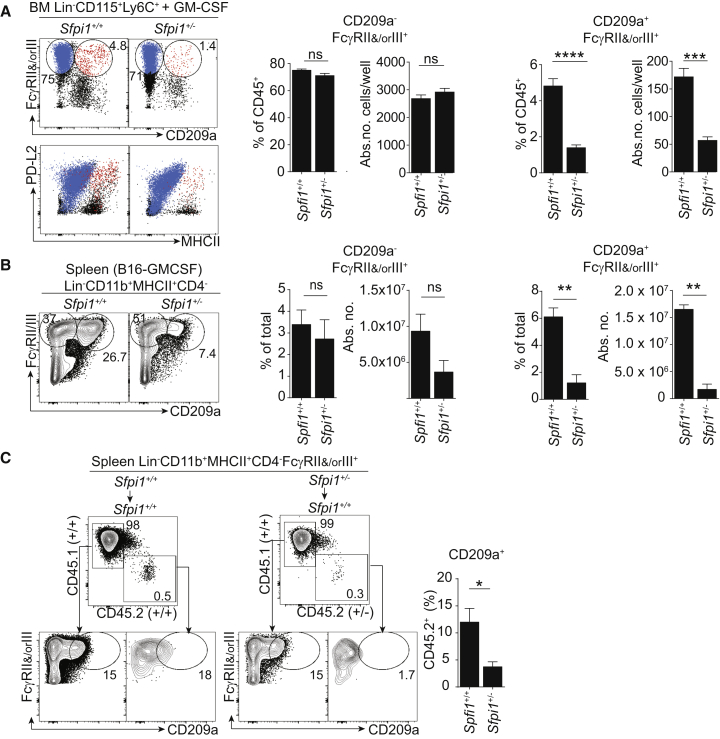
Finally, we investigated whether a higher expression of PU.1 is required for the differentiation of GM-CSF-induced moDCs. The generation of PD-L2hiCD86hiMHCIIhi moDCs (Barthélémy et al., 2015, Gao et al., 2013) from BM GM-CSF cultures was lower in Sfpi1+/− cultures than in WT cultures (Figure S7A). WT, but not Sfpi1+/− Ly6Chi, monocytes generated PD-L2+CD209a+ moDCs in vitro (Figure 7A). Analyzing B16-GM-CSF-engrafted Sfpi1+/+ or Sfpi1+/− mice, we found that the expansion of CD209a+ moDCs (but not FcγRII+ and/or FcγRIII+ CD209a− macrophages) was selectively decreased in Sfpi1+/− mice (Figure 7B).
Figure 7.
PU.1 Promotes the Generation of GM-CSF-Dependent PD-L2+CD209a+ moDCs
(A) FcγRII and/or FcγRIII and CD209a expression in GM-CSF culture of BM Lin−CD115+Ly6C+ cells from Sfpi1+/+ or Sfpi1+/− mice. FcγRII+ and/or FcγRIII+CD209a− (blue) and FcγRII+ and/or FcγRIII+CD209a+ (red) cells are overlaid on flow cytometry staining of PDL2 (CD273) and MHCII. The percentage and absolute number of DAPI−CD45+ cells of each indicated population of Sfpi1+/+ or Sfpi1+/− BM are shown. Data represent two similar experiments.
(B) Quantification of FcγRII+ and/or FcγRIII+CD209a− and FcγRII+ and/or FcγRIII+CD209a+ cells in Lin−CD11b+MHCII+CD4− splenocytes from B16-GM-CSF-bearing Sfpi1+/+or Sfpi1+/− mice. Data represent six mice.
(C) PU.1 promotes the generation of CD209a+ moDCs by a cell-intrinsic mechanism. Shown is the adoptive transfer of CD45.2+Sfpi1+/+ or Sfpi1+/− BM into B16-GM-CSF-engrafted CD45.1+ congenic recipients, as well as CD209a and FcγRII and/or FcγRIII expression on Lin−CD11b+MHCII+CD4− cells of recipient CD45.1+ or donor CD45.2+Sfpi1+/+ or Sfpi1+/− BM. The percentage of CD209a+ cells inside the donor Lin−CD11b+MHCII+CD4−FcγRII+ and/or FcγRIII+CD45.2+ cells is quantified (n = 3 mice per group in two identical experiments). Data represent the mean ± SEM (∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005; Student’s t test).
Please also refer to Figure S7.
We wondered whether PU.1 is required at the cell-autonomous level for the generation of CD209a+ moDCs. We adoptively transferred Sfpi1+/+ or Sfpi1+/− CD45.2+ BM into CD45.1+ B16-GM-CSF-injected recipients. Unlike Sfpi1+/+BM, Sfpi1+/− BM did not generate CD209a+ moDCs (Figure 7C). Control populations such as B cells and granulocytes were generated as efficiently upon transfer of either Sfpi1+/+ or Sfpi1+/− BM (Figure S7B). We conclude that PU.1 selectively controls the differentiation of GM-CSF-dependent CD209a+ moDCs by a cell-intrinsic mechanism.
Discussion
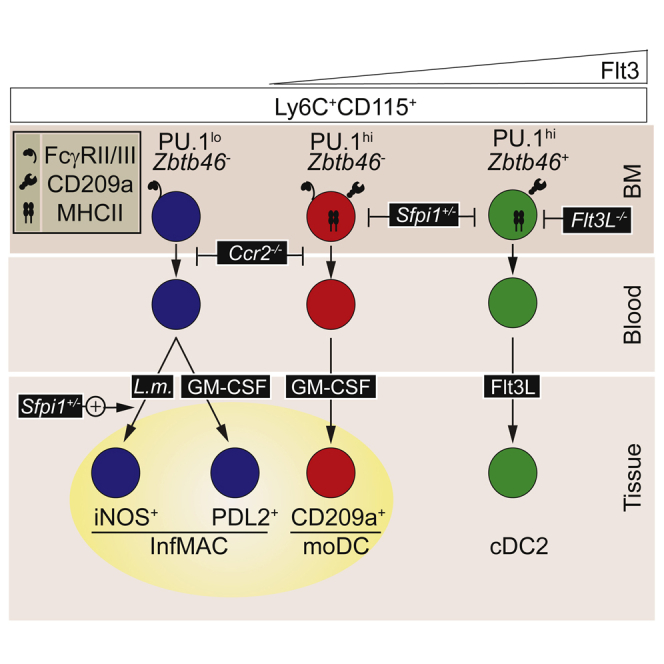
Depending on the study, monocyte-derived inflammatory cells (distinct from the cDC lineage) are termed inflammatory macrophages (Bain et al., 2013, Tamoutounour et al., 2012) or moDCs (Cheong et al., 2010, Plantinga et al., 2013, Zigmond et al., 2012). Here, we report that distinct monocyte subsets give rise to iNOS+ inflammatory macrophages and CD209a+ moDCs. Indeed, we have shown that a subset of monocytes (R2; CCR2 dependent and not labeled in Zbtb46Cre x RosalslYFP mice) contain MHCII+ cells that are distinct from BTBD4+ pre-DCs. These cells develop into CD209a+ moDCs upon exposure to GM-CSF both in vitro and in vivo. Conversely, MHCII− monocytes (R1) possess the progenitor function for iNOS+ inflammatory macrophages. This supports the view that inflammatory macrophages and moDCs are ontogenically distinct populations. In support of this hypothesis, fate mapping of iNOS+ inflammatory macrophages did not show an efficient conversion to the moDC phenotype. In addition, single-cell analysis of monocytes (R2; CD11c−) identified a mixed transcriptional program characterized by the expression of MHCII genes (downstream activation of CIITA by pI promoter) and DC-related genes such as Flt3, Cd209a, or Kmo, for example. These cells might originate from cMoPs (through Flt3 acquisition), from CDPs (via loss of DC-specific commitment), or from earlier myeloid-primed progenitors (LMPP or GMPs). Additional experiments are needed to address this. Of note, we have shown that R3 cells (CD11c+Flt3+CD115+BTBD4+SiglecH−Ly6C+) aligning with previously described cDC2-commited pre-cDCs (pre-DC2s) (Schlitzer et al., 2015, Tussiwand et al., 2015) are diverse in terms of MHCII or CD209a expression, for example. The functional relevance of pre-DC2 heterogeneity for the generation of multiple cDC2 subsets (Lewis et al., 2011, Tussiwand et al., 2015) remains to be addressed.
An important question that arises is the transcriptional mechanism driving the steady-state differentiation of Ly6C+ monocytes into a small sub-population with a distinct potential to generate moDCs or inflammatory macrophages. PU.1 is a lineage-determining TF essential for hematopoietic stem cells and has multiple roles in the myeloid lineage (Dakic et al., 2005, DeKoter and Singh, 2000, McKercher et al., 1996, Scott et al., 1994). PU.1 cooperates with multiple other TFs to shape the enhancer landscape of tissue-resident macrophages (Lavin et al., 2014, Norris et al., 2014). Here, we report that Sfpi1 haploinsufficiency promotes the generation of iNOS+ macrophages during L.m. infection. PU.1-dependent negative regulation of iNOS+ macrophages might constitute a regulatory mechanism limiting iNOS-dependent immunopathology. How does PU.1 downregulate the production of iNOS? PU.1 is known to upregulate multiple miRNAs, including miR-146 and miR-155, which in turn negatively regulate innate sensing through the regulation of TRAF6, IRAK4, and STAT1, for example (Ghani et al., 2011, Jurkin et al., 2010). Indeed, Ly6C+ monocytes from mir146−/− mice are hyper-responsive to microbial stimulation (Etzrodt et al., 2012). Higher amounts of PU.1 might be needed to induce mir146 and limit anti-microbial responses. Further experiments are needed to assess the relevance of miRNAs downstream of PU.1-dependent regulation of innate sensing.
We have found that, in addition to having a regulatory effect on microbicidal iNOS+ macrophages, the highest amounts of PU.1 selectively promote the generation of GM-CSF-dependent moDCs in vitro and in vivo. This could be explained by (1) the reduction of moDC precursors (MHCII+ R2 monocytes) in naive Sfpi1+/− mice and (2) an effect on moDC terminal differentiation. In support of the latter, overexpression of PU.1 promotes the differentiation of DC-like cells (Bakri et al., 2005), and inducible ablation of Sfpi1 prevents the differentiation of DCs (Carotta et al., 2010). In this context, PU.1 cooperation with TFs IRF4 and IRF8 could be relevant to explain the role of PU.1 in moDC differentiation from monocytes. PU.1 can bind to Ets binding sites on its own, but PU.1 also cooperates with IRF4 or IRF8 at Ets-IRF composite response elements called EICEs (Brass et al., 1999). IRF4 is known to be involved in the control of CIITA promoter pI in GM-CSF moDCs or CD11b+ DCs (Gao et al., 2013, Tamura et al., 2005, Vander Lugt et al., 2014). In addition, PU.1 might boost the expression of growth factor receptors required for the development of moDCs (e.g., CSFR2A; DeKoter et al., 1998).
In conclusion, our results shed light on the readiness of inflammatory monocyte subsets for distinct and specialized developmental programs activated in inflammatory conditions. Importantly, PU.1 amounts segregate the transcriptional programs of microbicidal iNOS+ macrophages or moDCs.
Experimental Procedures
Mice
All mice used were between 6 and 12 weeks old and were matched for age and sex in all experiments. They were maintained under specific-pathogen-free conditions in accordance with the UK Animals (Scientific Procedures) Act of 1986.
Cell Isolation and Fluorescence-Activated Cell Sorting
For preparation of BM cell suspensions, the bones of both hind limbs (two tibia and two femurs) were flushed with ice-cold fluorescence-activated cell sorting (FACS) buffer (PBS (Life technologies) with 1% BSA (Apollo Scientific Ltd) and 2 mM EDTA (Life Technologies). Spleens were collected, cut into small pieces, and incubated with collagenase D (Roche) and DNaseI (Roche) in Hank’s balanced salt solution (GE Healthcare) and 5% fetal bovine serum (Life technologies) for 20 min; they were further macerated through 100 μm cell strainers (BD Falcon). Red blood cells were lysed with 2 ml of Ack lysis buffer (Life Technologies), incubated for 2 min at room temperature, and then diluted with FACS buffer. After centrifugation, cells were either re-suspended in an antibody cocktail in FACS buffer or permeabilized and fixed for intracellular staining and analyzed by flow cytometry with FlowJo software (TreeStar). For cell sorting, see Supplemental Experimental Procedures.
In Vitro GM-CSF Cultures
Total BM or 104 sorted Ly6ChiCD115+ monocytes (total or subsets R1–R3) were cultured in 20 ng/ml of GM-CSF in complete RPMI with 6,000 live MS-5 cells as “feeders,” which were plated on the same day. Analyzed cells were pre-gated to be DAPI− and CD45+.
In Vitro L.m. Infections
Primary cells infected with L.m. were in vivo cultured overnight at an MOI of 0.01, 0.1, 1, or 10 (as indicated in Figure 5) in complete RPMI medium supplemented with macrophage colony-stimulating factor (MCSF, 20 ng/ml; Peprotech), GM-CSF (3 ng/ml; Peprotech), and human Flt3L (100 ng/ml; CellDex). BMDMs were derived by culture of whole BM in RPMI supplemented with culture medium from L-929 cells.
B16-GM-CSF Tumor Experiments
B16-GM-CSF tumor cells were checked for viability with Trypan Blue, and 1.5 × 105 to 3 × 105 live cells were injected subcutaneously (see Supplemental Experimental Procedures).
Infection
4 × 103 to 5 × 103 WT colony-forming units (CFU) (Listeria) or 106 ΔActA mutant CFU of L.m. (ΔActA Listeria) were injected intravenously into sterile PBS (see Supplemental Experimental Procedures).
Microarray
Cells sorted by flow cytometry were collected in complete RPMI and pelleted, lysed in Buffer RLT (RNeasy Kit, QIAGEN), and frozen at −80°C until they were processed for RNA. The NuGEN Ovation Pico WTA V2 Kit was used to process 1 ng RNA per sample into cDNA amplified by single-primer isothermal amplification. The Encore Biotin Module (NuGEN) was used to fragment and label the cDNA with biotin. Hybridization cocktails were prepared as recommended by NuGEN and hybridized to Affymetrix Mouse Gene 1.0 ST arrays overnight. Arrays were washed and stained with Affymetrix Fluidics Station FS450 and the GeneChip Hybridization, Wash, and Stain Kit and scanned by the GeneChip Scanner 3000 7G with Autoloader. Raw data files (DAT and CEL) were generated in Affymetrix GeneChip Command Console software and are available at GEO: GSE90471. Data were analyzed with GenePattern software (Broad Institute), and pre-DC and CDP datasets were obtained from public databases.
t-SNE Analysis
Single-cell analysis using the t-SNE algorithm was done on flow cytometry data in the online platform provided by Cytobank (see Supplemental Experimental Procedures).
Single-Cell qPCR
Single cells were sorted by flow cytometry, cDNA was amplified with the CellsDirect One-Step qRT-PCR Kit (ThermoFisher), and qPCR was run on a BioMark HD (Fluidigm) with the help of Taqman probes (Life Technologies) for the genes indicated in Figure 2. The 45 targeted genes were analyzed against an average of three housekeeping genes: Hprt, ActB, and Gapdh. Analysis was done with the help of Gene-E software (Broad Institute) (see Supplemental Experimental Procedures).
qPCR
qPCR was carried out in duplicate for samples sorted from three independent sorting experiments. Primers used for testing Zbtb46 were 5′-AGAGAGCACATGAAGCGACA-3′ (forward) and 5′-CTGGCTGCAGACATGAACAC-3′ (reverse). Results were normalized against β-actin: 5′-ATGCTCCCCGGGCTGTAT-3′ (forward) and 5′-CATAGGAGTCCTTCTGACCCATTC-3′ (reverse).
Adoptive Transfer
For adoptive transfer of cells sorted by flow cytometry, 3.3 × 105 cells of each population were collected in complete RPMI, centrifuged, resuspended in 120 μl of RPMI alone, and injected intravenously into each CD45.1+ congenic recipient. For whole-BM transfers, single-cell suspensions of BM were depleted of red blood cells by exposure to RBC lysis buffer (Life technologies) and counted. 40 × 106 whole-BM cells from Sfpi1+/+ or Sfpi1+/− mice were transferred into B16-GM-CSF-treated congenic CD45.1+ mice on day 9 and analyzed on day 11 after tumor injection. For L.m.-infected recipients, 20 × 106 whole-BM cells were transferred into congenic CD45.1+ recipients 2 hr before intravenous L.m. infection.
Statistical Analysis
Data were analyzed for statistical significance by unpaired Student’s t tests. Differences were considered significant at p < 0.05 (∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005; ∗∗∗∗p < 0.00005; ns, non-significant).
Author Contributions
S.M. performed most of the experiments (with help from R.P., D.M., and G.A. for some) and designed the figures. T.P. performed the single-cell multiplex qPCR; R.G. provided expertise for the design of these experiments. J.D.A., S.H., and A.-M.L.-D. provided help for the analysis of MHCII expression and its transcriptional regulation and performed some experiments addressing the role of CIITA. J.L., M.C.N., and M.P.L. provided tools and expertise for the analysis of Zbtb46 expression and the fate mapping of Zbtb46-expressing cells. G.L., A.B., and F.G. provided reagents and expertise with the fate mapping of iNOS-expressing cells. J.Y.H and S.A.Q provided reagents and expertise with the B16-GM-CSF melanoma model. E.L.G, E.G.-M, C.E.J.-G., E.G.-P., H.G., and F.G. provided mice models, expertise for the analysis of monocyte populations, and helpful discussions. The manuscript was written by P.G. and S.M. Experiments were designed by P.G. in collaboration with S.M.
Acknowledgments
P.G. is a CNRS investigator. S.M. is funded by King’s Overseas Research Studentships. The research was supported by the MRC (MR/K01241X/1), BBSRC (BB/M029735/1), and King’s Health Partners. We thank Dr. Loredana Saveanu, Prof. Caetano Reis e Sousa, and Dr. Julie Helft for reagents and insightful discussions. All flow cytometry work was performed within the NIHR Biomedical Research Centre based at Guy’s and St. Thomas’ NHS Foundation Trust and King’s College London; we especially thank Prabhjoat Singh Chana for help at the facility and Nedyalko Petrov for assistance with t-SNE analysis. We would like to thank Matthew Arno and Erick Nasser at the Genomics Centre of King’s College London for performing the microarray experiments and providing technical support. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
Published: December 20, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2016.12.001.
Accession Numbers
The accession number for the microarray data reported in this paper is GEO: GSE90471.
Supplemental Information
Genes have a fold change ≥ 2 and p value ≤ 0.05. Related to Figure 1F.
References
- Auffray C., Fogg D., Garfa M., Elain G., Join-Lambert O., Kayal S., Sarnacki S., Cumano A., Lauvau G., Geissmann F. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 2007;317:666–670. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- Bain C.C., Scott C.L., Uronen-Hansson H., Gudjonsson S., Jansson O., Grip O., Guilliams M., Malissen B., Agace W.W., Mowat A.M. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013;6:498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakri Y., Sarrazin S., Mayer U.P., Tillmanns S., Nerlov C., Boned A., Sieweke M.H. Balance of MafB and PU.1 specifies alternative macrophage or dendritic cell fate. Blood. 2005;105:2707–2716. doi: 10.1182/blood-2004-04-1448. [DOI] [PubMed] [Google Scholar]
- Barthélémy O., Degrell P., Berman E., Kerneis M., Petroni T., Silvain J., Payot L., Choussat R., Collet J.P., Helft G. Sex-related differences after contemporary primary percutaneous coronary intervention for ST-segment elevation myocardial infarction. Arch. Cardiovasc. Dis. 2015;108:428–436. doi: 10.1016/j.acvd.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Brass A.L., Zhu A.Q., Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G., Lee J., Zhou Y.J., Schreiber J.J., Keler T., Puhr S., Anandasabapathy N., Schlesinger S., Caskey M., Liu K., Nussenzweig M.C. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J. Exp. Med. 2015;212:401–413. doi: 10.1084/jem.20141441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin L.M., Stamatiades E.G., Auffray C., Hanna R.N., Glover L., Vizcay-Barrena G., Hedrick C.C., Cook H.T., Diebold S., Geissmann F. Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal. Cell. 2013;153:362–375. doi: 10.1016/j.cell.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotta S., Dakic A., D’Amico A., Pang S.H., Greig K.T., Nutt S.L., Wu L. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Cheong C., Matos I., Choi J.H., Dandamudi D.B., Shrestha E., Longhi M.P., Jeffrey K.L., Anthony R.M., Kluger C., Nchinda G. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakic A., Metcalf D., Di Rago L., Mifsud S., Wu L., Nutt S.L. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J. Exp. Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daro E., Pulendran B., Brasel K., Teepe M., Pettit D., Lynch D.H., Vremec D., Robb L., Shortman K., McKenna H.J. Polyethylene glycol-modified GM-CSF expands CD11b(high)CD11c(high) but notCD11b(low)CD11c(high) murine dendritic cells in vivo: a comparative analysis with Flt3 ligand. J. Immunol. 2000;165:49–58. doi: 10.4049/jimmunol.165.1.49. [DOI] [PubMed] [Google Scholar]
- DeKoter R.P., Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- DeKoter R.P., Walsh J.C., Singh H. PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J. 1998;17:4456–4468. doi: 10.1093/emboj/17.15.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranoff G., Jaffee E., Lazenby A., Golumbek P., Levitsky H., Brose K., Jackson V., Hamada H., Pardoll D., Mulligan R.C. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc. Natl. Acad. Sci. USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzrodt M., Cortez-Retamozo V., Newton A., Zhao J., Ng A., Wildgruber M., Romero P., Wurdinger T., Xavier R., Geissmann F. Regulation of monocyte functional heterogeneity by miR-146a and Relb. Cell Rep. 2012;1:317–324. doi: 10.1016/j.celrep.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg D.K., Sibon C., Miled C., Jung S., Aucouturier P., Littman D.R., Cumano A., Geissmann F. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- Gao Y., Nish S.A., Jiang R., Hou L., Licona-Limón P., Weinstein J.S., Zhao H., Medzhitov R. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 2013;39:722–732. doi: 10.1016/j.immuni.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., Immunological Genome Consortium Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F., Jung S., Littman D.R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- Ghani S., Riemke P., Schönheit J., Lenze D., Stumm J., Hoogenkamp M., Lagendijk A., Heinz S., Bonifer C., Bakkers J. Macrophage development from HSCs requires PU.1-coordinated microRNA expression. Blood. 2011;118:2275–2284. doi: 10.1182/blood-2011-02-335141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna R.N., Shaked I., Hubbeling H.G., Punt J.A., Wu R., Herrley E., Zaugg C., Pei H., Geissmann F., Ley K., Hedrick C.C. NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ. Res. 2012;110:416–427. doi: 10.1161/CIRCRESAHA.111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettinger J., Richards D.M., Hansson J., Barra M.M., Joschko A.C., Krijgsveld J., Feuerer M. Origin of monocytes and macrophages in a committed progenitor. Nat. Immunol. 2013;14:821–830. doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]
- Jakubzick C., Gautier E.L., Gibbings S.L., Sojka D.K., Schlitzer A., Johnson T.E., Ivanov S., Duan Q., Bala S., Condon T. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. 2013;39:599–610. doi: 10.1016/j.immuni.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkin J., Schichl Y.M., Koeffel R., Bauer T., Richter S., Konradi S., Gesslbauer B., Strobl H. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J. Immunol. 2010;184:4955–4965. doi: 10.4049/jimmunol.0903021. [DOI] [PubMed] [Google Scholar]
- Kurotaki D., Osato N., Nishiyama A., Yamamoto M., Ban T., Sato H., Nakabayashi J., Umehara M., Miyake N., Matsumoto N. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. 2013;121:1839–1849. doi: 10.1182/blood-2012-06-437863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlet C., Tamoutounour S., Henri S., Luche H., Ardouin L., Grégoire C., Malissen B., Guilliams M. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J. Immunol. 2012;188:1751–1760. doi: 10.4049/jimmunol.1102744. [DOI] [PubMed] [Google Scholar]
- Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Breton G., Oliveira T.Y., Zhou Y.J., Aljoufi A., Puhr S., Cameron M.J., Sékaly R.P., Nussenzweig M.C., Liu K. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J. Exp. Med. 2015;212:385–399. doi: 10.1084/jem.20141442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeibundGut-Landmann S., Waldburger J.M., Reis e Sousa C., Acha-Orbea H., Reith W. MHC class II expression is differentially regulated in plasmacytoid and conventional dendritic cells. Nat. Immunol. 2004;5:899–908. doi: 10.1038/ni1109. [DOI] [PubMed] [Google Scholar]
- Lewis K.L., Caton M.L., Bogunovic M., Greter M., Grajkowska L.T., Ng D., Klinakis A., Charo I.F., Jung S., Gommerman J.L. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., Nussenzweig M. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschko J., Schreiber H.A., Rieke G.J., Esterházy D., Meredith M.M., Pedicord V.A., Yao K.H., Caballero S., Pamer E.G., Mucida D., Nussenzweig M.C. Absence of MHC class II on cDCs results in microbial-dependent intestinal inflammation. J. Exp. Med. 2016;213:517–534. doi: 10.1084/jem.20160062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N., Gillessen S., Wilson S.B., Sheehan C., Mihm M., Dranoff G. Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 2000;60:3239–3246. [PubMed] [Google Scholar]
- MacMicking J.D., Nathan C., Hom G., Chartrain N., Fletcher D.S., Trumbauer M., Stevens K., Xie Q.W., Sokol K., Hutchinson N. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell. 1995;81:641–650. doi: 10.1016/0092-8674(95)90085-3. [DOI] [PubMed] [Google Scholar]
- McKenna H.J., Stocking K.L., Miller R.E., Brasel K., De Smedt T., Maraskovsky E., Maliszewski C.R., Lynch D.H., Smith J., Pulendran B. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- McKercher S.R., Torbett B.E., Anderson K.L., Henkel G.W., Vestal D.J., Baribault H., Klemsz M., Feeney A.J., Wu G.E., Paige C.J., Maki R.A. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- Meredith M.M., Liu K., Darrasse-Jeze G., Kamphorst A.O., Schreiber H.A., Guermonprez P., Idoyaga J., Cheong C., Yao K.H., Niec R.E., Nussenzweig M.C. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med. 2012;209:1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., Immunological Genome Consortium Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- Naik S.H., Perié L., Swart E., Gerlach C., van Rooij N., de Boer R.J., Schumacher T.N. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- Norris P.C., Gosselin D., Reichart D., Glass C.K., Dennis E.A. Phospholipase A2 regulates eicosanoid class switching during inflammasome activation. Proc. Natl. Acad. Sci. USA. 2014;111:12746–12751. doi: 10.1073/pnas.1404372111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig M.C., Steinman R.M., Gutchinov B., Cohn Z.A. Dendritic cells are accessory cells for the development of anti-trinitrophenyl cytotoxic T lymphocytes. J. Exp. Med. 1980;152:1070–1084. doi: 10.1084/jem.152.4.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., Manz M.G. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- Plantinga M., Guilliams M., Vanheerswynghels M., Deswarte K., Branco-Madeira F., Toussaint W., Vanhoutte L., Neyt K., Killeen N., Malissen B. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Sathe P., Metcalf D., Vremec D., Naik S.H., Langdon W.Y., Huntington N.D., Wu L., Shortman K. Lymphoid tissue and plasmacytoid dendritic cells and macrophages do not share a common macrophage-dendritic cell-restricted progenitor. Immunity. 2014;41:104–115. doi: 10.1016/j.immuni.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Satpathy A.T., Kc W., Albring J.C., Edelson B.T., Kretzer N.M., Bhattacharya D., Murphy T.L., Murphy K.M. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 2012;209:1135–1152. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., Sivakamasundari V., Chen J., Sumatoh H.R., Schreuder J., Lum J., Malleret B., Zhang S., Larbi A., Zolezzi F. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 2015;16:718–728. doi: 10.1038/ni.3200. [DOI] [PubMed] [Google Scholar]
- Schraml B.U., van Blijswijk J., Zelenay S., Whitney P.G., Filby A., Acton S.E., Rogers N.C., Moncaut N., Carvajal J.J., Reis e Sousa C. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell. 2013;154:843–858. doi: 10.1016/j.cell.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Schreiber H.A., Loschko J., Karssemeijer R.A., Escolano A., Meredith M.M., Mucida D., Guermonprez P., Nussenzweig M.C. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J. Exp. Med. 2013;210:2025–2039. doi: 10.1084/jem.20130903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E.W., Simon M.C., Anastasi J., Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Serbina N.V., Pamer E.G. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat. Immunol. 2006;7:311–317. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- Serbina N.V., Salazar-Mather T.P., Biron C.A., Kuziel W.A., Pamer E.G. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- Steinman R.M., Cohn Z.A. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J. Exp. Med. 1973;137:1142–1162. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderkötter C., Nikolic T., Dillon M.J., Van Rooijen N., Stehling M., Drevets D.A., Leenen P.J. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J. Immunol. 2004;172:4410–4417. doi: 10.4049/jimmunol.172.7.4410. [DOI] [PubMed] [Google Scholar]
- Tamoutounour S., Henri S., Lelouard H., de Bovis B., de Haar C., van der Woude C.J., Woltman A.M., Reyal Y., Bonnet D., Sichien D. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur. J. Immunol. 2012;42:3150–3166. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- Tamura T., Tailor P., Yamaoka K., Kong H.J., Tsujimura H., O’Shea J.J., Singh H., Ozato K. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J. Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- Tussiwand R., Everts B., Grajales-Reyes G.E., Kretzer N.M., Iwata A., Bagaitkar J., Wu X., Wong R., Anderson D.A., Murphy T.L. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 2015;42:916–928. doi: 10.1016/j.immuni.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Lugt B., Khan A.A., Hackney J.A., Agrawal S., Lesch J., Zhou M., Lee W.P., Park S., Xu M., DeVoss J. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat. Immunol. 2014;15:161–167. doi: 10.1038/ni.2795. [DOI] [PubMed] [Google Scholar]
- Varol C., Landsman L., Fogg D.K., Greenshtein L., Gildor B., Margalit R., Kalchenko V., Geissmann F., Jung S. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J. Exp. Med. 2007;204:171–180. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskow C., Liu K., Darrasse-Jèze G., Guermonprez P., Ginhoux F., Merad M., Shengelia T., Yao K., Nussenzweig M. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E., Varol C., Farache J., Elmaliah E., Satpathy A.T., Friedlander G., Mack M., Shpigel N., Boneca I.G., Murphy K.M. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37:1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes have a fold change ≥ 2 and p value ≤ 0.05. Related to Figure 1F.