Abstract
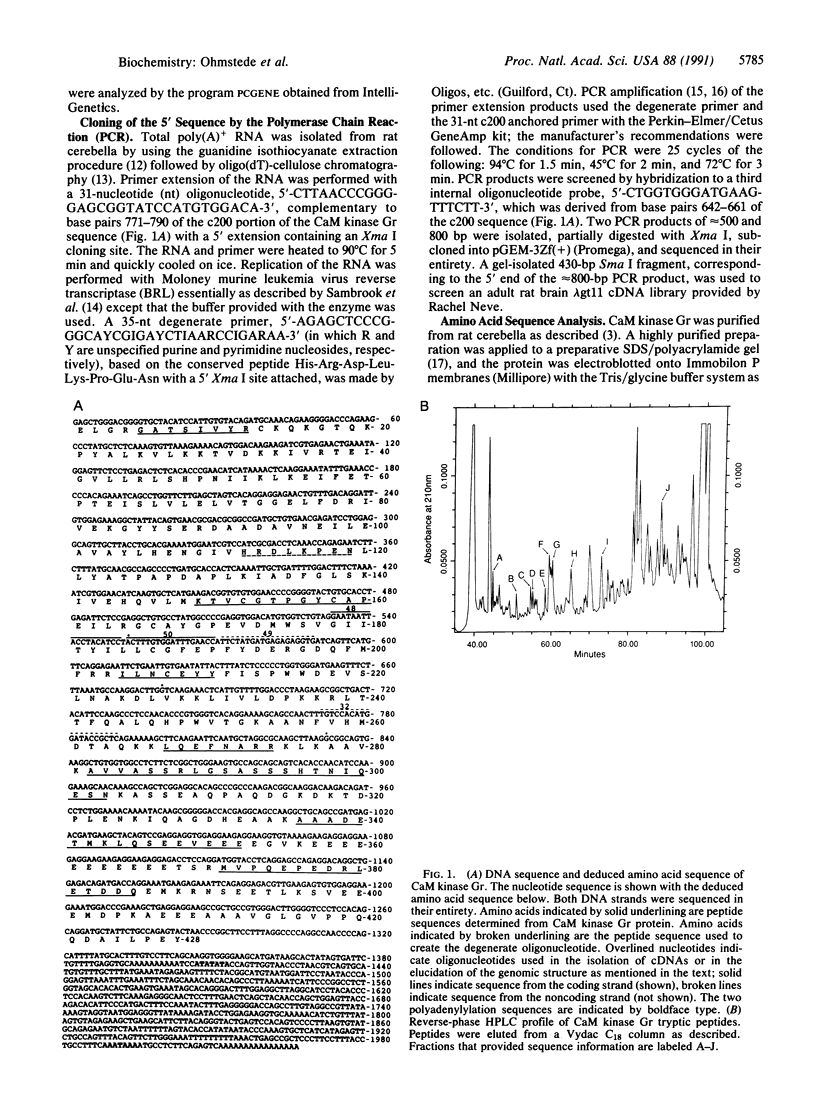
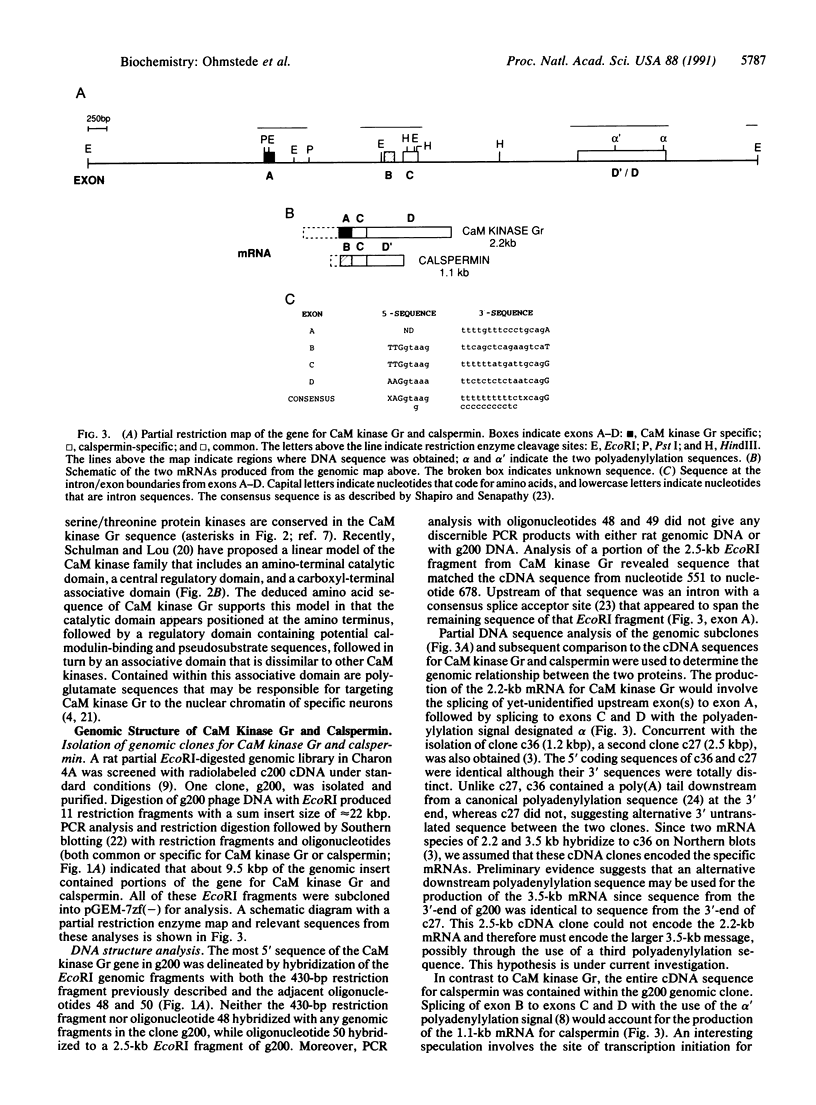
Ca2+/calmodulin-dependent protein kinase enriched in cerebellar granule cells (CaM kinase Gr) is a neuronal calmodulin-dependent protein kinase whose purification and partial cloning from rat brain has been described. A combination of the polymerase chain reaction and cDNA library screening was used to determine the DNA sequence that encodes most of the remaining polypeptide sequence. The deduced amino acid sequence was confirmed by comparison with the peptide sequence from purified CaM kinase Gr. Analysis of this sequence indicated the presence of potential catalytic, regulatory, and association domains with 42% overall homology to the alpha subunit of another neuronal Ca2+/calmodulin-dependent protein kinase, CaM kinase II. The degree of homology within the catalytic domain was 58% with conservation of all invariant amino acids. The portion of sequence that extended from the hypothesized calmodulin-binding domain to the carboxyl terminus of the protein was identical at both the amino acid and nucleotide level to the noncatalytic, calmodulin-binding protein calspermin from rat testis. Screening a genomic library with a portion of the cDNA for CaM kinase Gr allowed the isolation of a genomic clone that contained at least 9 kilobases (kb) of the gene for CaM kinase Gr. Analysis of the sequence revealed that the coding sequences for calspermin were contained within the CaM kinase Gr gene and that alternative splicing of internal exons may lead to the formation of the two different proteins, CaM kinase Gr and calspermin.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Chelly J., Hamard G., Koulakoff A., Kaplan J. C., Kahn A., Berwald-Netter Y. Dystrophin gene transcribed from different promoters in neuronal and glial cells. Nature. 1990 Mar 1;344(6261):64–65. doi: 10.1038/344064a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Dorai T., Wang L. H. An alternative non-tyrosine protein kinase product of the c-src gene in chicken skeletal muscle. Mol Cell Biol. 1990 Aug;10(8):4068–4079. doi: 10.1128/mcb.10.8.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C. Anionic regions in nuclear proteins. J Cell Biol. 1987 Oct;105(4):1479–1482. doi: 10.1083/jcb.105.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S. K., Quinn A. M., Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988 Jul 1;241(4861):42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Jensen K. F., Ohmstede C. A., Fisher R. S., Olin J. K., Sahyoun N. Acquisition and loss of a neuronal Ca2+/calmodulin-dependent protein kinase during neuronal differentiation. Proc Natl Acad Sci U S A. 1991 May 1;88(9):4050–4053. doi: 10.1073/pnas.88.9.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. F., Ohmstede C. A., Fisher R. S., Sahyoun N. Nuclear and axonal localization of Ca2+/calmodulin-dependent protein kinase type Gr in rat cerebellar cortex. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2850–2853. doi: 10.1073/pnas.88.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin C. R., Kapiloff M. S., Durgerian S., Tatemoto K., Russo A. F., Hanson P., Schulman H., Rosenfeld M. G. Molecular cloning of a brain-specific calcium/calmodulin-dependent protein kinase. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5962–5966. doi: 10.1073/pnas.84.16.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairn A. C., Hemmings H. C., Jr, Greengard P. Protein kinases in the brain. Annu Rev Biochem. 1985;54:931–976. doi: 10.1146/annurev.bi.54.070185.004435. [DOI] [PubMed] [Google Scholar]
- Ohmstede C. A., Jensen K. F., Sahyoun N. E. Ca2+/calmodulin-dependent protein kinase enriched in cerebellar granule cells. Identification of a novel neuronal calmodulin-dependent protein kinase. J Biol Chem. 1989 Apr 5;264(10):5866–5875. [PubMed] [Google Scholar]
- Ono T., Slaughter G. R., Cook R. G., Means A. R. Molecular cloning sequence and distribution of rat calspermin, a high affinity calmodulin-binding protein. J Biol Chem. 1989 Feb 5;264(4):2081–2087. [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf S. J., Horn G. T., Erlich H. A. Direct cloning and sequence analysis of enzymatically amplified genomic sequences. Science. 1986 Sep 5;233(4768):1076–1078. doi: 10.1126/science.3461561. [DOI] [PubMed] [Google Scholar]
- Schulman H., Lou L. L. Multifunctional Ca2+/calmodulin-dependent protein kinase: domain structure and regulation. Trends Biochem Sci. 1989 Feb;14(2):62–66. doi: 10.1016/0968-0004(89)90045-5. [DOI] [PubMed] [Google Scholar]
- Schulman H. The multifunctional Ca2+/calmodulin-dependent protein kinase. Adv Second Messenger Phosphoprotein Res. 1988;22:39–112. [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]



