Abstract
Both the 3D solution and the 2D interface play important roles in bioanalysis. For the former, reactions can be carried out adequately; while for the latter, interfering substance can be eliminated simply through wash. It is a challenge to integrate the advantages of solution-based assays and the interface-based assays. Here, we report an immuno-NRCA (netlike rolling circle amplification) strategy, which integrates immunoassay with NRCA for the ultrasensitive detection of tumor biomarker, by taking the assay of a tumour marker as an example. In this strategy, immunoreactions occur on interface, while the target-induced signal amplification can be completed totally in solution. As a result, this system has the merits of both solution- and interface-based assays. The whole procedure of this novel strategy is similar to the conventional ELISA, inheriting the usability. But in comparison with ELISA, the performance is greatly improved. The detection limit can be lowered to 5.5 fg/L, making it possible to detect the target tumour marker in one drop of blood. Also, in comparison with established immuno-PCR method, which integrates immunoassay with the commonly used nucleic acid amplification approach, this system has no requirement for thermal cycler owing to the isothermal amplification, and it has the ability to retain the immunoreactivities. So, the new immunoassay method proposed in this study may have more feasible applications in the future.
Keywords: tumor marker, immunoassay, isothermal amplification, netlike rolling circle amplification, ELISA.
Introduction
Currently, most of the available analytical methods for the detection of biomolecules can be subsumed under two broad categories: the solution-based assays and the interface-based assays. For the former, some well-known methods have been widely applied, e.g. polymerase chain reaction (PCR) for gene analysis 1 and enzyme-based colorimetric detection of small biomolecules like glucose 2-4. A large number of new methods are also being developed, e.g. gene analysis based on isothermal nucleic acid amplification and nanoparticle-based colorimetric assays 5-7. Generally speaking, solution-based assays use UV-vis (especially visible, which is known as "colorimetric") or fluorescent signals. Therefore, a main advantage of these methods is their easy-to-see signals. Another main advantage is the highly efficient signal amplification. Nucleic acid amplification and enzyme (or their mimics)-based catalysis show maximum benefit in the three-dimensional solution system. As a result, solution-based assays can provide excellent detection sensitivity 8-9. In the case of the interface-based assays, enzyme linked immunosorbent assay (ELISA) 10, DNA and protein chips 11, electrochemical biosensors 12, and etc., are usually referred. Because interfering substance can be eliminated simply through washing while the target can be captured onto the detection interface through specific molecular recognition, the most important advantage of the interface-based assays is their applicability for complex samples, e.g. serum and cell extraction.
It is a pity that the advantages of solution-based assays and interface-based assays usually cannot be integrated. In a solution-based assay, the co-existed interfering substance may affect the signal readout, resulting in false positive/negative signals. So, pre-separation is usually required, while a complex sample lies ahead. In an interface-based assay, the detection system usually suffers from low sensitivity. Though a variety of interfacial signal amplification strategies has been developed, the efficiency cannot rival that in solution-based assays. Taking the well-known ELISA for example, the catalytic reactivity of the immobilized enzymes is restricted, because the immobilized enzymes cannot react with the substrates through Brownian movement and a rate-limiting process, i.e. the diffusion of the substrates/products between the solution and the interface, should also been considered in the catalytic kinetics. While only tiny amounts of enzymes are recruited in the case of a low concentration of the target, the restriction on the catalytic reactivity becomes apparent.
In the last decades, a great deal of efforts has been made to develop novel analytical methods to integrate the advantages of solution-based assays and interface-based assays. A variety of biosensing strategies 13, amplification strategies 14, nanomaterials 15 and molecular labels 16 are involved. However, in most cases, the reactions in solution still partially rely on interface (either solid surface or nanomaterials surface) 17-18. A typical exception is that in 1992, Sano et. al. developed an immuno-PCR technique, in which the solution- and interface-based assays were well integrated 19. Like ELISA, in this system, antigen-antibody reactions also occur on interface. That is, the advantages of interface-based assays can be retained. But instead of labeling with enzyme, the antibody is labelled with a DNA fragment, which can be amplified by PCR. Unlike the interface-confined catalysis in ELISA, the immuno-PCR is launched from the interface but completed totally in solution. As a result, the exponential amplification of PCR can maximize the performance, leading to a highly improved sensitivity in comparison with ELISA. Nevertheless, the requirement for thermal cycler and the damage to the antibody-antigen under high temperature have restricted the widespread adoption of immuno-PCR.
Herein, on the basis of our previously developed netlike rolling circle amplification technique (NRCA) 20, we propose an immuno-NRCA strategy for immuno-assays. A tumour marker, i.e. platelet derived growth factor (PDGF), is selected as a representative target. In this strategy, the events of antigen-antibody reactions on interface can be transferred to highly efficient isothermal nucleic acid amplification in solution. Therefore, this system has the advantages of both solution- and interface-based assays. That is, this system can be applied for complex samples without pretreatment, and has an ultrahigh sensitivity in the meantime. Benefited from the ultrahigh sensitivity, only one drop of blood is required in the assay. Also, because of the isothermal amplification, this system has no requirement for thermal cycler, making it cost-efficient and easy-operated.
Materials and Methods
All oligonucleotides used in this research (Table S1) were synthesized by TaKaRa Biotechnology Co., Ltd. (Dalian, China) with HPLC purification. Deoxynucleotide solution mixture (dNTPs) and 6 × loading buffer were also obtained from the TaKaRa Biotechnology. Nb. BsrDI nicking enzyme, Bst DNA polymerase large fragment, and their corresponding buffer were purchased from New England Biolabs Inc. SanPrep Column PCR Product Purification Kit was purchased from Sangon Biotechnology Co., Ltd. (Shanghai, China). Streptavidin was purchased from Sigma Chemical Co. (St. Louis, MO). SYBR Green I was from Solarbio Technology Co., Ltd. Human platelet-derived growth factor (PDGF) ELISA Kit was purchased from Sangon Biotechnology Co., Ltd. (Shanghai, China). Fetal bovine serum (FBS). Other reagents were all of analytical grade and used as received. All solutions were prepared with Milli-Q water (18.2 MΩ cm-1) from a Milli-Q purification system (Millipore Corp, Milford, MA)
Isothermal amplification reactions were performed on a ThermoMixer C (Eppendorf China Ltd.). Gel electrophoresis was conducted using a Wide Mini-Sub Cell GT Cell and a Gel Doc XR Imaging System (Bio-Rad, US). The products of NRCA were also observed directly by an ex situ Agilent 5500 atomic force microscope system (Santa Clara, CA). Fluorescence intensity was measured using a SpectraMax M3 Multi-Mode Microplate Reader with SoftMax Pro software (Molecular Devices, Sunnyvale, CA, USA).
Immune reaction on the microplate. 50 μL volumes of serial dilutions of each test antigen were added to the capture antibody-coated well of the microplate. Negative control wells were filled with 50 μL of blocking buffer. The microplate was incubated at 37 °C for 1 h, the antigen solution was removed and the wells were washed 3 times. Next, 100 μL of biotinylated detection antibody (anti-PDGF antibodies) conjugate was added to the test wells, and the microplate was incubated at 37 °C for 1 h. Then, the conjugate solutions were removed and the wells were washed 3 times. After that, 50μL of 1 nM streptavidin was added to the test wells and incubated at 37 °C for 1 h to form antibody-biotin- streptavidin. The plates were washed 3 times again and 100 μL of 100 nM biotin-labeled primer 1 was added and incubated at 37 °C for 1 h. After the plate was washed and then the conjugate-primer 1 was prepared for the following NRCA.
NRCA reaction. The NRCA reaction was conducted in a 50 μL reaction mixture containing 1 × polymerase buffer, 1 μM Primer 2, 100 nM circular probe, 400 μM dNTPs, 10 U Nb. BsrDI nicking enzyme, and 8 U Bst. DNA polymerase. The reaction was allowed to proceed at 65 °C for 1 h and terminate by deactivating the enzymes at 95 °C for 10 min.
Gel electrophoresis analysis. Agarose gel electrophoresis was performed for the characterization of the products of immuno-NRCA. 5 μL of the products of immuno-NRCA with 1 μL 6 × loading buffer was loaded onto a 2% non-denaturing agarose gel. The electrophoresis experiments were carried out in 1 × Tris-acetate-EDTA (TAE) at 80 V for 30 min. Subsequently, the gel was stained with SYBR Green I for 30 min. The imaging of the gel was performed using a Gel Doc XR Imaging System.
Also, horseradish peroxidase (HRP) labeled anti-human PDGF antibody stored at -20 °C was thawed and diluted and diluted 1:1 with 2 × sample buffer, and loaded onto a 13 % non-denaturing polyacrylamide gel. Gel electrophoresis were done in mini protean gel assembly (BIO-RAD Inc, CA, USA) using 1 × Tris glycine running buffer at a voltage of 100 for 3 h on the ice. The gel was taken out of the cating frame and was stained with Coomassie brilliant blue for 2 h. Subsequently, gel image was captured using Gel Doc XR Imaging System.
Visualization of NRCA products. The products of immuno-NRCA was firstly purified using SanPrep Column PCR Product Purification Kit to exclude the interference of enzymes, dNTPs, and etc. Subsequently, the DNA fragments dissolved in 10 mM MgCl2 were spread onto a fresh mica slice (Yunfeng Co. Ltd., China) and allowed to incubate for 20 min under room temperature. The mica slice was then rinsed with double distilled water and blown completely dried with nitrogen gas. Morphological analysis of the NRCA products on the mica slice was achieved by using an ex situ Agilent 5500 AFM system. Samples were imaged at a scan rate of 0.5-1 Hz in a tapping mode. AFM tips with resonant frequency in a range 160-260 kHz were used. Images were acquired at a resolution of 512 × 512 pixels.
Fluorescent detection. For the detection of different concentration of the antigen, fluorescence spectrum analysis was adopted. 50 μL of NRCA products and 1 μL of 20 × SYBR Green I were added into 50 μL of double distilled water, followed by incubation at room temperature for 10 min. Fluorescence spectra were obtained using a SpectraMax M3 Multi-Mode Microplate Reader. The excitation wavelength was 470 nm; and the spectra were measured in the range from 500 nm to 650 nm. The fluorescence intensity was recorded at the emission wavelength of 520 nm. Fluorescent images were acquired using a Gel Doc XR Imaging System. The fluorescence detection for the interface of the NRCA, HRCA and RCA were obtained using an objective (20 ×, NA 1.3) on a confocal laser scanning fluorescence microscope setup consisting of Axio Scope, Axio Imager and Axio observer inverted microscope with a Carl Zeiss LSM710 confocal scanning system. An Ar+ laser (488 nm) was used as the excitation source for the detection of SYBER Green I, and a bandpass filter of 500-550 nm was used for fluorescence detection.
ELISA. In the conventional ELISA experiments, 50 μL volumes of serial dilutions of each test antigen were added to the capture antibody-coated well of the microplate. The microplate was incubated at 37 °C for 1 h and then washed 3 times. 100 μL of the biotin-conjugated anti-PDGF detection antibody was added and incubated at 37 °C for 1 h with shaking. The plate was washed again and 50 μL of the streptavidin-conjugated HRP solution was added and incubated at 37 °C for 1 h. After incubation, the plate was washed, and then 50 μL of the substrate A and B were added. After the plate was incubated for another 15 min at 37 °C, 50 μL of stop solution was added to stop the reaction, and the plate was read using an ELISA plate reader at 450 nm.
Detection of PDGF in real serum samples. Serum from patients who suffered lung cancer were provided by Shanghai Pulmonary Hospital. Serum samples were collected from sixty lung carcinoma patients and healthy individuals. As for the detection of PDGF, about 3 mL blood was required, which were stepwise diluted to about 10 ~ 20-fold in conventional ELISA experiments as mentioned above. Only 10 μL serum of each sample was used for the immuno-NRCA assay, which were stepwise diluted to about 4 ~ 5 × 106-fold. 50 μL volumes of serial dilutions of serum were added to the capture antibody-coated well of the microplate. The microplate was incubated at 37 °C for 1 h, the antigen solution was removed and the wells were washed 3 times. Next, 100 μL of biotinylated detection antibody (anti-PDGF antibodies) conjugate was added to the test wells, and the microplate was incubated at 37 °C for 1 h. Then, the conjugate solutions were removed and the wells were washed 3 times. After that, 50 μL of 1 nM streptavidin was added to the test wells and incubated at 37 °C for 1 h to form antibody-biotin- streptavidin. The plates were washed 3 times again and 100 μL of 100 nM biotin-labeled primer 1 was added and incubated at 37 °C for 1 h. After the plate was washed and then the conjugate-primer 1 was prepared for the following NRCA. The NRCA reaction was conducted in a 50 μL reaction mixture containing 1 × polymerase buffer, 1 μM Primer 2, 100 nM circular probe, 400 μM dNTPs, 10 U Nb. BsrDI nicking enzyme, and 8 U Bst. DNA polymerase. The reaction was allowed to proceed at 65 °C for 1 h and terminate by deactivating the enzymes at 95 °C for 10 min. Fluorescence spectrum analysis was adopted. 50 μL of NRCA products and 1 μL of 20 × SYBR Green I were added into 50 μL of double distilled water, followed by incubation at room temperature for 10 min. Fluorescence spectra were obtained using a SpectraMax M3 Multi-Mode Microplate Reader.
Results and Discussion
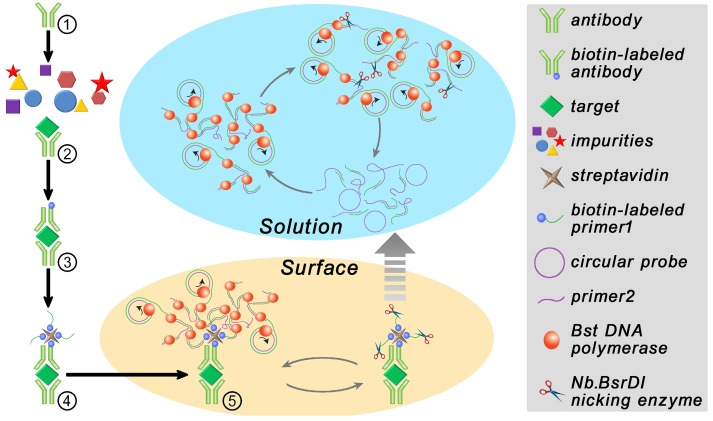
The procedure and principle of the proposed immuno-NRCA is shown in Scheme 1. The first four steps are similar to that in ELISA. That is (1) An anti-PDGF antibody-coated microplate is prepared. (2) Add sample into the plate well and incubate for a while to allow the specific binding of the target PDGF onto the interface; (3) After washing, biotin-labeled antibody is added; (4) After washing, streptavidin and a biotin-labeled DNA primer 1 (biotin-labeled HRP instead in ELISA) are added successively. In our system, the next fifth step is the core step, in which a DNA Primer 2, a circular probe, dNTPs, DNA polymerase and DNA nicking enzyme are added simultaneously. Three interdependent reactions occur in this system: (1) DNA polymerase extends the primer 1 using the circular probe as the template. This reaction is well known as the RCA; (2) In the presence of a reverse primer 2, ramified extension and strand displacement proceed simultaneously, resulting in the HRCA. (3) The nicking enzyme scissors the extended primer 1 into fragments and releases them into the solution. After this, these three reactions can cycle in solution independent from the interface. As a result, the cubic amplification (named as NRCA, including primer extension, strand displacement and nicking reaction) in solution is expected to maximize the signal amplification performance. Here, this detection system integrating immuno-assays with NRCA is termed as immuno-NRCA.
Scheme 1.
Schematic presentation of the principle of immuno-NRCA assay.
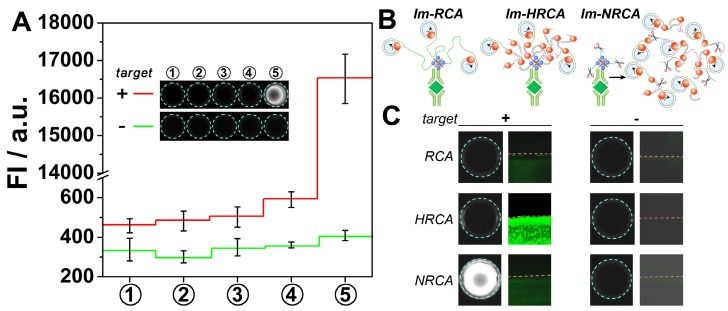
Proof-of-concept experiments were first carried out to verify the above scheme. SYBER Green I, a commonly used fluorescent molecule for quantitative analysis of DNA, is adopted to check whether large amounts of DNA can be produced in solution. As is shown Fig. 1A, in the presence of the target PDGF, no apparent fluorescent signals can be observed for the first four steps. However, after the cubic amplification is launched in the fifth step, a large leap of the fluorescent signal can be observed. The fluorescence intensity increases almost forty times, showing bright kelly under UV light (presenting bright white in the figure by using a monochrome camera). While in the absence of PDGF, no apparent signal changes can be observed all through the experiments. This result suggests the expected PDGF-induced DNA amplification. During the DNA amplification in the fifth step, three oligonucleotides are involved. Results also show that in the absence of any of them, the signal can't be amplified adequately (Fig. S1), suggesting the necessity of each of these oligonucleotides in the amplification. While the oligonucleotides are changed to another rationally designed set, the PDGF-induced DNA amplification in solution can be also observed (Fig. S2, the rightmost column). However, if these two unrelated oligonucleotides sets are mashed up, the amplified signals almost disappear (Fig. S2, the middle six columns). This result suggests that the cubic amplification relies on the combination of three dependent oligonucleotides, i.e. primer 1, circular probe, and primer 2. The sequences are adjustable; and different oligonucleotides sets can work independently, which may allow the detection of different targets simultaneously.
Figure 1.
(A) Fluorescent signals and overall appearance under UV-light (inset) obtained from each step of immuno-NRCA in the presence and absence of PDGF, respectively. (B) Schematic presentation of immuno-RCA, -HRCA and -NRCA. (C) Comparison of the performance of immuno-NRCA with immuno-RCA, and -HRCA in the presence and absence of PDGF. The left columns show the photographs of the overall appearance under UV-light. The right columns are the confocal fluorescence images of the interface.
Im-RCA and Im-HRCA are also conducted as comparison (Fig. 1B). In these two cases, no apparent fluorescence can be observed under UV light (Fig. 1C, the left graphs). The surficial fluorescence is also monitored using a laser scanning confocal microscope. A drop of solution with or without PDGF is adopted as the sample to cover only a little circle on the well, so as to produce a clear edge under the confocal microscope. As is showing in Fig. 1C (the middle graphs), if PDGF is presented in the drop of sample (the lower parts under the red dash lines), green fluorescence can be observed in all the three cases. It is also noted the surficial fluorescence of Im-HRCA is much brighter than the others. In control experiments, while PDGF is absent, no apparent surficial fluorescence can be observed for all the three cases. The confocal microscopic results as well as the photographic results under UV light agree with the principles. Briefly, in the case of Im-RCA and Im-HRCA, DNA amplification is confined only on the 2D interface. So, the fluorescence intensity is not homogeneous and high enough to produce a recordable signal under a 3D recorder. Nevertheless, while using a sensitive 2D recorder (confocal microscopy here), Im-RCA and especially Im-HRCA can produce recognizable signal. For Im-HRCA, hyperbranched double stranded DNA is produced, which is favorable for the intercalate of the fluorescent molecule (SYBR Green I). So, much bright fluorescence can be observed. In the case of immuno-NRCA, most of the amplified DNA has been scissored and released into the solution, retaining only short fragments on the interface. So, in contrast with the faint fluorescence on interface, the overall fluorescence under UV light is much brighter. In view of the availability of the signal, the direct observation of immuno-NRCA has a great advantage over the others.
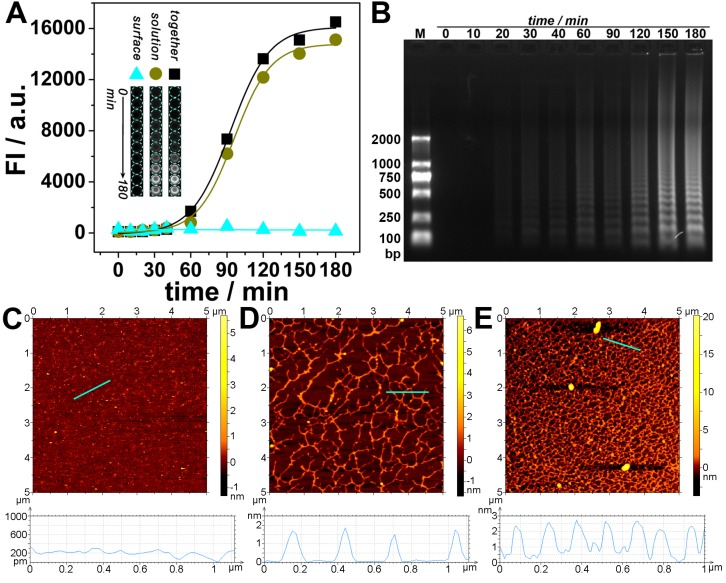
Experiments have been conducted to further confirm the transfer of DNA amplification from interface to solution. The solution is separated from the interface and transferred to a new well, while the interface is reperfusion with equal amount of buffer solution. As is shown in Fig. 2A, along with the amplification time, the fluorescence of the interface keeps almost unchanged, whereas the fluorescence of the solution as well as the total fluorescence increases and reaches a plateau at about 180 min. This time is also adopted as an optimized amplification time for the following experiments. Fig. 2A also shows that the fluorescence of the solution dominates and is approximately the same as that of the total fluorescence. Electrophoretic and AFM results confirm that a large amount of amplified DNA has been produced in the solution (Fig. 2B~E); and the amount depends on the amplification time.
Figure 2.
(A) Fluorescent signals and overall appearance under UV-light (inset) obtained from interface, solution, and both together respectively after different amplified reaction times. (B) Agarose gel electrophoretic analysis of immuno-NRCA products in solution. (C-E) AFM images of immuno-NRCA products in solution. The reaction times of NRCA are 0 (C), 60 (D) and 180 (E) min, respectively. The heights of the positions marked with cyan lines are shown under the corresponding AFM images.
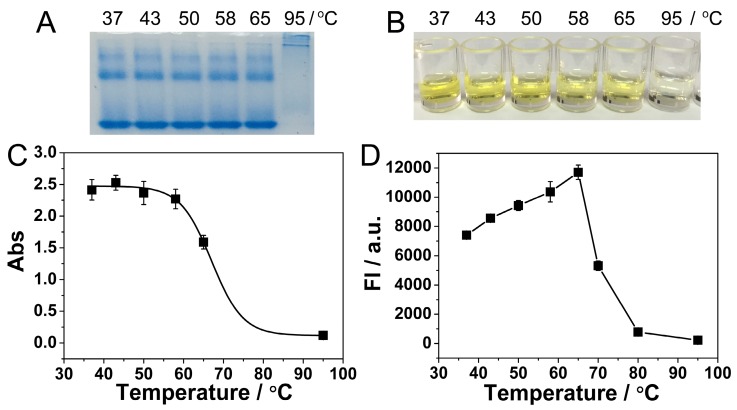
Another advantage of our immuno-NRCA system is the isothermal amplification and no requirements for thermal cycling. However, the optimum temperature for Nb. BsrDI nicking enzyme and Bst DNA polymerase is 65 °C, a relatively high temperature for immunoreactions. Considering the stability of the detection system and the reutilization of the antigen-antibody, a suitable temperature that can balance the DNA amplification and the reactivity of antigen-antibody should be found. As is shown in the protein electrophoretic pattern in Fig. 3A, the bands of anti-PDGF antibody remain unchanged in a temperature range from 37 to 65 °C. While 95 °C, a temperature for PCR, is applied, the bands miss, suggesting the denaturation of the antibody. ELISA is also conducted to study the impact of temperature on the reactivity of immunoreaction. During ELISA, PDGF is incubated with its antibody at different temperature, and then the enzyme-catalytic color reaction is conducted. As is shown in Fig. 3B and C, the color becomes lighter and the absorbance becomes lower with the increase of temperature. Especially at 95 °C, no apparent color or absorbance can be observed, suggesting the absolute loss of the immunoreactivity. At 65 °C, the immunoreactivity also loses about 1/3. Fig. 3D shows the impact of temperature on NRCA. With the increase of temperature, the efficiency of NRCA increases and reaches a plateau at 65 °C, and then decreases sharply. From Fig. 3C and 3D, it can be concluded that there is a trade-off in the range from 37 to 65 °C: with the increase of temperature, the efficiency of NRCA increases but the immunoreactivity decreases. After comprehensive consideration, 58 °C is adopted as an optimum temperature for the immuno-NRCA system. Under this temperature, the immunoreaction retains about 94% of its maximum activity at 37 °C (Fig. 3C), and the NRCA exerts 90% of its maximum efficiency at 65 °C (Fig. 3D).
Figure 3.
(A) Polyacrylamide gel electrophoretic patterns of the anti-PDGF antibody, which has been pre-incubated at different temperature for 3 hours. Photograph (B) and absorption at a wavelength of 450 nm (C) of ELISA detection of 10 ng/L PDGF. During the ELISA, the PDGF was allowed to react with the antibody at dirrerent temperatures. (D) Relationship between the fluorescence intensity of NRCA products and the reaction temperature. The NRCA was conducted independent of the immunoreaction, using 100 μL of 100 nM primer 1-1 as the trigger directly.
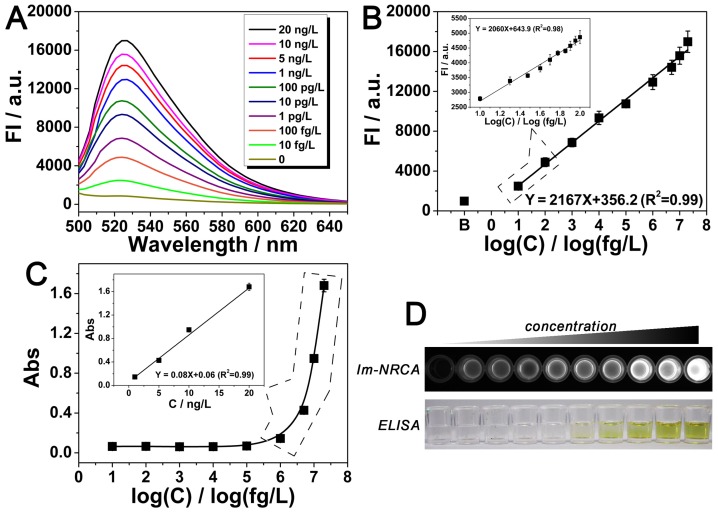
Detection of the target PDGF using the immuno-NRCA is then performed. As is shown in Fig. 4A and B, the fluorescence intensity increases with the PDGF concentration. In a large range from 10 fg/L to 20 ng/L, there is a linear relationship between the fluorescence intensity and the logarithmic concentration of PGDF. The detection limit is calculated to be 5.5 fg/L (defined as S/N = 3). Considering that the physiological concentration of PDGF in serum usually fluctuates in about one order of magnitude, we also narrow the concentration of PDGF to 10~100 fg/L, and study the detailed relationship between the signal and the concentration. As is shown in the inset of Fig. 4B, in this narrow range, a linear relationship can be also obtained. In comparison with the conventional ELISA method which just reaches a detection limit of 0.13 ng/L (LOD = 3SD / k, LOD: detection limit, SD: the standard deviation of blank sample, k: the slope of the fitting curve) (Fig. 4C and D), immuno-NRCA possesses a much better sensitivity. To further investigate the specificity of the proposed detection method, four kinds of potential interfering substances: bovine serum albumin (BSA), nuclear factor kappa-B (NF-κB), vascular endothelial growth factor (VEGF) and thrombin (Tb) are adopted. As is shown in Fig. S3, significant difference between the target and the interfering substances can be observed, suggesting a good specificity.
Figure 4.
(A) Emission spectra of the dye-intercalated products of immuno-NRCA in the presence of different concentrations of the target PDGF. The excitation wavelength was fixed at 470 nm. (B) Relationship between the fluorescence intensity at 520 nm of (A) and the corresponding concentration of the target protein. Inset: fluorescence intensity vs. PDGF concentration in a narrow concentration range from 10 fg/L to 100 fg/L. (C) Detection of PDGF using the conventional ELISA. The relationship between the UV-vis aborbance at 450 nm and the corresponding concentration of PDGF is presented. (D) Comparison of the overall appearance of the detection using immuno-NRCA and ELISA, respectively. The concentration of PDGF from left to right: 0, 10 fg/L, 100 fg/L, 1 pg/L, 10 pg/L, 100 pg/L, 1 ng/L, 5 ng/L, 10 ng/L, 20 ng/L.
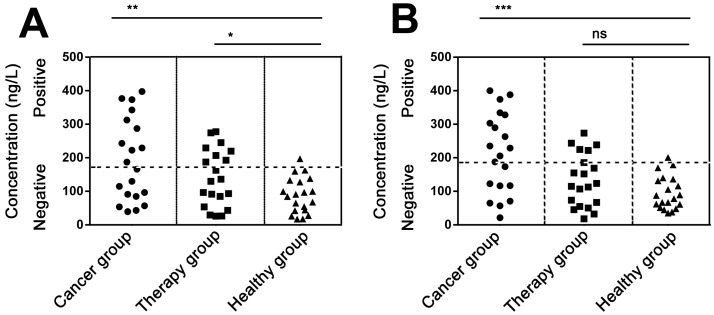
We next try to detect PDGF in real serum samples. In experiments, serum samples were collected from sixty lung carcinoma patients and healthy individuals. Only 10 μL serum of each sample was used for the immuno-NRCA assay. As is shown in Fig. 5A and B, we detect the concentration of serum PDGF using the conventional ELISA and our immuno-NRCA, respectively. The detailed data have been shown in the supporting information (Table S2). Similar results can be obtained by using these two methods. Results show that the mean concentration is higher in lung carcinoma patients (192.4 ng/L using ELISA and 214.1 ng/L using immuno-NRCA) than in patients who has been treated (140.3 ng/L using ELISA and 133.1 ng/L using immuno-NRCA) as well as those healthy controls (87.6 ng/L using ELISA and 95.4 ng/L using immuno-NRCA). Using the cut-off level of 192.8 ng/ml for ELISA and 195.1 ng/ml for immuno-NRCA, corresponding to the mean + 2SD of PDGF in healthy controls, ca. 50% (45% using ELISA and 55% using immuno-NRCA) of lung carcinoma patients and ca. 30% (35% using ELISA and 25% using immuno-NRCA) of treated patients show positive PDGF levels. The diagnostic and prognostic role of serum PDGF shown here is in accordance with literatures 21-22.
Figure 5.
Quantification of PDGF in the serum of lung carcinoma patients without treatment (cancer group), with chemotherapy or radiotherapy (therapy group), and healthy individuals (healthy group) using (A) ELISA and (B) immuno-NRCA, respectively. The positive signals are set to be on or above the healthy threshold (the horizontal dash line), which is defined as the mean + 2SD of PDGF in healthy individuals. (*P < 0.05, **P < 0.01, ***P < 0.001, ns: no significant difference by Student's t-test).
Finally, we would like to make a short discussion. The immuno-NRCA we developed here shows considerable usability in comparison with ELISA. The 96-wells ELISA plates as well as the antibody used in ELISA can be retained only with the replacement of the HRP catalytic system by the NRCA system. But in comparison with ELISA, the sensitivity of immuno-NRCA has been greatly improved for about 20 000 folds, making it possible to detect the target in trace amount of samples. The increased sensitivity should be ascribed to two points: (1) The cubic nucleic acids amplification catalyzed by cascade enzymatic reactions is much more efficient than the single enzyme-based amplification in ELISA; (2) The target-induced signal amplification can be completed totally in solution, avoiding the limits of interface-based assays. In some ways, the immuno-NRCA has common with the existing immuno-PCR technique. For example, they both combine immunoreactions with DNA amplification, and integrate the advantages of solution- and interface-based assays. The improvement of the sensitivity of immuno-PCR can also reach as high as 100 000 folds, while in comparison with ELISA 23. Nevertheless, there are significant advantages of immuno-NRCA over the immuno-PCR. Owing to the isothermal amplification, no thermal cycler is required, making the immuno-NRCA method cost-efficient and easy-operated. In addition, during the immuno-NRCA, the antibody and antigen can retain the immunoreactivity, whereas during the immuno-PCR, they are denaturized under the high temperature.
Conclusion
In summary, we have successfully combined isothermal NRCA technique with immunoassays to develop a novel immuno-NRCA system for the detection of PDGF. It is the first time that solution-based assays and interface-based assays are integrated and conducted under isothermy. Owing to the transfer of the DNA amplification from interface to solution, the signal amplification performance can be maximized to result in an ultrahigh sensitivity. The detection limit is improved to 5.5 fg/L, almost 20 000 folds lower than the conventional ELISA (0.13 ng/L). Detection of PDGF in trace amount of serum samples is thereby achieved. Because of the universality of immunoreactions, this novel immunoassay can also work as a versatile platform for the detection of a variety of antigens or antibodies, especially those with ultralow abundance.
Supplementary Material
Supplementary tables S1-S2 and figures S1-S3.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 21575088, 21235003,) and the Natural Science Foundation of Shanghai (14ZR1416500).
Abbreviations
- Immuno-NRCA
Immuno-netlike rolling circle amplification
- Im-HRCA
Immuno-hyperbranched rolling circle amplification
- Im-RCA
Immuno-rolling circle amplification
- ELISA
Enzyme-linked immunosorbent assay
- PCR
Polymerase chain reaction
- PDGF
Platelet derived growth factor
- dNTPs
Deoxynucleotide solution mixture
- FBS
Fetal bovine serum
- HRP
horseradish peroxidase
References
- 1.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro:the polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 1986;51:263–73. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 2.Chen S, Hai X, Chen XW, Wang JH. In situ growth of silver nanoparticles on graphene quantum dots for ultrasensitive colorimetric detection of H₂O₂ and glucose. Anal Chem. 2014;86:6689–94. doi: 10.1021/ac501497d. [DOI] [PubMed] [Google Scholar]
- 3.Yuan J, Cen Y, Kong XJ, Wu S, Liu CL, Yu RQ, Chu X. MnO2-nanosheet-modified upconversion nanosystem for sensitive turn-on fluorescence detection of H2O2 and glucose in blood. ACS Appl Mater Interfaces. 2015;7:10548–55. doi: 10.1021/acsami.5b02188. [DOI] [PubMed] [Google Scholar]
- 4.Zhou C, Zhao J, Pang D, Zhang Z. Enzyme-induced metallization as a signal amplification strategy for highly sensitive colorimetric detection of avian influenza virus particles. Anal Chem. 2014;86:2752–9. doi: 10.1021/ac404177c. [DOI] [PubMed] [Google Scholar]
- 5.Cheng F, He Y, Xing X, Tan D, Lin Y, Pang D. et al. A gold nanoparticle-based label free colorimetric aptasensor for adenosine deaminase detection and inhibition assay. Analyst. 2015;140:1572–7. doi: 10.1039/c4an02070g. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Li F, Chen H, Ma Y, Qi S, Chen X, Zhou L. AuNPs colorimetric sensor for detecting platelet-derived growth factor-BB based on isothermal target-triggering strand displacement amplification. Sens Actuators B Chem. 2015;207:748–55. [Google Scholar]
- 7.Shi C, Liu Q, Zhou M, Zhao H, Yang T, Ma C. Nicking endonuclease-mediated isothermal exponential amplification for double-stranded DNA detection. Sens Actuators B Chem. 2016;222:221–5. [Google Scholar]
- 8.Wei L, Wang X, Wu D, Li C, Yin Y, Li G. Proximity ligation-induced assembly of DNAzyme for simple and cost-effective colorimetric detection of protein with high sensitivity. Chem Commun. 2016;52:5633–6. doi: 10.1039/c6cc00205f. [DOI] [PubMed] [Google Scholar]
- 9.Zou P, Liu Y, Wang H, Wu J, Zhu F, Wu H. G-quadruplex DNAzyme-based chemiluminescence biosensing platform based on dual signal amplification for label-free and sensitive detection of protein. Biosens Bioelectron. 2016;79:29–33. doi: 10.1016/j.bios.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry. 1971;9:871–4. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 11.Lathwal S, Sikes HD. Assessment of colorimetric amplification methods in a paper-based immunoassay for diagnosis of malaria. Lab Chip. 2016;16:1374–82. doi: 10.1039/c6lc00058d. [DOI] [PubMed] [Google Scholar]
- 12.Huang H, Bai W, Dong C, Guo R, Liu Z. An ultrasensitive electrochemical DNA biosensor based on graphene/Au nanorod/polythionine for human papillomavirus DNA detection. Biosens Bioelectron. 2015;68:442–6. doi: 10.1016/j.bios.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 13.Sang S, Wang Y, Feng Q, Wei Y, Ji J, Zhang W. Progress of new label-free techniques for biosensors: a review. Crit Rev Biotechnol. 2016;36:465–81. doi: 10.3109/07388551.2014.991270. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115:12491–545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 15.Chinen AB, Guan CM, Ferrer JR, Barnaby SN, Merkel TJ, Mirkin CA. Nanoparticle probes for the detection of cancer biomarkers, cells, and tissues by fluorescence. Chem Rev. 2015;115:10530–74. doi: 10.1021/acs.chemrev.5b00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng J, Yang R, Shi M, Wu C, Fang X, Li Y, Li J, Tan W. Rationally designed molecular beacons for bioanalytical and biomedical applications. Chem Soc Rev. 2015;44:3036–55. doi: 10.1039/c5cs00020c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong B, Akter R, Han OH, Rhee CK, Rahman MA. Increased electrocatalyzed performance through dendrimer-encapsulated gold nanoparticles and carbon nanotube-assisted multiple bienzymatic labels: highly sensitive electrochemical immunosensor for protein detection. Anal Chem. 2013;85:1784–91. doi: 10.1021/ac303142e. [DOI] [PubMed] [Google Scholar]
- 18.Zhu X, Feng C, Ye Z, Chen Y, Li G. Fabrication of magneto-controlled moveable architecture to develop reusable electrochemical biosensors. Sci Rep. 2014;4:4169. doi: 10.1038/srep04169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sano T, Smith CL, Cantor CR. Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science. 1992;258:120–2. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 20.Zhu X, Feng C, Zhang B, Tong H, Gao T, Li G. A netlike rolling circle nucleic acid amplification technique. Analyst. 2015;140:74–8. doi: 10.1039/c4an01711k. [DOI] [PubMed] [Google Scholar]
- 21.Gui R, Huang ZY, Liu J, Li HW, Nie XM. Expression of platelet-derived growth factor-BB in patients with lung cancer. Chin J Blood Transfusion. 2009;10:810–2. [Google Scholar]
- 22.Feng X, Sun Y, Xia A. Clinical significance of the combined determination of NSE, CYFRA21-1, PDGF-BB and D-D for the patients with lung cancer. Laboratory Medicine. 2011;26:741–5. [Google Scholar]
- 23.Niemeyer CM, Adler M, Wacker R. Immuno-PCR: high sensitivity detection of proteins by nucleic acid amplification. Trends Biotechnol. 2005;23:208–16. doi: 10.1016/j.tibtech.2005.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables S1-S2 and figures S1-S3.