Abstract
Mutation of the PARK2 gene can promote both Parkinson's Disease and cancer, yet the underlying mechanisms of how PARK2 controls cellular physiology is incompletely understood. Here, we show that the PARK2 tumor suppressor controls the apoptotic regulator BCL-XL and modulates programmed cell death. Analysis of approximately 10,000 tumor genomes uncovers a striking pattern of mutual exclusivity between PARK2 genetic loss and amplification of BCL2L1, implicating these genes in a common pathway. PARK2 directly binds to and ubiquitinates BCL-XL. Inactivation of PARK2 leads to aberrant accumulation of BCL-XL both in vitro and in vivo, and cancer-specific mutations in PARK2 abrogate the ability of the ubiquitin E3 ligase to target BCL-XL for degradation. Furthermore, PARK2 modulates mitochondrial depolarization and apoptosis in a BCL-XL-dependent manner. Thus, like genes at the nodal points of growth arrest pathways such as p53, the PARK2 tumor suppressor is able to exert its antiproliferative effects by regulating both cell cycle progression and programmed cell death.
Introduction
Genetic alterations in PARK2 are common across many cancers as well as in juvenile Parkinson's Disease [1], [2], [3], [4], [5]. In human malignancies, PARK2 is a tumor suppressor that is mutated and/or deleted, with copy number loss being the dominant mode of alteration [1], [6], [7]. The PARK2 gene encodes a ubiquitin E3 ligase that can target proteins for degradation through the ubiquitin-proteasome system [8], [9]. Loss of PARK2 can lead to acceleration of tumorigenesis or enhanced tumor aggressiveness [6], [7], [10], [11].
As a tumor suppressor, PARK2 may have pleiotropic effects. Recent studies have shown that PARK2 is an important regulator of cell cycle progression [3]. PARK2 controls the cell cycle by acting as a master regulator of multiple G1/S cyclins. Using systems mainly geared towards examining neural biology, several studies have shown that PARK2 plays a role in mitochondrial maintenance and turnover [12], [13], [14]. However, the full extent of PARK2’s functions and how inactivation of PARK2 promotes tumor growth is unknown.
It is well known that the BCL-2 family of proteins is central to regulation of apoptosis [15], [16]. These proteins govern mitochondrial outer membrane permeabilization (MOMP) and can either be pro-apoptotic (i.e., BAX, BAK, PUMA) or anti-apoptotic (i.e., BCL-2, BCL-XL). The anti-apoptotic functions of BCL-XL include: prevention of activation of pro-apoptotic factors, enhancement of bioenergetic efficiency, prevention of mitochondrial permeability transition channel activity, protection from mitochondrial outer membrane permeabilization to pro-apoptotic factors, and improvement in the rate of vesicular trafficking [17]. In cancer, BCL-XL is involved in a tumor cell's ability to escape programmed cell death. Overexpression of BCL-XL occurs in diseases such as non-small-cell lung cancer (NSCLC) [18], [19], breast cancer [1], giant-cell tumors of the bone [20], and lymphomas [16], [21], [22], [23]. How BCL-XL levels are controlled is still not completely understood.
In the present study, we demonstrate that PARK2 tumor suppressor plays important roles in regulating cell death via modulating the stability of BCL-XL protein. More specifically, our in silico results of pan-cancer analyses show that deletions involving the PARK2 gene are significantly anti-correlated with focal amplifications of the gene encoding BCL-XL. Our further in vitro and in vivo experiments reveal that PARK2 binds to and ubiquitinates BCL-XL protein to control apoptosis.
Results
Genetic Evidence Demonstrates the PARK2 Tumor Suppressor Gene is Integrally Involved in BCL-XL Regulation
One approach to determine the function of a genetic alteration is to identify alterations that are mutually exclusive with it, since patterns of genetic alterations can be used to delineate epistasis and define biological pathways [24], [25]. The availability of large numbers of genome-wide data from many cancer types allows the in silico inference of relationships between different somatic lesions [4], [26]. Previously, we showed that PARK2 deletion was frequent across human malignancies [3]. Here, we examined copy number data from 10,844 patient samples across 33 cancer types (TCGA data) as previously described [4] and identified a number of genetic lesions that were significantly anti-correlated with PARK2 loss (Supplementary Table 1). Figure 1A shows the first order anti-correlation relationships shared between PARK2 loss and other copy number alterations. These instances of anti-correlation were identified after rigorously controlling for tumor lineage and for overall levels of genomic disruption, both of which can confound correlation analyses [4], [27]. PARK2 loss is significantly associated with amplification of CCNE1 (encodes cyclin E1) and amplification of CCND1 (cyclin D1), as previously described [3], as well as amplification of BRD4.
Figure 1.
Pan-cancer analysis implicates PARK2 in the regulation of program cell death via BCL-XL. (A) Network of significant regions of amplification and deletion that anti-correlate with deletions of PARK2. (B) Significance of anticorrelation between PARK2 deletion and amplification of BCL2L1. The graph represents the distribution of co-occurrences of PARK2 deletion and BCL2L1 amplification in our lineage-controlled permutations. The red line represents the observed number of co-occurrences. Blue line represents the predicted number of co-occurrences in the absence of a correlative relationship, as calculated by multiple permutations. P values that correspond to these graphs are provided in Supplementary Table 2.
Pan-cancer analysis implicates PARK2 in the regulation of program cell death via BCL-XL. (A) Network of significant regions of amplification and deletion that anti-correlate with deletions of PARK2. (B) Significance of anticorrelation between PARK2 deletion and amplification of BCL2L1. The graph represents the distribution of co-occurrences of PARK2 deletion and BCL2L1 amplification in our lineage-controlled permutations. The red line represents the observed number of co-occurrences. Blue line represents the predicted number of co-occurrences in the absence of a correlative relationship, as calculated by multiple permutations. P values that correspond to these graphs are provided in Supplementary Table 2.
Interestingly, PARK2 deletion was significantly anti-correlated with focal amplifications of BCL2L1, which is a known oncogene that encodes the anti-apoptotic protein BCL-XL. Our analysis, which used a lineage control to detect anti-correlations between peak regions of significance, found strong anti-correlation between amplification at the loci containing BCL2L1, CCND1, and 11q14.1, as well as between the presence of amplification of either of these regions and PARK2 loss (Figure 1A and Supplementary Table 1). The anti-correlation between PARK2 loss and BCL2L1 amplification was highly statistically significant and was much stronger than would be expected in the absence of selective pressure (Figure 1B). The genetic relationship between PARK2 and BCL2L1 suggests that these genes are functionally related. One possibility is that, if PARK2 loss serves a similar function as copy gains of BCL2L1, this redundancy should lead to anti-correlation of these events in tumors [28], [29], this relationship is especially strong in tumor lineages that where alterations of PARK2 and BCL2L1 are frequent (i.e., ovarian, breast cancer) (Supplementary Table 2). Such a pattern of mutual exclusivity is consistent with PARK2 and BCL-XL functioning in a common pathway.
The PARK2 Ubiquitin E3 Ligase Interacts with BCL-XL Protein
Although our genetic data implicate a functional relationship between PARK2 and BCL-XL, the mechanistic basis underlying this relationship is not well understood. To investigate whether BCL-XL and PARK2 associate with each other, we performed co-immunoprecipitation experiments using lysates from several different human cell lines (A549, T202, and HEK 293 T). BCL-XL protein associates with PARK2 under both normal and apoptotic conditions (Figure 2A and Supplementary Figure 1). Next, we verified the interaction between PARK2 and BCL-XL by expressing these proteins in combination using the baculovirus system and characterizing their association. As expected, PARK2 interacted with BCL-XL but not non-specific antibody control, as assessed by immunoprecipitation (Figure 2B).
Figure 2.
PARK2 interacts with BCL-XL. (A) Immunoprecipitation assays showing binding of PARK2 to BCL-XL endogenously. (B) Recapitulation of PARK2 binding associations using baculovirus. Sf9 cells were co-transduced with different combinations of PARK2 and BCL-XL genes as indicated. Associated proteins were co-purified using immunoprecipitation. (C) Cancer-specific mutations of PARK2 decrease affinity of PARK2 for BCL-XL. A549 cells were transfected with vector only (pcDNA3.1), WT PARK2 (Flag-tagged) or one of three mutant PARK2 cDNAs (Flag-tagged). After treatment with MG132 (25 μM), cell lysates were subjected to immunoprecipitation with antibody to Flag followed by western blotting.
Cancer-specific mutations have been found in the PARK2 gene [6], [7], [30]. To determine whether these mutations affect the ability of PARK2 to bind BCL-XL, we used site-directed mutagenesis to introduce these mutations into PARK2 cDNA. These mutant cDNAs were then expressed in cells to generate mutant PARK2 protein. Co-immunoprecipitation assays showed that all three PARK2 mutants tested had a decreased ability to associate with BCL-XL (Figure 2C). Together, these data demonstrate that PARK2 normally associates with BCL-XL and mutations of PARK2 found in cancer help abrogate this association.
PARK2 Regulates Degradation of BCL-XL Protein
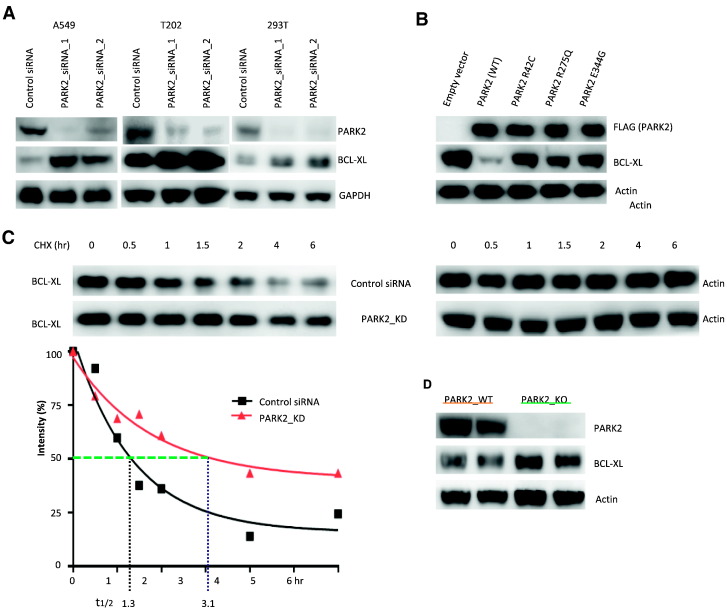
To determine the functional significance of the interaction of BCL-XL and PARK2, we knocked down PARK2 in a number of human cell lines and analyzed BCL-XL levels. Depletion of PARK2 with two different siRNAs but not control caused accumulation of BCL-XL protein (Figure 3A). Furthermore, overexpression of PARK2 wild-type cDNA led to reduced BCL-XL, an effect not seen with PARK2 harboring cancer-specific mutations Figure 3B). Furthermore, PARK2 altered the stability of BCL-XL protein. Cycloheximide chase experiments indicated a clear change in the stability of BCL-XL when PARK2 was inactivated. The half-life time of BCL-XL was increase from 1.3 hr. to 3.1 hr. when PARK2 was knocked down, showing that PARK2 controls the steady-state levels of BCL-XL (Figure 3C). We also used PARK2 knockout mouse to examine the effects of PARK2 on BCL-XL expression levels (The Jackson Laboratory, USA). We observed that, in liver, BCL-XL protein levels in PARK2-deficit mice were higher than that in PARK2 wild-type mice (Figure 3D). Levels were not changed in some other tissues examined, indicating that other factors may also influence BCL-XL levels.
Figure 3.
BCL-XL protein level is regulated by the PARK2 ubiquitin ligase. (A) Knockdown of PARK2 results in increased BCL-XL levels. Cells indicated were transfected with scrambled siRNA control or PARK2 siRNAs and western blotting were performed with the antibodies indicated. Representative results shown from triplicates. (B) Expression of wild-type but not mutant PARK2 leads to lower levels of BCL-XL protein. A549 cells were transfected with vector encoding wild-type or mutant Flag-tagged PARK2 as indicated. (C) PARK2 regulates the half-life of BCL-XL. Time course showing levels of BCL-XL at various time points following pulse treatment with cyclohexamide (10 μM). PARK2 knockdown increased BCL-XL half-life in A549 cells. (D) PARK2 deficit results in higher expression of BCL-XL protein in mouse liver. Livers were prepared from 3–4 month old PARK2 knockout and wild type mice. Lysates were made for western blotting.
PARK2 Regulates BCL-XL Stability Through Proteasome-Mediated Degradation
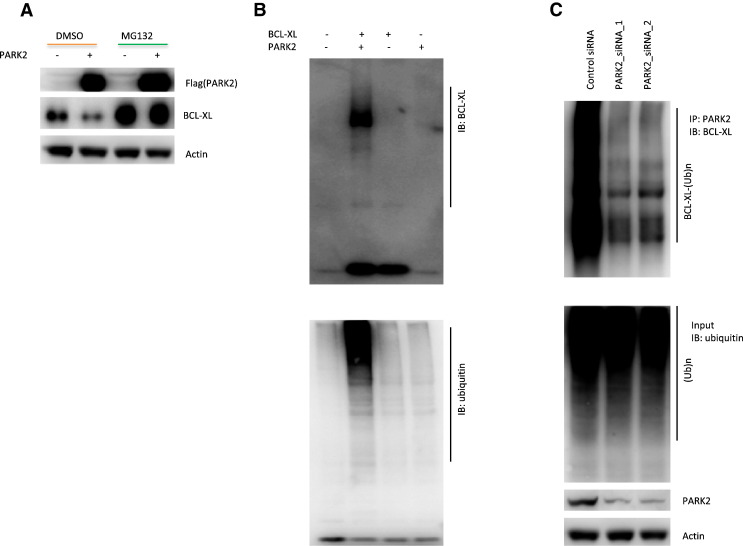
How does PARK2 control BCL-XL protein levels? PARK2 is a ubiquitin E3 ligase, an enzyme that can promote degradation of its substrates through the ubiquitin proteasome system. In cells, we found BCL-XL levels were reduced by overexpression of PARK2, but this catalytic activity was blocked by the proteasome inhibitor MG132, suggesting degradation of BCL-XL protein by PARK2 is dependent on the proteasome system (Figure 4A). To determine whether the PARK2 E3 ligase can directly ubiquitinate BCL-XL, we used in vitro ubiquitination assays with recombinant BCL-XL and PARK2. Our results indicate that BCL-XL is directly ubiquitinated by PARK2 in the presence of the ubiquitin -activating enzyme E1 and the ubiquitin-conjugating enzyme E2 (Figure 4B). In A549 cells, PARK2 knockdown results in decrease in ubiquitination of BCL-XL (Figure 4C).
Figure 4.
PARK2 controls BCL-XL degradation through ubiquitin proteasome system. (A) PARK2-mediated BCL-XL degradation is mediated by the proteasome. BCL-XL levels are shown in A549 cells transfected with pcDNA3.1 empty vector or Flag tagged PARK2, in the presence or absence of proteasomal inhibitor MG132 (25 μM). (B) PARK2 E3 ubiquitin ligase ubiquitinates BCL-XL in vitro. Recombinant proteins BCL-XL and PARK2 were incubated in Hepes buffer, pH 8.0 containing E1, E2, ubiquitin, and Mg2+-ATP. Reactions were cultured for 10 min at 30°C, then 30–60 min at 37°C, and last terminated by adding loading buffer. (C) Knockdown of PARK2 decreases ubiquitination of BCL-XL protein. A549 cells were transfected with two independent PARK2 siRNAs or scrambled siRNA control. PARK2’s E3 ligase for ubiquitinating BCL-XL were examined as previously described [3].
PARK2 Regulates BCL-XL to Control Apoptosis and Mitochondrial Depolarization
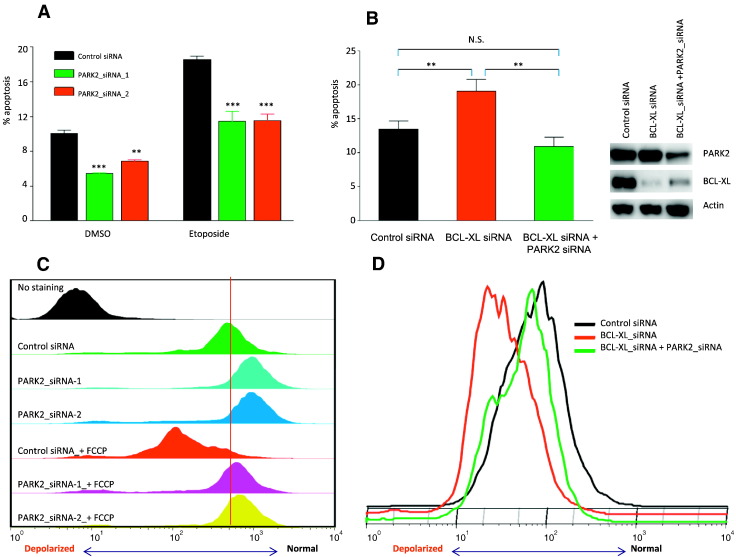
Because BCL-XL is a known modulator of apoptosis, we examined the effects of PARK2 on this process. When A549 cells were treated with PARK2 siRNA, we found that the apoptotic rate was decreased, pointing to a pro-apoptotic activity of PARK2 (Figure 5A). A possible rationale is that depletion of PARK2 causes accumulation of BCL-XL, which can be anti-apoptotic. Consistent with this hypothesis, knockdown of BCL-XL promoted the frequency of apoptosis, an effect that was abrogated by depletion of PARK2 (Figure 5B).
Figure 5.
Effects of PARK2 regulating BCL-XL on apoptosis and MOMP. (A) PARK2 depletion inhibits apoptotic rate. A549 cells were transfected with different PARK2 siRNAs or scrambled siRNA as control. Cells were treated with 100 μM Etoposide for 18 h, as indicated. Cells were stained with FITC and PI and performed by flow cytometry. Histograms show total apoptosis rates (%) obtained from flow cytometer at 48 h post transfection. Error bars show 1 standard deviation (SD). **P < .01, t-test. (B) PARK2 regulates BCL-XL to control programmed cell death. Scramble siRNA, BCL-XL siRNA, and BCL-XL siRNA + PARK2 siRNA were used to treat A549 cells, respectively. Cells were stained with FITC and PI, and performed with FACS analysis. Histograms show total apoptosis rates (%) (Left). Error bars show 1 standard deviation (SD). **P < .01, t-test. Western blots show the expression of proteins indicated (Right). (C) PARK2 depletion inhibits depolarization of mitochondrial outer membrane. A549 cells transfected with different PARK2 siRNAs or scrambled siRNA were stained with TMRE and analyzed by flow cytometry. 20 μM FCCP was used to induce depolarization of mitochondrial outer membrane. FACS profiles represent TMRE uptake (reflective of mitochondrial membrane potential). Leftward peak shifts indicate decreased TMRE, whereas rightward peak shifts indicate increased TMRE uptake. The vertical red line indicates the position of the major population of scramble control RNA cells. Cell sample without being stained with TMRE was taken as negative control. (D) PARK2 abrogates BCL-XL control of mitochondrial outer membrane integrity. A549 cells were treated same as above in (C). Cells were stained with TMRE, and analyzed with FACS analysis. All of the above results are representative data from at least three independent experiments.
During programmed cell death, BCL-XL increases the release of ATP through enhanced voltage-dependent anion channel (VDAC) opening and decreases mitochondrial outer membrane permeabilization [31], [32]. If PARK2 acts on apoptotic pathways, we hypothesize that these effects may be apparent at the mitochondria. Therefore, we measured the effects of PARK2 on mitochondrial membrane potential using the tetramethyl rhodamine ethyl ester (TMRE) staining assay. We observed that PARK2 depletion helped to prevent cells from undergoing mitochondrial depolarization (Figure 5C). Importantly, we also found that PARK2 knockdown inhibited mitochondrial depolarization induced by BCL-XL knockdown (Figure 5D). These results show that PARK2 plays an important roles in maintaining mitochondrial membrane polarization via regulation the anti-apoptotic BCL-XL protein.
Discussion
Our study presents both pan-cancer genetic and biochemical data demonstrating that PARK2 is an important regulator of programmed cell death. Apoptosis involves a cascade of events that can be divided into three phases: (1) initiatory events that induce changes in mitochondrial function; (2) a decisional stage leading to the release of pro-apoptotic proteins from the intermembrane space of mitochondria such as cytochrome c; and (3) activation of effector proteases and endonucleases [33]. Any one of these stages can be interrupted by proteins with anti-apoptotic functions. Recent studies have linked PARK2 and PINK1 in a pathway critical for the maintenance of mitochondrial integrity and function. Expression of mitochondrial PINK1 is required for the recruitment of PARK2 to the dysfunctional mitochondria and for selective elimination by PARK2 [34]. It has been suggested that PARK2 can promote apoptotic activity [35], [36], [37], but the mechanism is unclear.
It is now well established that members of the BCL-2 family proteins play a central role in dictating the onset of apoptosis through regulation of the permeability of the mitochondrial outer membrane [38]. BCL-XL has been shown to counteract the pro-apoptotic functions of BAX and BAD by preventing their translocation from the cytosol to the mitochondria. BCL-XL antagonizes BAK or BAX by binding to their BH3 domains, and members of the BH3-only proteins can relieve this interaction by binding to BCL-XL [39]. Recent work showed that PINK1 interacts with, and phosphorylates BCL-XL, but the PINK1–BCL-XL interaction is independent from the role of the PINK1-PARK2 axis in regulating mitochondrial degradation [40]. Moreover, PARK2 has been shown to monoubiquitinate BCL-2, enhancing BCL-2-Beclin-1 interaction [41]. PARK2 can sensitize cells to apoptosis induced by mitochondrial depolarization by promoting degradation of MCL-1 [42]. In this study, we found PARK2 regulates BCL-XL, a key member of the pro-survival BCL-2 family proteins. Our results show that PARK2 regulates mitochondrial outer membrane permeability and apoptosis by controlling the stability of BCL-XL protein. Pan-cancer genetics suggests that this particular interaction is critical across a large number of human cancers.
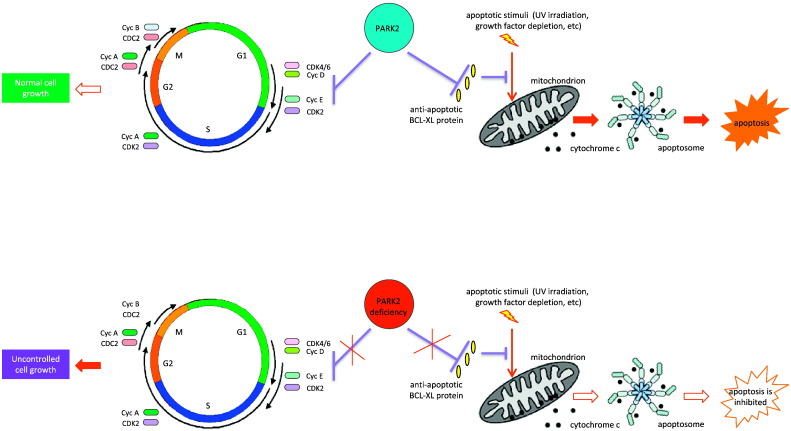
Our findings are important for understanding how apoptosis dysregulation leads to cancer formation. Impaired apoptosis is pivotal for tumor development, and altered expression of molecular determinants of apoptosis, such as the BCL-2 family proteins, contributes to oncogenesis (Figure 6). Anti-apoptotic BCL-2 family proteins such as BCL-2 and BCL-XL are frequently overexpressed in cancers [43], [44]. Many pro-apoptotic members of the BCL-2 family have demonstrated tumor-suppression activity in mouse models of cancer. Conversely, overexpression of pro-survival BCL-2 family members promotes tumorigenesis in humans and in mouse models. Since PARK2 can regulate apoptosis via BCL-2-like proteins, deletion of PARK2 may be a root cause of anti-apoptotic BCL-2 family protein overexpression in cancer.
Figure 6.
Model of coordinate regulation of appropriate cell growth and apoptosis by PARK2 ubiquitin E3 ligase. PARK2 controls cyclin D and E stability to orchestrate cell cycle, and PARK2 controls BCL-XL anti-apoptotic protein level to regulate cell death.
Strategies to target BCL-2 family proteins are in development. For example, BH3-nimetic drugs that directly activate apoptosis in cancer cells by binding and inhibiting pro-survival BCL-2 family members are in development. Such compounds include ABT-263/navitoclax and ABT-199/venetoclax and are currently undergoing testing for the treatment of a variety of both hematological and solid cancers. Based upon our findings, it is tantalizing to hypothesize that PARK2 genetic status may be a molecular determinant of response to these compounds.
In summary, through pan-cancer analysis, we discovered that deletions involving the PARK2 gene are significantly anti-correlated with focal amplifications of the gene encoding BCL-XL. We used multiple approaches to show that PARK2 helps regulate apoptosis via the BCL-XL protein. Our findings shed light on the wide effects the PARK2 tumor suppressor can have on cancer cells and uncovers a critical mechanistic dependency between PARK2 and BCL-XL.
Funding
This work was supported by the National Institutes of Health (RO1 NS086875–01 and R01 CA188228) (T.A.C. and R.B.), the MSKCC Brain Tumor Center (T.A.C.), the Sontag Foundation (T.A.C. and R.B.), the Pediatric Low-Grade Astrocytoma Foundation (R.B.), the Frederick Adler Fund (T.A.C.), and the Paine Webber Chair (T.A.C.).
The authors have no relevant disclosures.
Materials and Methods
Genomic Analyses
GISTIC 2.0 analysis [1] was performed on the copy number profiles of 10,844 primary tumor samples spanning 33 lineages obtained from the TCGA Copy Number Portal [4]. To determine significant regions that anti-correlate with one another, we performed a permutation test that maintained the overall SCNA distribution, event structure and controlled for each of the 33 lineages in the dataset [4]. Benjamini-Hochberg was used to correct for false discovery rate [45] and anti-correlations with FDR p-value less than or equal to 0.25 were considered significant.
Cell Culture
Cell lines were obtained from the American Type Culture Collection (ATCC) and cultured using the recommended media supplemented with 10% FBS (Invitrogen) and penicillin–streptomycin at 37°C in 5% CO2. Specifically, F12-K (ATCC, Cat. 30–2004) was used to culture A549 cells, DMEM (Invitrogen) was used to culture HEK 293 T cells, and MEM + NEAA (Invitrogen) was used to culture T202 cells. Grace's Insect Cell Culture Medium (Invitrogen) was used for Sf9 insect cells.
Mouse Administration
Homozygous PARK2 knockout mice were purchased from The Jackson Laboratory. Mice were in a C57BL/6 background. Mice experiments were performed with MSKCC Institutional Animal Care and Use Committee approval.
SiRNA Knockdown and Plasmid Transfection
siRNAs to PARK2 and BCL-XL were obtained from Invitrogen and Integrated DNA Technologies. The targeted sequences are listed in Supplementary Table 3. siRNAs were transfected into cells in antibiotic-free medium using Lipofectamine RNAiMAX (Invitrogen), medium was changed after 4–6 hours (h), and cells were harvested after 48 h. For exogenous expression, cells were transfected with Lipofectamine (Invitrogen) in serum- and antibiotic-free medium. Medium was changed after 4–6 h, and cells were harvested after 48 h.
Baculovirus Expression in Insect Cells
Full length cDNAs of PARK2 and BCL-XL were cloned into pENTR-D-TOPO (Invitrogen). All constructs were verified by Sanger sequencing. The LR reaction was performed between the entry clone and the C-terminal BaculoDirect™ linear DNA (Invitrogen) to generate recombinant baculovirus DNA as directed by the manufacturer. Sf9 insect cells were co-infected with PARK2 and BCL-XL baculoviruses and grown to produce proteins. Cells were harvested and lysed for co-immunoprecipitation in buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 10% glycerol.
Immunoprecipitation
Cells were harvested and lysed in lysis buffer (50 mM Tris, pH 7.4, 250 mM NaCl, 5 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 1% Nonidet P40 (NP40), 0.02% NaN3) containing protease inhibitors. Lysates were clarified by centrifugation at 12,000 × g for 10 min, and protein concentration was determined using a BCA assay kit (Thermo). For immunoprecipitation assays on endogenous proteins, samples were incubated with antibodies (10 μl of anti-BCL-XL (Cell signaling, 2762); 10 μl of anti-PARK2 (4211,Cell Signaling); or 3.3 μl non-specific mouse IgG (Invitrogen)). Proteins were precipitated by using protein A-sepharose beads that had been blocked with 3% powdered milk. For Flag-tagged construct transfected samples, EZview Red ANTI-FLAG M2 Affinity Gel (Sigma) was used according to manufacturer's protocol. The beads were washed four times with lysis buffer and then mixed with 2× Laemmli sample buffer. Western blotting was performed using standard methods.
Protein Stability and Half-Life Analysis
Protein stability and half-life was measured using a standard cycloheximide release assay [46]. For analysis of BCL-XL decay following protein synthesis inhibition, after 48 h of transfection, cells were treated with cycloheximide (Sigma) at a final concentration of 10 μg/ml (time 0). Cell extracts from each time point were analyzed by Western blotting. Protein intensities were quantified and measured using ImageJ software (Research Services Branch, NIH).
Ubiquitination Assay
In vivo ubiquitination assays were performed as previously described [3]. In brief, A549 cells were transfected with different PARK2 siRNAs or scrambled siRNA control for 24 h, then with plasmid encoding hemagglutinin-tagged ubiquitin (HA-Ub) for 48 h. After treatment with MG132 (25 μM), cell lysates were prepared and subjected to immunoprecipitation with antibody to PARK2 followed by western blotting. In vitro ubiquitination assays were performed using a commercial ubiquitination kit (Boston Biochem) per the manufacturer's protocol. Recombinant proteins of human BCL-XL and PARK2 were purchased from Origene Technologies Inc. and Boston Biochem, respectively.
Annexin V-FITC Apoptosis Assay
The presence of apoptosis in siRNA-silenced A549 cells was detected using the Annexin V-FITC Apoptosis Detectioin Kit (Clontech Laboratories, Inc.) according to the manufacturer's protocol. Briefly, cells were trypsinized, rinsed and resuspended with Binding Buffer, and then incubated with Annexin V and Propidium Iodide at room temperature in the dark. Flow cytometry was performed using FACS Calibur laser flow cytometer (Becton Dickinson Immunocytometry Systems). Ten thousand events were acquired for each sample. Results were quantified using FlowJo software.
Detection of Mitochondrial Membrane Potential
Using the TMRE-Mitochondrial Membrance Potential AssayKt (Abcam), mitochondrial membrane potential was assessed using the fluorescent dye tetramethyl rhodamine ethyl ester (TMRE). Cells were loaded with 100 nM TMRE for 30 min in PBS buffer containing 0.2% BSA. At the end of the incubation, cells were trypsinized, washed, and resuspended. Flow cytometry was performed using FACS Calibur laser flow cytometer (Becton Dickinson Immunocytometry System). Ten thousand events were acquired for each sample. Results were quantified using FlowJo software.
Antibodies
Antibodies used included those against Flag (Sigma, F7425), Actin (Sigma, A2066), PARK2 (Cell signaling, 4211 and 2132), GAPDH (Cell Signaling, 2118), and BCL-XL (Cell signaling, 2762).
Statistical Analysis
All experiments were performed at least in triplicate. Statistical significance was evaluated using either t-tests or ANOVA as appropriate.
The following are the supplementary data related to this article.
BCL-XL protein associates with PARK2 under both normal and apoptotic conditions.
Pan-Cancer Analyses of Significant SCNA and PARK2 Anticorrelates.
Anti-Correlation between PARK2 and BCL-XL in Different Tumor Lineages.
PARK2 and BCL-XL siRNA Target Sequences.
References
- 1.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiong D, Wang Y, Kupert E, Simpson C, Pinney SM, Gaba CR, Mandal D, Schwartz AG, Yang P, de Andrade M. A recurrent mutation in PARK2 is associated with familial lung cancer. Am J Hum Genet. 2015;96:301–308. doi: 10.1016/j.ajhg.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong Y, Zack TI, Morris LG, Lin K, Hukkelhoven E, Raheja R, Tan IL, Turcan S, Veeriah S, Meng S. Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nat Genet. 2014;46:588–594. doi: 10.1038/ng.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denefle P, Wood NW. Association between early-onset Parkinson's disease and mutations in the Parkin gene. N Engl J Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 6.Lin DC, Xu L, Chen Y, Yan H, Hazawa M, Doan N, Said JW, Ding LW, Liu LZ, Yang H. Genomic and functional analysis of the E3 ligase PARK2 in glioma. Cancer Res. 2015;75:1815–1827. doi: 10.1158/0008-5472.CAN-14-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, Janakiraman M, Schultz N, Hanrahan AJ, Pao W. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K. Familial Parkinson disease gene product, Parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 9.Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G. Structure of Parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 10.Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, Luo F, Cantley LC, Wyllie AH, Adams DJ. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci U S A. 2010;107:15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, Hu W, Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norris KL, Hao R, Chen LF, Lai CH, Kapur M, Shaughnessy PJ, Chou D, Yan J, Taylor JP, Engelender S. Convergence of Parkin, PINK1, and alpha-synuclein on stress-induced mitochondrial morphological remodeling. J Biol Chem. 2015;290:13862–13874. doi: 10.1074/jbc.M114.634063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W, Xu H, Wang Z, Mao Y, Yuan L, Luo W, Cui Z, Cui T, Wang XL, Shen YH. PINK1-Parkin-mediated mitophagy protects mitochondrial integrity and prevents metabolic stress-induced endothelial injury. PLoS One. 2015;10:e0132499. doi: 10.1371/journal.pone.0132499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 16.Delbridge AR, Strasser A. The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ. 2015;22:1071–1080. doi: 10.1038/cdd.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonas EA, Porter GA, Alavian KN. Bcl-xL in neuroprotection and plasticity. Front Physiol. 2014;5:355. doi: 10.3389/fphys.2014.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karczmarek-Borowska B, Filip A, Wojcierowski J, Smolen A, Korobowicz E, Korszen-Pilecka I, Zdunek M. Estimation of prognostic value of Bcl-xL gene expression in non-small cell lung cancer. Lung Cancer. 2006;51:61–69. doi: 10.1016/j.lungcan.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith LT, Mayerson J, Nowak NJ, Suster D, Mohammed N, Long S, Auer H, Jones S, McKeegan C, Young G. 20q11.1 amplification in giant-cell tumor of bone: Array CGH, FISH, and association with outcome. Genes Chromosomes Cancer. 2006;45:957–966. doi: 10.1002/gcc.20354. [DOI] [PubMed] [Google Scholar]
- 21.Delbridge AR, Grabow S, Bouillet P, Adams JM, Strasser A. Functional antagonism between pro-apoptotic BIM and anti-apoptotic BCL-XL in MYC-induced lymphomagenesis. Oncogene. 2015;34:1872–1876. doi: 10.1038/onc.2014.132. [DOI] [PubMed] [Google Scholar]
- 22.Kelly PN, Grabow S, Delbridge AR, Strasser A, Adams JM. Endogenous Bcl-xL is essential for Myc-driven lymphomagenesis in mice. Blood. 2011;118:6380–6386. doi: 10.1182/blood-2011-07-367672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly PN, Grabow S, Delbridge AR, Adams JM, Strasser A. Prophylactic treatment with the BH3 mimetic ABT-737 impedes Myc-driven lymphomagenesis in mice. Cell Death Differ. 2013;20:57–63. doi: 10.1038/cdd.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boone J, Borel Rinkes IH, van Hillegersberg R. Robot-assisted thoracolaparoscopic esophagolymphadenectomy for esophageal cancer. Surg Endosc. 2007;21:2342–2343. doi: 10.1007/s00464-007-9604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nijhawan D, Zack TI, Ren Y, Strickland MR, Lamothe R, Schumacher SE, Tsherniak A, Besche HC, Rosenbluh J, Shehata S. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842–854. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 29.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 30.Hu HH, Kannengiesser C, Lesage S, Andre J, Mourah S, Michel L, Descamps V, Basset-Seguin N, Bagot M, Bensussan A. PARKIN inactivation links Parkinson's disease to melanoma. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djv340. [DOI] [PubMed] [Google Scholar]
- 31.Vander Heiden MG, Li XX, Gottleib E, Hill RB, Thompson CB, Colombini M. Bcl-xL promotes the open configuration of the voltage-dependent anion channel and metabolite passage through the outer mitochondrial membrane. J Biol Chem. 2001;276:19414–19419. doi: 10.1074/jbc.M101590200. [DOI] [PubMed] [Google Scholar]
- 32.Gottlieb E, Armour SM, Thompson CB. Mitochondrial respiratory control is lost during growth factor deprivation. Proc Natl Acad Sci U S A. 2002;99:12801–12806. doi: 10.1073/pnas.202477599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroemer G, Reed JC. Mitochondrial control of cell death. Nat Med. 2000;6:513–519. doi: 10.1038/74994. [DOI] [PubMed] [Google Scholar]
- 34.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, Lee S, Peng Y, Bunker E, Giaime E, Shen J, Zhou Z, Liu X. PINK1 triggers autocatalytic activation of Parkin to specify cell fate decisions. Curr Biol. 2014;24:1854–1865. doi: 10.1016/j.cub.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee K, Lee MH, Kang YW, Rhee KJ, Kim TU, Kim YS. Parkin induces apoptotic cell death in TNF-alpha-treated cervical cancer cells. BMB Rep. 2012;45:526–531. doi: 10.5483/bmbrep.2012.45.9.104. [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara M, Marusawa H, Wang HQ, Iwai A, Ikeuchi K, Imai Y, Kataoka A, Nukina N, Takahashi R, Chiba T. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27:6002–6011. doi: 10.1038/onc.2008.199. [DOI] [PubMed] [Google Scholar]
- 38.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 40.Arena G, Gelmetti V, Torosantucci L, Vignone D, Lamorte G, De Rosa P, Cilia E, Jonas EA, Valente EM. PINK1 protects against cell death induced by mitochondrial depolarization, by phosphorylating Bcl-xL and impairing its pro-apoptotic cleavage. Cell Death Differ. 2013;20:920–930. doi: 10.1038/cdd.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen D, Gao F, Li B, Wang H, Xu Y, Zhu C, Wang G. Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J Biol Chem. 2010;285:38214–38223. doi: 10.1074/jbc.M110.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carroll RG, Hollville E, Martin SJ. Parkin sensitizes toward apoptosis induced by mitochondrial depolarization through promoting degradation of Mcl-1. Cell Rep. 2014;9:1538–1553. doi: 10.1016/j.celrep.2014.10.046. [DOI] [PubMed] [Google Scholar]
- 43.Walensky LD. BCL-2 in the crosshairs: tipping the balance of life and death. Cell Death Differ. 2006;13:1339–1350. doi: 10.1038/sj.cdd.4401992. [DOI] [PubMed] [Google Scholar]
- 44.Sasi N, Hwang M, Jaboin J, Csiki I, Lu B. Regulated cell death pathways: new twists in modulation of BCL2 family function. Mol Cancer Ther. 2009;8:1421–1429. doi: 10.1158/1535-7163.MCT-08-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 46.Magal SS, Jackman A, Ish-Shalom S, Botzer LE, Gonen P, Schlegel R, Sherman L. Downregulation of Bax mRNA expression and protein stability by the E6 protein of human papillomavirus 16. J Gen Virol. 2005;86:611–621. doi: 10.1099/vir.0.80453-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BCL-XL protein associates with PARK2 under both normal and apoptotic conditions.
Pan-Cancer Analyses of Significant SCNA and PARK2 Anticorrelates.
Anti-Correlation between PARK2 and BCL-XL in Different Tumor Lineages.
PARK2 and BCL-XL siRNA Target Sequences.