Abstract
Congenital anomalies of the kidneys and urinary tract (CAKUT) are the leading cause of CKD in children, featuring a broad variety of malformations. A monogenic cause can be detected in around 12% of patients. However, the morphologic clinical phenotype of CAKUT frequently does not indicate specific genes to be examined. To determine the likelihood of detecting causative recessive mutations by whole-exome sequencing (WES), we analyzed individuals with CAKUT from 33 different consanguineous families. Using homozygosity mapping and WES, we identified the causative mutations in nine of the 33 families studied (27%). We detected recessive mutations in nine known disease–causing genes: ZBTB24, WFS1, HPSE2, ATRX, ASPH, AGXT, AQP2, CTNS, and PKHD1. Notably, when mutated, these genes cause multiorgan syndromes that may include CAKUT as a feature (syndromic CAKUT) or cause renal diseases that may manifest as phenocopies of CAKUT. None of the above monogenic disease–causing genes were suspected on clinical grounds before this study. Follow-up clinical characterization of those patients allowed us to revise and detect relevant new clinical features in a more appropriate pathogenetic context. Thus, applying WES to the diagnostic approach in CAKUT provides opportunities for an accurate and early etiology–based diagnosis and improved clinical management.
Keywords: CAKUT, monogenic disease, WES
Congenital anomalies of the kidneys and urinary tract (CAKUT) are the leading cause of CKD in children, accounting for >40% of patients.1 CAKUT features a broad variety of structural malformations, such as renal agenesis, renal hypodysplasia, multicystic dysplastic kidney, hydronephrosis, ureteropelvic junction obstruction, megaureter, ureter duplex, vesicoureteral reflux (VUR), and posterior urethral valves.2,3 CAKUT may appear as an isolated feature or part of systemic conditions that encompass extrarenal manifestations2,3; >200 recessive and dominant monogenic syndromes have been described involving renal or urinary anomalies as one of their features.4 Recently, it was shown that a monogenic cause can be detected in approximately 12% of patients.2,5–8 The documented mode of inheritance in most published pedigrees is compatible with an autosomal dominant inheritance with variable expressivity and incomplete penetrance.2 However, several CAKUT–causing genes with autosomal recessive mode of inheritance have been recently identified.5,9,10 Nevertheless, in the clinical practice, the genetic basis of most patients with CAKUT is elusive because of a very weak genotype-phenotype correlation. To determine the likelihood of detecting causative recessive mutations in CAKUT by whole-exome sequencing (WES), we have analyzed individuals with CAKUT from 33 consanguineous families. We used homozygosity mapping combined with WES11 to detect recessive monogenic disease–causing genes of CAKUT. Using this strategy, we identified the causative mutations in nine of the 33 families studied (27%). In all nine families, we established a specific etiologic genetic diagnosis that was not made on the basis of the patient’s clinical findings.
We have previously shown that recessive-causing mutations can be identified in a high percentage of individuals if (1) consanguinity of the parents is present and (2) a combination of homozygosity mapping with WES is applied.11,12 We, therefore, selected, of approximately 700 families with the diagnosis of CAKUT that were referred to us from worldwide sources, 33 unrelated individuals from consanguineous families for homozygosity mapping and WES. All patients were referred to us by a pediatric nephrologist who made a clinical diagnosis of CAKUT on the basis of renal imaging studies. Mutations in all genes known to be mutated in isolated CAKUT in humans were excluded previously in this cohort.6 In addition, a clinical diagnosis of a known monogenic syndrome had not been previously made in any of the patients.
We used homozygosity mapping in combination with WES as previously described11,12 to detect recessive homozygous disease–causing mutations. We also performed a separate analysis using individuals’ WES data for X-linked diseases. All families had evidence of consanguinity, with an average proportion of the genome exhibiting homozygosity by descent of 290 Mb (range =34–546 Mb). This is, on average, compatible with a first cousin marriage of the parents of the affected individual. After WES, genetic variants were filtered to retain only homozygous nonsynonymous and splice variants found in homozygous by descent regions. Next, we used strict criteria for allele calling of causative recessive mutations as shown in Supplemental Material and in line with proposed guidelines.13
After homozygosity mapping and WES, we detected recessive mutations in nine known disease–causing genes: WFS1, ZBTB24, HPSE2, ATRX, ASPH, AGXT, AQP2, CTNS and PKHD1 (Table 1, Supplemental Material). Interestingly, five of these genes (ZBTB24, WFS1, HPSE2, ATRX, and ASPH) if mutated cause multiorgan syndromes that may include CAKUT as one of their features (syndromic CAKUT) (Table 1, syndromes with CAKUT). Four other genes (AGXT, AQP2, CTNS, and PKHD1) cause, if mutated, a kidney disease that may represent a phenocopy of CAKUT (Table 1, phenocopies with CAKUT).
Table 1.
Disease-causing mutations and clinical phenotypes in nine individuals from 33 consanguineous families with CAKUT
| Individual | Origin | Causative Gene | Nucleotide Alteration | Alteration in Coding Sequence | Zygosity | Continuous AA Sequence Conservation | PP2a SIFT | Renal Phenotype | Extrarenal Phenotype | Syndrome Name (OMIM No.)b |
|---|---|---|---|---|---|---|---|---|---|---|
| Syndromes with CAKUT | ||||||||||
| A3857 | India | WFS1 | c.2452C>Tc | p.Arg818Cys | Homo | Xenopus tropicalis | 0.97 (PD) Del | Neurogenic bladder, bilateral ureterohydronephrosis, ESRD | Growth retardation and spina bifida | Wolfram syndrome (222300) |
| A3835 | India | ZBTB24 | c.1457G>A | p.Gly486Asp | Homo | Danio rerio | 1 (PD) Del | Bilateral VUR | Intellectual disability, deafness, growth retardation | ICF-2 syndrome (614069) |
| B268 | Macedonia | HPSE2 | c.457C>Tc | p.Arg153* | Homo | Truncating | Left UVJ obstruction, left renal hypodysplasia | Distorted facial expression with smilingd | Urofacial Syndrome 1 (236730) | |
| B407 | Saudi Arabia | ATRX | c.477_478insA | p.Arg160Thrfs*29 | Hemi | Truncating | Bilateral VUR | Global developmental delay, seizures, and undescended testis | ATRX (301040) | |
| A3813 | India | ASPH | c.1301G>T | p.Gly434Val | Homo | Danio rerio | 1 (PD) Del | VUR and CKD | Lens subluxation and hyperhomocystinemia | Traboulsi syndrome (601552) |
| Phenocopies of CAKUT | ||||||||||
| A3871 | India | AGXT | c.524+1G>A | Splice | Homo | Splice mutatione | Bilateral renal hypodysplasia and ESRDf | Growth retardation | Hyperoxaluria type 1 (259900) | |
| A3879 | India | AQP2 | c.377C>Tc | p.Thr126Met | Homo | Drosophila | 0.99 (PD) Del | Bilateral renal dysplasiag | Brain calcifications and growth retardation | NDI (125800) |
| A3913 | India | CTNS | c.809_811del | p.Ser270del | Homo | Caenorhabditis elegans | n/a | Bilateral VUR and polyuriah | Hepatomegaly,d growth retardation, and skeletal deformities | Cystinosis (219800) |
| A3772 | Kuwait | PKHD1 | c.4870C>Tc | p.Arg1624Trp | Homo | Gallus gallus | 0.99 (PD) Del | Renal hypodysplasia and ESRD | Gallbladder stones, dilated segmental intrahepatic biliary radiclesd | PKHD1 (263200) |
AA, amino acid; PP2, PolyPhen2; SIFT, sorting intolerant from tolerant; OMIM, online mMendelian inheritance in man; Homo, homozygous; PD, probably damaging; Del, deleterious; ICF-2, immunodeficiency–centromeric instability facial anomalies; UVJ, ureterovesical junction obstruction; Hemi, hemizygous; ATRX, α-thalassemia/mental retardation syndrome X-linked; NDI, nephrogenic diabetes insipidus; n/a, not applicable; PKHD1, polycystic kidney and hepatic disease 1.
All mutations had a PP2 and SIFT score of functional deleteriousness (http://genetics.bwh.harvard.edu/pph2 and http://sift.jcvi.org/).
This mutation has been previously reported as disease causing.
This information was obtained after the genetic diagnosis by gene–specific direct phenotypic characterization (Figure 1).
This mutation is predicted to change conserved adenine, thymine donor site and be deleterious using three different in silico splice prediction software (MaxEnt, NNSPLICE, and HSF).
After the genetic diagnosis, a revision of the patient’s renal ultrasound revealed multiple calcification.
After the genetic diagnosis, a water deprivation test as well as a vasopressin stimulation test revealed that the patient has nephrogenic diabetes insipidus.
After the genetic diagnosis, the patient’s primary nephrologist reported that the patient has unexplained Fanconi syndrome and hepatomegaly.
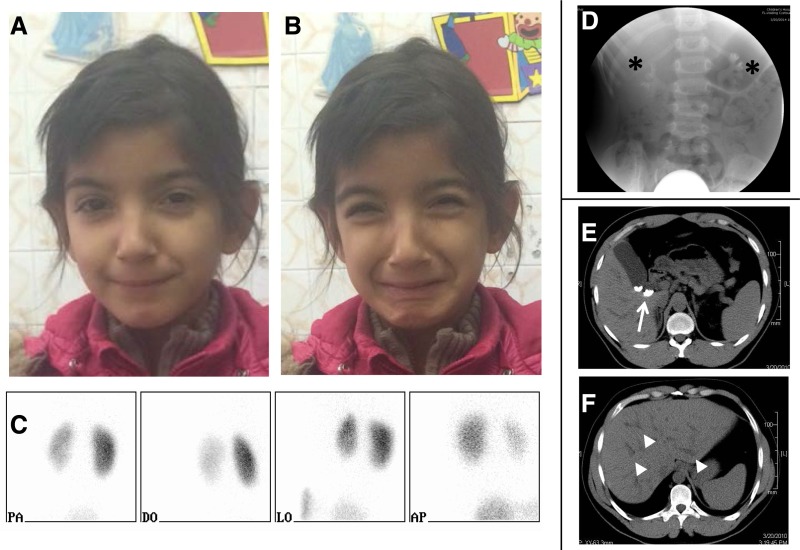
In five patients, we identified recessive mutations that cause syndromic CAKUT (Table 1, syndromes with CAKUT). Patient A3857 had ESRD secondary to neurogenic bladder and recurrent urinary tract infections. This patient presented with CAKUT and harbors a previously reported missense mutation in the gene WFS1 responsible to Wolfram syndrome, a diagnosis that had not been made in this patient (Table 1, syndromes with CAKUT).14 Wolfram syndrome is a neurodegenerative disease that can be associated with variable clinical manifestations, including renal structural abnormalities. These abnormalities include urethral sphincter disturbances, neurogenic bladders, VUR, and hydronephrosis. As a result, recurrent urinary tract infections are a common finding.14 It is estimated that around 58% of patients with Wolfram syndrome have urinary tract abnormalities.15 This patient had no additional typical syndromic findings, such as diabetes mellitus, deafness, and diabetes insipidus. However, the patient had spina bifida, which is a common finding in Wolfram syndrome.16 In patient A3853, we identified a highly conserved missense mutation affecting a zinc finger coding domain of the gene ZBTB24. The disease phenotype included bilateral VUR, intellectual disability, growth retardation, and deafness. Mutations in this gene lead to the rare syndrome immunodeficiency–centromeric instability facial anomalies, a diagnosis that was not established previously in this patient.17 This syndrome features pleiotropic clinical presentations, which include immunodeficiency of variable extent, developmental delay, intellectual disability, and mild facial dimorphism. Immunodeficiency–centromeric instability facial anomalies has also been reported to include congenital malformations, including CAKUT.18 Patient B268 presented with isolated CAKUT and was found to harbor a previously reported truncating mutation in the gene HPSE2.19 This gene is responsible for urofacial syndrome.19,20 This syndrome, also known as Ochoa syndrome, is characterized by typical distorted facial expression with smiling or laughing expression as well as bladder malfunctions leading to voiding dysfunction, recurrent urinary tract infections, and other accompanying features of CAKUT. After establishment of the molecular genetic diagnosis, we contacted the primary nephrologist who originally recruited this patient at a very young age and asked him to look for the typical facial grimacing associated with this syndrome. In a follow-up clinical examination, the patient was found to exhibit the syndrome–typical distorted facial expression with smiling (Figure 1, A–C). This highlights the difficulty in diagnosing subtle phenotypes, such as facial finding, at a very young age and the benefits of early molecular genetic diagnosis.
Figure 1.
Renal imaging and clinical findings in selected patients with disease-causing mutations. (A and B) Photographs of an affected girl (B268) with the HPSE2 mutation show characteristic grimace on smiling. (C) Dimercaptosuccinic acid scan at different views of the patient (B268) with the HPSE2 mutation shows differential renal function. Left kidney, 30%; right kidney, 70%. (D) Voiding cystourethrogram imaging of patient B407 with the ATRX mutation shows bilateral grade 3 VUR (black asterisks). (E and F) Abdominal computed tomography scans of patient A3772 with the PKHD1 mutation show gallbladder stones (white arrow) and dilated segmental intrahepatic biliary radicles (white arrowheads).
Patient B407 presented with CAKUT and nonspecific extrarenal features of seizures and global developmental delay. He was found to harbor a novel truncating mutation in the gene ATRX affecting the plant homeodomain–like domain. ATRX was originally described as a severe form of X–linked mental retardation associated with urogenital abnormalities, facial dysmorphism, and α-thalassemia.21 Later, the clinical spectrum was extended to clinical phenotypes without α-thalassemia, predominantly among patients harboring mutations affecting the protein plant homeodomain–like domain, similar to our patients.22 Similarly, our patient with VUR (Figure 1D) did not have α-thalassemia. Lastly, patient A3813 was found to harbor a novel conserved missense mutation in the gene ASPH. He presented with VUR and lens dislocation. This gene was recently reported to be mutated in the genetic cause of Traboulsi syndrome, which features characteristics finding of lens dislocation.23 Although CAKUT has not been previously reported as part of this syndrome, it is plausible that VUR is a feature of the syndrome, because ASPH is highly expressed in the lower urinary tract (http://www.proteinatlas.org and http://www.gudmap.org/index.html).
Surprisingly, in four individuals, we identified mutations in genes known to cause nonsyndromic kidney diseases (i.e., AGXT, AQP2, NDI, CTNS, and PKHD1) (Table 1, phenocopies with CAKUT). This shows that these diseases apparently represent clinical phenocopies of CAKUT. In our follow-up analysis of all four patients, we were able to contact the recruiting physician to further deepen the phenotypic characterization in light of having detected the disease–defining causative monogenic mutations.
Patient A3871 presented with hypertension. Subsequent evaluation revealed that he had ESRD and bilateral small kidneys, initially suspected to be the result of congenital bilateral renal dysplasia. Identification of a homozygous splice site mutation in AGXT (Table 1, phenocopies with CAKUT) enabled us to establish the unexpected etiologic diagnosis of hyperoxaluria type 1. Recessive mutations of AGXT have been reported previously to cause a nephronophthisis-like phenotype12 and therefore, may also phenocopy renal cystic ciliopathies. Both conditions, CAKUT and nephronophthisis, can ultrasonographically present similarly with features of echogenic kidneys.24 In a follow-up analysis with the referring physician, a revision of the renal ultrasound of this patient showed multiple calcification in addition to small kidneys, cortical echogenicity, and loss of corticomedullary differentiation. The former may represent calcium oxalate depositions, which in retrospect, coincide with the genetic diagnosis. This finding is in line with previous reports showing that primary hyperoxaluria has a heterogeneous clinical phenotype,25 often leading to a delayed diagnosis.25,26 Moreover, in the setting of early childhood ESRD, a diagnosis of hyperoxaluria can be confused with bilateral congenital renal hypodysplasia, and therefore, a molecular genetic analysis for those patients is important.
Patient A3879 was referred to our attention because of bilateral renal hypodysplasia evident by renal ultrasound at 1 year of age. Postnatally, he had hypoxic ischemic encephalopathy and secondary brain calcifications. We identified a previously reported missense mutation in AQP2 (Table 1, phenocopies with CAKUT). Review of his medical history revealed that he initially presented with fever and polyuria, supporting the diagnosis of his overlooked congenital NDI. The AQP2 mutation that we identified was previously described in two brothers with congenital NDI who originate from the same geographic region as our patient and therefore, may represent a founder mutation.27 The role of AQP2 mutations as phenocopying CAKUT is evident in the Aqp2 null mouse model, which presents with neonatal mortality from kidney failure, hydronephrosis, and renal tubular abnormalities.28,29 This association is thought to be caused by urinary overflow obstruction, leading to secondary renal damage. Alternatively, as recently suggested, AQP2 may play an additional role during kidney development as an integrin binding membrane protein that promotes cell migration and epithelial morphogenesis.30 Follow-up analysis with the referring physician revealed that the patient has a urine concentration defect with urine osmolality of 180 mosM and a loss of rise in urine osmolality after water deprivation test and vasopressin stimulation.
In patient A3913 who presented with hydronephrosis, VUR, growth retardation, and skeletal deformities, we identified a previously reported pathogenic mutation that causes cystinosis (Table 1, phenocopies with CAKUT).31 Follow-up analysis with the referring physician revealed that the patient had renal Fanconi syndrome and hepatomegaly. Both findings further support the genetic diagnosis of cystinosis. Surprisingly, the patient’s corneal examination was normal, showing no corneal cystine crystal formation. Nonetheless, this incomplete clinical presentation has been described in cystinosis before, leading to a missed diagnosis of cystinosis.32,33
Finally, in patient A3772, who was diagnosed with CAKUT, we detected a previously reported disease–causing missense mutation in the gene PKHD1.34 Specifically and similar to in our patient, this mutation has been reported to lead to a late-onset phenotype. Follow-up analysis with the referring physician revealed that the patient has had gallbladder stones, dilated segmental intrahepatic biliary radicles, and mild common bile duct dilation (Figure 1, E and F). These anomalies (also known as Caroli disease) are associated with PKHD1 mutations,35 thereby confirming the clinical diagnosis in retrospect.
In this study, we examined a group of 33 unrelated consanguineous families with CAKUT for the presence of autosomal recessive disease–causing mutations. We identified nine different causative mutated genes in nine of 33 individuals (27%). In five families, we found predominantly CAKUT phenotype in genes known to cause syndromic CAKUT if mutated. In four families, we found mutations in genes that apparently may phenocopy CAKUT. None of the above disease–causing mutations were suspected on clinical grounds before this study, and affected patients were not clinically distinguished from other patients with CAKUT. In all patients available for follow-up, a molecular genetic diagnosis led to an unequivocal etiology–based diagnosis.
Our study highlights several important conclusions for the diagnosis of CAKUT. First, it shows that, for individuals with CAKUT, clinical diagnosis, renal ultrasound, and other renal imaging studies represent relatively blunt diagnostic tools, which may not be sufficient to establish the correct diagnosis. In this setting, WES offers a powerful and noninvasive tool for a precise, unequivocal, etiology–based diagnosis. Second, individuals with mutations in genes that cause syndromic CAKUT can initially present as isolated CAKUT or with a predominant CAKUT phenotype. As a result, pediatric nephrologists, who may be the first clinicians to be approached, should be aware that there are >200 different multiorgan syndromes that can have CAKUT as one of their features.4 Third, the diagnosis of CAKUT in the context of existing CKD may be challenging. Patients may present late in the CKD course with bilateral small kidneys, a common pathway for numerous other CKD etiologies that cannot easily be distinguished. Fourth, rare genetically heterogeneous syndromes probably are responsible for a significant number of cases of CAKUT in children but very difficult to diagnose, especially at early disease stages and in patients with nontypical or incomplete disease presentation.
In retrospect, the clinical courses of most respective patients were typical for the gene found to be mutated. Nonetheless, the primary physicians had not fully established the diagnosis clinically at the time of study. In each patient, the clinical phenotype still suggested multiple causative genes. Evaluation of WES data for the candidate genes in question is now considered the gold standard for molecular genetic diagnostics. This takes into account that, today, WES is not more expensive than sequencing of candidate gene panels.
Our study shows that, for patients from consanguineous families, by using the WES approach, an etiologic diagnosis can be made in a very high percentage of patients. The advent of WES in consanguineous families often leads to correction of a diagnosis made on clinical grounds, because monogenic mutations allow the clinician to arrive at an unequivocal etiologic diagnosis. This approach may reveal a syndrome or kidney disease that otherwise would not be suspected solely from the patient’s clinical presentation. In some cases, such as primary hyperoxaluria, for example, this would have very important therapeutic consequences.
Concise Methods
Study Participants
After informed consent, we obtained clinical data, blood samples, and pedigrees from individuals with CAKUT. Approval for research on humans was obtained from the Boston Children’s Hospital and the University of Michigan Institutional Review Board. The diagnosis of CAKUT was made by (pediatric) nephrologists on the basis of standardized clinical and renal ultrasound criteria. Clinical data were obtained using a standardized questionnaire (http://www.renalgenes.org). Mutations in the following 17 genes known to be mutated in isolated CAKUT in humans were excluded by high–throughput multiplex PCR and next generation sequencing6 before this study: BMP4, BMP7, CDC5L, CHD1L, EYA1, GATA3, HNF1B, PAX2, RET, ROBO2, SALL1, SIX1, SIX2, SIX5, SOX17, UMOD, and UPK3A. HNF1B deletions were excluded by quantitative PCR in individuals with the CAKUT phenotype of renal hypodysplasia.6
Homozygosity Mapping
For genome–wide homozygosity mapping of the 33 families, GeneChip HumanMapping 250,000 StyI Array (Affymetrix, Santa Clara, CA) was used. Genomic DNA samples were hybridized and scanned using the manufacturer’s standard protocol at the University of Michigan Core Facility (www.michiganmicroarray.com). Regions of homozygosity were identified using GENEHUNTER 2.11 and ALLEGRO with a disease allele frequency of 0.0001 and marker allele frequencies of white descent. Genetic regions of homozygosity by descent (homozygosity peaks) were plotted across the genome as candidate regions for recessive genes as previously described.11,12
Whole-Exome Resequencing
WES was performed using genomic DNA isolated from blood lymphocytes and later processed using Agilent SureSelect Human Exome Capture Arrays (Life Technologies, Carlsbad, CA) with next generation sequencing on an Illumina sequencing platform at the Broad Institute (Cambridge, MA).
Mutation Calling
Sequence reads were mapped to the human reference genome assembly (National Center for Biotechnology Information build 37/hg19; www.genome.ucsc.edu) using CLC Genomics Workbench (version 6.5.1) software (CLC Bio, Aarhus, Denmark) as previously described.12 Mutation calling was performed by a team of clinician scientists who had knowledge of the clinical phenotypes and pedigree structure as well as experience with homozygosity mapping and exome evaluation as previously described.11,12
Disclosures
None.
Supplementary Material
Acknowledgments
A.V. is a recipient of the Fulbright Post-Doctoral Scholar Award for 2013. A.V. is also supported by grants from the Manton Center Fellowship Program, Boston Children’s Hospital, and the Mallinckrodt Research Fellowship Award. F.H. was supported by the Howard Hughes Medical Institute and is the Warren E. Grupe Professor of Pediatrics. This research was supported by National Institutes of Health grant R01DK088767 (to F.H.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2015080962/-/DCSupplemental.
References
- 1.The EMMES Corporation : North American Pediatric Renal Transplant Cooperative Study (NAPRTCS) 2008 Annual Report, Rockville, MD, The EMMES Corporation, 2008 [Google Scholar]
- 2.Vivante A, Kohl S, Hwang DY, Dworschak GC, Hildebrandt F: Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans. Pediatr Nephrol 29: 695–704, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soliman NA, Ali RI, Ghobrial EE, Habib EI, Ziada AM: Pattern of clinical presentation of congenital anomalies of the kidney and urinary tract among infants and children. Nephrology (Carlton) 20: 413–418, 2015 [DOI] [PubMed] [Google Scholar]
- 4.Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O’Toole JF, Helou J, Attanasio M, Utsch B, Sayer JA, Lillo C, Jimeno D, Coucke P, De Paepe A, Reinhardt R, Klages S, Tsuda M, Kawakami I, Kusakabe T, Omran H, Imm A, Tippens M, Raymond PA, Hill J, Beales P, He S, Kispert A, Margolis B, Williams DS, Swaroop A, Hildebrandt F: Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet 37: 282–288, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Kohl S, Hwang DY, Dworschak GC, Hilger AC, Saisawat P, Vivante A, Stajic N, Bogdanovic R, Reutter HM, Kehinde EO, Tasic V, Hildebrandt F: Mild recessive mutations in six Fraser syndrome-related genes cause isolated congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol 25: 1917–1922, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang DY, Dworschak GC, Kohl S, Saisawat P, Vivante A, Hilger AC, Reutter HM, Soliman NA, Bogdanovic R, Kehinde EO, Tasic V, Hildebrandt F: Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int 85: 1429–1433, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vivante A, Kleppa MJ, Schulz J, Kohl S, Sharma A, Chen J, Shril S, Hwang DY, Weiss AC, Kaminski MM, Shukrun R, Kemper MJ, Lehnhardt A, Beetz R, Sanna-Cherchi S, Verbitsky M, Gharavi AG, Stuart HM, Feather SA, Goodship JA, Goodship TH, Woolf AS, Westra SJ, Doody DP, Bauer SB, Lee RS, Adam RM, Lu W, Reutter HM, Kehinde EO, Mancini EJ, Lifton RP, Tasic V, Lienkamp SS, Jüppner H, Kispert A, Hildebrandt F: Mutations in TBX18 cause dominant urinary tract malformations via transcriptional dysregulation of ureter development. Am J Hum Genet 97: 291–301, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang DY, Kohl S, Fan X, Vivante A, Chan S, Dworschak GC, Schulz J, van Eerde AM, Hilger AC, Gee HY, Pennimpede T, Herrmann BG, van de Hoek G, Renkema KY, Schell C, Huber TB, Reutter HM, Soliman NA, Stajic N, Bogdanovic R, Kehinde EO, Lifton RP, Tasic V, Lu W, Hildebrandt F: Mutations of the SLIT2-ROBO2 pathway genes SLIT2 and SRGAP1 confer risk for congenital anomalies of the kidney and urinary tract. Hum Genet 134: 905–916, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humbert C, Silbermann F, Morar B, Parisot M, Zarhrate M, Masson C, Tores F, Blanchet P, Perez MJ, Petrov Y, Khau Van Kien P, Roume J, Leroy B, Gribouval O, Kalaydjieva L, Heidet L, Salomon R, Antignac C, Benmerah A, Saunier S, Jeanpierre C: Integrin alpha 8 recessive mutations are responsible for bilateral renal agenesis in humans. Am J Hum Genet 94: 288–294, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saisawat P, Kohl S, Hilger AC, Hwang DY, Yung Gee H, Dworschak GC, Tasic V, Pennimpede T, Natarajan S, Sperry E, Matassa DS, Stajić N, Bogdanovic R, de Blaauw I, Marcelis CL, Wijers CH, Bartels E, Schmiedeke E, Schmidt D, Märzheuser S, Grasshoff-Derr S, Holland-Cunz S, Ludwig M, Nöthen MM, Draaken M, Brosens E, Heij H, Tibboel D, Herrmann BG, Solomon BD, de Klein A, van Rooij IA, Esposito F, Reutter HM, Hildebrandt F: Whole-exome resequencing reveals recessive mutations in TRAP1 in individuals with CAKUT and VACTERL association. Kidney Int 85: 1310–1317, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hildebrandt F, Heeringa SF, Rüschendorf F, Attanasio M, Nürnberg G, Becker C, Seelow D, Huebner N, Chernin G, Vlangos CN, Zhou W, O’Toole JF, Hoskins BE, Wolf MT, Hinkes BG, Chaib H, Ashraf S, Schoeb DS, Ovunc B, Allen SJ, Vega-Warner V, Wise E, Harville HM, Lyons RH, Washburn J, Macdonald J, Nürnberg P, Otto EA: A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet 5: e1000353, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gee HY, Otto EA, Hurd TW, Ashraf S, Chaki M, Cluckey A, Vega-Warner V, Saisawat P, Diaz KA, Fang H, Kohl S, Allen SJ, Airik R, Zhou W, Ramaswami G, Janssen S, Fu C, Innis JL, Weber S, Vester U, Davis EE, Katsanis N, Fathy HM, Jeck N, Klaus G, Nayir A, Rahim KA, Al Attrach I, Al Hassoun I, Ozturk S, Drozdz D, Helmchen U, O’Toole JF, Attanasio M, Lewis RA, Nürnberg G, Nürnberg P, Washburn J, MacDonald J, Innis JW, Levy S, Hildebrandt F: Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int 85: 880–887, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C: Guidelines for investigating causality of sequence variants in human disease. Nature 508: 469–476, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gómez-Zaera M, Strom TM, Rodríguez B, Estivill X, Meitinger T, Nunes V: Presence of a major WFS1 mutation in Spanish Wolfram syndrome pedigrees. Mol Genet Metab 72: 72–81, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Barrett TG, Bundey SE, Macleod AF: Neurodegeneration and diabetes: UK nationwide study of Wolfram (DIDMOAD) syndrome. Lancet 346: 1458–1463, 1995 [DOI] [PubMed] [Google Scholar]
- 16.Hadidy AM, Jarrah NS, Al-Till MI, El-Shanti HE, Ajlouni KM: Radiological findings in Wolfram syndrome. Saudi Med J 25: 638–641, 2004 [PubMed] [Google Scholar]
- 17.de Greef JC, Wang J, Balog J, den Dunnen JT, Frants RR, Straasheijm KR, Aytekin C, van der Burg M, Duprez L, Ferster A, Gennery AR, Gimelli G, Reisli I, Schuetz C, Schulz A, Smeets DF, Sznajer Y, Wijmenga C, van Eggermond MC, van Ostaijen-Ten Dam MM, Lankester AC, van Tol MJ, van den Elsen PJ, Weemaes CM, van der Maarel SM: Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet 88: 796–804, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weemaes CM, van Tol MJ, Wang J, van Ostaijen-ten Dam MM, van Eggermond MC, Thijssen PE, Aytekin C, Brunetti-Pierri N, van der Burg M, Graham Davies E, Ferster A, Furthner D, Gimelli G, Gennery A, Kloeckener-Gruissem B, Meyn S, Powell C, Reisli I, Schuetz C, Schulz A, Shugar A, van den Elsen PJ, van der Maarel SM: Heterogeneous clinical presentation in ICF syndrome: Correlation with underlying gene defects. Eur J Hum Genet 21: 1219–1225, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daly SB, Urquhart JE, Hilton E, McKenzie EA, Kammerer RA, Lewis M, Kerr B, Stuart H, Donnai D, Long DA, Burgu B, Aydogdu O, Derbent M, Garcia-Minaur S, Reardon W, Gener B, Shalev S, Smith R, Woolf AS, Black GC, Newman WG: Mutations in HPSE2 cause urofacial syndrome. Am J Hum Genet 86: 963–969, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al Badr W, Al Bader S, Otto E, Hildebrandt F, Ackley T, Peng W, Xu J, Li J, Owens KM, Bloom D, Innis JW: Exome capture and massively parallel sequencing identifies a novel HPSE2 mutation in a Saudi Arabian child with Ochoa (urofacial) syndrome. J Pediatr Urol 7: 569–573, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weatherall DJ, Higgs DR, Bunch C, Old JM, Hunt DM, Pressley L, Clegg JB, Bethlenfalvay NC, Sjolin S, Koler RD, Magenis E, Francis JL, Bebbington D: Hemoglobin H disease and mental retardation: A new syndrome or a remarkable coincidence? N Engl J Med 305: 607–612, 1981 [DOI] [PubMed] [Google Scholar]
- 22.Badens C, Lacoste C, Philip N, Martini N, Courrier S, Giuliano F, Verloes A, Munnich A, Leheup B, Burglen L, Odent S, Van Esch H, Levy N: Mutations in PHD-like domain of the ATRX gene correlate with severe psychomotor impairment and severe urogenital abnormalities in patients with ATRX syndrome. Clin Genet 70: 57–62, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Patel N, Khan AO, Mansour A, Mohamed JY, Al-Assiri A, Haddad R, Jia X, Xiong Y, Mégarbané A, Traboulsi EI, Alkuraya FS: Mutations in ASPH cause facial dysmorphism, lens dislocation, anterior-segment abnormalities, and spontaneous filtering blebs, or Traboulsi syndrome. Am J Hum Genet 94: 755–759, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildebrandt F, Benzing T, Katsanis N: Ciliopathies. N Engl J Med 364: 1533–1543, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopp K, Cogal AG, Bergstralh EJ, Seide BM, Olson JB, Meek AM, Lieske JC, Milliner DS, Harris PC Rare Kidney Stone Consortium : Phenotype-genotype correlations and estimated carrier frequencies of primary hyperoxaluria. J Am Soc Nephrol 26: 2559–2570, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandrile G, van Woerden CS, Berchialla P, Beck BB, Acquaviva Bourdain C, Hulton SA, Rumsby G OxalEurope Consortium : Data from a large European study indicate that the outcome of primary hyperoxaluria type 1 correlates with the AGXT mutation type. Kidney Int 86: 1197–1204, 2014 [DOI] [PubMed] [Google Scholar]
- 27.Mulders SM, Knoers NV, Van Lieburg AF, Monnens LA, Leumann E, Wühl E, Schober E, Rijss JP, Van Os CH, Deen PM: New mutations in the AQP2 gene in nephrogenic diabetes insipidus resulting in functional but misrouted water channels. J Am Soc Nephrol 8: 242–248, 1997 [DOI] [PubMed] [Google Scholar]
- 28.Rojek A, Füchtbauer EM, Kwon TH, Frøkiaer J, Nielsen S: Severe urinary concentrating defect in renal collecting duct-selective AQP2 conditional-knockout mice. Proc Natl Acad Sci U S A 103: 6037–6042, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang B, Zhao D, Qian L, Verkman AS: Mouse model of inducible nephrogenic diabetes insipidus produced by floxed aquaporin-2 gene deletion. Am J Physiol Renal Physiol 291: F465–F472, 2006 [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Rice W, Gu Z, Li J, Huang J, Brenner MB, Van Hoek A, Xiong J, Gundersen GG, Norman JC, Hsu VW, Fenton RA, Brown D, Lu HA: Aquaporin 2 promotes cell migration and epithelial morphogenesis. J Am Soc Nephrol 23: 1506–1517, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Attard M, Jean G, Forestier L, Cherqui S, van’t Hoff W, Broyer M, Antignac C, Town M: Severity of phenotype in cystinosis varies with mutations in the CTNS gene: Predicted effect on the model of cystinosin. Hum Mol Genet 8: 2507–2514, 1999 [DOI] [PubMed] [Google Scholar]
- 32.Soliman NA, El-Baroudy R, Rizk A, Bazaraa H, Younan A: Nephropathic cystinosis in children: An overlooked disease. Saudi J Kidney Dis Transplant 20: 436–442, 2009 [PubMed] [Google Scholar]
- 33.Emma F, Nesterova G, Langman C, Labbé A, Cherqui S, Goodyer P, Janssen MC, Greco M, Topaloglu R, Elenberg E, Dohil R, Trauner D, Antignac C, Cochat P, Kaskel F, Servais A, Wühl E, Niaudet P, Van’t Hoff W, Gahl W, Levtchenko E: Nephropathic cystinosis: An international consensus document. Nephrol Dial Transplant 29[Suppl 4]: iv87–iv94, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onuchic LF, Furu L, Nagasawa Y, Hou X, Eggermann T, Ren Z, Bergmann C, Senderek J, Esquivel E, Zeltner R, Rudnik-Schöneborn S, Mrug M, Sweeney W, Avner ED, Zerres K, Guay-Woodford LM, Somlo S, Germino GG: PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats. Am J Hum Genet 70: 1305–1317, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Courcet JB, Minello A, Prieur F, Morisse L, Phelip JM, Beurdeley A, Meynard D, Massenet D, Lacassin F, Duffourd Y, Gigot N, St-Onge J, Hillon P, Vanlemmens C, Mousson C, Cerceuil JP, Guiu B, Thevenon J, Thauvin-Robinet C, Jacquemin E, Rivière JB, Michel-Calemard L, Faivre L: Compound heterozygous PKHD1 variants cause a wide spectrum of ductal plate malformations. Am J Med Genet A 167: 3046–3053, 2015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.