Abstract
IgA nephropathy (IgAN) is the most common form of primary GN and an important cause of kidney failure. Characteristically, patients with IgAN have increased serum levels of undergalactosylated IgA1 (gd-IgA1). To assess the degree to which serum gd-IgA1 levels are genetically determined in healthy individuals, we determined serum IgA and gd-IgA1 levels by ELISA in a sample of 148 healthy female twins, including 27 monozygotic and 47 dizygotic pairs. Using the classic twin model, we found the heritability of serum gd-IgA1 and IgA levels to be 80% (95% confidence interval, 66% to 89%) and 46% (95% confidence interval, 15% to 69%), respectively. These data indicate that serum gd-IgA1 levels are highly heritable. Elucidating the genetic basis of this heritability will be important in understanding the pathogenesis of IgAN.
Keywords: IgA nephropathy, human genetics, gd-IgA1, glycosylation
IgA nephropathy (IgAN) is the most frequently diagnosed type of GN in the world.1 The course of disease is complex and not yet fully understood; the prognosis is variable. Some patients have a very mild form of the disease that requires little to no treatment. However, others have progressive disease with up to 50% of patients developing ESRD within 20 years of diagnosis. IgAN recurs in approximately 50%–60% of transplanted patients, indicating an important contribution of extrarenal factors to pathogenesis.2–5 A great deal of evidence exists to support a significant genetic contribution to IgAN.5–9 The incidence of IgAN varies geographically, being most prevalent in East Asian populations and less prevalent in European and African populations.10,11 Six genome-wide association studies have collectively identified 20 distinct loci associated with IgAN. Interestingly, most of these loci are shared with other immune-related diseases.12–17 One associated single-nucleotide polymorphism has been located within the ST6GAL1 gene.16 ST6GAL1 encodes ST6 β-galactosamide α-2,6-sialyltranferase 1, a glycosyltransferase. However, none of the loci are specifically associated with genes involved in IgA1 glycosylation. One of the hallmarks of IgAN is the presence of increased amounts of circulating undergalactosylated IgA1 antibodies. Usually, glycans on the hinge region of IgA1 terminate with galactose. In IgAN patients an increased proportion of IgA1 glycans terminate in N-acetylgalactosamine or sialylated N-acetylgalactosamine.5,18 This type of IgA1 is termed galactose-deficient IgA1 (gd-IgA1). gd-IgA1 has an established role in the development of IgAN. In the proposed four-hit hypothesis of IgAN pathogenesis, an increase in gd-IgA1 triggers the production of antiglycan autoantibodies.8,19,20 This leads to the formation of immune complexes that, under this hypothesis, may deposit in the kidney and cause kidney injury.21,22 Levels of gd-IgA1 are elevated in IgAN patients, regardless of ethnicity or age.23,24,25
Studies of familial IgAN have provided heritability estimates for gd-IgA1 between 54% and 76%.25–27 One difficulty of these studies is that at-risk relatives tend to show increased gd-IgA1 levels, biasing the heritability estimates which have been suggested to strongly depend on the gd-IgA1 levels of the index IgAN case.26 In order to understand the genetic contribution to gd-IgA1 levels in IgAN patients, it is first necessary to understand the genetic contribution to gd-IgA1 levels in healthy individuals. The classic twin model allows the estimation of the environmental and genetic contribution to phenotypic variation. We assessed the heritability of serum gd-IgA1 and IgA levels in a randomly ascertained sample of 148 healthy female twins from the TwinsUK cohort consisting of 47 dizygotic and 27 monozygotic pairs. All individuals were white females. The mean age was 56.9 years (range, 27.1–84.8; SD=13). Phenotypic characteristics are summarized in Table 1.
Table 1.
Phenotypic details of the 148 white female individuals in the study sample
| Mean | SD | First Quantile | Third Quantile | |
|---|---|---|---|---|
| Monozygotic (n=54) | ||||
| Age, yr | 52.73 | 12.16 | 45.14 | 62.86 |
| IgA, mg/ml | 3.22 | 0.65 | 2.81 | 3.54 |
| gd-IgA1, AU | 0.54 | 0.16 | 0.42 | 0.66 |
| Dizygotic (n=94) | ||||
| Age, yr | 59.28 | 12.98 | 48.91 | 69.76 |
| IgA, mg/ml | 3.13 | 0.77 | 2.62 | 3.67 |
| gd-IgA1, AU | 0.53 | 0.15 | 0.43 | 0.63 |
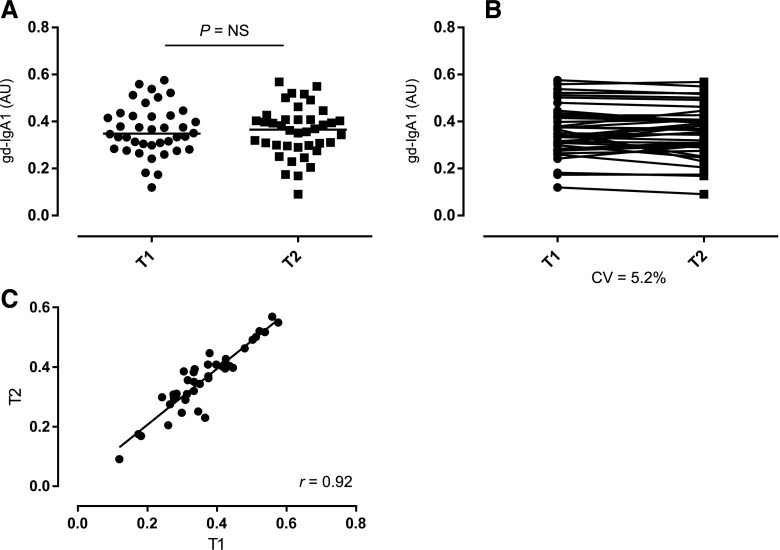
We measured IgA and gd-IgA1 levels in serum by ELISA. To demonstrate consistency among multiple measurements of both IgA and gd-IgA1 we designed a fully crossed experiment, where 58 randomly selected control samples were assessed on 3 testing days. We assessed the intrarater reliability of each experiment by calculating the intraclass correlation coefficient (ICC).28 The ICC for the IgA assay was 0.74 (95% confidence interval [95% CI], 0.63 to 0.83) and the ICC for the gd-IgA1 assay was 0.89 (95% CI, 0.73 to 0.95), showing a good-to-excellent reproducibility.29 We next assessed serum IgA and gd-IgA1 levels in our twin cohort. The mean serum IgA level was 3.16 mg/ml (range, 0.91–5.41; SD=0.73). The mean serum gd-IgA1 level was 0.54 absorbance units (AU) (range, 0.21–0.89; SD=0.15). For both parameters the data were normally distributed (P>0.05, Shapiro–Wilk normality test). To control for potential batch effects analyses were carried out using the plate-adjusted residuals for both traits and age at assessment was included in all models. To determine longitudinal stability of gd-IgA1, we analyzed gd-IgA1 levels in two samples from each individual (n=40 individuals). The samples were collected 5 years apart. There was no difference in Helix aspersa agglutinin (HAA) binding of the paired samples over time (Figure 1, r=0.92, P<0.001). This data demonstrated longitudinal stability of gd-IgA1 in the twins and is consistent with previous studies.30
Figure 1.
Stability of gd-IgA1 levels over time. Unpaired (A) and paired (B) scatterplots show gd-IgA1 levels determined at two time points, T1 and T2 (5 years apart), for individual samples (n=40 individuals with paired samples from each; bars represent group median). There was no difference in gd-IgA1 levels in the paired samples over time (paired t-test, P=NS), with a good correlation between T1 and T2 (C) (r=0.92, P<0.001 [Pearson correlation]). AU, absorbance units; CV, coefficient of variation.
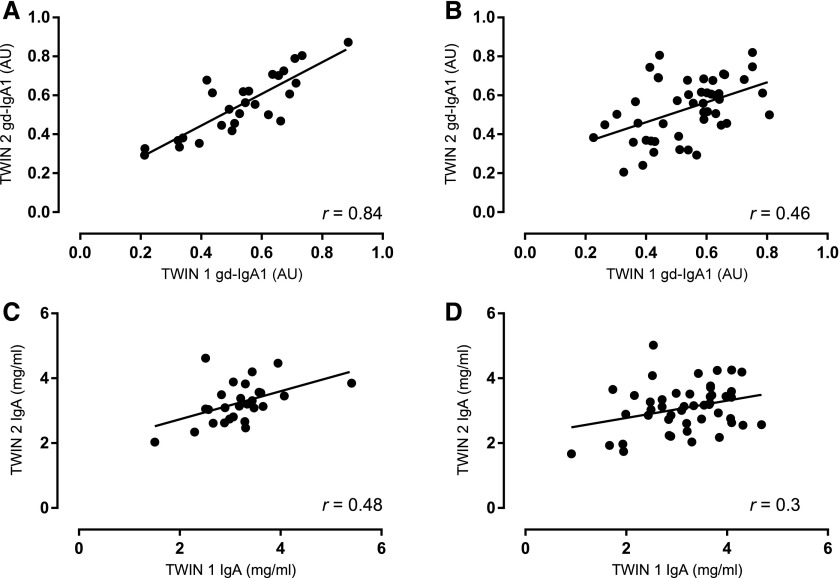
We fitted three different nested genetic models to the data using OpenMX: (1) the E model, which assumes that the phenotypic variability in the population is determined only by the environment; (2) the AE model, which assumes that both additive genetic effects and the environment play a role; and (3) the ACE model that includes an additional component for the common shared familial environment. The AE model was the best-fitting model for the estimation of both gd-IgA1 and IgA level heritability (Akaike information criterion [AIC]ACE=−485.6, AICAE=−487.5 and AICACE=11.0 AICAE=9.0, respectively). Additive genetic effects accounted for 80.4% (95% CI, 65.6% to 88.7%) of the variance of gd-IgA1 levels and individual-specific environmental effects explained the remaining 19.6% (95% CI, 11.3% to 34.4%) of the variance. Additive genetic effects accounted for 46.3% (95% CI, 15.2% to 68.6%) of the variance of IgA levels, and individual-specific environmental effects explained the remaining 53.7% (95% CI, 31.4% to 84.8%) of the variance. These data show that, unlike serum IgA, serum gd-IgA1 is highly heritable. This is confirmed by the fact that, in contrast to serum IgA levels, the correlation of gd-IgA1 between monozygotic twins (r=0.84) was much higher than the correlation between dizygotic twins (r=0.46; Figure 2). Our analyses suggest that the variability of gd-IgA1 levels in the healthy general population is strongly determined by genes with additive effects on the trait, whereas the individual environment (lifestyle, exposure) plays a much smaller role. Analogously, no effects due to common environmental or lifestyle factors that are shared within each family were identified in our sample. Overall, our data show that circulating gd-IgA1 is highly heritable (80.4%) in a healthy population, indicating that serum gd-IgA1 levels are under strong genetic control. Heritability of gd-IgA1 has previously been demonstrated in studies based on families ascertained through the presence of IgA nephropathy or Henoch–Schonlein Purpura, although estimates were sometimes dependent on the gd-IgA1 levels of the index case.25–27 An increased level of gd-IgA1 is associated with IgAN and is considered to be the ‘first hit’ in the proposed disease pathogenesis model.8,20 Notably, asymptomatic first-degree relatives of IgAN patients have high gd-IgA1 levels, suggesting that additional factors (‘hits’) are required for IgAN to develop. Consistent with previous studies, serum IgA levels showed low heritability (46.3%) in our cohort.31,32 In conclusion, our study found gd-IgA1 levels to be a highly heritable trait in the general population, with a heritability estimate of about 80%. Disentangling the genetic component underlying gd-IgA1 variability may help the identification of genetic risk factors for IgAN susceptibility.
Figure 2.
Twin-by-twin scatterplots. Serum gd-IgA1 levels in monozygotic (MZ) (A) and dizygotic (DZ) (B) twins and of serum IgA levels in MZ (C) and DZ (D) twins. The levels of gd-IgA1 were more correlated in MZ twins than in DZ twins; intrapair correlations were 0.84 (P<0.001) and 0.46 (P=0.001), respectively. These data suggest gd-IgA1 level is highly heritable. Conversely, intrapair correlations of IgA level were 0.48 (P=0.01) and 0.30 (P=0.04), indicative of IgA level being less heritable than gd-IgA1 level. AU, absorbance units.
Concise Methods
Sample Cohort
The TwinsUK adult twin registry includes about 12,000 subjects, predominately white females, unselected for any specific disease, recruited from all over the United Kingdom from 1992. Individuals from the TwinsUK cohort have been shown to have similar disease and lifestyle characteristics to the general population.33 St. Thomas’ Hospital Research Ethics Committee approved this study, and all twins provided informed written consent. The sample used for this study was randomly ascertained among healthy twin pairs. All of the data are available upon request from the Twin Research Unit website (www.twinsuk.ac.uk/data-access/submission-procedure).
Measurement of Serum IgA
Serum IgA levels were measured by ELISA.23 MaxiSorb immunoplates (Nunc; Life Technologies, Carlsbad, CA) were coated overnight at 4°C with 3 µg/ml F(ab’)2 fragment goat antihuman IgA (Jackson ImmunoResearch Laboratories, West Grove, PA) in coating buffer (0.05 M carbonate-bicarbonate buffer, pH 9.6). Between each incubation step, plates were washed three times with washing buffer (PBS and 0.1% Tween 20). Plates were blocked for 1 hour at room temperature with carbofree (Vector Labs). Samples were diluted 1:80,000 in carbofree and incubated at room temperature for 2 hours. Detection was carried out for 1 hour at room temperature using F(ab’)2 fragment biotinylated goat anti-human IgA1 (Jackson ImmunoResearch Laboratories), followed by Extravidin-HRP. ELISAs were developed using TMB substrate (BD Biosciences, San Jose, CA) and absorbance was measured at 450 nm. A standard curve was produced on each plate using serial dilutions of purified IgA1 (Abcam, Inc., Cambridge, MA) from 100 ng/ml to 1.56 ng/ml.
Measurement of gd-IgA1
Levels of serum gd-IgA1 were measured using a lectin-based ELISA.34 MaxiSorb immunoplates were coated overnight at 4°C with polyclonal rabbit antihuman IgA (Dako) diluted 1:1000 in coating buffer (0.05 M carbonate-bicarbonate buffer, pH 9.6). Between each incubation step, plates were washed four times with washing buffer (PBS and 0.1% Tween 20, 0.355 M sodium chloride). Plates were blocked for 1 hour at room temperature with carbofree (Vector Laboratories, Burlingame, CA) and samples were diluted 1:100 in PBS (to ensure saturation of IgA) and incubated overnight at 4°C. Helix aspersa agglutinin-biotin (Sigma-Aldrich, St. Louis, MO) diluted 1:1000 in PBS was added to each well for 90 min at room temperature, followed by poly-streptavidin HRP (Pierce, Rockford, IL) diluted 1:10,000, also for 90 min at room temperature. ELISAs were developed using TMB substrate (BD Biosciences) and absorbance was measured at 450 nm. Three control samples were run on each plate (“low,” “medium,” and “high”) to test for inter- and intra-assay variation.
Heritability Estimation
We used OpenMX (http://openmx.psyc.virginia.edu, version 2.2.4) to estimate the contribution of additive genetic, shared, and individual-specific environmental effects on serum IgA and gd-IgA1 level variation (ACE model).35 We also compared the ACE model with the most parsimonious AE model, which does not include the effect of common environmental influences, assuming that all familial aggregation results from additive genetic effects, and against the E model that assumes all variability to be determined by the environment. The models were compared using AIC in order to determine which model attained the best goodness-of-fit in the most parsimonious way. In the analyses the age at serum level collection was included as a covariate.
Disclosures
None
Acknowledgments
We acknowledge support by the National Institute for Health Research Biomedical Research Centre based at Imperial College Healthcare National Health Service Trust and Imperial College London. The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health research, or the Department of Health.
This work was supported by funding from the Medical Research Council (MR/K01353X/1). M.C.P. is a Wellcome Trust Senior Fellow in Clinical Science (fellowship WT082291MA). TwinsUK is funded by the Wellcome Trust, the Medical Research Council, the European Union and the National Institute for Health Research–funded BioResource, the Clinical Research Facility, and the Biomedical Research Centre based at Guy's and St Thomas' National Health Service Foundation Trust in partnership with King's College London.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Lai KN: Pathogenesis of IgA nephropathy. Nat Rev Nephrol 8: 275–283, 2012 [DOI] [PubMed] [Google Scholar]
- 2.Berger J, Yaneva H, Nabarra B, Barbanel C: Recurrence of mesangial deposition of IgA after renal transplantation. Kidney Int 7: 232–241, 1975 [DOI] [PubMed] [Google Scholar]
- 3.Schena FP: A retrospective analysis of the natural history of primary IgA nephropathy worldwide. Am J Med 89: 209–215, 1990 [DOI] [PubMed] [Google Scholar]
- 4.Bumgardner GL, Amend WC, Ascher NL, Vincenti FG: Single-center long-term results of renal transplantation for IgA nephropathy. Transplantation 65: 1053–1060, 1998 [DOI] [PubMed] [Google Scholar]
- 5.Kiryluk K, Novak J: The genetics and immunobiology of IgA nephropathy. J Clin Invest 124: 2325–2332, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiryluk K, Julian BA, Wyatt RJ, Scolari F, Zhang H, Novak J, Gharavi AG: Genetic studies of IgA nephropathy: past, present, and future. Pediatr Nephrol 25: 2257–2268, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiryluk K, Novak J, Gharavi AG: Pathogenesis of immunoglobulin A nephropathy: recent insight from genetic studies. Annu Rev Med 64: 339–356, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyatt RJ, Julian BA: IgA nephropathy. N Engl J Med 368: 2402–2414, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Magistroni R, D’Agati VD, Appel GB, Kiryluk K: New developments in the genetics, pathogenesis, and therapy of IgA nephropathy. Kidney Int 88: 974–989, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall YN, Fuentes EF, Chertow GM, Olson JL: Race/ethnicity and disease severity in IgA nephropathy. BMC Nephrol 5: 10, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiryluk K, Li Y, Sanna-Cherchi S, Rohanizadegan M, Suzuki H, Eitner F, Snyder HJ, Choi M, Hou P, Scolari F, Izzi C, Gigante M, Gesualdo L, Savoldi S, Amoroso A, Cusi D, Zamboli P, Julian BA, Novak J, Wyatt RJ, Mucha K, Perola M, Kristiansson K, Viktorin A, Magnusson PK, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Boland A, Metzger M, Thibaudin L, Wanner C, Jager KJ, Goto S, Maixnerova D, Karnib HH, Nagy J, Panzer U, Xie J, Chen N, Tesar V, Narita I, Berthoux F, Floege J, Stengel B, Zhang H, Lifton RP, Gharavi AG: Geographic differences in genetic susceptibility to IgA nephropathy: GWAS replication study and geospatial risk analysis. PLoS Genet 8: e1002765, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feehally J, Farrall M, Boland A, Gale DP, Gut I, Heath S, Kumar A, Peden JF, Maxwell PH, Morris DL, Padmanabhan S, Vyse TJ, Zawadzka A, Rees AJ, Lathrop M, Ratcliffe PJ: HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol 21: 1791–1797, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, Sanna-Cherchi S, Men CJ, Julian BA, Wyatt RJ, Novak J, He JC, Wang H, Lv J, Zhu L, Wang W, Wang Z, Yasuno K, Gunel M, Mane S, Umlauf S, Tikhonova I, Beerman I, Savoldi S, Magistroni R, Ghiggeri GM, Bodria M, Lugani F, Ravani P, Ponticelli C, Allegri L, Boscutti G, Frasca G, Amore A, Peruzzi L, Coppo R, Izzi C, Viola BF, Prati E, Salvadori M, Mignani R, Gesualdo L, Bertinetto F, Mesiano P, Amoroso A, Scolari F, Chen N, Zhang H, Lifton RP: Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43: 321–327, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu XQ, Li M, Zhang H, Low HQ, Wei X, Wang JQ, Sun LD, Sim KS, Li Y, Foo JN, Wang W, Li ZJ, Yin XY, Tang XQ, Fan L, Chen J, Li RS, Wan JX, Liu ZS, Lou TQ, Zhu L, Huang XJ, Zhang XJ, Liu ZH, Liu JJ: A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet 44: 178–182, 2011 [DOI] [PubMed] [Google Scholar]
- 15.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, Fischman C, Snyder HJ, Appel G, Izzi C, Viola BF, Dallera N, Del Vecchio L, Barlassina C, Salvi E, Bertinetto FE, Amoroso A, Savoldi S, Rocchietti M, Amore A, Peruzzi L, Coppo R, Salvadori M, Ravani P, Magistroni R, Ghiggeri GM, Caridi G, Bodria M, Lugani F, Allegri L, Delsante M, Maiorana M, Magnano A, Frasca G, Boer E, Boscutti G, Ponticelli C, Mignani R, Marcantoni C, Di Landro D, Santoro D, Pani A, Polci R, Feriozzi S, Chicca S, Galliani M, Gigante M, Gesualdo L, Zamboli P, Battaglia GG, Garozzo M, Maixnerová D, Tesar V, Eitner F, Rauen T, Floege J, Kovacs T, Nagy J, Mucha K, Pączek L, Zaniew M, Mizerska-Wasiak M, Roszkowska-Blaim M, Pawlaczyk K, Gale D, Barratt J, Thibaudin L, Berthoux F, Canaud G, Boland A, Metzger M, Panzer U, Suzuki H, Goto S, Narita I, Caliskan Y, Xie J, Hou P, Chen N, Zhang H, Wyatt RJ, Novak J, Julian BA, Feehally J, Stengel B, Cusi D, Lifton RP, Gharavi AG: Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 46: 1187–1196, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Foo JN, Wang JQ, Low HQ, Tang XQ, Toh KY, Yin PR, Khor CC, Goh YF, Irwan ID, Xu RC, Andiappan AK, Bei JX, Rotzschke O, Chen MH, Cheng CY, Sun LD, Jiang GR, Wong TY, Lin HL, Aung T, Liao YH, Saw SM, Ye K, Ebstein RP, Chen QK, Shi W, Chew SH, Chen J, Zhang FR, Li SP, Xu G, Tai ES, Wang L, Chen N, Zhang XJ, Zeng YX, Zhang H, Liu ZH, Yu XQ, Liu JJ: Identification of new susceptibility loci for IgA nephropathy in Han Chinese. Nat Commun 6: 7270, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saka S, Hirawa N, Oka A, Yatsu K, Hirukawa T, Yamamoto R, Matsusaka T, Imai E, Narita I, Endoh M, Ichikawa I, Umemura S, Inoko H: Genome-wide association study of IgA nephropathy using 23 465 microsatellite markers in a Japanese population. J Hum Genet 60: 573–580, 2015 [DOI] [PubMed] [Google Scholar]
- 18.Novak J, Rizk D, Takahashi K, Zhang X, Bian Q, Ueda H, Ueda Y, Reily C, Lai LY, Hao C, Novak L, Huang ZQ, Renfrow MB, Suzuki H, Julian BA: New Insights into the Pathogenesis of IgA Nephropathy. Kidney Dis (Basel) 1: 8–18, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki H, Fan R, Zhang Z, Brown R, Hall S, Julian BA, Chatham WW, Suzuki Y, Wyatt RJ, Moldoveanu Z, Lee JY, Robinson J, Tomana M, Tomino Y, Mestecky J, Novak J: Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest 119: 1668–1677, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki H, Kiryluk K, Novak J, Moldoveanu Z, Herr AB, Renfrow MB, Wyatt RJ, Scolari F, Mestecky J, Gharavi AG, Julian BA: The pathophysiology of IgA nephropathy. J Am Soc Nephrol 22: 1795–1803, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomana M, Novak J, Julian BA, Matousovic K, Konecny K, Mestecky J: Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J Clin Invest 104: 73–81, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novak J, Tomana M, Matousovic K, Brown R, Hall S, Novak L, Julian BA, Wyatt RJ, Mestecky J: IgA1-containing immune complexes in IgA nephropathy differentially affect proliferation of mesangial cells. Kidney Int 67: 504–513, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Moldoveanu Z, Wyatt RJ, Lee JY, Tomana M, Julian BA, Mestecky J, Huang WQ, Anreddy SR, Hall S, Hastings MC, Lau KK, Cook WJ, Novak J: Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int 71: 1148–1154, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Shimozato S, Hiki Y, Odani H, Takahashi K, Yamamoto K, Sugiyama S: Serum under-galactosylated IgA1 is increased in Japanese patients with IgA nephropathy. Nephrol Dial Transplant 23: 1931–1939, 2008 [DOI] [PubMed] [Google Scholar]
- 25.Hastings MC, Moldoveanu Z, Julian BA, Novak J, Sanders JT, McGlothan KR, Gharavi AG, Wyatt RJ: Galactose-deficient IgA1 in African Americans with IgA nephropathy: serum levels and heritability. Clin J Am Soc Nephrol 5: 2069–2074, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gharavi AG, Moldoveanu Z, Wyatt RJ, Barker CV, Woodford SY, Lifton RP, Mestecky J, Novak J, Julian BA: Aberrant IgA1 glycosylation is inherited in familial and sporadic IgA nephropathy. J Am Soc Nephrol 19: 1008–1014, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiryluk K, Moldoveanu Z, Sanders JT, Eison TM, Suzuki H, Julian BA, Novak J, Gharavi AG, Wyatt RJ: Aberrant glycosylation of IgA1 is inherited in both pediatric IgA nephropathy and Henoch-Schönlein purpura nephritis. Kidney Int 80: 79–87, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallgren KA: Computing Inter-Rater Reliability for Observational Data: An Overview and Tutorial. Tutor Quant Methods Psychol 8: 23–34, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cicchetti DV: Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol Assess 6: 284–290, 1994 [Google Scholar]
- 30.Hastings MC, Moldoveanu Z, Suzuki H, Berthoux F, Julian BA, Sanders JT, Renfrow MB, Novak J, Wyatt RJ: Biomarkers in IgA nephropathy: relationship to pathogenetic hits. Expert Opin Med Diagn 7: 615–627, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viktorin A, Frankowiack M, Padyukov L, Chang Z, Melén E, Sääf A, Kull I, Klareskog L, Hammarström L, Magnusson PK: IgA measurements in over 12 000 Swedish twins reveal sex differential heritability and regulatory locus near CD30L. Hum Mol Genet 23: 4177–4184, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frankowiack M, Kovanen RM, Repasky GA, Lim CK, Song C, Pedersen NL, Hammarström L: The higher frequency of IgA deficiency among Swedish twins is not explained by HLA haplotypes. Genes Immun 16: 199–205, 2015 [DOI] [PubMed] [Google Scholar]
- 33.Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, MacGregor AJ: Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res 4: 464–477, 2001 [DOI] [PubMed] [Google Scholar]
- 34.Smith AC, de Wolff JF, Molyneux K, Feehally J, Barratt J: O-glycosylation of serum IgD in IgA nephropathy. J Am Soc Nephrol 17: 1192–1199, 2006 [DOI] [PubMed] [Google Scholar]
- 35.Verweij KJ, Mosing MA, Zietsch BP, Medland SE: Estimating heritability from twin studies. Methods Mol Biol 850: 151–170, 2012 [DOI] [PubMed] [Google Scholar]