ABSTRACT
The DosR regulon, a set of 48 genes normally expressed in Mycobacterium tuberculosis under conditions that inhibit aerobic respiration, is controlled via the DosR-DosS/DosT two-component system. While the regulon requires induction in most M. tuberculosis isolates, for members of the Beijing lineage, its expression is uncoupled from the need for signaling. In our attempts to understand the mechanistic basis for this uncoupling in the Beijing background, we previously reported the identification of two synonymous single-nucleotide polymorphisms (SNPs) within the adjacent Rv3134c gene. In the present study, we have interrogated the impact of these SNPs on dosR expression in wild-type strains, as well as a range of dosR-dosS-dosT mutants, for both Beijing and non-Beijing M. tuberculosis backgrounds. In this manner, we have unequivocally determined that the C601T dosR promoter SNP is the sole requirement for the dramatic shift in the pattern of DosR regulon expression seen in this globally important lineage. Interestingly, we also show that DosT is completely nonfunctional within these strains. Thus, a complex series of evolutionary steps has led to the present-day Beijing DosR phenotype that, in turn, potentially confers a fitness advantage in the face of some form of host-associated selective pressure.
IMPORTANCE Mycobacterium tuberculosis strains of the Beijing lineage have been described as being of enhanced virulence compared to other lineages, and in certain regions, they are associated with the dramatic spread of multidrug-resistant tuberculosis (TB). In terms of trying to understand the functional basis for these broad epidemiological phenomena, it is interesting that, in contrast to the other major lineages, the Beijing strains all constitutively overexpress members of the DosR regulon. Here, we identify the mutational events that led to the evolution of this unique phenotype. In addition, our work highlights the fact that important phenotypic differences exist between distinct M. tuberculosis lineages, with the potential to impact the efficacy of diagnosis, vaccination, and treatment programs.
KEYWORDS: Tuberculosis, DosR regulon, strain variation, evolution, Mycobacterium tuberculosis
INTRODUCTION
Despite being an ancient disease, human tuberculosis (TB) resulting from infection with Mycobacterium tuberculosis still remains a leading cause of death worldwide as a consequence of multiple contributing factors, including drug resistance, HIV coinfection, and inadequate access to high-quality health care (1). Furthermore, recent evidence suggests that the level of genetic diversity that exists among distinct M. tuberculosis isolates was previously underestimated and has the potential to impact pathogenicity, as well as to limit the effectiveness of current diagnostic and therapeutic interventions (2–4).
The identification of specific single-nucleotide polymorphisms (SNPs) and/or large-sequence polymorphisms (LSPs) has enabled the classification of M. tuberculosis isolates into 7 major strain lineages (lineages 1 to 7) that show a degree of geographic restriction (5, 6). From an epidemiological perspective, it is particularly interesting to note that the incidence of TB resulting from the major Beijing subbranch of lineage 2 (also known as the East Asian lineage) is reported to be increasing disproportionately in multiple settings, and it is often associated with the spread of multidrug-resistant (MDR) M. tuberculosis (7–12). This has led to frequent speculation that strains belonging to the Beijing lineage possess unique phenotypic attributes that confer an increased ability to transmit and/or cause disease, as well as an increased capacity to acquire antibiotic resistance (10, 11). Indeed, it has recently been demonstrated that members of this lineage acquire resistance to multiple antibiotics in vitro at a rate that is up to 10-fold higher than that of lineage 4 strains (including the laboratory strain M. tuberculosis H37Rv), due to an increased basal mutation frequency. In the same paper, modeling simulations used by Ford et al. indicated that the probability of de novo MDR arising in patients infected with Beijing strains is more than 20-fold higher than for non-Beijing strain infections (13). Although the exact mechanism(s) underlying this enhanced mutation rate has yet to be defined, we and other groups have suggested that perturbations in regulatory, metabolic, or structural pathways that exist in the Beijing strains potentially account for an alteration in antibiotic sensitivity/tolerance (2, 11, 13, 14). In turn, this could provide a mechanism by which an increased proportion of cells are able to survive drug exposure, thereby increasing the potential for the acquisition of specific resistance mutations (15).
One striking example of a regulatory perturbation unique to the Beijing lineage involves the DosR regulon, a 48-member regulon that includes a diverse array of genes involved in anaerobic respiration, metabolism (nucleotide, carbon, and lipid), and stress responses, and which is controlled through a two-component signaling system comprising the transcription factor DosR and one of two sensor kinases, DosS or DosT (16, 17). The DosR regulon normally serves as the primary mechanism by which M. tuberculosis responds to changes in the redox environment as a result of hypoxia or nitric oxide (NO) exposure (18–21). These signals are encountered as part of the host granulomatous response and are proposed to be among the most critical stimuli for the development of dormancy and latent TB (22–24). Mechanistic links between DosR and other “master regulators,” such as WhiB3 and the PhoPR two-component regulatory system, have also recently been described, suggestive of a wider stress-associated regulatory network linking hypoxia and redox adaptation (DosR-WhiB3), with cell wall lipid biosynthesis (WhiB3-PhoP) (25, 26). In contrast to the other M. tuberculosis lineages, the Beijing lineage constitutively overexpress members of the DosR regulon, a phenotype we first described in 2007 (14, 27) and that has since been independently confirmed in multiple laboratories (28, 29). Thus, the DosR regulon is always activated in the Beijing isolates, even in the absence of the aforementioned external redox signals.
In 2010, we reported several Beijing-specific mutations within the dosR-dosS-dosT two-component regulatory system that coincide with the appearance of the constitutive DosR phenotype (30). The one on which we initially focused as being a potential contributor to the Beijing DosR phenotype was a frameshift mutation in the DosT sensor kinase gene. We originally hypothesized that the C-terminal kinase domain may have been constitutively active in the absence of the upstream regulatory sequence. However, gene complementation assays employing both wild-type and mutant versions of dosT in diverse strain backgrounds suggested that the mutation in dosT was not directly responsible for the constitutive DosR regulon phenotype (30). Next, we investigated a novel 350-kb gene duplication that was identified in members of the most recently evolved Beijing strains (14, 31). The presence of this duplication, which includes a second copy of the Rv3134c-dosR-dosS operon, does increase the extent of DosR regulon expression; however, gene disruption studies coupled with the finding that it is not uniformly present among all Beijing strains highlight the fact that the duplication is not the underlying cause of the constitutive DosR phenotype (14, 32). Furthermore, recent work by our laboratory has shown that this large chromosomal duplication event is rapidly selected for as a consequence of in vitro growth (32).
In the present study, we have reinvestigated the two SNPs we identified in the coding region of the Rv3134c gene that lies immediately adjacent to dosR and is included as part of the dosR-dosS operon. We originally discounted these SNPs on account of the fact that they are both synonymous and are located quite some distance upstream (>200 bp) of the dosR start codon (30). Herein, using a variety of approaches, we comprehensively demonstrate that one of these SNPs is the sole cause of the constitutive DosR overexpression phenotype that typifies the widespread Beijing lineage. Our data also serve to emphasize the dramatic effect that the evolution of a single regulatory mutation can have on shaping lineage-specific gene expression in M. tuberculosis. In turn, these alterations in gene expression, in the absence of environmental signaling, might contribute to the reputed epidemiologic success of the Beijing strains.
RESULTS
DosR regulon is the major feature that distinguishes gene expression between Beijing and non-Beijing strains.
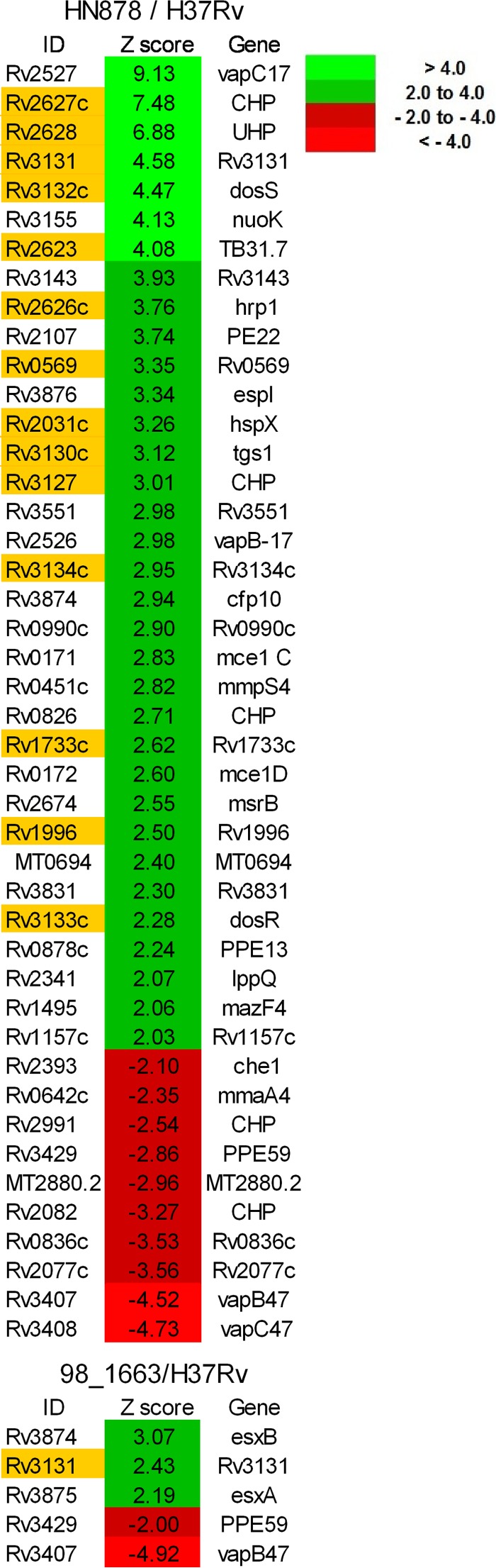
In order to compare the gene expression profiles of a “modern” Beijing isolate versus a “pre-Beijing” lineage 2 (East Asian) isolate, we performed whole-genome microarray analyses with HN878 (14, 33) and 98_1663 (5) with reference to the H37Rv (ATCC) laboratory strain. HN878 is a modern Beijing isolate belonging to the region of difference 142 (RD142) Beijing subbranch, while 98_1663 is an ancestral lineage 2 strain lacking the RD207 deletion that is responsible for the characteristic Beijing spoligotyping profile (5, 34, 35). In contrast, H37Rv is a non-Beijing lineage 4 strain. When HN878, 98_1663, and H37Rv were grown under standard in vitro conditions, the ancestral isolate displayed an overall pattern of gene expression very similar to that of H37Rv. We identified only 5 genes that were differentially expressed, although the pattern of expression of these genes appears to be shared with the modern HN878 Beijing isolate (Fig. 1). Of particular note, members of the ESX-1 type VII secretion system that are critical for full virulence of M. tuberculosis (36–39) were significantly overexpressed in both of these isolates compared to H37Rv, including the esxA (esat-6), esxB (cfp-10), and espI genes. It should be noted that for the HN878 arrays, the relative expression level of esxA (+1.9) fell just below the Z-score threshold of >2.0. Although there were other findings of potential interest in these comparisons, for example, the VapBC-17 and VapBC-47 toxin-antitoxin complexes and certain mce (mammalian cell entry) genes were differentially regulated compared to H37Rv, from the point of view of the present study, the most striking observation was the very large influence that DosR appears to have on shaping the differential gene expression profile of HN878 compared to both H37Rv and the 98_1663 (pre-Beijing) isolate. Indeed, of the 34 genes that we reproducibly detected as being overexpressed in HN878 relative to H37Rv via microarray, >40% (14/34) were members of the DosR regulon (Fig. 1). Included in this list are dosR and dosS, encoding the main regulatory components of the regulon. Other examples of note include tgs1, encoding a triacylglycerol synthase whose expression has been associated with triglyceride accumulation under hypoxic conditions (40); hspX, encoding an α-crystallin-like chaperonin that is inducible under stress conditions, including hypoxia (41); hrp1, encoding a protein of unknown function that is reportedly able to be used to discriminate between latent and active TB infection (42, 43); and multiple genes from the Rv2623 to Rv2630 cluster that have recently been described as being upregulated by isoniazid (INH) treatment, specifically within infected macrophages (44).
FIG 1.
The DosR regulon is the major feature that distinguishes gene expression between Beijing and non-Beijing strains. Heatmaps of the microarray-based expression profiles comparing Beijing isolate HN878 and pre-Beijing isolate 98_1663 to the Euro-American strain H37Rv are shown. Data represent the mean from three arrays for each comparison. Only genes whose expression differs from that in H37Rv by a Z-score of more than ±2-fold are indicated. Values in green represent genes that are significantly overexpressed relative to H37Rv, while those in red indicate genes that are significantly repressed relative to H37Rv. Genes highlighted in yellow are members of the DosR regulon.
Overall, these findings reaffirm and expand upon our initial description of the constitutive overexpression of the DosR regulon in the Beijing lineage that was based on comparative real-time PCR analysis of a select set of genes (14, 27, 30). Next, as a step toward developing the tools necessary to understand the evolution and functional significance of this dramatic shift in the pattern of DosR regulon gene expression, we continued our ongoing efforts aimed at identifying the underlying molecular basis of this Beijing-specific phenotype.
Beijing-specific dosR promoter region confers constitutive overexpression of dosR.
In our 2010 publication that documents the dosT frameshift mutation, we also reported the identification of three synonymous SNPs within the 3.9-kb region surrounding dosR that includes Rv3131, dosS, and Rv3134c (30). Two of these SNPs were termed C507G and C601T, based on their position within the Rv3134c coding region. The C507G SNP was present in both the ancestral East Asian (95_1848) and modern Beijing strains (HN878 and W210) that were sequenced, while the C601T SNP was specific to the modern Beijing strains. We also identified a single G1068A substitution within dosS of the modern Beijing strains. As indicated above, we initially discounted the C507G and C601T SNPs on account of the fact that they are synonymous and are located >200 bp upstream of the dosR start codon. However, after eliminating the dosT mutation as the root cause of the Beijing constitutive DosR phenotype, we decided to look more closely at these SNPs on account of their relative proximity to the T1 and T2 transcription start sites (TSS) that had previously been reported for dosR (45, 46). We postulated that SNPs within this same region potentially impact dosR transcription.
To start, we sequenced the region of Rv3134c containing the C507G and C601T SNPs from representatives of each of the 6 major M. tuberculosis lineages that had been described at that time, including 3 to 5 representatives of each of the 5 RD-based subgroups of the East Asian lineage/lineage 2. Of these 5 subgroups, groups 2 to 5 represent the Beijing sublineage (27, 47, 48). Interestingly, the C507G SNP is restricted to the phylogenetically related East African/Indian (lineage 3) and East Asian lineages, while the C601T SNP is unique to the Beijing sublineage of the East Asian lineage (Fig. 2A). Thus, as with the dosT frameshift mutation (30), the presence of the C601T SNP correlates precisely with the occurrence of the constitutive DosR regulon phenotype. Meanwhile, the East African/Indian strains that bear only the C507G SNP display a level of dosR expression that is equivalent to that in H37Rv (see Fig. S1 in the supplemental material).
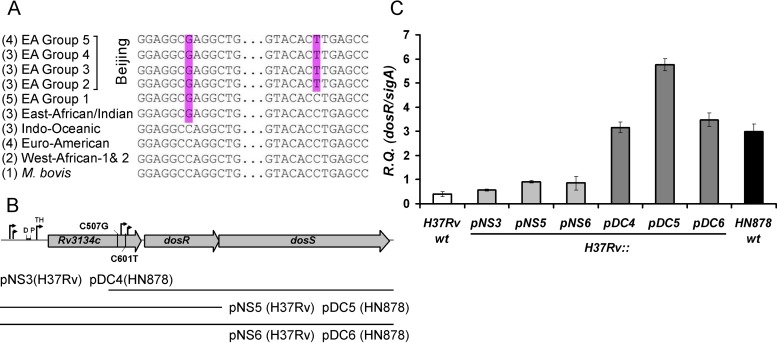
FIG 2.
The Beijing-specific dosR promoter region confers constitutive overexpression of dosR. (A) Partial sequence alignment of nucleotides 501 to 612 of the Rv3134c gene (nucleotide positions 3500444 to 3500555; numbering based on H37Rv). A total of 31 M. tuberculosis complex isolates were analyzed, 18 of which belong to the East Asian lineage. Of these, 13 were from the Beijing sublineage (East Asian [EA] groups 2 to 5). The positions of the C507G (left) and C601T (right) synonymous SNPs are highlighted. Only the C601T SNP is unique to the Beijing lineage. (B) Genomic organization of the Rv3134c-dosR-dosS operon. The approximate positions of the C507G and C601T SNPs found in the Rv3134c gene of M. tuberculosis Beijing isolates are shown. Arrows indicate previously identified promoter sequences, while D and P refer to distal and proximal binding sites, respectively, of the DosR-dependent hypoxic TH promoter (45, 46). A schematic representation of the inserts present in the various plasmids introduced into wild-type H37Rv is shown: pNS3 and pDC4 contain a 3,359-bp fragment that includes Rv3134c truncated at nucleotide 305, along with the complete dosR and dosS gene sequences; pNS5 and pDC5 contain a 1,883-bp fragment that includes 392 bp upstream of the Rv3134c gene, along with the complete Rv3134c and dosR gene sequences; and pNS6 and pDC6 contain a 4,049-bp fragment that includes 392 bp upstream of the Rv3134c gene in addition to the Rv3134c, dosR, and dosS genes. The pNS3, pNS5, and pNS6 plasmids contain the 507C and 601C H37Rv allele, while pDC4, pDC5, and pDC6 contain the 507G and 601T HN878 (Beijing) allele. (C) qRT-PCR analysis of dosR expression in H37Rv transformed with the plasmids indicated in panel B. For reference, wild-type H37Rv and HN878 are also included. Results are shown as relative quantities (R.Q.), using sigA as the normalizing gene. The error bars represent standard deviations calculated as indicated in Materials and Methods.
To further investigate the specific contribution of the aforementioned SNPs to the expression of dosR within the Beijing strain background, we generated a series of plasmid constructs capable of integrating at the nonessential mycobacteriophage L5 attachment site (attB) and that harbor distinct segments of the Rv3134c-dosR-dosS operon (Fig. 2B). Two versions of each of these segments were cloned: a wild-type non-Beijing version based on the H37Rv sequence (pNS series) and a Beijing version based on the sequence found in HN878 (pDC series). The pNS3 and pDC4 plasmids contain dosR and dosS, along with the 3′ region of Rv3134c that includes the C507G and C601T SNPs, in the case of pDC4. This region also includes the T1 and T2 TSS that have been reported to partially drive basal dosR expression (45, 46). Plasmids pNS5 and pDC5 contain full-length Rv3134c and dosR (without dosS), in addition to the promoter region of Rv3134c that includes both DosR-dependent (TH) and DosR-independent promoter elements (45, 46). Finally, pNS6 and pDC6 contain the full-length Rv3134c-dosR-dosS operon, including the Rv3134c promoter region. All six of these constructs were transformed into wild-type H37Rv, and the expression of dosR was analyzed via quantitative real-time PCR (qRT-PCR) and normalized against expression of the sigA gene encoding the housekeeping sigma factor. In a comparison of the pNS and pDC plasmids, which differ only by the presence of the C507G and C601T SNPs present in the pDC series, there was a very striking effect of the Beijing-specific sequence (pDC4, pDC5, and pDC6) on dosR expression, even when placed in the non-Beijing (H37Rv) background (Fig. 2C). Indeed, the pNS-transformed strains all behaved much like the parental H37Rv strain (albeit with slightly elevated dosR expression due to the presence of a second dosR copy). On the other hand, all the pDC-transformed H37Rv strains exhibited dosR expression levels that bore a remarkable resemblance to the wild-type HN878 strain. In one case (pDC5), dosR expression exceeded the level observed for HN878. This is likely attributable to the relatively small size of the plasmid insert in this case, allowing for more efficient transcription. Together, these data are highly suggestive that the C507G and/or C601T SNPs are directly contributing to the development of the constitutive dosR phenotype displayed by the Beijing strains (including HN878).
C601T promoter SNP is sufficient to cause the Beijing DosR phenotype.
To further refine our understanding of the mechanistic requirements necessary for the constitutive overexpression of dosR, we repeated several of the above-mentioned transformations within the dosR knockout (KO) background, for both the H37Rv (non-Beijing) and HN878 (Beijing) strains. In this manner, all dosR expression was derived from the exogenous copy of dosR contained on the integrated plasmid. In addition, several new plasmid constructs were generated to test the specific role of the individual C601T SNP, as well as the role of dosR itself on the evolution of the Beijing phenotype (Fig. 3A).
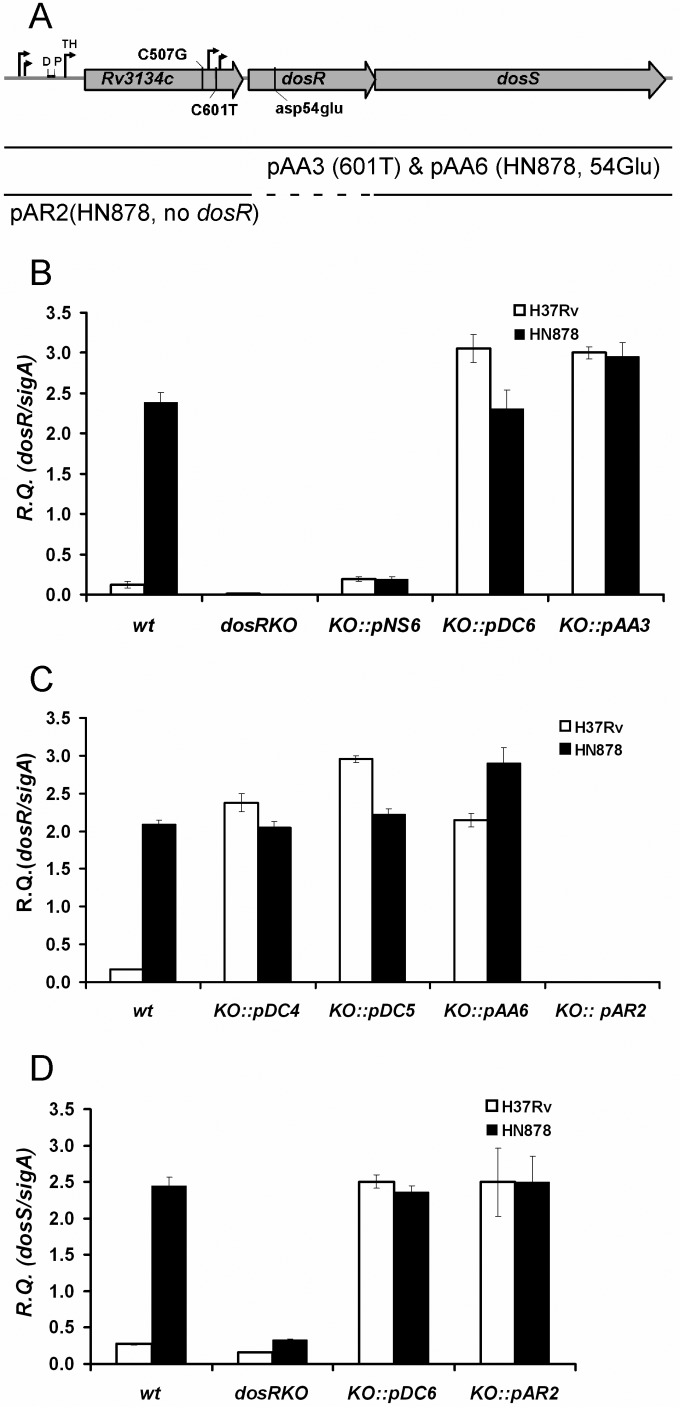
FIG 3.
The C601T promoter SNP is sufficient to cause the Beijing DosR phenotype. (A) Genomic organization of the Rv3134c-dosR-dosS operon, as in Fig. 2. The position of the Asp54Glu mutation (pAA6) is also indicated. A schematic representation of the inserts included in the plasmids used to transform dosR KO mutants of H37Rv and HN878 is shown. pAA3 and pAA6 contain a 4,049-bp fragment that includes 392 bp upstream of the Rv3134c gene in addition to the Rv3134c, dosR, and dosS genes. pAA3 contains a 507C (H37Rv) and 601T (HN878) hybrid allele (i.e., it has only the C601T SNP), while pAA6 contains the 507G and 601T HN878 allele along with a T/A transversion at position 161 of dosR that replaces Asp54 with Glu54. This renders DosR nonphosphorylatable. pAR2 contains a 3,393-bp fragment that incorporates 392 bp upstream of Rv3134c in addition to the Rv3134c and dosS genes (dashed line, no dosR). It is also based on the 507G and 601T HN878 allele. The pNS6, pDC6, pDC4, and pDC5 plasmids are all described in the legend to Fig. 2. (B to D) qRT-PCR analysis of dosR (B and C) and dosS (D) expression in wild-type, dosR KO mutant, and complemented strains. Results are shown as relative quantities (R.Q.), using sigA as the normalizing gene. The error bars represent standard deviation.
In both the dosR KO backgrounds, the effect of introducing the Beijing version of the Rv3134c-dosR-dosS operon was again very striking. In a comparison of the pNS6 and pDC6 plasmids, which differ only in the presence of the C507G and C601T SNPs (as described above), we observed an approximately 10-fold increase in dosR expression for both the H37Rv and HN878 transformants (Fig. 3B). Moreover, when the pAA3 plasmid (a hybrid containing the H37Rv allele at position 507 and the HN878 allele at position 601) was introduced into both dosR KO mutant backgrounds, the level of dosR expression was virtually identical to that seen for wild-type HN878 as well as the pDC6-containing strains. This important result clearly demonstrated that the presence of the Beijing-specific C601T promoter SNP is sufficient to drive the constitutive overexpression of dosR within the Beijing lineage. Indeed, we were able to recreate the same phenotype in the non-Beijing (H37Rv) background just by introducing this single mutation (Fig. 3B).
Next, we investigated the impact of dosR activation status, as well as the complete absence of dosR, on transcription arising from the Beijing promoter region. This was done in order to determine if dosR itself has any role to play in activating transcription in the presence of the C601T mutation. Plasmid pAA6 is a derivative of pDC6 (containing both C507G and C601T SNPs) that has an Asp-to-Glu mutation at position 54 of DosR that renders it nonphosphorylatable; hence, it is unable to be activated by either the DosS or DosT sensor kinase (16). As seen in Fig. 3C, the level of dosR expression for both the H37Rv and HN878 dosR KO strains transformed with pAA6 was not substantially different from that of either wild-type HN878 or the pDC transformants. From this, we conclude that activated DosR is not responsible for mediating transcription from the novel Beijing dosR promoter region. Likewise, pAR2 is a derivative of pDC6 that lacks dosR entirely. As expected, the KO strains transformed with this plasmid are devoid of dosR expression (Fig. 3C). However, the level of dosS expression in these strains was identical to that of wild-type HN878 or the pDC6 transformants (Fig. 3D). Hence, the C601T SNP is able to exert its effect on transcription completely independently of the DosR protein. Last, given that the level of dosS expression in the pDC6-containing strains was approximately 10-fold higher than in the dosR KOs (Fig. 3D), yet the level of dosR in the pDC5 and pDC6 transformants was equivalent (Fig. 3B and C), we deduce that the C601T SNP is also able to function independently of dosS, a fact we later confirmed in the HN878 ΔdosR ΔdosS mutant strain background that has no residual dosS expression (data not shown). Thus, the presence of the C601T promoter SNP is the sole feature of the dosR operon that is required for initiating the unique dosR phenotype displayed by the Beijing lineage.
Constitutive DosR regulon phenotype of the Beijing lineage does not impact virulence within the mouse model of infection.
To begin to interrogate the Beijing DosR phenotype from a functional perspective, we performed a series of aerosol mouse infections in order to examine the impact of constitutive (Beijing) versus inducible (non-Beijing) DosR overexpression on bacterial growth within the lungs and spleens of infected animals, as well as its impact on animal survival. Note that several prior studies had already validated the mouse as being a suitable model for distinguishing Beijing and non-Beijing M. tuberculosis strains on the basis of animal survival (49, 50). These low-dose aerosol infections (10 to 20 CFU/lung) were carried out for the following strains: wild-type HN878 (Beijing), HN878 in which both dosR and dosS were disrupted (dosRS KO mutant), and the HN878 dosRS KO mutant complemented with either pNS6 (non-Beijing Rv3134c-dosR-dosS operon) or pDC6 (Beijing Rv3134c-dosR-dosS operon). Thus, the only difference between the complemented strains is the presence of the two dosR promoter SNPs (C507G and C601T). The expression of dosR, as well as an independent marker of DosR regulon activation, Rv3130c (tgs1), was examined in these strains in order to confirm their expected phenotypes prior to use (Fig. S2).
In a comparison of the growth of wild-type HN878 to that of the double-knockout mutant (dosRS KO) in the lungs of infected mice, we observed an approximately 1-log10 reduction in CFU for the strain lacking both dosR and dosS between weeks 4 and 9 postinfection (Fig. 4A). However, as the infection progressed beyond 9 weeks, this difference in CFU seemed to resolve. By 28 weeks postinfection, there was no longer any detectable CFU difference. Interestingly, the reduction in bacterial CFU seen between 4 and 9 weeks with the double-KO mutant was not observed in the spleens (Fig. 4B). This suggests that any defect attributable to the loss of DosR and DosS may be specific to the lungs, at least within the context of this mouse model. In terms of the animal survival study (12 mice/group) that was halted after 9 months, disrupting both dosR and dosS in the HN878 background had no statistically significant effect on the median survival time of these mice compared to the wild-type group (236 versus 269 days, respectively) (Fig. 4C).
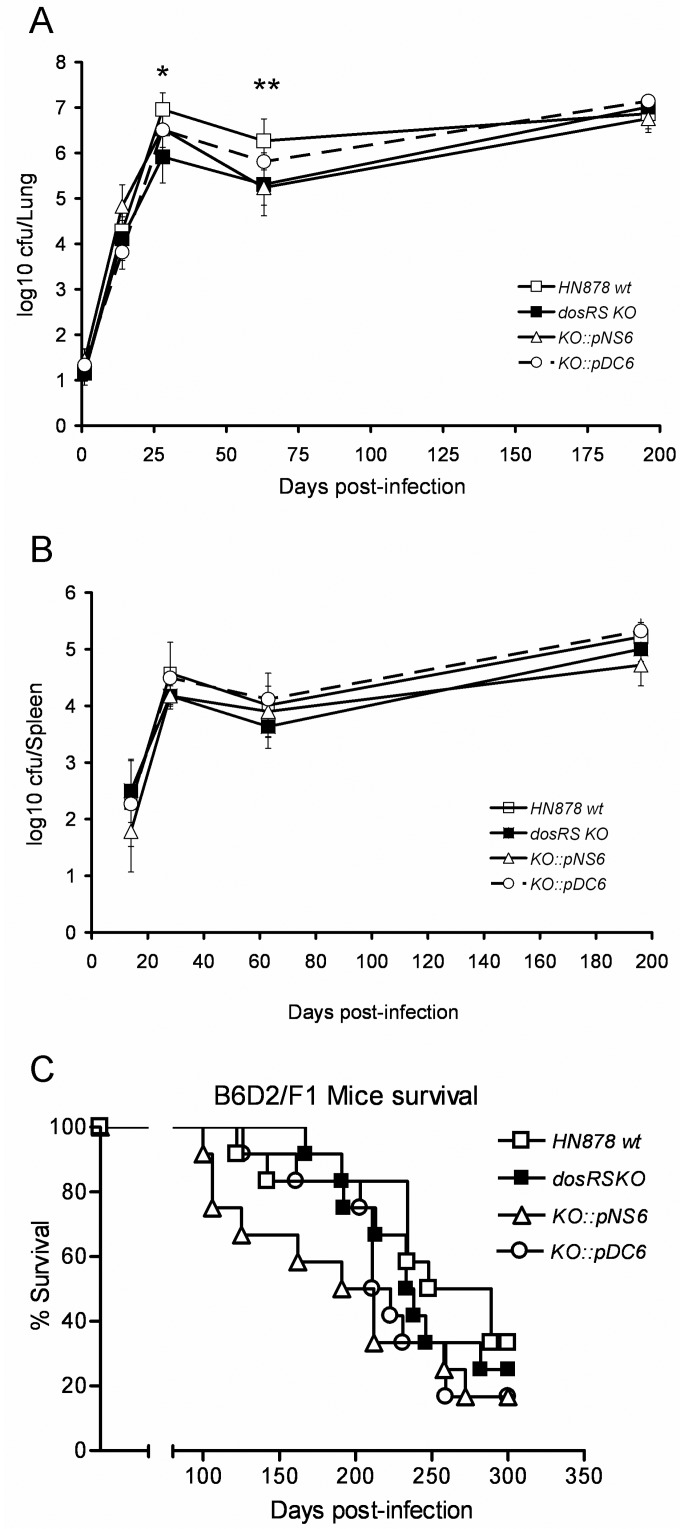
FIG 4.
The constitutive DosR regulon phenotype of the Beijing lineage does not impact virulence within the mouse model of infection. (A and B) B6D2/F1 mice were infected via aerosol with wild-type (wt) HN878, an HN878 dosRS KO mutant, HN878 dosRS KO::pNS6 (H37Rv allele), and HN878 dosRS KO::pDC6 (HN878 allele). Bacterial numbers were monitored at the indicated times postinfection by harvesting lungs (A) and spleens (B) of infected mice. Results are expressed as the average log10 CFU obtained from five mice at each time point. Error bars represent the standard deviation. *, P = 0.047 for the HN878 wt versus the HN878 dosRS KO mutant at 28 days postinfection; **, P = 0.009 for the HN878 wt versus the HN878 dosRS KO mutant at 63 days postinfection. (C) Survival curve for 12 mice per group infected with the same strains as those for panels A and B. Analysis of these data was carried out using the Kaplan-Meier method, and a log rank test was used to determine the statistical significance of observed differences in survival (GraphPad Prism, version 3.0; GraphPad Software, CA). No significant survival differences were observed between the 4 strains tested in this case.
Although the pDC6-complemented strain was more closely in line with the lung CFU of the wild-type HN878 at 4 and 9 weeks postinfection than the pNS6 complement, the most plausible interpretation of the data presented is that there is no biologically relevant difference between the two strains (Fig. 4A). The same observation can be made from the spleen CFU data (Fig. 4B). For the animal survival experiments, the median survival times of the pNS6 and pDC6 groups were 202 and 217 days, respectively (Fig. 4C). Again, there was no significant difference between the two groups. Although there was a trend early on toward the pNS6-infected animals dying more rapidly, this was not sustained.
Overall, we conclude that in the low-dose aerosol model of infection employing C57BL6/J × DBA/2J F1 mice, there is no impact on virulence for strains that constitutively overexpress dosR along with members of the DosR regulon.
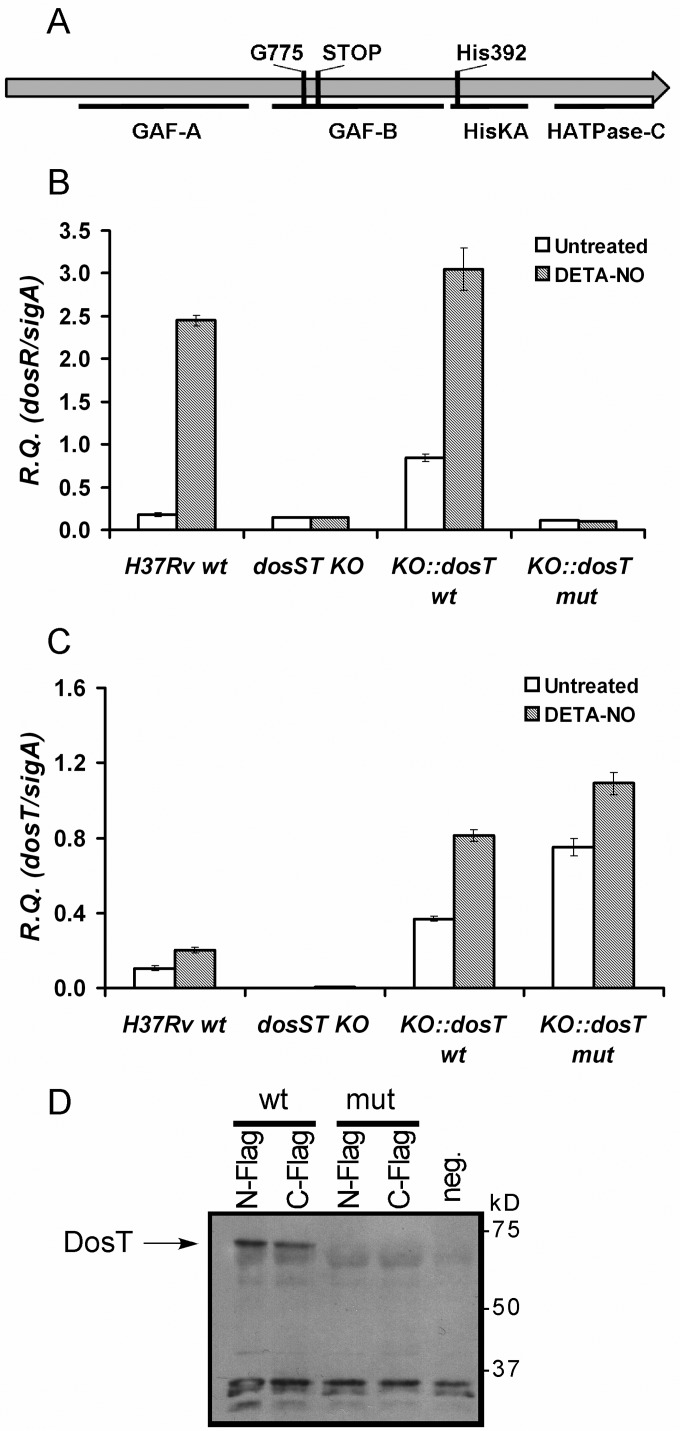
DosT sensor kinase is completely nonfunctional in the Beijing lineage.
We have previously examined the effect of the Beijing-specific dosT frameshift mutation (Fig. 5A) on dosR expression and activation through a series of gene complementation assays carried out in wild-type Beijing and non-Beijing strain backgrounds (30). These initial data suggested that the mutation in dosT was not the direct cause of the constitutive dosR overexpression seen in the Beijing lineage. Our subsequent examination of the C601T dosR promoter mutation (see above) confirms this observation. However, a clear interpretation regarding the effect of the dosT frameshift mutation was difficult in the aforementioned study due to the fact that the strains examined were diploid for dosT (wild-type and mutant copies) and also retained functional DosS. To conclusively demonstrate whether expression of the Beijing dosT gene has any effect on dosR expression and/or the ability of DosR to be activated in the presence of nitric oxide (NO), herein, we generated an H37Rv strain in which both dosS and dosT were disrupted (dosST KO mutant) prior to carrying out the complementation assays with wild-type or mutant dosT. Thus, the strains analyzed in this case expressed only the wild-type or mutant (Beijing) version of dosT, in the complete absence of the DosS sensor kinase.
FIG 5.
The DosT sensor kinase is completely nonfunctional in the M. tuberculosis Beijing lineage. (A) A schematic representation of the DosT protein showing the relative position of each of its conserved functional domains (http://www.rcsb.org/pdb/protein/P9WGK1) is shown. The approximate location of the frameshift mutation (G775) identified within dosT of Beijing strains that is predicted to result in the introduction of a premature termination codon (STOP) is indicated, as is the autophosphorylation site at residue His392 (17). (B and C) qRT-PCR analysis of dosR (B) and dosT (C) expression in the H37Rv wild type, the H37Rv dosST KO mutant, and the H37Rv dosST KO mutant complemented with the dosT gene from either H37Rv (wt) or HN878 (mut). Results are shown as relative quantities (R.Q.), using sigA as the normalizing gene. The error bars represent standard deviations. Gray bars indicate samples prepared from cultures treated with the NO donor diethylenetriamine-nitric oxide (DETA-NO) for 2 h prior to RNA extraction. (D) Western immunoblotting using anti-FLAG M2 peroxidase to probe reduced protein extracts prepared from M. smegmatis transformants harboring N- or C-terminal FLAG epitope-tagged mutant (mut; Beijing) or wild-type (wt; non-Beijing) DosT. The negative control (neg.) corresponds to protein extracts from M. smegmatis carrying wild-type dosT (i.e., with no FLAG epitope). The arrow indicates the position of the full-length DosT protein. Molecular mass markers are indicated along the right.
Expression of dosT containing the frameshift mutation had no impact on the basal level of dosR expression that was observed in the parental dosST KO mutant strain (Fig. 5B). Similarly, there was no increase in dosR expression following treatment with diethylenetriamine-nitric oxide (DETA-NO), a compound that spontaneously releases NO when in solution. In contrast, expression of wild-type dosT resulted in dosR levels that were slightly above those in wild-type H37Rv under both treated and nontreated conditions. This can be explained by the fact that dosT was being overexpressed by the hsp60 promoter in these strains compared to the normal levels seen in H37Rv (Fig. 5C). Overall, these data confirm our original finding that the Beijing dosT gene is not functionally contributing to the constitutive DosR regulon phenotype.
In our previous study, we also speculated that a truncated version of DosT may be expressed in an active form that lacks the N-terminal GAF regulatory domain (Fig. 5A) (51). To look at what forms of the DosT protein are produced as a result of the Beijing frameshift mutation, in the present study, we have expressed N- and C-terminal FLAG epitope-tagged DosT (mutant and wild type) in Mycobacterium smegmatis, followed by Western immunoblotting of total protein lysates with anti-FLAG antibodies. When wild-type DosT was expressed with a FLAG epitope located at either the N or C terminus, we readily detected the predicted 63-kDa DosT fusion protein (Fig. 5D). However, in the case of the mutant dosT, we did not observe any novel protein species distinct from those appearing in the negative control (a plasmid bearing wild-type dosT without any FLAG epitope). Therefore, it appears as if dosT bearing the Beijing frameshift mutation is either no longer expressed, or any translation product that arises is rapidly being degraded. This finding is consistent with the gene expression data described above (Fig. 5B), indicating that the mutated dosT gene is nonfunctional.
To ascertain the effect that a loss in DosT sensor kinase function has on the cell in terms of its ability to induce the DosR regulon, we took our H37Rv dosRST triple-KO mutant and transformed it with the integrative pNS6 and pDC6 plasmids containing the non-Beijing and Beijing versions of the Rv3134c-dosR-dosS operon, respectively (see Fig. 2B). We then compared the expression of dosR and Rv3130c (tgs1; a marker of DosR regulon induction) in untreated in vitro cultures versus those treated for 2 h with 150 μM DETA-NO. With respect to the expression of dosR and Rv3130c, it is clear that the loss of DosT had no detrimental impact on inducibility when the C601T SNP was present (Fig. 6). However, when the wild-type non-Beijing promoter was used (pNS6), there was an approximately 2-fold reduction in the maximal levels of dosR and Rv3130c expression that were attained following induction with NO. This is likely attributable to the fact that the “resting” levels of the two genes are 18 to 20 times higher when the Beijing SNP is present. In the presence of DETA-NO, the level of induction of Rv3130c when dosR was expressed from the C601T SNP-containing promoter was 9-fold, versus 75-fold when the SNP was absent (Fig. 6B). For dosR itself, there appeared to be no further induction upon DETA-NO addition in the pDC6-complemented strain, compared to a 9-fold induction for the pNS6-complement and a 1.3-fold induction in the case of wild-type HN878 (Fig. 6A).
FIG 6.
The dosT defect in Beijing isolates has no impact on the functionality of the DosR regulon. (A and B) qRT-PCR analysis of dosR (A) and Rv3130c (B) expression in H37Rv and HN878 wild-type strains (wt), as well as the H37Rv dosTRS triple-KO mutant complemented with either pNS6 (harboring the Rv3134c promoter and complete Rv3134c-dosR-dosS operon from H37Rv) (Fig. 2) or pDC6 (harboring the Rv3134c promoter and complete Rv3134c-dosR-dosS operon from HN878) (Fig. 2). Results are shown as relative quantities (R.Q.), using sigA as the normalizing gene. The error bars represent standard deviation as per Materials and Methods. Gray bars indicate samples prepared from cultures treated with the NO donor diethylenetriamine-nitric oxide (DETA-NO) for 2 h prior to RNA extraction.
To summarize, the frameshift in dosT renders it a nonfunctional pseudogene in the Beijing lineage. However, as a result of the associated dosR promoter mutation, this dosT defect appears to have no impact on the functionality of the DosR regulon within this same set of strains.
DISCUSSION
While we were in the process of conducting this study, Rose et al. published an article that combined M. tuberculosis genome sequence data with transcriptomics (RNA sequencing [RNA-seq]) in order to identify lineage-specific mutations that are associated with phenotypic diversity at the level of gene expression (29). In this manner, these authors identified the same C3500149T synonymous SNP in the dosR promoter region (referred to herein as the C601T SNP), which they showed correlated with a novel dosR transcript, as well as a strong overall enhancement of the DosR regulon response in the two Beijing strains they analyzed. While our data certainly confirm the predictions made by Rose et al., in the present study, we have also gone one step further and set out to prove a causal relationship between the SNP we identified through sequence analysis (30) (Fig. 2A) and the constitutive Beijing DosR phenotype via a combination of traditional molecular approaches. In this manner, we have effectively satisfied the molecular Koch's postulates (52).
Rose et al. also suggest that the C3500149T SNP generates a novel −10 TANNNT consensus motif, thereby effectively creating a new TSS within the dosR promoter region from which transcription is activated via the housekeeping SigA σ factor (29, 53–55). Although this seems a reasonable assumption, it has not been confirmed experimentally. Thus, it remains possible that a transcription factor other than SigA is involved in promoting transcription from this Beijing-specific promoter region, or if SigA is involved, it may be doing so in the presence of an additional transcriptional activator (56).
Aside from an early defect in bacterial replication in the dosRS double-KO mutant around weeks 4 to 9 that coincides with the early phases of development of the adaptive immune response, the results of our mouse aerosol infections do not point to any substantial benefit of the Beijing DosR regulon phenotype in vivo. However, as other research groups have demonstrated, the standard mouse model of infection may not be suitable for teasing apart differences between strains based solely around DosR expression (57–60). The granuloma-like cellular aggregations that form in the normal mouse lung appear not to develop areas of hypoxia; thus, any benefit to the bacteria that may arise due to continuous DosR regulon expression during the early phases of hypoxia development, for example, may not be observable within this model (61, 62). In this regard, rabbits, guinea pigs, monkeys or the Kramnik mouse model may be more informative in future efforts aimed at comparing the effect of the Beijing versus non-Beijing DosR phenotypes on pathogenesis (57, 59, 60, 62). The early delayed-growth phenotype seen with the dosRS KO mutant suggests that this strain may be at a disadvantage early on after T-cell-mediated immunity develops in the lung. This finding is interesting in light of reports that indicate that T cells specific to several DosR regulon antigens are generated during the course of natural infection, and in some cases, these antigens may be recognized more strongly in latently infected individuals (42, 43). Thus, given the apparent evolutionary benefit to M. tuberculosis that maintaining host recognition of critical T-cell antigens provides, the loss of the DosR regulon antigens in the double-KO mutant may render it more susceptible to prolonged inflammatory attack as a result of the diminished T-cell repertoire and, possibly, a delay in the resulting granulomatous response (63). This hypothesis awaits further investigation.
In our original 2007 description of constitutive DosR regulon overexpression within the Beijing lineage, we indicated a connection between the DosR phenotype and the marked triacylglyceride (TAG) accumulation we had also observed in these strains. This connection was made primarily on the basis of tgs1 (Rv3130c), a triacylglycerol synthase, being overexpressed as part of the DosR regulon (27). In addition, TAG had been reported to accumulate in H37Rv under hypoxia and NO treatment, both of which induce the DosR regulon (40, 64). However, as part of the present study, we determined that constitutive dosR expression is not the underlying cause of the TAG accumulation seen in Beijing strains. For example, when the entire dosR-dosS-dosT system was knocked out in the Beijing background, the accumulation of TAG was unaffected (Fig. S3A). Similarly, when dosR-dosS was overexpressed in H37Rv from the pDC6 plasmid bearing the Beijing dosR promoter, no increase in TAG synthesis was detectable via thin-layer chromatography (Fig. S3A). However, an HN878 Δtgs1 mutant we have generated does accumulate slightly less TAG than the parental strain, suggestive of a partial role for tgs1 in Beijing TAG synthesis that is independent of DosR (Fig. S3B). Thus, although both DosR regulon activation and TAG synthesis are associated with conditions leading to growth arrest in non-Beijing M. tuberculosis strains, these two phenotypes are seemingly uncoupled from the aforementioned signals within the Beijing lineage (i.e., they occur natively) and appear to be independent of each other. Moreover, neither phenotype has any noticeable impact on the in vitro (aerobic) growth rate of Beijing strains, nor does reconstruction of the constitutive Beijing dosR phenotype in the non-Beijing H37Rv background (data not shown). Thus, while dosR activation and TAG synthesis may well be markers of dormancy or nonreplicative persistence, for example, when M. tuberculosis is placed under hypoxic conditions, by themselves, they are not sufficient to induce these physiological states. A similar observation has been made previously by Minch et al. (65). Our data also serve to reinforce the point that there is the potential for considerable metabolic flexibility among the different circulating M. tuberculosis lineages. Thus, understanding the metabolic flux of one particular strain, including a long-term lab-adapted strain, such as H37Rv, does not guarantee that we understand the individual metabolic nuances of all strains. We and others have previously highlighted the importance of considering the impact of M. tuberculosis genetic and metabolic diversity in the generation of future treatment and diagnostic programs (2, 4, 28, 66, 67).
Given the potential energetic cost associated with the dramatic shift in the pattern of DosR regulon expression observed for the Beijing lineage shown herein (Fig. 1) and elsewhere (27–30), it seems reasonable to expect that the evolution and maintenance of this phenotype must confer a significant survival or fitness advantage to the bacteria in response to some form of host-associated selective pressure. Presumably, the promoter mutation responsible for the constitutive dosR overexpression phenotype occurred early on in the evolution of the Beijing lineage, as it is conserved across all of the Beijing sublineages in the strains we have examined to date (Fig. 2a). Furthermore, based on the whole-genome sequences of 358 unique lineage 2 M. tuberculosis strains representing 15 countries (most within the East Asia region, including Russia), Luo et al. have independently determined that the C601T SNP in Rv3134c is phylogenetically informative for defining strains that belong to the Beijing lineage (12). Although the published estimates of exactly when the most recent Beijing sublineages evolved vary widely (from 8,000 to 200 years ago), in any case, they appear to have origins that are well before the modern era of antibiotics, Mycobacterium bovis BCG vaccination, or HIV (10, 12). Thus, the original selection event(s) that led to the Beijing promoter mutation is not related to any of these recently applied evolutionary forces. However, this certainly does not preclude the possibility that the Beijing DosR phenotype provides an added fitness benefit to these strains in the modern context of anti-TB drugs or the BCG vaccine. Therefore, it is theoretically possible that the constitutive overexpression of DosR has contributed to the recent global expansion of the Beijing lineage from its ancestral home in East Asia to eastern Europe and other parts of the globe (7, 8, 10, 12). Indeed, the recent associations between the Beijing lineage and drug resistance are most often recorded for countries in which the lineage has recently emerged (8, 11). The potential role of DosR in this process is intriguing in light of two recent reports: first, Ford et al. have demonstrated an up to 10-fold increase in the in vitro mutation rate of Beijing compared to non-Beijing (lineage 4) strains in response to multiple front-line anti-TB drugs (13). Second, Liu et al. have very recently shown that a large component of the transcriptional response of intracellular M. tuberculosis toward drug treatment is mediated via dosR (44). Liu et al. also showed that there is considerable overlap between the intracellular response of M. tuberculosis to anti-TB drugs and the stress conditions encountered as a result of immune activation. Thus, within the intracellular milieu, both antibiotic treatment and immune activation converge at the level of DosR regulon activation. In the same study, immune activation of M. tuberculosis-infected cells was tied to the development of antibiotic tolerance (44). Hence, it is of great interest to investigate the possibility that constitutive DosR regulon expression in the Beijing background contributes to either drug tolerance or resistance. This is also interesting in light of the precedent in the literature that implicates the WhiB3 and WhiB7 transcription factors in modifying M. tuberculosis drug susceptibility. Like DosR, both of these transcription factors are known to regulate the cellular response to redox stress (56, 68).
As with the dosR promoter mutation, the frameshift mutation that renders dosT nonfunctional is present across all sublineages (30). Although the relative timing of the two events is still unknown, we feel that the evolution of the dosT mutation most likely represents a compensatory event that occurred secondary to the initial dosR promoter mutation, and it reflects the fact that maintaining a functional DosT kinase in this context serves no functional purpose. This is entirely consistent with previous suggestions that DosT acts as the “first responder” in terms of oxygen or NO sensing but plays no further role once DosS is induced by virtue of being a member of the DosR regulon and also part of the Rv3134c-dosR-dosS operon (69–72). In the context of the Beijing DosR phenotype, the early activatory role normally attributed to DosT appears to have been supplanted through the constitutive overexpression of dosR.
In summary, we have comprehensively demonstrated that the C601T dosR promoter mutation is the sole factor underlying the constitutive dosR overexpression phenotype unique to the Beijing M. tuberculosis lineage. In turn, this correlates with a dramatic shift in the overall pattern of DosR regulon gene expression within these strains that, bearing in mind the high energetic cost likely associated with maintaining this phenotype, almost certainly affords some form of survival or fitness advantage within the environment of the host. Other host-adaptive regulatory mutations described within the wider M. tuberculosis complex, such as those in the PhoPR and SigK systems, have been associated with broad evolutionary shifts linked to specific host species (73, 74). It will be intriguing in the future to determine if the lineage-specific DosR trait detailed herein also represents a bacterial adaptation to a specific host population or host-associated selective pressure. In addition, given the pleiotropic nature of the proteins encoded within the DosR regulon, it is undoubtedly important that we understand the impact of this phenotype, along with other examples of strain variation, on the long-term success of TB diagnosis, vaccination, and treatment programs.
MATERIALS AND METHODS
Chemicals, bacterial strains, and culture conditions.
All chemicals were supplied by Sigma-Aldrich, Inc., unless otherwise noted. Escherichia coli NEB 5-alpha (NEB) used for cloning was cultured in Luria-Bertani broth or agar (Difco). When necessary, ampicillin (100 μg · ml−1), kanamycin (50 μg · ml−1), hygromycin (200 μg · ml−1; Wisent), or gentamicin (5 μg · ml−1; Invitrogen) was added to the medium. M. smegmatis mc2155 and M. tuberculosis strains were grown in Middlebrook 7H9 broth (Difco) supplemented with 10% albumin-dextrose-catalase (ADC; 8.1 g · liter−1 NaCl, 50 g · liter−1 bovine serum albumin [BSA] Fraction V [Calbiochem], 20 g · liter−1 glucose), 0.2% glycerol, and 0.05% Tween 80 or on Middlebrook 7H11 agar (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC) enrichment (as per ADC, plus 0.6 ml · l−1 oleic acid, 3.6 mM NaOH). Kanamycin (25 μg · ml−1), hygromycin (50 μg · ml−1), or gentamicin (10 μg · ml−1) was used as necessary. Treatment of M. tuberculosis with NO was accomplished by adding the NO donor diethylenetriamine-nitric oxide (DETA-NO) to early log-phase cultures (optical density at 600 nm [OD600], 0.2 to 0.3) at a final concentration of 150 μM and incubating for 2 h at 37°C with constant mixing prior to RNA extraction. Control cultures were grown under identical conditions in the absence of DETA-NO.
M. tuberculosis H37Rv was originally obtained from the American Type Culture Collection (ATCC 27294). The majority of clinical M. tuberculosis isolates used in this study were originally reported as part of an earlier molecular epidemiology investigation (48) in which isolates were classified according to the presence of large-sequence polymorphisms (LSPs) (47). Isolate 98_1663 was kindly provided by S. Gagneux (Swiss Tropical and Public Health Institute, Basel, Switzerland) and P. Small (Stony Brook University, Stony Brook, NY). Strain HN878 was originally obtained from J. Musser (Methodist Hospital Research Institute, Houston, TX) (75). The HN878-27 subclone derived in our laboratory (14) lacks the 350-kb genomic duplication generated in vitro by HN878 and certain other Beijing isolates, and it is referred to here as HN878 (32).
General nucleic acid techniques.
Taq polymerase and dinucleoside triphosphates (dNTPs) were obtained from Thermo Scientific. PCR was carried out according to standard protocols (76), except, where necessary, 5 or 10% dimethyl sulfoxide (DMSO) was also included in the reaction mixtures. The primers used in this study are shown in Table S1 in the supplemental material. M. tuberculosis knockout (KO) strains were generated by homologous recombination using the system developed by Pelicic et al. (77). The plasmids used in this study are listed in Table 1. Specific details of how these plasmids were constructed are described in the supplemental material. The gene arrangement of each KO strain around the site of homologous recombination was confirmed by PCR, and their gene expression phenotypes were confirmed by qRT-PCR. The absence of the 350-kb duplication that can occur in strains belonging to groups 3 to 5 of the East Asian lineage was confirmed by PCR, as previously described (14, 32).
TABLE 1.
Plasmids used in this study
| Name | Insert |
|---|---|
| pNS3 | Truncated Rv3134c-dosR-dosS from H37Rv in pMV306-Kan; 507C plus 601C allele |
| pNS5 | Rv3134c promoter-Rv3134c-dosR from H37Rv in pMV306-Kan; 507C plus 601C allele |
| pNS6 | Rv3134c promoter-Rv3134c-dosR-dosS from H37Rv in pMV306-Kan; 507C plus 601C allele |
| pDC4 | Truncated Rv3134c-dosR-dosS from HN878 in pMV306-Kan; 507G plus 601T allele |
| pDC5 | Rv3134c promoter-Rv3134c-dosR from HN878 in pMV306-Kan; 507G plus 601T allele |
| pDC6 | Rv3134c promoter-Rv3134c-dosR-dosS from HN878 in pMV306-Kan; 507G plus 601T allele |
| pAA3 | Rv3134c promoter-Rv3134c-dosR-dosS from HN878 in pMV306-Kan; 507C plus 601T hybrid allele |
| pAA6 | Rv3134c promoter-Rv3134c-dosR-dosS from HN878 in pMV306-Kan; 507G plus 601T, DosR (D54E) |
| pAR2 | Rv3134c promoter-Rv3134c-dosS from HN878 in pMV306-Kan; 507G plus 601T allele (no dosR) |
| pdosT-wt | dosT from H37Rv in pMV361-Gent |
| pdosT-mut | dosT from HN878 (G775 frameshift mutation) in pMV361-Gent |
| pN-FLAGdosT-wt | 5′ FLAG-wild-type dosT in pMV361-Kan |
| pC-FLAGdosT-wt | Wild-type dosT-3′ FLAG in pMV361-Kan |
| pN-FLAGdosT-mut | 5′ FLAG-mutant dosT (G775 frameshift mutation) in pMV361-Kan |
| pC-FLAGdosT-mut | Mutant dosT (G775 frameshift mutation)-3′ FLAG in pMV361-Kan |
| pPR23ΔdosR+S | dosR-kan-dosS in pPR23; used for generating M. tuberculosis dosR-dosS KO mutants |
| pPR23ΔdosT/Hyg | dosT-hyg-dosT in pPR23; used for generating M. tuberculosis dosT KO mutants |
| pΔdosSv2.0 | dosS-kan-dosS in pPR23; used for generating M. tuberculosis dosS KO mutants |
| pPDM15 | dosR-hyg-dosR in pPR23; used for generating M. tuberculosis dosR KO mutants |
| pPDM4 | tgs1-hyg-tgs1 in pPR23; used for generating M. tuberculosis tgs1 KO mutants |
Sequence analysis of the dosR promoter region.
A 647-bp fragment was generated by PCR using primers Rv3134c-M-F and dosR-B. This region corresponds to nucleotides 3499869 to 3500516 of the genome, numbered according to the H37Rv sequence provided in TubercuList (http://tuberculist.epfl.ch/) (78). This sequence harbors 574 bp at the 3′ end of Rv3134c and 46 bp at the 5′ end of dosR. Sanger sequencing of the reaction products was carried out at the McGill University and Génome Québec Innovation Centre.
Microarray analysis.
Whole-genome transcriptome profiling was performed by microarray analysis of RNA, according to protocols previously described (79). In brief, 4 μg of RNA was used in the preparation of cDNA labeled with either Cy3 or Cy5-NHS esters (GE Healthcare) via the aminoallyl indirect labeling method (80) in the presence of SuperScript III reverse transcriptase (Invitrogen). The M. tuberculosis whole-genome microarrays used in these experiments were kindly provided by F. McIntosh and M. Behr (McGill University) and are composed of 70-bp oligonucleotides (TB Array-Ready oligo set; Operon) printed in duplicate. The mean fluorescence values for each spot, minus the surrounding background, were used in the analysis. Spots flagged as misrepresentative or those with a negative value following background correction were excluded from further analysis. The fluorescence intensity ratios (Cy3/Cy5 or Cy5/Cy3) were calculated for each spot, log10 transformed, and then normalized with respect to the mean value of all fluorescence ratios. A Z-score [Z = (log10 fluorescence ratio − mean)/standard deviation] was calculated for each spot, and the Z-scores for each gene were averaged across all replicates. Two independent biological samples were analyzed for each M. tuberculosis strain (a total of 3 arrays for each comparison).
qRT-PCR.
RNA was purified from cultures with an OD600of 0.2 to 0.3, as previously described (18). Primer pairs DosR-RT-6-f and DosR-RT-6-r, dosTSYBR-F2 and dosTSYBR-R2, Rv3130c-F and Rv3130c-R, and DosS-F2 and DosS-R2 (Table S1) were used to quantify the expression of dosR, dosT, Rv3130c, and dosS, respectively, while primers SigA1-F and SigA1-R were used to quantify sigA for normalization purposes. The procedures used for first-strand cDNA synthesis and qRT-PCR were as described by Domenech and Reed (79). The relative standard curve method was used for quantification. Standard deviations (SD) were calculated according to the following formula as specified by Applied Biosystems (81): SD = (cv)(X), where X is the ratio of the mean values of the nominator (n) and denominator (d). cv is the coefficient of variation (standard deviation normalized to the mean values for each gene) calculated from the following formula: . Three biological replicates were each analyzed in triplicate.
Mouse infections.
Prior to infection, well-dispersed liquid cultures were adjusted to an OD600 of 0.4 and stored at −70°C as 10% (vol/vol) glycerol stocks. Inocula were prepared by diluting these stocks to 4 × 106 CFU/ml in phosphate-buffered saline (PBS)–Tween 80 (0.05%). Eight-week-old B6D2/F1 mice (The Jackson Laboratory) were infected using a Lovelace nebulizer (In-Tox) for 10 min. Bacteria were enumerated at 1, 14, 28, 63, and 196 days postinfection (5 mice per time point) by homogenizing the lungs and spleens of infected mice in 1 ml of 7H9 medium and plating 10-fold serial dilutions on 7H11-OADC medium containing PANTA antibiotic mixture (BD; 12 U/ml polymyxin B, 1.2 μg/ml amphotericin B, trimethoprim, and azlocillin, and 4.8 μg/ml nalidixic acid) to avoid contamination of the plates. Twelve mice were left in each group for the animal survival experiments. Survival proportions were calculated using the Kaplan-Meier method (82), and the Mann-Whitney test was used to determine the statistical significance of the observed survival differences (GraphPad Prism version 3.0; GraphPad Software, CA). All animal experiments were approved and carried out in accordance with the guidelines and regulations of the Animal Care Committee of McGill University.
Protein techniques.
Crude protein extracts from M. smegmatis mc2155 transformants were prepared by bead-beating, according to the procedure described by Parish and Wheeler (83). Samples were added to an equal volume of 2× reducing sample buffer (containing 8 M urea), boiled, and then electrophoresed in 10% acrylamide–bis-acrylamide (29:1) (Bio-Rad) SDS-PAGE, according to standard procedures (76). The monoclonal anti-FLAG M2–horseradish peroxidase (HRP) clone M2 (Sigma) and the Amersham ECL detection system (GE Healthcare) were used for Western blotting, according to the manufacturer's instructions.
Lipid analysis.
Apolar lipids were extracted as previously described (84). The presence of phthiocerol dimycocerosate (PDIM) in all M. tuberculosis strains used in this study was confirmed by radiolabeling cultures with 0.1 μCi/ml of [1-14C]propionic acid (1 mCi/37 MBq; American Radiolabeled Chemicals), and extracted lipids were analyzed by thin-layer chromatography (TLC) (silica gel 60 plates; EM Science) run three times in hexanes/ethyl acetate (98:2 [vol/vol]) (79). Triacylglyceride (TAG) content was analyzed by radiolabeling with 1 μCi/ml of [14C(U)]glycerol (1 mCi/37 MBq; American Radiolabeled Chemicals) and analyzed as described above (27). TLC plates were visualized using a Storm 840 PhosphorImager (Molecular Dynamics).
Accession number(s).
The data associated with this paper have been deposited in NCBI's Gene Expression Omnibus and are accessible with GEO series accession no. GSE83677.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anya Rog for her technical assistance throughout this project. We are also grateful to Marcel Behr and Fiona McIntosh (Research Institute of the McGill University Health Centre) for providing access to several of the M. tuberculosis strains and also the microarrays used in this study.
We declare no conflicts of interest.
This work was supported by Canadian Institutes of Health Research (CIHR) grant MOP115133 to M.B.R.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00696-16.
REFERENCES
- 1.Harries AD, Dye C. 2006. Tuberculosis. Ann Trop Med Parasitol 100:415–431. [DOI] [PubMed] [Google Scholar]
- 2.Köser CU, Feuerriegel S, Summers DK, Archer JA, Niemann S. 2012. Importance of the genetic diversity within the Mycobacterium tuberculosis complex for the development of novel antibiotics and diagnostic tests of drug resistance. Antimicrob Agents Chemother 56:6080–6087. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, Gagneux S. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol 6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coscolla M, Gagneux S. 2014. Consequences of genomic diversity in Mycobacterium tuberculosis. Semin Immunol 26:431–444. doi: 10.1016/j.smim.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comas I, Homolka S, Niemann S, Gagneux S. 2009. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, Yeboah-Manu D, Bothamley G, Mei J, Wei L, Bentley S, Harris SR, Niemann S, Diel R, Aseffa A, Gao Q, Young D, Gagneux S. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowley D, Govender D, February B, Wolfe M, Steyn L, Evans J, Wilkinson RJ, Nicol MP. 2008. Recent and rapid emergence of W-Beijing strains of Mycobacterium tuberculosis in Cape Town, South Africa. Clin Infect Dis 47:1252–1259. doi: 10.1086/592575. [DOI] [PubMed] [Google Scholar]
- 8.European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis. 2006. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg Infect Dis 12:736–743. doi: 10.3201/eid1205.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanekom M, van der Spuy GD, Streicher E, Ndabambi SL, McEvoy CR, Kidd M, Beyers N, Victor TC, van Helden PD, Warren RM. 2007. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J Clin Microbiol 45:1483–1490. doi: 10.1128/JCM.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, Willery E, Blum MG, Rusch-Gerdes S, Mokrousov I, Aleksic E, Allix-Beguec C, Antierens A, Augustynowicz-Kopec E, Ballif M, Barletta F, Beck HP, Barry CE III, Bonnet M, Borroni E, Campos-Herrero I, Cirillo D, Cox H, Crowe S, Crudu V, Diel R, Drobniewski F, Fauville-Dufaux M, Gagneux S, Ghebremichael S, Hanekom M, Hoffner S, Jiao WW, Kalon S, Kohl TA, Kontsevaya I, Lillebaek T, Maeda S, Nikolayevskyy V, Rasmussen M, Rastogi N, Samper S, Sanchez-Padilla E, Savic B, Shamputa IC, Shen A, Sng LH, Stakenas P, Toit K, Varaine F, Vukovic D, et al. . 2015. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet 47:242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parwati I, van Crevel R, van Soolingen D. 2010. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis 10:103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- 12.Luo T, Comas I, Luo D, Lu B, Wu J, Wei L, Yang C, Liu Q, Gan M, Sun G, Shen X, Liu F, Gagneux S, Mei J, Lan R, Wan K, Gao Q. 2015. Southern East Asian origin and coexpansion of Mycobacterium tuberculosis Beijing family with Han Chinese. Proc Natl Acad Sci U S A 112:8136–8141. doi: 10.1073/pnas.1424063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. 2013. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet 45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domenech P, Kolly GS, Leon-Solis L, Fallow A, Reed MB. 2010. Massive gene duplication event among clinical isolates of the Mycobacterium tuberculosis W/Beijing family. J Bacteriol 192:4562–4570. doi: 10.1128/JB.00536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ. 2016. Within-host evolution of bacterial pathogens. Nat Rev Microbiol 14:150–162. doi: 10.1038/nrmicro.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol 48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini DK, Malhotra V, Tyagi JS. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett 565:75–80. doi: 10.1016/j.febslet.2004.02.092. [DOI] [PubMed] [Google Scholar]
- 18.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc Natl Acad Sci U S A 98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med 198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saini DK, Malhotra V, Dey D, Pant N, Das TK, Tyagi JS. 2004. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150:865–875. doi: 10.1099/mic.0.26218-0. [DOI] [PubMed] [Google Scholar]
- 21.Boon C, Dick T. 2002. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J Bacteriol 184:6760–6767. doi: 10.1128/JB.184.24.6760-6767.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wayne LG, Sohaskey CD. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev Microbiol 55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 23.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med 198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi L, Jung YJ, Tyagi S, Gennaro ML, North RJ. 2003. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc Natl Acad Sci U S A 100:241–246. doi: 10.1073/pnas.0136863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, Gomes A, Rustad T, Dolganov G, Glotova I, Abeel T, Mahwinney C, Kennedy AD, Allard R, Brabant W, Krueger A, Jaini S, Honda B, Yu WH, Hickey MJ, Zucker J, Garay C, Weiner B, Sisk P, Stolte C, Winkler JK, Van de Peer Y, Iazzetti P, Camacho D, Dreyfuss J, Liu Y, Dorhoi A, Mollenkopf HJ, Drogaris P, Lamontagne J, Zhou Y, Piquenot J, Park ST, Raman S, Kaufmann SH, Mohney RP, Chelsky D, Moody DB, Sherman DR, Schoolnik GK. 2013. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalo Asensio J, Maia C, Ferrer NL, Barilone N, Laval F, Soto CY, Winter N, Daffe M, Gicquel B, Martin C, Jackson M. 2006. The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J Biol Chem 281:1313–1316. doi: 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- 27.Reed MB, Gagneux S, Deriemer K, Small PM, Barry CE III. 2007. The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J Bacteriol 189:2583–2589. doi: 10.1128/JB.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Homolka S, Niemann S, Russell DG, Rohde KH. 2010. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog 6:e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose G, Cortes T, Comas I, Coscolla M, Gagneux S, Young DB. 2013. Mapping of genotype-phenotype diversity among clinical isolates of Mycobacterium tuberculosis by sequence-based transcriptional profiling. Genome Biol Evol 5:1849–1862. doi: 10.1093/gbe/evt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fallow A, Domenech P, Reed MB. 2010. Strains of the East Asian (W/Beijing) lineage of Mycobacterium tuberculosis are DosS/DosT-DosR two-component regulatory system natural mutants. J Bacteriol 192:2228–2238. doi: 10.1128/JB.01597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiner B, Gomez J, Victor TC, Warren RM, Sloutsky A, Plikaytis BB, Posey JE, van Helden PD, Gey van Pittius NC, Koehrsen M, Sisk P, Stolte C, White J, Gagneux S, Birren B, Hung D, Murray M, Galagan J. 2012. Independent large scale duplications in multiple M. tuberculosis lineages overlapping the same genomic region. PLoS One 7:e26038. doi: 10.1371/journal.pone.0026038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domenech P, Rog A, Moolji JU, Radomski N, Fallow A, Leon-Solis L, Bowes J, Behr MA, Reed MB. 2014. Origins of a 350-kilobase genomic duplication in Mycobacterium tuberculosis and its impact on virulence. Infect Immun 82:2902–2912. doi: 10.1128/IAI.01791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manca C, Tsenova L, Barry CE III, Bergtold A, Freeman S, Haslett PA, Musser JM, Freedman VH, Kaplan G. 1999. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J Immunol 162:6740–6746. [PubMed] [Google Scholar]
- 34.Gagneux S, Burgos MV, DeRiemer K, Encisco A, Munoz S, Hopewell PC, Small PM, Pym AS. 2006. Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS Pathog 2:e61. doi: 10.1371/journal.ppat.0020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsolaki AG, Gagneux S, Pym AS, Goguet de la Salmoniere YO, Kreiswirth BN, Van Soolingen D, Small PM. 2005. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J Clin Microbiol 43:3185–3191. doi: 10.1128/JCM.43.7.3185-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guérin attenuation. J Infect Dis 187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, Eisenberg D, Russell RG, Derrick SC, Collins FM, Morris SL, King CH, Jacobs WR Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guérin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A 100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol 51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bitter W, Houben EN, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao LY, Liu J, Gey van Pittius NC, Pym AS, Rubin EJ, Sherman DR, Cole ST, Brosch R. 2009. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog 5:e1000507. doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sirakova TD, Dubey VS, Deb C, Daniel J, Korotkova TA, Abomoelak B, Kolattukudy PE. 2006. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology 152:2717–2725. doi: 10.1099/mic.0.28993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan Y, Crane DD, Barry CE III. 1996. Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial alpha-crystallin homolog. J Bacteriol 178:4484–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peña D, Rovetta AI, Hernandez Del Pino RE, Amiano NO, Pasquinelli V, Pellegrini JM, Tateosian NL, Rolandelli A, Gutierrez M, Musella RM, Palmero DJ, Gherardi MM, Iovanna J, Chuluyan HE, Garcia VE. 2015. A Mycobacterium tuberculosis dormancy antigen differentiates latently infected bacillus Calmette-Guérin-vaccinated individuals. EBioMedicine 2:882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leyten EM, Lin MY, Franken KL, Friggen AH, Prins C, van Meijgaarden KE, Voskuil MI, Weldingh K, Andersen P, Schoolnik GK, Arend SM, Ottenhoff TH, Klein MR. 2006. Human T-cell responses to 25 novel antigens encoded by genes of the dormancy regulon of Mycobacterium tuberculosis. Microbes Infect 8:2052–2060. doi: 10.1016/j.micinf.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Tan S, Huang L, Abramovitch RB, Rohde KH, Zimmerman MD, Chen C, Dartois V, VanderVen BC, Russell DG. 2016. Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo. J Exp Med 213:809–825. doi: 10.1084/jem.20151248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bagchi G, Chauhan S, Sharma D, Tyagi JS. 2005. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology 151:4045–4053. doi: 10.1099/mic.0.28333-0. [DOI] [PubMed] [Google Scholar]
- 46.Chauhan S, Tyagi JS. 2008. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J Bacteriol 190:4301–4312. doi: 10.1128/JB.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, Hilty M, Hopewell PC, Small PM. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed MB, Pichler VK, McIntosh F, Mattia A, Fallow A, Masala S, Domenech P, Zwerling A, Thibert L, Menzies D, Schwartzman K, Behr MA. 2009. Major Mycobacterium tuberculosis lineages associate with patient country of origin. J Clin Microbiol 47:1119–1128. doi: 10.1128/JCM.02142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE III, Freedman VH, Kaplan G. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc Natl Acad Sci U S A 98:5752–5757. doi: 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE III. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 51.Sardiwal S, Kendall SL, Movahedzadeh F, Rison SC, Stoker NG, Djordjevic S. 2005. A GAF domain in the hypoxia/NO-inducible Mycobacterium tuberculosis DosS protein binds haem. J Mol Biol 353:929–936. doi: 10.1016/j.jmb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 52.Falkow S. 1988. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis 10(Suppl 2):S274–276. [DOI] [PubMed] [Google Scholar]
- 53.Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, Aebersold R, Young DB. 2013. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep 5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newton-Foot M, Gey van Pittius NC. 2013. The complex architecture of mycobacterial promoters. Tuberculosis (Edinb) 93:60–74. doi: 10.1016/j.tube.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 55.Hu Y, Coates AR. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol 181:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burian J, Ramon-Garcia S, Sweet G, Gomez-Velasco A, Av-Gay Y, Thompson CJ. 2012. The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J Biol Chem 287:299–310. doi: 10.1074/jbc.M111.302588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Converse PJ, Karakousis PC, Klinkenberg LG, Kesavan AK, Ly LH, Allen SS, Grosset JH, Jain SK, Lamichhane G, Manabe YC, McMurray DN, Nuermberger EL, Bishai WR. 2009. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect Immun 77:1230–1237. doi: 10.1128/IAI.01117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, Bishai WR. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med 200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gautam US, McGillivray A, Mehra S, Didier PJ, Midkiff CC, Kissee RS, Golden NA, Alvarez X, Niu T, Rengarajan J, Sherman DR, Kaushal D. 2015. DosS is required for the complete virulence of Mycobacterium tuberculosis in mice with classical granulomatous lesions. Am J Respir Cell Mol Biol 52:708–716. doi: 10.1165/rcmb.2014-0230OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehra S, Foreman TW, Didier PJ, Ahsan MH, Hudock TA, Kissee R, Golden NA, Gautam US, Johnson AM, Alvarez X, Russell-Lodrigue KE, Doyle LA, Roy CJ, Niu T, Blanchard JL, Khader SA, Lackner AA, Sherman DR, Kaushal D. 2015. The DosR regulon modulates adaptive immunity and is essential for Mycobacterium tuberculosis persistence. Am J Respir Crit Care Med 191:1185–1196. doi: 10.1164/rccm.201408-1502OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, Lee EG, Park SK, Raleigh JA, Cho SN, McMurray DN, Flynn JL, Barry CE III. 2008. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun 76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gautam US, Mehra S, Kaushal D. 2015. In-vivo gene signatures of Mycobacterium tuberculosis in C3HeB/FeJ mice. PLoS One 10:e0135208. doi: 10.1371/journal.pone.0135208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S. 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE. 2004. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol 186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Minch K, Rustad T, Sherman DR. 2012. Mycobacterium tuberculosis growth following aerobic expression of the DosR regulon. PLoS One 7:e35935. doi: 10.1371/journal.pone.0035935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohen T, Colijn C, Murray M. 2008. Modeling the effects of strain diversity and mechanisms of strain competition on the potential performance of new tuberculosis vaccines. Proc Natl Acad Sci U S A 105:16302–16307. doi: 10.1073/pnas.0808746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gagneux S, Small PM. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis 7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 68.Saini V, Cumming BM, Guidry L, Lamprecht DA, Adamson JH, Reddy VP, Chinta KC, Mazorodze JH, Glasgow JN, Richard-Greenblatt M, Gomez-Velasco A, Bach H, Av-Gay Y, Eoh H, Rhee K, Steyn AJ. 2016. Ergothioneine maintains redox and bioenergetic homeostasis essential for drug susceptibility and virulence of Mycobacterium tuberculosis. Cell Rep 14:572–585. doi: 10.1016/j.celrep.2015.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Honaker RW, Leistikow RL, Bartek IL, Voskuil MI. 2009. Unique roles of DosT and DosS in DosR regulon induction and Mycobacterium tuberculosis dormancy. Infect Immun 77:3258–3263. doi: 10.1128/IAI.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim MJ, Park KJ, Ko IJ, Kim YM, Oh JI. 2010. Different roles of DosS and DosT in the hypoxic adaptation of mycobacteria. J Bacteriol 192:4868–4875. doi: 10.1128/JB.00550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sousa EHS, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. 2007. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci 16:1708–1719. doi: 10.1110/ps.072897707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sivaramakrishnan S, Ortiz de Montellano PR. 2013. The DosS-DosT/DosR mycobacterial sensor system. Biosensors 3:259–282. doi: 10.3390/bios3030259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Broset E, Martin C, Gonzalo-Asensio J. 2015. Evolutionary landscape of the Mycobacterium tuberculosis complex from the viewpoint of PhoPR: implications for virulence regulation and application to vaccine development. mBio 6(5):e01289-15. doi: 10.1128/mBio.01289-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Veyrier F, Said-Salim B, Behr MA. 2008. Evolution of the mycobacterial SigK regulon. J Bacteriol 190:1891–1899. doi: 10.1128/JB.01452-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, Whittam TS, Musser JM. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A 94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 77.Pelicic V, Jackson M, Reyrat JM, Jacobs WR Jr, Gicquel B, Guilhot C. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lew JM, Kapopoulou A, Jones LM, Cole ST. 2011. TubercuList–10 years after. Tuberculosis (Edinb) 91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 79.Domenech P, Reed MB. 2009. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: implications for virulence studies. Microbiology 155:3532–3543. doi: 10.1099/mic.0.029199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu J, Othman MI, Farjo R, Zareparsi S, MacNee SP, Yoshida S, Swaroop A. 2002. Evaluation and optimization of procedures for target labeling and hybridization of cDNA microarrays. Mol Vis 8:130–137. [PubMed] [Google Scholar]
- 81.Applied Biosystems. 2005. Real-time PCR systems: Applied Biosystems 7900HT fast real-time PCR system and 7300/7500 real-time PCR systems. Chemistry guide. Part no. 4348358, re E. Applied Biosystems, Foster City, CA: http://www3.appliedbiosystems.com/cms/groups/mcb_marketing/documents/generaldocuments/cms_041440.pdf. [Google Scholar]
- 82.Kaplan EL, Meier P. 1958. Nonparametric estimation for imcomplete observations. J Am Stat Assoc 53:457–481. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- 83.Parish T, Wheeler PR. 1998. Preparation of cell-free extracts from mycobacteria. Methods Mol Biol 101:77–89. [DOI] [PubMed] [Google Scholar]
- 84.Slayden RA, Barry CE III. 2001. Analysis of the lipids of Mycobacterium tuberculosis. Methods Mol Med 54:229–245. doi: 10.1385/1-59259-147-7:229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.