ABSTRACT
RNase Y is a major endoribonuclease that plays a crucial role in mRNA degradation and processing. We study the role of RNase Y in the Gram-positive anaerobic pathogen Clostridium perfringens, which until now has not been well understood. Our study implies an important role for RNase Y-mediated RNA degradation and processing in virulence gene expression and the physiological development of the organism. We began by constructing an RNase Y conditional knockdown strain in order to observe the importance of RNase Y on growth and virulence. Our resulting transcriptome analysis shows that RNase Y affects the expression of many genes, including toxin-producing genes. We provide data to show that RNase Y depletion repressed several toxin genes in C. perfringens and involved the virR-virS two-component system. We also observe evidence that RNase Y is indispensable for processing and stabilizing the transcripts of colA (encoding a major toxin collagenase) and pilA2 (encoding a major pilin component of the type IV pili). Posttranscriptional regulation of colA is known to be mediated by cleavage in the 5′ untranslated region (5′UTR), and we observe that RNase Y depletion diminishes colA 5′UTR processing. We show that RNase Y is also involved in the posttranscriptional stabilization of pilA2 mRNA, which is thought to be important for host cell adherence and biofilm formation.
IMPORTANCE RNases have important roles in RNA degradation and turnover in all organisms. C. perfringens is a Gram-positive anaerobic spore-forming bacterial pathogen that produces numerous extracellular enzymes and toxins, and it is linked to digestive disorders and disease. A highly conserved endoribonuclease, RNase Y, affects the expression of hundreds of genes, including toxin genes, and studying these effects is useful for understanding C. perfringens specifically and RNases generally. Moreover, RNase Y is involved in processing specific transcripts, and we observed that this processing in C. perfringens results in the stabilization of mRNAs encoding a toxin and bacterial extracellular apparatus pili. Our study shows that RNase activity is associated with gene expression, helping to determine the growth, proliferation, and virulence of C. perfringens.
KEYWORDS: Clostridium perfringens, RNase Y, posttranscriptional regulation, toxin regulation, type IV pili, ribonuclease, toxin gene
INTRODUCTION
RNA degradation and turnover are essential processes in all organisms, and bacterial mRNAs are rapidly degraded to facilitate quick adaptation to changing environments (1). In addition to transcriptional control, posttranscriptional control is also important for gene regulation. RNA regulation without translation requires smaller amounts of energy and permits more rapid control of expression than regulation exerted through protein factors (2). Ribonucleases (RNases) are critical factors that determine transcript instability and function in posttranscriptional gene regulation. RNases are broadly divided into endo- or exoribonuclease classes. The former class of enzymes plays an important role in initiating degradation, while the latter mediates further transcript degradation (1).
RNases have been well studied in Bacillus subtilis, and roles for RNases in the principle mRNA decay pathway of this bacterium have been predicted (3). RNase Y is the major endoribonuclease that cleaves transcripts; the cleaved fragments are then degraded by the 5′ to 3′ exoribonuclease activity of RNase J1 and/or the 3′ exoribonuclease activity of polynucleotide phosphorylase (PNPase), RNase PH, RNase R, and YhaM (4–6). RNase Y affects mRNA turnover and normal proliferation in B. subtilis (7). The important role of RNase Y in bacterial pathogenesis is highlighted by the fact that RNase Y is conserved in the majority of Firmicutes and also affects mRNA degradation and gene expression in the Gram-positive pathogens Staphylococcus aureus and Streptococcus pyogenes (8–10).
C. perfringens is a Gram-positive spore-forming anaerobic bacterium that produces numerous extracellular enzymes and toxins (11). This organism is a causative agent of gas gangrene, food poisoning, and antibiotic-associated diarrhea (AAD). Rapid gene activation and repression are important for regulating virulence in C. perfringens, since virulence-associated gene expression is observed to peak during the exponential phase (12). The two-component system (TCS) VirR/VirS regulates a number of toxin genes (13, 14). The response regulator, VirR, is phosphorylated and activated by a sensor kinase, VirS, inducing target gene expression (15, 16). The transcription of a small RNA (sRNA), VR-RNA, is activated by VirR/VirS; this sRNA is an effector molecule and directly or indirectly regulates >100 genes (17). Taken together, the VirR/VirS-VR-RNA regulatory cascade is important for the regulation of virulence gene expression in C. perfringens. Type IV pili (TFP) are virulence factors for host-microbe interactions and are also involved in host cell adherence, motility, and biofilm formation (18–20). Bacterial pathogens quickly respond to environmental stimuli and possess multiple strategies in order to rapidly regulate genes to achieve effective infection (21). The rapid degradation of mRNA and RNA regulation affect the regulation of virulence factors in C. perfringens. It is not clearly unknown, however, whether mRNA decay and processing are involved in gene expression and pathogenesis of this organism.
A putative major RNase, RNase Y, is highly conserved in Clostridia and is predicted to be encoded by CPE1672 (here called the rny gene) in C. perfringens, based on sequence similarity. In this study, we constructed an RNase Y-depleted strain of C. perfringens to understand its importance for growth and virulence factor expression in C. perfringens. In the RNase Y-depleted strain, we observed a drastic attenuation of expression of toxin genes regulated by the VirR/VirS TCS cascade and a decrease in the expression of a major secreted toxin. Notably, transcripts of the collagenase gene colA were processed and accumulated in the presence of RNase Y, which suggests the involvement of RNase Y-dependent cleavage of colA transcripts in colA regulation. Additionally, RNase Y was involved in the processing and accumulation of pilA2 mRNA (encoding a TFP pilin in the TFP biosynthesis operon, pilD-pilB2-pilC2-pilA2). These results show that RNase Y affects TFP component expression and host cell attachment. Transcriptome analysis of the RNase Y-depleted strain revealed the effects of RNase Y on the expression of a number of mRNAs. Thus, RNase Y plays a critical role in RNA degradation and gene expression in C. perfringens.
RESULTS
RNase Y is important for the growth of Clostridium perfringens cells.
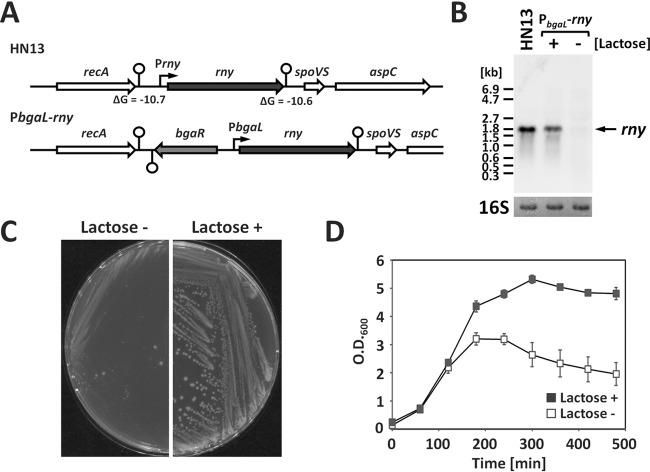
We compared the sequence of the rny gene encoding RNase Y of B. subtilis with clostridial genome sequences and found that the rny gene is highly conserved in Clostridia (see Fig. S1 in the supplemental material). This suggests that the protein product of the rny gene in Clostridia also exhibits endoribonuclease activity and is involved in RNA degradation and processing. We attempted to construct a null mutant of the rny gene via double crossover to analyze RNase Y function in C. perfringens. However, construction was not successful. Despite the potential for low efficiency in electroporation with our method for homologous recombination with linear DNA fragments, we assumed that RNase Y was important for cell proliferation in C. perfringens. Due to this assumption, we then constructed a conditional knockdown rny strain by replacing the native rny promoter region with the lactose-inducible bgaL promoter (22, 23) (Fig. 1A). In the resultant strain, the rny promoter was replaced with the bgaR gene and the bgaL promoter, and we induced transcription of the rny gene by adding lactose to the culture medium (Fig. 1B). rny expression in response to lactose was stable and reproducible in different experiments (Fig. S2). We observed that for the rny conditional knockdown strain, cell growth on agar plates was drastically inhibited in the absence of lactose compared to that in the presence of lactose, which suggested the importance of RNase Y for C. perfringens cell growth (Fig. 1C).
FIG 1.
RNase Y is important for the growth of C. perfringens. (A) Schematics of the rny locus in HN13 and the rny conditional knockdown strain. The transcriptional start sites and terminators are represented by bent arrows and stem-loop structures, respectively. (B) Northern blot analysis of the rny gene. Each lane contained 2 μg of total RNA isolated from the culture at the mid-exponential phase. The rny conditional knockdown strain was cultured with or without 1 mM lactose to induce rny gene expression under the bgaL promoter. 16S rRNA stained with methylene blue on the blots is indicated at the bottom as a loading control. (C) Lactose addition is required for growth of the rny conditional knockdown strain on GAM/2 plates. The rny conditional knockdown strain was anaerobically grown for 12 h on GAM/2 plates with or without 1 mM lactose under anaerobic conditions. (D) Effects of the inducer on cell growth in liquid culture. rny conditional knockdown strains were grown overnight in PGY medium containing 1 mM lactose, and the cells were washed twice with GAM broth. The cells were inoculated with and cultured in GAM broth with or without 1 mM lactose at 37°C.
RNase Y affects global gene expression.
To determine the role of RNase Y in gene expression, we compared the transcriptome profiles of the rny conditional knockdown strain in the presence or absence of the inducer. The rny conditional knockdown strain was precultured in PGY medium containing 1 mM lactose, and the cells were washed twice with GAM broth. The washed cells were inoculated in GAM broth with or without lactose, and the optical densities of the culture were plotted (Fig. 1D). The growth rate of the strain both with and without lactose was comparable at mid-exponential phase; however, the absence of the inducer inhibited growth during the late-exponential or stationary phase (Fig. 1D). To account for the effects of these different growth rates exerted by the two culture conditions, we isolated total RNA from the cultures at mid-exponential phase (120 min; optical density at 600 nm [OD600] of 1.0 to 2.0). Microarray analysis revealed the effects of RNase Y depletion on the mRNA levels of over 400 genes. The expression of 235 and 188 genes significantly decreased and increased, respectively (the log2 ratio of the expression levels in the rny conditional knockdown strain with or without the inducer was less than −1 or greater than 1, and the P value was less than 0.05) (Fig. S3 and Tables S2 and S3). This indicates that RNase Y is involved in mRNA turnover and affects global gene expression in C. perfringens.
RNase Y is involved in destabilizing specific mRNAs.
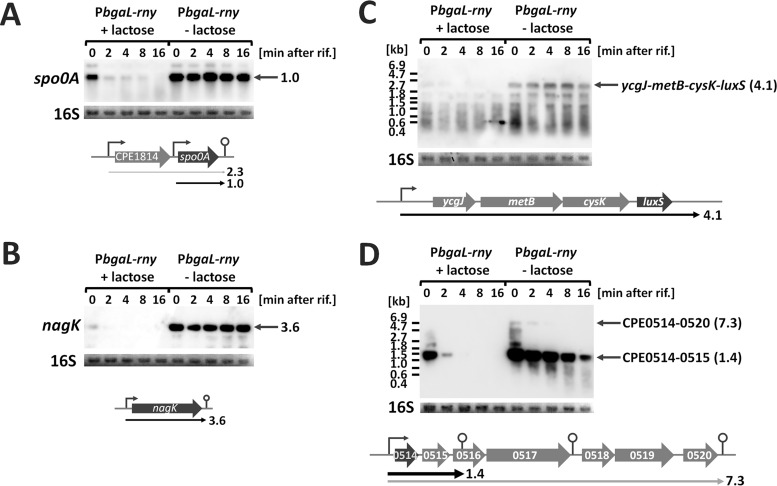
According to our transcriptome analysis, a number of mRNAs accumulated under RNase Y-depleted conditions, suggesting these mRNAs are direct targets for degradation by RNase Y (Table S2). To analyze the effects of RNase Y on mRNA half-life, we selected the following genes that accumulated when RNase Y was depleted: spo0A, encoding a sporulation master regulator (24); monocistronic nagK, encoding the μ-toxin hyaluronidase (25); the tetracistronic operon ycgJ-metB-cysK-luxS, encoding the AI-2-producing enzyme LuxS (26); and the long operon CPE0514-CPE0520, which encodes putative extracellular proteins and the two-component system (27). The monocistronic spo0A and nagK mRNAs were highly stable under RNase Y-depleted conditions, although they were rapidly degraded after the addition of rifampin in the presence of the inducer lactose (Fig. 2A and B). In addition, the 4.1-kb ycgJ-metB-cysK-luxS mRNA was unstable and undetectable 2 min after rifampin addition in the presence of the inducer but was stable and detectable at 16 min when RNase Y was depleted (Fig. 2C). We detected the 1.4-kb CPE0514-CPE0515 bicistronic mRNA and the 7.3-kb CPE0514-CPE0520 polycistronic mRNA and observed stabilization of the bicistronic mRNA upon RNase Y depletion (Fig. 2D). This shows that in C. perfringens RNase Y is involved in the degradation of at least four and possibly more transcripts.
FIG 2.
RNase Y affects the stability of specific transcripts in vivo. Half-lives of spo0A (A), nagK (B), ycgJ-metB-cysK-luxS (C), and CPE0514-CPE0520 (D) mRNAs. rny conditional knockdown strains were cultured with or without 1 mM lactose to mid-exponential phase at 37°C. Total RNA was isolated prior to or after the addition of rifampin (rif; 200 μg ml−1). In each lane, 2 μg of total RNA was loaded and hybridized with spo0A-, nagK-, luxS-, or CPE0514-specific probes. The lengths of the transcripts detected are indicated in kilobases. 16S rRNA stained with methylene blue on the blots is indicated at the bottom as a loading control. Schematics of genetic maps are shown below. The predicted transcriptional start sites and terminators are indicated by bent arrows and stem-loop structures, respectively. Arrows represent the mRNAs detected by Northern blotting.
RNase Y impacts toxin gene expression through the VirR/VirS two-component system.
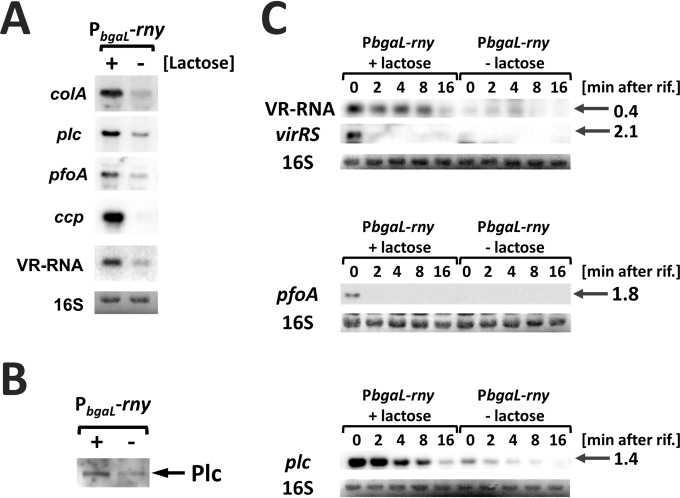
The expression of several major toxin genes, specifically plc, pfoA, colA, ccp, and cpb2, was decreased under RNase Y-depleted conditions (Fig. 3A and Table 1; Table S3). RNase Y depletion also decreased the amount of extracellular secreted alpha-toxin, a phospholipase C (Fig. 3B). Furthermore, expression of the vrr gene, which encodes a small regulatory RNA, VR-RNA, decreased under RNase Y-depleted conditions (Fig. 3A). We postulated that RNase Y affects toxin production via the VirR/VirS–VR-RNA cascade because toxin genes were either directly or indirectly regulated by the VirR/VirS–VR-RNA cascade (17). We examined the mRNA half-life of the plc gene, which encodes the alpha-toxin phospholipase C, the virRS gene, which encodes the response regulator VirR, and the sensor kinases VirS and VR-RNA. RNase Y depletion negatively affected the amount of each transcript but demonstrated little impact on the stability of plc, virRS mRNA, and VR-RNA (Fig. 3B). Northern blots and transcriptome analysis (the log2 ratio with/without inducer was −2.1) indicated a decrease in virRS mRNA under RNase Y-depleted conditions (Fig. 3B). The virRS transcript was highly unstable; we were unable to detect it 2 min after the addition of rifampin with or without the inducer lactose. Thus, RNase Y affects virRS gene expression at the transcriptional level, not at the posttranscriptional level. The expression of the VirR direct target gene pfoA, whose promoter sequence contains the VirR protein-binding box (15), also decreased. However, transcript stability was not affected by RNase Y depletion (Fig. 3C).
FIG 3.
RNase Y affects the expression of toxin genes through the VirR/VirS–VR-RNA cascade. (A) The expression of virulence-associated genes was affected by RNase Y. Total RNAs (2 μg) isolated from cells cultured for 2 h at 37°C were used for Northern blotting of colA, plc, pfoA, ccp, and VR-RNA. 16S rRNA stained with methylene blue on the blots is indicated at the bottom as a loading control. (B) Western blotting of Plc proteins in the culture supernatant. Extracellular proteins equivalent to 0.01 OD600 units were separated by SDS-PAGE and were probed with anti-Plc antisera. (C) Half-lives of virRS mRNA and VirR/VirS target gene mRNAs in rny conditional knockdown strains. rny conditional knockdown strains were cultured with or without 1 mM lactose to mid-exponential phase at 37°C. Total RNA was isolated prior to or after the addition of rifampin (200 μg ml−1). In each lane, 2 μg of total RNA was loaded and hybridized with VR-RNA-, plc-, vir-, or pfoA-specific probes. The lengths of the detected transcripts are indicated in kilonucleotides on the right. 16S rRNA stained with methylene blue on the blots is indicated at the bottom as a loading control.
TABLE 1.
Effect of RNase Y on virulence-associated genesa
| Locus | Gene | Product name | Expression level log2 ratiob | P value |
|---|---|---|---|---|
| Loci with increased expression under RNase Y-depleted condition | ||||
| CPE0030 | hlyA | Hemolysin-related protein | 1.99 | 0.065 |
| CPE0452 | entC | Enterotoxin | 1.88 | 0.019 |
| CPE1234 | nagJ | Hyaluronidase | 3.44 | 0.007 |
| CPE1279 | nagK | Hyaluronidase | 2.61 | 0.001 |
| Loci with decreased expression under RNase Y-depleted condition | ||||
| CPE0036 | plc | Phospholipase C | −2.88 | 0.101 |
| CPE0163 | pfoA | Perfringolysin O | −3.38 | 0.049 |
| CPE0173 | colA | Collagenase | −3.10 | 0.087 |
| CPE0846 | ccp | Alpha-clostripain | −5.75 | 0.097 |
| CPE0957 | vrr | Hypothetical protein CPE0957 | −2.49 | 0.044 |
| CPE1231 | Surface protein | −1.07 | 0.008 | |
| CPE1258 | entA | Putative enterotoxin | −2.81 | 0.052 |
| CPE1500 | virS | Sensor histidine kinase VirS | −1.57 | 0.046 |
| CPE1818 | hlyD | Hemolysin A | −1.04 | 0.004 |
| PCP17 | cpb2 | Beta2-toxin | −2.30 | 0.356 |
| PCP57 | cna | Collagen adhesin | −1.27 | 0.163 |
| Unaffected loci | ||||
| CPE0191 | nagH | Hyaluronidase | −0.18 | 0.083 |
| CPE0378 | Myosin cross-reactive antigen family protein | −0.02 | 0.838 | |
| CPE0437 | hlyB | Hemolysin | −0.38 | 0.131 |
| CPE0553 | nanJ | Exo-alpha-sialidase | −0.41 | 0.285 |
| CPE0606 | entD | Enterotoxin | −0.66 | 0.050 |
| CPE0725 | nanI | Exo-alpha-sialidase | −0.59 | 0.048 |
| CPE0737 | Putative fibronectin-binding protein | −0.70 | 0.004 | |
| CPE0881 | nagI | Hyaluronidase | 0.58 | 0.001 |
| CPE1354 | entB | Enterotoxin | 0.43 | 0.034 |
| CPE1474 | hlyC | Hemolysin III | −0.85 | 0.072 |
| CPE1501 | virR | DNA-binding response regulator, LytTR family | −0.85 | 0.048 |
| CPE1523 | nagL | Hyaluronidase | 0.04 | 0.647 |
| CPE1847 | Fibronectin-binding protein | 0.52 | 0.046 | |
| CPE1915 | hlyE | Hemolysin III | −0.76 | 0.218 |
Virulence-associated genes were predicted by Shimizu et al. (45).
Log2 ratio of expression in the rny conditional knockdown strain without the inducer to expression with the inducer.
RNase Y is responsible for colA primary transcript processing.
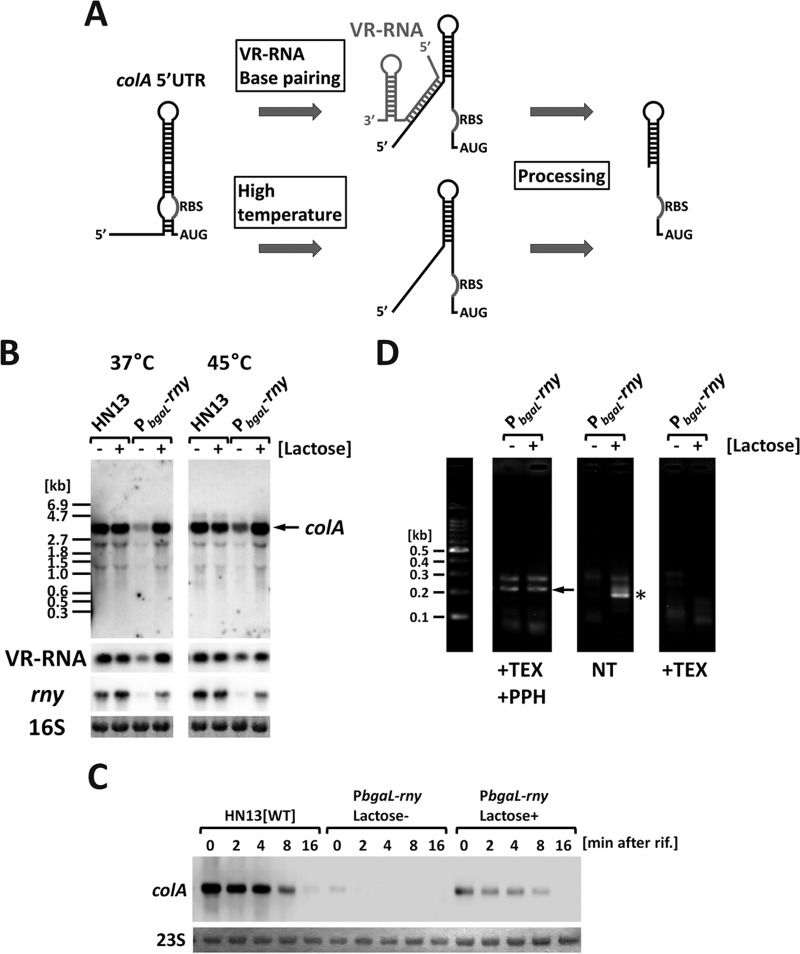
We previously observed base pairing between VR-RNA and the colA primary transcript, resulting in a partial melting of the stem-loop structure in the 5′ untranslated region (5′UTR) of the transcript (28). The primary colA transcript is unstable because the stem-loop structure masks the ribosome binding site (RBS), thereby inhibiting translation. Partial melting of the colA 5′UTR stem-loop structure via VR-RNA–colA base pairing induces cleavage of the colA mRNA 5′UTR; this induced cleavage activates translation and stabilizes the transcript (28) (Fig. 4A). The RNase responsible for this cleavage was not previously identified. As described above, expression of colA mRNA decreased after RNase Y depletion (Fig. 4B). Thus, RNase Y is potentially involved in colA primary mRNA cleavage.
FIG 4.
colA 5′UTR cleavage requires RNase Y. (A) A model of posttranscriptional regulation in the colA mRNA 5′UTR. The long stem-loop structure in the colA 5′UTR is partially melted by VR-RNA base pairing or high temperatures, and this structural change induces cleavage. The colA RBS in the processed transcripts is free from the RNA secondary structure, and translation was stimulated. (B) Northern blotting of colA mRNA in the rny conditional knockdown strain. Total RNA was isolated from cultures grown to mid-exponential phase at 37 or 45°C with or without lactose, and 2 μg of RNA was dissolved in denaturing agarose. VR-RNA and colA and rny mRNA were detected using DIG-labeled specific DNA probes. 16S rRNA stained with methylene blue on the blots is indicated at the bottom as a loading control. (C) The half-life of colA mRNA was affected by RNase Y. rny conditional knockdown strains or HN13 was cultured with or without 1 mM lactose to mid-exponential phase at 45°C. Total RNA was isolated prior to or after the addition of rifampin (200 μg ml−1). In each lane, 2 μg of total RNA was loaded and hybridized with colA-specific probes. 16S rRNA stained with methylene blue on the blots is indicated at the bottom as a loading control. (D) 5′RLM-RACE analysis of colA transcripts. Total RNA isolated from rny conditional knockdown strains cultured at 45°C with or without lactose was treated with or without terminal-dependent exonuclease (TEX) and pyrophosphatase (PPH) to distinguish primary transcripts from processed transcripts. The PCR products were resolved on a 2% agarose gel and stained with ethidium bromide. Asterisks and arrows show PCR products derived from processed and primary transcripts (estimated product sizes are 182 and 244 bp), respectively.
RNase Y depletion represses colA expression, which potentially is a secondary effect that results from decreased amounts of VR-RNA because the amount of VR-RNA decreased under RNase Y-depleted conditions. However, to exclude this possibility, we analyzed colA expression at 45°C. At higher temperatures, the stem-loop structure is partially melted, which can induce colA 5′UTR processing without VR-RNA (29) (Fig. 4A). If colA repression under RNase Y-depleted conditions is a secondary effect of VR-RNA repression, RNase Y depletion should have no effect on colA expression at 45°C. However, because RNase Y depletion reduced colA expression even at 45°C, we can infer that the repression of colA due to RNase Y depletion is independent of VR-RNA (Fig. 4B). Moreover, we observed stabilization of colA mRNA through the induction of RNase Y expression (Fig. 4C).
We employed 5′ RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) to discriminate between primary and processed colA transcripts, which enabled us to determine whether RNase Y affects colA primary transcript processing. Treatment with 5′-dependent exoribonuclease (TEX) and pyrophosphatase (PPH) eliminates processed transcripts and makes the primary transcripts detectable by PCR. The PCR products were detected in both the presence and absence of the inducer (Fig. 4D, +TEX +PPH). However, to detect the processed transcripts we used the total RNA that was not treated with PPH and TEX for PCR. In this case, the processed transcripts were amplified when we induced RNase Y expression (Fig. 4D, NT). This PCR product was not amplified when the RNAs were treated with TEX, indicating that the PCR product corresponds to the processed transcripts (Fig. 4D, TEX). Based on these results, RNase Y is responsible for the cleavage and stabilization of colA mRNA.
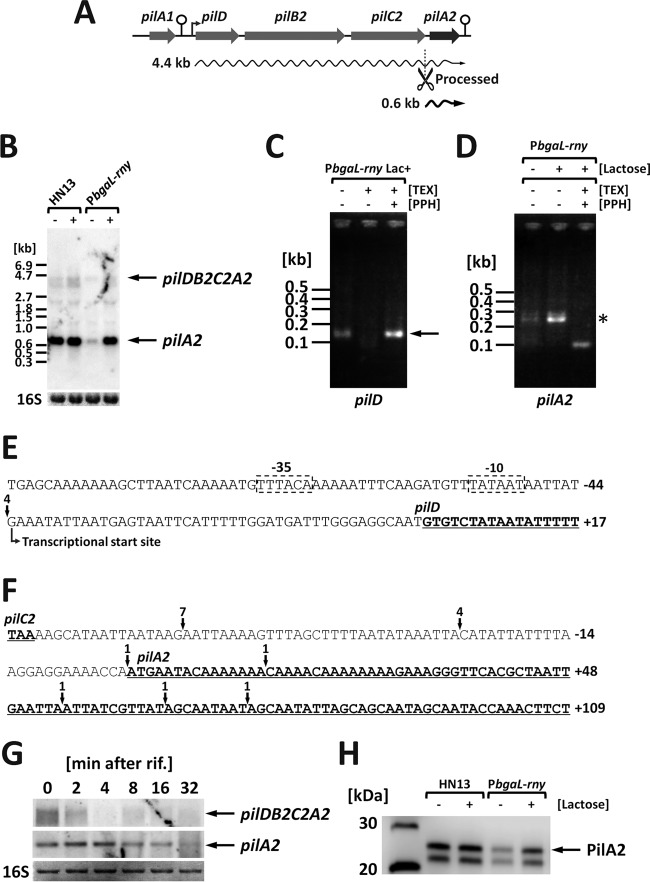
Effect of RNase Y on the expression of a type IV pilus component, pilA2.
In C. perfringens, the type IV pili (TFP) are involved in motility, attachment to eukaryotic cells, and biofilm formation, suggesting an important role for TFP in pathogenesis (18, 19, 30). A TFP biosynthesis operon, pilD-pilB2-pilC2-pilA2, encodes the PilD peptidase and PilA2 pilin, a major component of TFP (Fig. 5A). The expression of pilA2 was drastically repressed under RNase Y-depleted conditions (the log2 ratio of the expression levels under RNase Y-depleted conditions was −2.65) (Table S3). Although we have predicted that there is only one promoter sequence upstream of pilD, we detected both a monocistronic pilA2 mRNA and polycistronic pilD-pilB2-pilC2-pilA2 mRNA by Northern blotting (20) (Fig. 5B). The amount of monocistronic pilA2 mRNA was reduced under RNase Y-depleted conditions (Fig. 5B), and therefore we hypothesized that RNase Y-dependent posttranscriptional cleavage occurs in polycistronic mRNA.
FIG 5.
Expression of the pilin-encoding pilA2 gene is regulated by RNase Y. (A) Schematic representation of the pilD-pilB2-pilC2-pilA2 operon. The predicted transcriptional start site and terminators are indicated by a bent arrow and stem-loop structures, respectively. Wavy lines represent the transcripts detected in the Northern blots. The scissor symbol indicates a putative processing site. (B) Northern blotting of pilA2. Total RNA was isolated from cultures grown to mid-exponential phase at 37°C with or without 1 mM lactose, and 2 μg of RNA was used for blotting and hybridization with a pilA2-specific probe. 16S rRNA stained with methylene blue on the blots is indicated at the bottom as a loading control. (C and D) Determination of the 5′ end of pilD (C) or pilA2 (D) mRNA. Total RNA isolated from rny conditional knockdown strain cells cultured at 37°C was left untreated or treated with TEX and PPH to distinguish primary transcripts from processed transcripts. PCR products amplified using primers for detection were resolved on a 2% agarose gel and stained with ethidium bromide. Asterisks and arrows show PCR products derived from the processed and primary transcripts, respectively. (E) Nucleotide sequence of the pilD promoter region. Putative −10 and −35 sequence regions are boxed in dashed lines. The pilD ORF is underlined. Arrows represent 5′-terminal sites of the pilD transcripts detected and of a number of clones obtained by 5′RLM-RACE. Numbers relative to the ATG start codon of pilD (A is +1) are shown to the right of the sequences. (F) Nucleotide sequence of the pilC2-pilA2 intergenic region. The pilC2 and pilA2 ORFs are underlined. Arrows represent the 5′-terminal sites of the pilA2 transcripts detected and of a number of clones obtained by 5′RLM-RACE. Numbers relative to the ATG start codon of pilA2 (A is +1) are shown to the right of the sequences. (G) The stability of the pilD-pilB2-pilC2-pilA2 polycistronic and pilA2 monocistronic transcripts. C. perfringens was grown to mid-exponential phase at 37°C. Total RNA was isolated prior to or after the addition of rifampin (200 μg ml−1). In each lane, 2 μg of total RNA was loaded and hybridized with pilA2-specific probes. 16S rRNA stained with methylene blue on the blots is indicated at the bottom as a loading control. (H) Western blotting of PilA2 proteins. Cell lysates (equivalent to 0.005 OD600 units) isolated from HN13 or rny conditional knockdown strains grown for 2 h in GAM broth at 37°C were separated by SDS-PAGE. PilA2 proteins were detected using anti-PilA2 antisera.
To determine whether monocistronic pilA2 mRNA is a processed product, the 5′ termini of the polycistronic pilD-pilB2-pilC2-pilA2 mRNA and the monocistronic pilA2 mRNA were detected by 5′RLM-RACE (Fig. 5C and D). Prior to RNA adaptor ligation, we treated the total RNA with pyrophosphatase (PPH). This allowed us to detect the PCR products amplified from the 5′ terminus of pilD mRNA, which indicates that the 5′ terminus of pilD mRNA is a transcription start site (Fig. 5C). Additionally, a typical σA consensus sequence was found in the region upstream of the predicted pilD transcriptional start site (Fig. 5E). However, the 250-bp PCR products amplified from the 5′ terminus of pilA2 mRNA were completely eliminated in the presence of TEX; this indicates that monocistronic pilA2 mRNA is a processed transcript (Fig. 5D and F). Although we detected a 100-bp band in the TEX- and PPH-treated lanes (Fig. 5D), we can justifiably assume that this is the result of nonspecific amplification; PCR products of less than 100 bp are too small to correspond to the monocistronic pilA2 transcripts, because we detected only monocistronic pilA2 transcripts of greater than 500 nucleotides by Northern blot analysis (Fig. S4).
To examine the effect of processing on mRNA, we compared the stability of the primary and processed mRNA in wild-type cells. We observed increased stability of the processed monocistronic pilA2 mRNA over the primary polycistronic transcripts (Fig. 5G). Moreover, the amount of PilA2 protein decreased under RNase Y-depleted conditions (Fig. 5H). Therefore, we reason that RNase Y is involved in the processing and stabilization of pilA2 mRNA, which is required for maximal PilA2 protein expression.
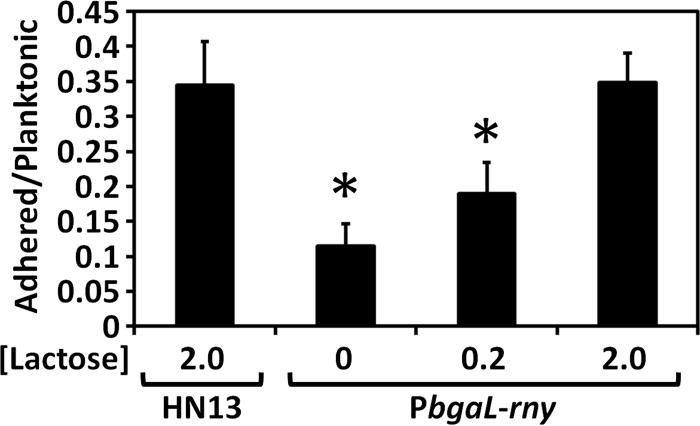
RNase Y may also play an important role in TFP biosynthesis and function. RNase Y likely affects cell-substrate attachment and biofilm formation in C. perfringens, because TFP are involved in host cell attachment and biofilm formation (19, 30). We cultured the C. perfringens strains in polystyrene microtiter plates and measured the amount of adhered cells and biofilm biomass. The attachment and biofilm formation by the RNase Y conditional knockdown strain to polystyrene surfaces were significantly decreased compared to those of the wild-type HN13 strain (Fig. 6). In the presence of lactose, these amounts increased to the same levels observed for the wild-type strain (Fig. 6). This shows that RNase Y activity has an important role in attachment and biofilm formation in C. perfringens.
FIG 6.
RNase Y depletion decreases attachment to polystyrene surface and biofilm formation. C. perfringens was grown with or without lactose for 24 h. Cell adhesion activity was calculated by measuring the OD600 of adherent or planktonic cells. Asterisks show statistically significant differences with respect to HN13 (P < 0.05). P values were calculated using a two-tailed unpaired t test with Welch's correction.
RNase Y is involved in the degradation of mRNAs of the protein secretion system.
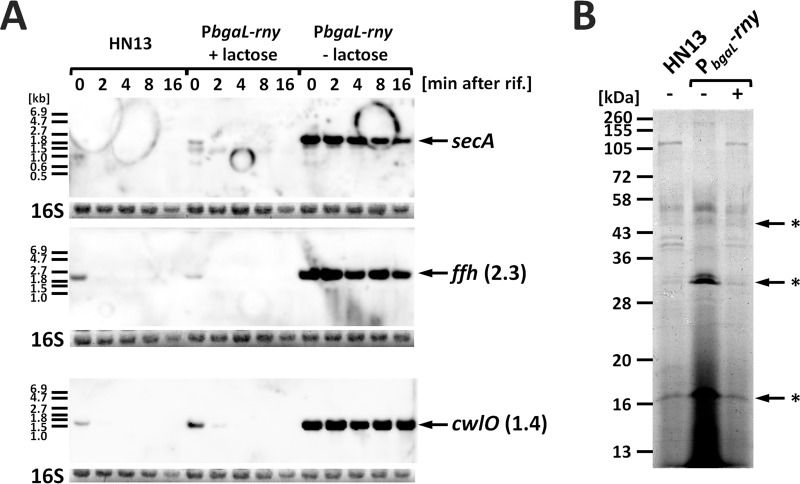
It is believed that C. perfringens secretes extracellular enzymes and toxins through a Sec-dependent pathway because these protein sequences are predicted to contain a signal peptide (31) (Table S4). Under RNase Y-depleted conditions, the expression of the ffh gene, which encodes a signal recognition particle protein, was highly upregulated according to our microarray analysis (Table S2). This observation led us to analyze Sec-dependent protein secretion in the rny conditional knockdown strain. We found that the secA and ffh mRNAs were stable when RNase Y was depleted but were unstable and thus undetectable 2 min after the addition of rifampin in the presence of RNase Y (Fig. 7A). This result is consistent with the transcriptome analysis and suggests the involvement of RNase Y in secA and ffh transcript turnover. The cwlO gene encodes a cell wall lytic endopeptidase, and in B. subtilis transcript stability is regulated by RNase Y (32). Northern blot analysis showed the cwlO transcript stability was increased under RNase Y-depleted conditions in C. perfringens (Fig. 7A). The 45-, 30-, and 15-kDa proteins have been predicted to be CwlO fragments and are the major extracellular proteins in this organism (22). We investigated the amounts of CwlO protein present in the culture supernatant of the rny conditional knockdown strain. The 45-, 30-, and 15-kDa proteins accumulated in the rny conditional knockdown strain cultured without lactose, suggesting the effects of RNase Y depletion on the Sec secretion system in C. perfringens (Fig. 7B).
FIG 7.
RNase Y affects the Sec secretion system. (A) rny conditional knockdown strains or HN13 were cultured with or without 1 mM lactose to mid-exponential phase at 37°C. Total RNA was isolated prior to or after the addition of rifampin (200 μg ml−1). In each lane, 2 μg of total RNA was loaded and hybridized with secA-, ffh-, and cwlO-specific probes. The lengths of the detected transcripts are indicated in kilobases on the right. 16S rRNA stained with methylene blue on the blots is indicated at the bottom as a loading control. (B) Extracellular protein expression in HN13 or rny conditional knockdown strains. Extracellular proteins equivalent to 0.04 OD600 units were prepared from the supernatant of the culture grown for 6 h at 37°C and were loaded in each lane. Arrows indicate CwlO proteins estimated based on molecular size.
DISCUSSION
RNase Y is a major endoribonuclease and plays an important role in mRNA turnover in C. perfringens. Cells are unable to grow normally under RNase Y-depleted conditions (Fig. 1), indicating the importance of RNase Y for the proliferation and physiological activity of C. perfringens. RNase Y depletion impacts a large number of genes in various functional classes, which suggests that combinatorial effects cause cell growth defects (see Fig. S3 in the supplemental material). In this study, we measured lactose-inducible RNase Y expression at the transcript level by Northern blotting. We cannot exclude the possibility that the amount of RNase Y protein was not accurately controlled by lactose. However, the stability of several transcripts was drastically affected as a function of the lactose content (Fig. 2 and 7). Thus, lactose plays a significant role in the regulation of RNase Y protein levels in the rny conditional knockdown strain. We attempted to inactivate rny using a gene replacement system but were unsuccessful, so it is still unclear whether RNase Y is essential for C. perfringens growth. The B. subtilis RNase Y deletion strain exhibits major defects in cell morphology and growth but is viable (33); however, to inactivate RNase Y in B. subtilis, high recombination efficiency appears to be required. In C. perfringens, we suspect that low transformation and recombination efficiency is the cause of our failure to generate an RNase Y deletion strain.
Transcriptome analysis revealed the global effects of RNase Y on gene expression, including toxin genes. We conducted a transcriptome analysis in the mid-exponential phase when the growth rate of the strain under both lactose-positive and -negative conditions were comparable. However, the downregulation of the toxin genes under the RNase Y-depleted condition might correlate with a decrease in growth rate and may not be specific to RNase Y. Meanwhile, we found that RNase Y depletion affected the expression of virulence factors under VirR/VirS TCS control. VirR/VirS–VR-RNA is the major regulatory cascade involved in toxin gene regulation; over 100 genes are controlled by the VirR/VirS–VR-RNA cascade, suggesting indirect effects of RNase Y on the expression of many genes through the VirR/VirS TCS (34). RNase Y depletion did not affect virR-virS mRNA stability, suggesting RNase Y affects virR-virS expression at the transcriptional level. The mRNA encoding the virR-virS gene regulator may be processed or degraded by RNase Y.
RNase Y is an endoribonuclease responsible for mRNA turnover in B. subtilis, S. aureus, and S. pyogenes (7–10, 35, 36). We suggest that this enzyme also plays an important role in RNA degradation in C. perfringens, because the expression of 188 genes was upregulated in our microarray experiment under RNase Y-depleted conditions. A Northern blot analysis of spo0A, nagK, ycgI-metB-cysK-luxS, CPE0514-CPE0515, secA, ffh, and cwlO also supported this hypothesis. The transcript of the cwlO homolog in B. subtilis is unstable and is cleaved by RNase Y in its 5′UTR, which initiates degradation of the transcript (32). In C. perfringens, cwlO transcripts are also highly unstable and are undetectable 2 min after the addition of rifampin. cwlO mRNA may be cleaved by RNase Y, triggering transcript degradation. In addition, several target genes directly regulated by RNase Y have been identified in B. subtilis. The glycolytic gapA operon is composed of the cggR-gapA-pgk-tpi-pgm-eno genes and is cleaved at a position upstream of the gapA open reading frame (ORF) by RNase Y in B. subtilis (37). In C. perfringens, the operon is conserved and composed of the cggR-gapC-pgk-tpiA-pgm genes. The longer gapC operon mRNAs were detected under RNase Y-depleted conditions, which suggests that the effects of RNase Y on gene expression are realized through mRNA cleavage in C. perfringens (Fig. S5).
The kappa-toxin collagenase is a major virulence factor and is encoded by the colA gene. The colA mRNA 5′UTR forms a long stem-loop structure that masks the RBS, thereby inhibiting translation and rendering the transcripts unstable (28). There are two ways to partially melt the secondary structure of the colA mRNA and induce processing in the 5′UTR: (i) base pairing of the sRNA VR-RNA or (ii) elevating temperatures (29). The processed transcripts are stable and translatable because the RBS is free from the secondary structure. However, the enzyme that cleaves colA transcripts is unknown. Under RNase Y-depleted conditions, we saw that colA transcripts became unstable even at higher temperatures. Additionally, we could not detect the processed transcript. From this we can induce that RNase Y is responsible for colA processing and stabilization and, furthermore, is coupled to sRNA- or temperature-dependent colA regulation. Endoribonuclease E (RNase E) is a major enzyme involved in RNA turnover in gamma proteobacteria such as Escherichia coli. RNase E is involved not only in maintaining steady-state mRNA levels but also in the sRNA-dependent regulation of specific mRNAs (38). In B. subtilis, RNase Y is thought to be the functional equivalent of RNase E and functions in concert with the RoxS sRNA in regulation of ppnKB and sucCD (39, 40). colA regulation by VR-RNA and RNase Y is the first example of trans-acting sRNAs and endoribonucleases functioning in concert in clostridia. RNase Y may be an important factor for sRNA-dependent posttranscriptional regulation in Clostridium and Bacillus (41, 42).
In this study, we also revealed the effects of RNase Y on the expression of the TFP gene pilA2, which encodes a pilin that is the primary component of TFP. The TFP biosynthesis operon consists of the pilD, pilB2, pilC2, and pilA2 genes, and these genes are cotranscribed from the promoter located upstream of the pilD gene. Northern blotting detected pilA2 monocistronic mRNA in addition to pilD-pilB2-pilC2-pilA2 polycistronic mRNA. Promoter prediction, the reporter assay, and 5′RLM-RACE indicated the absence of any promoter immediately upstream of the pilA2 gene (20) (Fig. 5). This indicates that pilA2 monocistronic mRNA is the processed product of primary polycistronic transcripts and consequently that RNase Y is required for the efficient processing of pilA2 mRNA.
The PilA2 pilin, a major component of TFP, polymerizes and forms a pilus structure (43). TFP activity and ability to rapidly respond to the environment is tightly controlled by the amount of pilA2 transcripts. We know that RNase Y-mediated pilA2 transcript processing actually controls pilA2 expression because pilD-pilB2-pilC2-pilA2 polycistronic mRNA is unstable, while processed pilA2 monocistronic mRNA is stable. The 5′-proximal RNA structure of the pilA2 monocistronic mRNA was predicted to form a stem-loop structure (data not shown), which might contribute to the stability of the transcript (44). We recently observed the accumulation of monocistronic pilA2 mRNA in cells grown at 37°C compared to 25°C, which highly suggests that PilA2 protein amounts and processing depends on temperature (20). In fact, it is known that TFP-mediated cell surface adhesion decreased at 25°C (20). Western blotting for glutathione S-transferase (GST) expressed from the pilD promoter revealed minimal effects of temperature on promoter activity (Fig. S6). RNase Y depletion decreased C. perfringens attachment to polystyrene surfaces and biofilm formation (Fig. 6). Therefore, the RNase Y-dependent cleavage of pilA2 is important for TFP response to environmental change and its relative functions in cell adhesion and biofilm formation.
RNase Y depletion also leads to the accumulation of mRNAs encoding two components of the Sec-dependent secretion system, SecA and Ffh. C. perfringens has no secretion system other than the Sec system (45). Therefore, the Sec secretion system is important for the physiology and proliferation of this organism. In B. subtilis, transcriptome analysis of three ribonucleases, RNase Y, RNase J1, and RNase III, has been performed, but the ribonucleases involved in regulating the Sec secretion components have not been identified (36). The results shown in Fig. 7 reveal the posttranscriptional effects of RNase Y on secA and ffh expression. The Sec system in C. perfringens must be rapidly regulated in response to the environment and requires RNase Y-dependent posttranscriptional degradation. The smaller secA mRNA was detected in the presence of RNase Y, as shown in Fig. 7. Thus, secA mRNA may be cleaved by RNase Y.
The major RNase RNase Y is involved in mRNA turnover and cleaves specific mRNAs. This RNase also affects toxin production and secretion, cell adhesion, and cell proliferation in C. perfringens. Furthermore, a number of sRNAs have been predicted in the genus Clostridium, and these sRNAs are potentially involved in physiological functions (42). RNase Y is more likely to have an important role in sRNA function, because this enzyme is involved in posttranscriptional colA regulation by VR-RNA, an sRNA in C. perfringens. Moreover, RNase Y activity affects the amounts of specific transcripts related to toxin, adherence, and protein secretion. Therefore, RNase Y is potentially a good target to control pathogenic, solventogenic, and cellulose-degrading clostridia.
MATERIALS AND METHODS
Bacterial strains and culturing conditions.
Clostridium perfringens strain 13 (45), HN13 (22), and derivative strains were cultured on Gifu anaerobic medium/2 (GAM/2) agar plates (Nissui Co., Japan) under anaerobic conditions using an Anaeropack system (Mitsubishi Gas Chemical Co. Inc., Tokyo, Japan) and PGY (3% proteose peptone no. 3, 2% glucose, 1% yeast extract, and 0.1% sodium thioglycolate) or GAM liquid broth supplemented with 1 mM lactose. Escherichia coli JM109 was cultured in LB medium supplemented with 50 μg ml−1 ampicillin or 25 μg ml−1 chloramphenicol.
Oligonucleotides.
The oligonucleotides used in this study are listed in Table S1 in the supplemental material.
Conditional knockdown strain construction.
We constructed the rny conditional knockdown strain using a galactose counterselection system and lactose-inducible promoter (22, 23). The upstream and downstream regions of the rny promoter region were amplified using primer pairs NOB-0642/NOB-0643 and NOB-0644/NOB-0645 and digested with SalI/SacI and BamHI/PstI, respectively. The bgaR ORF and bgaL promoter region were amplified using primers NOB-0488/NOB-0489 and digested with SacI/BamHI. These fragments were ligated and cloned into the SalI/PstI site of pCM-GALK (22), and the resulting plasmid was introduced into C. perfringens HN13 via electroporation. Agar plates containing 1 mM lactose with or without 20 μg/ml chloramphenicol were used for isolation of the strains. We confirmed the generated strain by PCR and DNA sequencing.
Northern blot analysis.
The rny conditional knockdown strains were cultured overnight in PGY medium containing 1 mM lactose. The cells were washed twice with GAM broth and inoculated in fresh GAM broth containing either 1 mM or no lactose to reach an OD600 of 0.1. The cultures were incubated for 2 h at 37°C or 1.5 h at 45°C to reach a mid-exponential growth phase. Total RNA was extracted from C. perfringens using the SV total RNA isolation system according to the manufacturer's instructions (Promega, Tokyo, Japan). A Northern blot analysis was performed as previously described (20). Digoxigenin (DIG)-labeled DNA probes were generated using DIG-High Prime (Roche), and template DNAs were amplified using primers that are listed in Table S1 or that were previously described (12).
Microarray analysis.
Transcriptome analysis of the RNase Y-depleted strain was performed using a custom C. perfringens DNA 15K array (TaKaRa Bio Inc., Japan), which contains the coding DNA sequences based on reference data for C. perfringens strain 13 from the National Center for Biotechnology Information (NCBI). We isolated the total RNA from the rny conditional knockdown strain grown with or without 1 mM lactose to mid-exponential phase and used the RNA for cDNA synthesis. Cy3-labeled cRNA was then synthesized via in vitro transcription using the Low Input Quick Amp WT labeling kit, one-color (Agilent Technologies, Tokyo, Japan). We hybridized the cRNAs to the custom DNA array and then scanned and analyzed the fluorescence using an Agilent DNA microarray scanner (G2565CA; Agilent Technologies). TaKaRa Bio Inc. supported probe labeling, hybridization, and partial data analysis. To determine fold ratios of gene expression, we compared the signal intensities of the rny conditional knockdown strains grown in the presence of lactose. The P values were determined using an unpaired Welch's t test.
5′RLM-RACE.
The 5′ ends of the transcripts were analyzed by 5′RLM-RACE as described previously, with some modifications (28). Total RNA (10 μg) was left untreated or was treated with 1 U of Terminator exonuclease in 20 μl of reaction buffer at 30°C for 1 h. We then purified the RNA with phenol-chloroform-isoamylalcohol (PCI) extraction and ethanol precipitation. RNA samples equivalent to 3 μg were left untreated or were treated with 5 U of RNA 5′ polyphosphatase in 10 μl of reaction buffer at 37°C for 1 h. We then incubated 2 μl of untreated or 5′ polyphosphatase-treated RNA (equivalent to 0.6 μg of RNA) with the 5′RACE adaptor and 5 U of T4 RNA ligase (Life Technologies, Tokyo, Japan) at 14°C for 16 h. The total RNA that ligated with the adaptor (equivalent to 0.12 μg of RNA) was used as a template for reverse transcription, which was performed with PrimeScript reverse transcriptase (TaKaRa Bio). We amplified cDNAs via nested PCR using the primers listed in Table S1. The amplified DNA products were cloned into E. coli using the pUC18 vector. The resulting vector sequences were determined by DNA sequencing using M13 primers.
Attachment and biofilm formation assay.
C. perfringens cells were inoculated from cultures grown in PGY into the wells of 24-well microtiter plates containing 1 ml of GAM broth with or without lactose. The cells were grown anaerobically for 24 h at 37°C. After removing culture supernatants and floating cells, we then washed the wells with 1 ml of phosphate-buffered saline (PBS). The residual cells adhering to the substratum were resuspended in 1 ml of PBS via thorough pipetting. OD600 readings were obtained for floating and adherent cells.
Western blot analysis.
Extracellular proteins equivalent to an OD600 of 0.01 were separated by SDS-PAGE and electroblotted onto polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 2.5% skim milk in Tris-buffered saline containing 0.05% Tween 20. Plc proteins were probed using anti-CPa (Clostridium perfringens alpha-toxin) and anti-rabbit IgG (GE Healthcare) antibodies diluted 1:1,000 and 1:20,000, respectively, in Can Get signal solution (Toyobo, Osaka, Japan). The bound antibodies were detected using Immunostar LD (Wako, Osaka, Japan). For PilA2 detection, we used an anti-PilA2-specific peptide (QANSKDYLVYPDGKKPYDL)-directed antibody (Sigma-Aldrich) diluted to 1:5,000.
Extraction of extracellular proteins.
C. perfringens HN13 and rny conditional knockdown strains were cultured overnight in TY-G1 medium (3% tryptone, 2% yeast extract, 1% glucose, and 0.1% sodium thioglycolate) containing 1 mM lactose. The cells were washed twice with TY-G1 medium and inoculated in fresh TY-G1 containing 1 mM or no lactose to reach an OD600 of 0.1. The cultures were incubated for 6 h at 37°C, and then the culture supernatants were collected via centrifugation at 5,000 × g for 10 min. The culture supernatants that passed through a 0.2-μm-pore-size membrane filter were incubated with 20% trichloroacetic acid (TCA). After washing with ice-cold acetone, precipitated proteins were dissolved in SDS sample buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 2.5% mercaptoethanol, 0.1% bromophenol blue).
Supplementary Material
ACKNOWLEDGMENTS
C. perfringens strain HN13 and pCM-GALK plasmid were kindly provided by Hirofumi Nariya (Department of Microbiology, Faculty of Medicine, Kagawa University). We thank Andrew S. Utada (University of Tsukuba) for a critical reading of the manuscript.
This work was financially supported by a Grant-in-Aid for Young Scientists (B) and ERATO of the Japan Science and Technology Agency (JST).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00703-16.
REFERENCES
- 1.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, Silva IJ, Viegas SC. 2010. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 2.Rochat T, Bouloc P, Repoila F. 2013. Gene expression control by selective RNA processing and stabilization in bacteria. FEMS Microbiol Lett 344:104–113. doi: 10.1111/1574-6968.12162. [DOI] [PubMed] [Google Scholar]
- 3.Lehnik-Habrink M, Lewis RJ, Mader U, Stulke J. 2012. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol Microbiol 84:1005–1017. doi: 10.1111/j.1365-2958.2012.08072.x. [DOI] [PubMed] [Google Scholar]
- 4.Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 2007. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 5.Liu B, Deikus G, Bree A, Durand S, Kearns DB, Bechhofer DH. 2014. Global analysis of mRNA decay intermediates in Bacillus subtilis wild-type and polynucleotide phosphorylase-deletion strains. Mol Microbiol 94:41–55. doi: 10.1111/mmi.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oussenko IA, Abe T, Ujiie H, Muto A, Bechhofer DH. 2005. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J Bacteriol 187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shahbabian K, Jamalli A, Zig L, Putzer H. 2009. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J 28:3523–3533. doi: 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laalami S, Bessieres P, Rocca A, Zig L, Nicolas P, Putzer H. 2013. Bacillus subtilis RNase Y activity in vivo analysed by tiling microarrays. PLoS One 8:e54062. doi: 10.1371/journal.pone.0054062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marincola G, Schafer T, Behler J, Bernhardt J, Ohlsen K, Goerke C, Wolz C. 2012. RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol Microbiol 85:817–832. doi: 10.1111/j.1365-2958.2012.08144.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Itzek A, Malke H, Ferretti JJ, Kreth J. 2013. Multiple roles of RNase Y in Streptococcus pyogenes mRNA processing and degradation. J Bacteriol 195:2585–2594. doi: 10.1128/JB.00097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rood JI, Cole ST. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev 55:621–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abe K, Obana N, Nakamura K. 2010. Effects of depletion of RNA-binding protein Tex on the expression of toxin genes in Clostridium perfringens. Biosci Biotechnol Biochem 74:1564–1571. doi: 10.1271/bbb.100135. [DOI] [PubMed] [Google Scholar]
- 13.Ba-Thein W, Lyristis M, Ohtani K, Nisbet IT, Hayashi H, Rood JI, Shimizu T. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J Bacteriol 178:2514–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohtani K, Hirakawa H, Tashiro K, Yoshizawa S, Kuhara S, Shimizu T. 2010. Identification of a two-component VirR/VirS regulon in Clostridium perfringens. Anaerobe 16:258–264. doi: 10.1016/j.anaerobe.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Cheung JK, Rood JI. 2000. The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J Bacteriol 182:57–66. doi: 10.1128/JB.182.1.57-66.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung JK, Awad MM, McGowan S, Rood JI. 2009. Functional analysis of the VirSR phosphorelay from Clostridium perfringens. PLoS One 4:e5849. doi: 10.1371/journal.pone.0005849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu T, Yaguchi H, Ohtani K, Banu S, Hayashi H. 2002. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol Microbiol 43:257–265. doi: 10.1046/j.1365-2958.2002.02743.x. [DOI] [PubMed] [Google Scholar]
- 18.Varga JJ, Nguyen V, O'Brien DK, Rodgers K, Walker RA, Melville SB. 2006. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Mol Microbiol 62:680–694. doi: 10.1111/j.1365-2958.2006.05414.x. [DOI] [PubMed] [Google Scholar]
- 19.Rodgers K, Arvidson CG, Melville S. 2011. Expression of a Clostridium perfringens type IV pilin by Neisseria gonorrhoeae mediates adherence to muscle cells. Infect Immun 79:3096–3105. doi: 10.1128/IAI.00909-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obana N, Nakamura K, Nomura N. 2014. A sporulation factor is involved in the morphological change of Clostridium perfringens biofilms in response to temperature. J Bacteriol 196:1540–1550. doi: 10.1128/JB.01444-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, Johansson J. 2010. RNAs: regulators of bacterial virulence. Nat Rev Microbiol 8:857–866. doi: 10.1038/nrmicro2457. [DOI] [PubMed] [Google Scholar]
- 22.Nariya H, Miyata S, Suzuki M, Tamai E, Okabe A. 2011. Development and application of a method for counterselectable in-frame deletion in Clostridium perfringens. Appl Environ Microbiol 77:1375–1382. doi: 10.1128/AEM.01572-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartman AH, Liu H, Melville SB. 2011. Construction and characterization of a lactose-inducible promoter system for controlled gene expression in Clostridium perfringens. Appl Environ Microbiol 77:471–478. doi: 10.1128/AEM.01536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang IH, Waters M, Grau RR, Sarker MR. 2004. Disruption of the gene (spo0A) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol Lett 233:233–240. doi: 10.1111/j.1574-6968.2004.tb09487.x. [DOI] [PubMed] [Google Scholar]
- 25.Obana N, Nakamura K. 2011. A novel toxin regulator, the CPE1446-CPE1447 protein heteromeric complex, controls toxin genes in Clostridium perfringens. J Bacteriol 193:4417–4424. doi: 10.1128/JB.00262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohtani K, Hayashi H, Shimizu T. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol Microbiol 44:171–179. doi: 10.1046/j.1365-2958.2002.02863.x. [DOI] [PubMed] [Google Scholar]
- 27.Yuan Y, Ohtani K, Yoshizawa S, Shimizu T. 2012. Complex transcriptional regulation of citrate metabolism in Clostridium perfringens. Anaerobe 18:48–54. doi: 10.1016/j.anaerobe.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Obana N, Shirahama Y, Abe K, Nakamura K. 2010. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5′ leader sequence. Mol Microbiol 77:1416–1428. doi: 10.1111/j.1365-2958.2010.07258.x. [DOI] [PubMed] [Google Scholar]
- 29.Obana N, Nomura N, Nakamura K. 2013. Structural requirement in Clostridium perfringens collagenase mRNA 5′ leader sequence for translational induction through small RNA-mRNA base pairing. J Bacteriol 195:2937–2946. doi: 10.1128/JB.00148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varga JJ, Therit B, Melville SB. 2008. Type IV pili and the CcpA protein are needed for maximal biofilm formation by the gram-positive anaerobic pathogen Clostridium perfringens. Infect Immun 76:4944–4951. doi: 10.1128/IAI.00692-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamane K, Bunai K, Kakeshita H. 2004. Protein traffic for secretion and related machinery of Bacillus subtilis. Biosci Biotechnol Biochem 68:2007–2023. doi: 10.1271/bbb.68.2007. [DOI] [PubMed] [Google Scholar]
- 32.Noone D, Salzberg LI, Botella E, Basell K, Becher D, Antelmann H, Devine KM. 2014. A highly unstable transcript makes CwlO D,L-endopeptidase expression responsive to growth conditions in Bacillus subtilis. J Bacteriol 196:237–247. doi: 10.1128/JB.00986-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Figaro S, Durand S, Gilet L, Cayet N, Sachse M, Condon C. 2013. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J Bacteriol 195:2340–2348. doi: 10.1128/JB.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banu S, Ohtani K, Yaguchi H, Swe T, Cole ST, Hayashi H, Shimizu T. 2000. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol Microbiol 35:854–864. doi: 10.1046/j.1365-2958.2000.01760.x. [DOI] [PubMed] [Google Scholar]
- 35.Lehnik-Habrink M, Schaffer M, Mader U, Diethmaier C, Herzberg C, Stulke J. 2011. RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol 81:1459–1473. doi: 10.1111/j.1365-2958.2011.07777.x. [DOI] [PubMed] [Google Scholar]
- 36.Durand S, Gilet L, Bessieres P, Nicolas P, Condon C. 2012. Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet 8:e1002520. doi: 10.1371/journal.pgen.1002520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Volker U, Stulke J. 2009. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics 8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackie GA. 2013. RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol 11:45–57. [DOI] [PubMed] [Google Scholar]
- 39.Lehnik-Habrink M, Newman J, Rothe FM, Solovyova AS, Rodrigues C, Herzberg C, Commichau FM, Lewis RJ, Stulke J. 2011. RNase Y in Bacillus subtilis: a natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J Bacteriol 193:5431–5441. doi: 10.1128/JB.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durand S, Braun F, Lioliou E, Romilly C, Helfer AC, Kuhn L, Quittot N, Nicolas P, Romby P, Condon C. 2015. A nitric oxide regulated small RNA controls expression of genes involved in redox homeostasis in Bacillus subtilis. PLoS Genet 11:e1004957. doi: 10.1371/journal.pgen.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irnov I, Sharma CM, Vogel J, Winkler WC. 2010. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res 38:6637–6651. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Indurthi DC, Jones SW, Papoutsakis ET. 2011. Small RNAs in the genus Clostridium. mBio 2:e00340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melville S, Craig L. 2013. Type IV pili in Gram-positive bacteria. Microbiol Mol Biol Rev 77:323–341. doi: 10.1128/MMBR.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bechhofer DH. 2009. Messenger RNA decay and maturation in Bacillus subtilis. Prog Mol Biol Transl Sci 85:231–273. doi: 10.1016/S0079-6603(08)00806-4. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci U S A 99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.