ABSTRACT
Staphylococcus aureus is a major human pathogen that causes infection in a wide variety of sites within the human body. Its ability to adapt to the human host and to produce a successful infection requires precise orchestration of gene expression. While DNA-dependent RNA polymerase (RNAP) is generally well characterized, the roles of several small accessory subunits within the complex have yet to be fully explored. This is particularly true for the omega (ω or RpoZ) subunit, which has been extensively studied in Gram-negative bacteria but largely neglected in Gram-positive counterparts. In Escherichia coli, it has been shown that ppGpp binding, and thus control of the stringent response, is facilitated by ω. Interestingly, key residues that facilitate ppGpp binding by ω are not conserved in S. aureus, and consequently, survival under starvation conditions is unaffected by rpoZ deletion. Further to this, ω-lacking strains of S. aureus display structural changes in the RNAP complex, which result from increased degradation and misfolding of the β′ subunit, alterations in δ and σ factor abundance, and a general dissociation of RNAP in the absence of ω. Through RNA sequencing analysis we detected a variety of transcriptional changes in the rpoZ-deficient strain, presumably as a response to the negative effects of ω depletion on the transcription machinery. These transcriptional changes translated to an impaired ability of the rpoZ mutant to resist stress and to fully form a biofilm. Collectively, our data underline, for the first time, the importance of ω for RNAP stability, function, and cellular physiology in S. aureus.
IMPORTANCE In order for bacteria to adjust to changing environments, such as within the host, the transcriptional process must be tightly controlled. Transcription is carried out by DNA-dependent RNA polymerase (RNAP). In addition to its major subunits (α2ββ′) a fifth, smaller subunit, ω, is present in all forms of life. Although this small subunit is well studied in eukaryotes and Gram-negative bacteria, only limited information is available for Gram-positive and pathogenic species. In this study, we investigated the structural and functional importance of ω, revealing key roles in subunit folding/stability, complex assembly, and maintenance of transcriptional integrity. Collectively, our data underline, for the first time, the importance of ω for RNAP function and cellular harmony in S. aureus.
KEYWORDS: RNA polymerase subunit omega, RpoZ, Staphylococcus aureus, gene regulation
INTRODUCTION
Transcription in all forms of life is a tightly controlled process, necessitated by the essentiality of correct temporal and spatial expression of genes for survival. All transcriptional activity within a cell is maintained by the DNA-dependent RNA polymerase (RNAP). This multiprotein complex is structurally and functionally similar in distant forms of life, displaying only minor variations in composition, e.g., the presence/absence of certain subunits (1, 2). In bacteria RNAP consists of four main subunits, i.e., two α (RpoA) subunits and one subunit each of β (RpoB) and β′ (RpoC), forming the α2ββ′ complex. Together they facilitate transcriptional elongation, but they require a σ factor to initiate the process. Interestingly, a large number of bacteria, and particularly the Firmicutes, possess several other accessory RNAP subunits (3–6). These are considerably smaller than the major subunits, ranging from 8.5 to 21.5 kDa, and include the δ (RpoE), ε (RpoY), and ω (RpoZ) subunits. Deletion of these subunits does not result in lethality for the cell, and thus their diminutive size and nonessential nature have resulted in their being classified as the “small accessory subunits” (reviewed in reference 6). Nevertheless, for the δ factor it has been shown that, in various species, deletion is accompanied by a deregulation of the transcriptional process, leading to decreased fitness and impaired virulence in pathogenic organisms (7, 8). While there is an extensive history of research for δ, ε has only recently been described as an RNAP subunit, with only a single study thus far performed in Bacillus subtilis (5), which suggested a role in phage immunity. While the presence of these two subunits is largely confined to the Firmicutes, homologs of ω, the smallest of the three subunits, can be found not only in bacteria but also in eukaryotes (RPB6) and archaea (RpoK) (6, 9). Although this conservation might suggest a vital role and perhaps similar function across widely different species, there are in fact marked differences in how this subunit influences cells across the various kingdoms (6, 10). Most strikingly is the observation that while it is accessory in bacteria, the ω subunit is essential in eukaryotic organisms (11).
The majority of studies on ω have been carried out in Gram-negative bacteria, with a focus on the model organism Escherichia coli (10). In this organism it has been shown that ω influences the transcriptional machinery, and thus the transcriptional process, in a variety of ways. Most notably, ω is known to interact with β′ to ensure correct folding of the subunit, as well as to facilitate docking to the α2β complex (9, 12–14). Accordingly, deletion of ω leads to misfolding as well as degradation of β′ (13, 15), which is also observed in Mycobacterium smegmatis (16). The crystal structure of ω in complex with the other RNAP subunits of both E. coli and Thermus aquaticus has been solved and confirms the binding of ω to β′ (17, 18). Interestingly, these structures also reveal species-specific differences in the interaction of these two proteins, again highlighting the heterogeneous nature of ω function in different organisms. In contrast, no such crystal structure is available for ω, or RNAP at large, in Gram-positive organisms.
Quite strikingly, in terms of functional difference, is the finding that in E. coli, the stringent response, and thus adaption to nutrient-limiting conditions, is dependent on ω. Specifically, the stringent response-inducing molecule ppGpp is recognized by and binds to the interface of β′ and ω (19–21), which in turn leads to the adjustment of transcriptional profiles to promote survival under nutrient-limiting conditions. While this role is true for E. coli, the ω protein in B. subtilis has been suggested to have no such role in the stringent response, due to an alternative mechanism of ppGpp recognition. Instead of binding to ω, the adaption to limiting conditions in B. subtilis is mediated by ppGpp-induced alterations of GTP concentrations within the cell (22, 23). Subsequently, these changes lead to alterations in gene expression, driven by the sensitivity of certain promoters to GTP availability as an initiating nucleotide. In line with this model, where onset of the stringent response does not require the interaction of ppGpp with the RNAP complex, is the observation that the conserved E. coli residues required for ppGpp binding to RNAP are largely absent in the B. subtilis ω and β′ subunits (20).
The final major function of ω described in the literature is a putative role in facilitating σ factor biding to the RNAP complex. For E. coli and cyanobacteria, it has been reported that depletion of ω can lead to increased binding of alterative σ factors and in turn to increased expression of genes within alternative σ factor regulons (24, 25). Again, structural differences within the ω-depleted RNAP have been implicated in this alteration of σ factor affinity for the complex. As with many of the other ω phenotypes, no in-depth studies have been performed in Gram-positive bacteria regarding this role, further underscoring the need to characterize this diverse protein.
For Gram-positive organisms, only a limited number of studies exist, detailing a few phenotypic effects resulting from the abrogation of ω activity. Indeed, none of these studies have unraveled the molecular basis for alterations in mutant strains, meaning that the role of ω in Gram-positive species is still relatively elusive. Those effects that have been detailed for rpoZ mutants include alterations in cell wall morphology, cell motility, protein secretion, and biofilm formation (16, 26–29). Importantly, the role of this subunit in the virulence of pathogenic species has yet to be evaluated. Therefore, in this study we explored the role of ω in Staphylococcus aureus, demonstrating that it is not involved in the stringent response but instead mediates structural integrity of the RNAP complex. Deletion of this factor leads to individual RNAP subunit degradation and an induction of cellular stress responses. The latter effect was characterized by global transcriptional analyses, revealing that a significant number of the observed changes were due, at least in part, to altered σ factor abundance in the RNAP complex. Finally, we demonstrate that deletion of rpoZ influences the ability of S. aureus to form biofilms, a process that mediates persistent infections and the capacity to resist antibiotic treatment. We suggest that collectively, our data underline the importance of ω for RNAP stability, function, and cellular physiology in S. aureus.
RESULTS
ω is cotranscribed with gmk and is highly expressed throughout growth.
Analysis of RNA sequencing data previously generated by our laboratory (31) reveals that the ω-encoding gene, rpoZ, is highly expressed during all phases of growth and is seemingly cotranscribed with its upstream gene, gmk, encoding a guanylate kinase (see Fig. S2A in the supplemental material). To validate the latter observation, we performed Northern blot analysis on RNA extracted from wild-type S. aureus cells grown to mid-exponential phase using rpoZ-specific probes. In so doing, a band of ∼1100 nucleotides (nt) was observed, indicating that rpoZ and gmk are indeed organized in a bicistronic operon (Fig. S2B). In order to study the role of ω in S. aureus, we next created an unmarked deletion mutant, removing the majority of the rpoZ-encoding gene and leaving gmk intact. Validation of this arrangement and that no deleterious effects were observed on gmk expression was obtained by RNA sequencing analysis of the mutant strain and by Northern blotting (Fig. S2A and B). RNA sequencing data for the ω-depleted strain showed the expected deletion of the rpoZ gene but confirmed that there were no unintended effects on transcript abundance for gmk (see Table S1 in the supplemental material).
The S. aureus ω and β′ proteins lack conserved residues required for ppGpp binding.
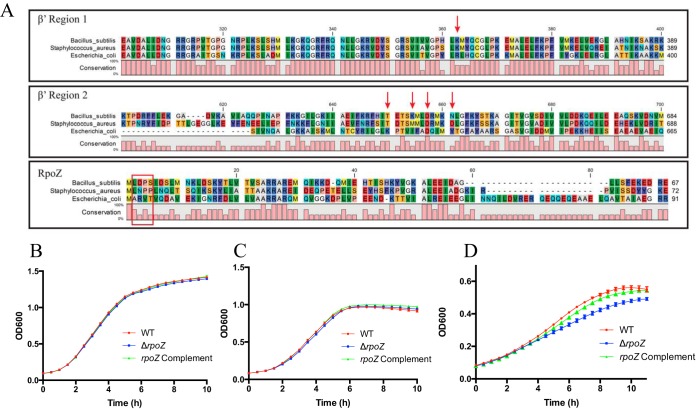
In E. coli the interaction of ppGpp with RNAP is mediated by conserved residues within the ω and β′ subunits, thus controlling the stringent response. Such residues are absent in B. subtilis, suggesting an alternate mechanism for stringent response regulation in Gram-positive organisms (20, 32). In corroboration of this, alignment analysis of ω and β′ subunits from E. coli, B. subtilis, and S. aureus (Fig. 1A) reveals that, although there is partial conservation of the β′ and ω proteins for all three organisms, the residues for ppGpp binding are almost entirely absent (a single aspartic acid is conserved in β′ region 2) at the primary sequence level and in the biochemical characteristics of each amino acid. This finding is further validated by the observation that growth of a ΔrpoZ strain is not impaired under standard, nutrient-limiting, or stringent response-inducing conditions. Specifically, in addition to growth in complex medium (tryptic soy broth [TSB]) (Fig. 1B), we investigated growth during amino acid limitation (Fig. 1C) as well as in amino acid-limiting medium completely depleted of valine and leucine (Fig. 1D). Importantly, the rpoZ mutant showed growth rates similar to those of the wild type in both TSB and amino acid-limiting medium and only a minor growth defect in medium devoid of valine and leucine. This defect, however, suggests an impaired ability to adapt to changing growth conditions rather than a stringent response defect, as stringent response-deficient mutants display a characteristic stalling of growth during the stringent response, rather than delayed growth (33). To further validate this finding, we assessed sensitivity to mupirocin of the rpoZ mutant alongside its parental strain and a codY mutant strain, as it has previously been shown that S. aureus mutants impaired in the stringent response display decreased resistance to this stringent response-inducing antibiotic (34). In so doing, we found that the ΔrpoZ strain has a mupirocin MIC (0.32 μg/ml) similar to that of the wild type, while the codY mutant has a 2-fold decrease in MIC (∼0.16 μg/ml), comparable to that of stringent response-deficient mutants documented elsewhere (34).
FIG 1.
Influence of the S. aureus ω subunit on survival during nutrient-limiting conditions. (A) Alignment of select RNAP subunits from E. coli, B. subtilis, and S. aureus, showing conservation and divergence of amino acids sequences for RpoC (β′) and RpoZ (ω). For E. coli, the conserved β′ (arrows) and ω (boxed) regions important for ppGpp binding are shown. (B to D) S. aureus growth under standard conditions (TSB) (B), in amino acid-limiting medium (CDM) (C), and in CDM completely depleted of valine and leucine (D). Error bars show the standard error of the mean (SEM).
Depletion of rpoZ leads to destabilization of the RNAP complex.
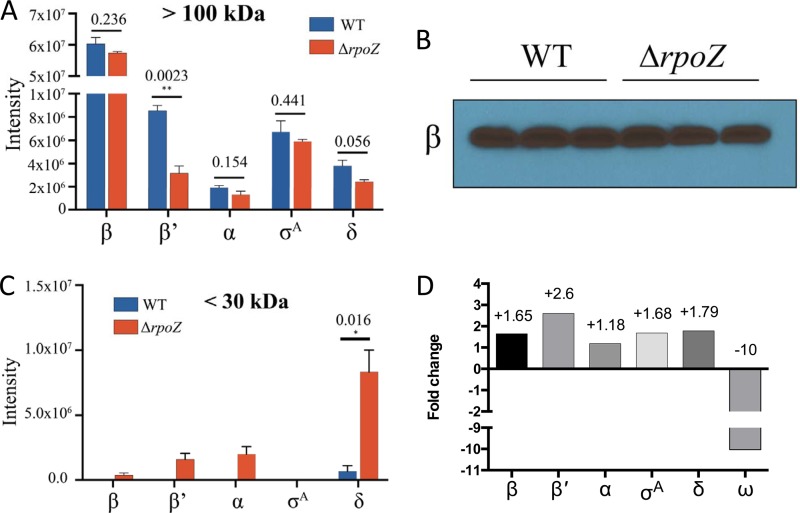
We next assessed whether ω has a role in maintaining RNAP complex integrity in S. aureus and, more precisely, in the folding of its subunits, as described for M. smegmatis and E. coli (13, 16). We isolated cytoplasmic proteins from exponentially growing cells and determined the abundances of the intact RNAP complex in the wild-type and mutant strains. This was achieved by concentrating fractions using a 100-kDa-cutoff filter, ensuring that the resulting retentate harbored only proteins of >100 kDa in size. As β and β′ are the only RNAP subunits larger than 100 kDa, any subunit found in the concentrated fraction must originate from protein in complex with other RNAP subunits. Consequently, β and β′ are expected to be present independently and in complex with RNAP and thus should demonstrate no alteration within the mutant even if complex instability is observed. As expected, we consistently observed equimolar amounts of β for both the wild type and the rpoZ mutant in tested samples (Fig. 2A), which was confirmed by Western blotting using a β-specific antibody (Fig. 2B). Conversely, we found a marked decrease in β′ abundance in the mutant compared to the parent in the same samples. Given the large size of β′, these results point not only toward a release of the subunit from the complex but also to its likely degradation in the absence of ω, as observed in other organisms. Similar to β′, but to a lesser extent, the α subunit, the housekeeping σ factor, σA, and the small subunit δ were also decreased in abundance in the mutant strain. In order to investigate the fate of each subunit, we performed additional concentration experiments using a 30-kDa-cutoff filter and again isolated proteins from the wild-type and rpoZ mutant strains (Fig. 2C). Interestingly, while only trace amounts of β were detected in these fractions (as expected), we observed significant levels of β′ in the rpoZ mutant strain, again suggesting β′ instability upon ω deletion. With regard to δ, which should always be found in the <30-kDa fraction, the amount of this protein was significantly higher in the ΔrpoZ strain than in the wild type. This would tend to indicate disassembly and release of this subunit from the RNAP complex upon ω depletion.
FIG 2.
Cells lacking the ω factor have altered RNAP composition and individual subunit stability. Free or complex-bound subunits were separated according to their molecular masses via size selection before analysis using mass spectrometry. (A) In the >100-kDA fraction only large RNAP subunits (β and β′) and subunits within the RNAP complex should be found. (B) Data were validated by Western blotting using a β subunit antibody. (C) Protein degradation in the ΔrpoZ strain is highlighted by the presence of larger subunits within the <30-kDa fraction. (D) Transcriptional changes as a cause of alterations in RNAP composition were excluded by analyzing expression of RNAP genes from RNA sequencing data sets. Shown are the fold changes for each RNAP gene in the mutant strain compared to the wild type. Where relevant, error bars show SEM. Statistical significance was determined using Student's t test (*, P < 0.05; **, P < 0.01).
To exclude the possibility that alterations in subunit abundance were caused by differential expression patterns for RNAP-encoding genes in the rpoZ-depleted strain, we analyzed RNA sequencing data sets of the USA300 parent and its corresponding rpoZ mutant (Fig. 2D). The rpoZ gene was included as an internal control for our data set, as a portion of the locus is still present in the mutant strain (Fig. S2A), and therefore an expression value is generated from the RNA sequencing experiment. Of note, all of the subunits were shown to have a moderate increase in expression in the mutant strain. Importantly, β′, which displayed the strongest decrease in protein abundance, showed the strongest upregulation, with a 2.6-fold increase in transcription. This perhaps suggests a possible attempt by the cell to counteract β′ degradation by enhancing transcriptional activity of the rpoC gene within the rpoZ mutant strain.
The δ and ω factors of RNAP have distinct and contrasting influences on S. aureus gene expression.
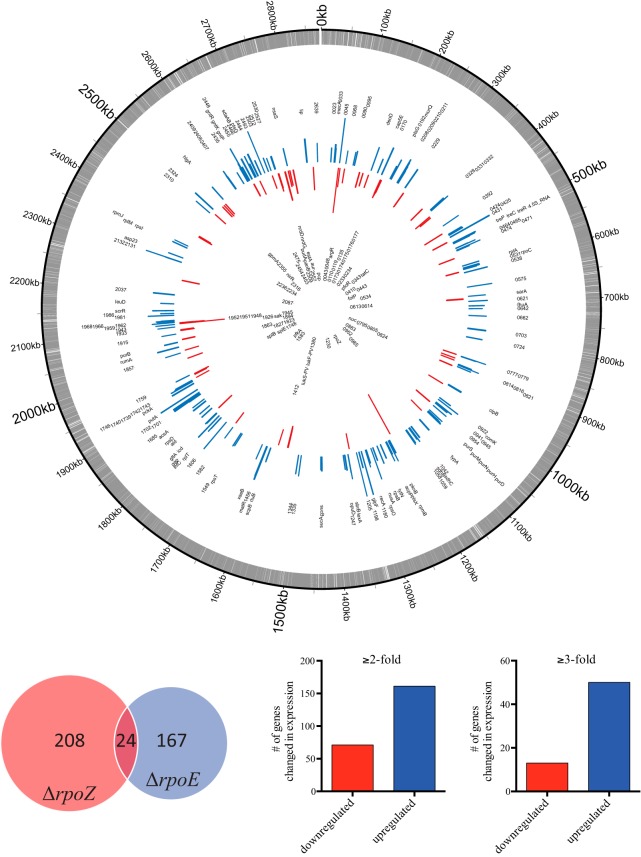
When further exploring the RNA sequencing comparison of the wild-type and rpoZ mutant strains, we observed 232 genes with altered expression at ≥2-fold and 63 genes at ≥3-fold (Fig. 3; Table S1). In order to validate these alterations, we performed quantitative reverse transcription-PCR (RT-qPCR) for a subset of differentially expressed genes (Fig. S3A and B), revealing similar changes in this and the RNA sequencing data sets. Since our RNAP composition studies showed that rpoZ deletion also results in δ depletion from RNAP, we next assessed whether removal of δ from the transcription complex was the driving force behind the observed changes. This is particularly important, as our previous studies have shown that δ is a key factor for maintaining transcriptional specificity within the S. aureus cell (8). Upon comparing RNA sequencing-derived transcriptional changes of the rpoE and rpoZ mutants, we found an overlapping regulon of only 24 genes (Fig. 3). Comparing these 24 genes to the overall number of genes changed in both mutants, only ∼10.5% (ΔrpoZ) or 12.5% (ΔrpoE) of each data set is identical. This low level of regulon overlap indicates that the changes seen in an rpoZ mutant are not the result of an inability of δ to exert its influence on RNAP.
FIG 3.
The δ and ω factors of RNAP have distinct and contrasting influences on S. aureus gene expression. (Top) Genomic map of altered transcription in the rpoZ mutant strain. The outermost circle (gray) represents annotations for the S. aureus USA300 genome. Blue (upregulation) and red (downregulation) bars show genes with differential expression in the rpoZ mutant (a maximum change of ±7-fold is displayed). Labels represent either gene names or USA300 gene numbers. (Bottom right) Number of genes changed in expression upon rpoZ depletion at ≥2-fold (232) or ≥3-fold (63). (Bottom left) Comparison of RNA sequencing data sets for ΔrpoE (δ) and ΔrpoZ (ω) strains. Shown are the number of genes within each regulon and the overlap between the two data sets (only ∼10% was detected).
ω influences σ factor recruitment to the S. aureus RNAP complex.
In E. coli and the cyanobacterium Synechocystis, it is known that depletion of ω causes alterations in σ factor abundance within the RNAP complex. Therefore, we next investigated if the multifaceted transcriptional changes of the rpoZ mutant were influenced by alterations in σ factor binding to the ω-less RNAP. Since σB is the major alternative σ factor in S. aureus, we compared transcriptome changes upon rpoZ deletion to those observed in a sigB mutant. This was facilitated using wild-type and sigB mutant RNA sequencing data sets previously produced in our lab in the SH1000 background. SH1000 was chosen for this study as it is known to have a σB overexpression phenotype. Subsequent comparison of these data sets detected an inverse relationship between the regulons of ω and σB. Specifically, for the 20 most strongly downregulated genes in the sigB mutant, the majority (70%) demonstrated increased expression upon rpoZ disruption (Fig. 4; Tables S2 and S3). Conversely, for the 20 most strongly upregulated genes in the sigB mutant, most (60%) were found to be decreased in expression in the ΔrpoZ strain.
FIG 4.
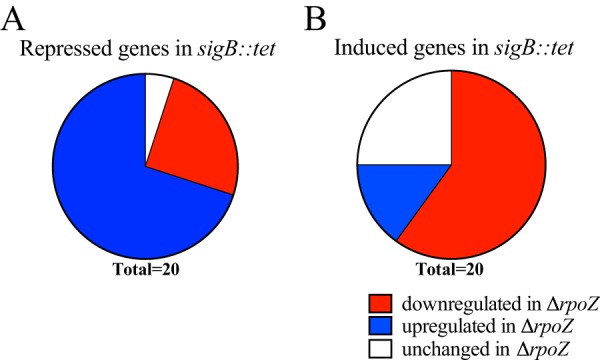
ω affects σ factor recruitment to the S. aureus RNAP complex. A comparison of RNA sequencing data sets from sigB::tet (σB) and ΔrpoZ strains is depicted. Shown are the most highly repressed (A) or highly induced (B) genes in the sigB::tet strain and their concomitant change in expression upon rpoZ deletion. Genes with a change of less than 10% between the rpoZ mutant and its parental strain were considered unchanged.
It is noted that the comparison of regulons between different strains is not without drawbacks. Our rationale was that by choosing a sigB-overexpressing strain such as SH1000, we could obtain better resolution for the identification of σB-dependent genes. Nevertheless, there remains the possibility that additional genomic differences between SH1000 and USA300 drive strain-specific expression patterns and thereby influence their corresponding σB regulons. However, the strongly correlated inverse relationship observed suggests that there are indeed higher levels of σB within the ω-lacking RNAP complex, which corroborates effects seen in other bacterial species. Accordingly, it appears that ω not only is required to maintain RNAP integrity but also plays a role in association of the complex with available σ factors, thereby influencing transcriptional stringency in S. aureus.
Depletion of rpoZ leads to decreased transcriptional specificity that influences multiple cellular processes within the S. aureus cell.
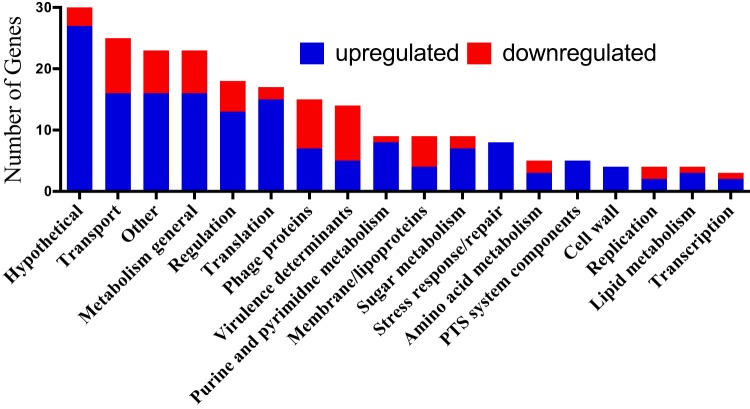
We next used functional clustering to investigate alterations in gene expression upon rpoZ disruption (Fig. 5). While a variety of cellular processes were shown to be affected by rpoZ deletion, we noted major changes in genes related to central dogma. Specifically, genes related to transcription and translation, as well as cellular processes connected to DNA, RNA, or protein synthesis, were strongly affected by the loss of ω. These include changes within purine and pyrimidine metabolism, enzymes for DNA replication and repair, and amino acid biosynthesis. Additionally, genes in more general ontological categories, such as genes for sugar uptake systems (phosphotransferase system [PTS]), the general stress response, regulators, transporters, and metabolism, displayed altered expression. Interestingly, the changes also negatively affect several well-characterized virulence determinants of S. aureus. These include aureolysin (−3.7-fold) and the Panton-Valentine leukocidin-encoding operon (LukSF, −3.4-fold and −3.1-fold, respectively). Lastly, a significant number of phage genes were altered in their expression. This point is particularly interesting as we have previously described this phenomenon to be one of the hallmarks of rpoE deletion (8); thus, these effects could result from its depletion from the RNAP complex. Therefore, although overlap of the δ and ω regulons is limited (as described above) and unlikely to be causative for the majority of transcriptional changes observed in the ΔrpoZ strain, it cannot be entirely excluded that a number of effects seen in the rpoZ mutant are driven, at least in part, by δ depletion from RNAP.
FIG 5.
Ontological grouping for genes altered in expression upon rpoZ disruption. The transcriptional changes from Fig. 3 were analyzed and organized into ontological groups.
Disruption of the ω subunit results in an impaired ability to circumvent multiple forms of environmental stress.
To explore the physiological outcome of these findings, we sought to determine if the mutant was impaired in resisting the impact of environmental stressors. In so doing, we observed that the mutant shows an augmented sensitivity toward various antibacterial compounds with unrelated mechanisms of action. These include increased sensitivity to triclosan (2-fold; a fatty acid biosynthesis inhibitor), erythromycin and pyrogallol (both ∼4-fold; a translation inhibitor and a reducing agent, respectively), and diamide (8-fold; creates disulfide stress). As these compounds all affect different cellular targets, it would appear that depletion of ω leads to widespread fitness defects, highlighting its global importance within the S. aureus cell.
The ω subunit of RNAP is required for biofilm formation in S. aureus.
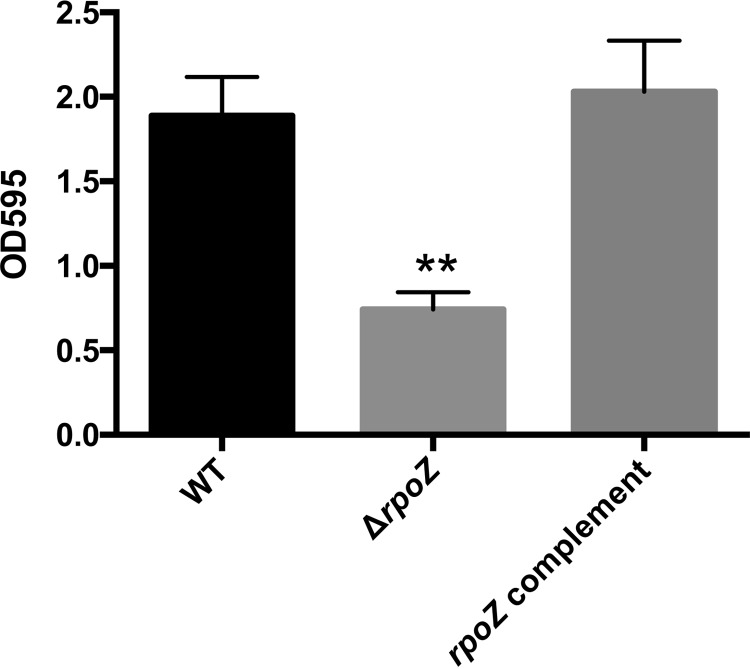
We next set out to investigate whether the observed changes in gene expression influence pathophysiologically relevant processes. Thus, we determined whether the formation of a bacterial biofilm, a key hallmark of S. aureus infection (35), was influenced by rpoZ depletion in vitro. Importantly, upon analysis we observed that loss of ω does indeed lead to an approximately 3-fold reduction in biofilm formation that could be restored via complementation in trans (Fig. 6). As biofilm formation is strongly influenced by extracellular proteins in S. aureus (36), we analyzed the secretomes of the wild-type and rpoZ mutant strains. Although the vast majority of proteins appeared to be unchanged in abundance, we noted a marked decrease in a major protein band at around 75 kDa in the mutant strain (see Fig. S4 in the supplemental material). In order to identify this protein, we excised the corresponding band from gels and performed mass spectrometric (MS) analysis. It was identified as two separate tributyrin lipases (the products of SAUSA300_0320 and SAUSA300_2603), with molecular masses of 76 and 77 kDa, respectively. Interestingly, a recent report on the role of lipase activity in S. aureus demonstrated that deleting these enzymes results in drastically impaired biofilm formation (37). This may provide some explanation for these findings, although given the complex process of biofilm formation in S. aureus, it is likely that other factors are also involved.
FIG 6.
The ω subunit of RNAP is required for biofilm formation in S. aureus. Biofilm formation by the wild-type, rpoZ mutant, and complemented strains was measured using a 24-well plate biofilm assay. Error bars show SEM, with statistical significance measured using Student's t test (**, P < 0.01).
DISCUSSION
The ω subunit of RNAP is a widely distributed protein that is found in all branches of life. While it is well studied in Gram-negative bacteria (10) and eukaryotes (9), a recent review by our group highlighted the need for ω to be examined in a wider range of species, and particularly Gram-positive bacteria (6). This is especially true since (i) the subunit has a variety of possible roles that have received only limited attention in Gram-positive species, including RNAP subunit folding (9, 12, 13, 15) and stability (16), complex assembly/stabilization (10, 13, 14), maintenance of σ factor specificity (24, 25), and ppGpp binding (19–21), and (ii) Gram-positive species, and particularly the Firmicutes, include several epidemiologically relevant human pathogens. To the latter point, an understanding of cellular factors that are required for transcription and to maintain transcriptional stringency is crucial in order to better comprehend the physiology of pathogens and thus potentially aid in counteracting and preventing the spread of disease. Here we describe the role of ω within S. aureus for the first time. Our study explores previously documented functions from other bacterial species, assessing its impact on the global transcriptome as well as its key role in fitness and, most importantly, virulence, which to our knowledge has not yet been examined.
It has previously been suggested that the stringent response in Firmicutes is independent of ppGpp recognition by ω (32). Here we demonstrate that under amino acid-limiting conditions, as well as during exposure to stringent response-inducing compounds, the ΔrpoZ strain does not exhibit typical stringent response deficiency phenotypes. This is consistent with findings in B. subtilis, where ppGpp, instead of directly binding to ω as in Gram-negative species, exerts its function indirectly. Instead, the generation of ppGpp from GTP and ATP (38) in B. subtilis leads to diminished GTP pools within the cell (39, 40), which in turn causes decreased transcription of GTP-sensitive promoters (i.e., those with GTP at position +1 [22, 41, 42]) and interferes with the role of GTP as a corepressor for CodY (43). These findings are mirrored in S. aureus, where CodY- and GTP-regulated genes are similarly influenced by the nutritional status of the cell in a ppGpp-dependent manner (33, 44). Therefore, we hypothesize that rather than resulting from a dysfunctional stringent response, the diminished ability of a ΔrpoZ strain to survive challenging conditions is a consequence of basal stress levels experienced by the cell upon loss of the subunit. This is supported by our findings that S. aureus lacking the rpoZ gene displays a reduced-growth phenotype not only when exposed to nutrient-limiting and stringent response-inducing conditions but also during growth at elevated temperatures (data not shown).
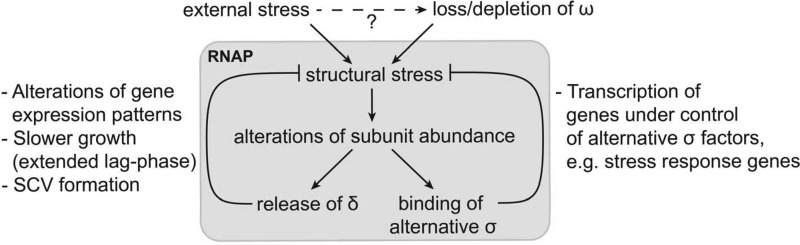
To investigate how a lack of ω affects the transcriptional process and integrity of RNAP, we performed complex stability analysis and determined that rpoZ deletion causes decreased levels of intact RNAP, perhaps as a result of reduced β′ within the complex. This is consistent with findings in other organisms, which describe a chaperone-like function for ω, assisting in the folding of β′ and its subsequent docking to RNAP (9, 10, 12–15). Indeed, the likely misfolding of β′ leads not only to degradation of the subunit but also to impaired assembly and/or dissolution of RNAP and the proteolysis of several other subunits. Importantly, in addition to this apparent failure of β′ to fully associate with RNAP, ω deletion leads to a jettisoning of δ from the transcription complex and an increase in alternative σ factor binding. Indeed, the finding that δ and σA are preferentially released from ω-less, and presumably structurally unstable, RNAP suggests that ω-deficient RNAP can provide a unique insight into the effects of stress on the transcription complex and, more broadly, the role of the accessory subunits. Based on the current literature, as well as our findings here, we propose a model whereby stress within the cell leads to RNAP instability, resulting in an alteration of subunits present within the transcription complex that specifically facilitates survival during unfavorable conditions (Fig. 7). In support of this, those sigma factors that have increased binding to RNAP upon ω deletion (σS in E. coli [24], σB/σF in cyanobacteria [25], and σB in S. aureus) are all major components of the general stress response in their respective organism (45–49). Furthermore, as with σ factors, the release of δ from RNAP has the potential for profound alterations in gene expression, as a recent publication demonstrates the ability of δ to selectively affect individual promoters (7, 8) via binding to specific promoter elements (50). As the disruption of rpoE results in slower bacterial growth in a variety of organisms (e.g., an extended lag phase [8, 51, 52]), removal of this subunit from RNAP may actually present a survival advantage during stress, redirecting resources away from division and toward repair and cellular maintenance. In agreement with this is a recent study with Streptococcus pneumoniae biofilms, where there appears to be selective pressure for rpoE mutations leading to small-colony variant (SCV) phenotypes (53). SCV formation is a common mechanism used by many different bacteria which would initially appear to present a defect in viability but in actuality results in enhanced survival (54) and antibiotic resistance and the development of persister cells (55). Interestingly, our group has observed similar effects in S. aureus, where δ-deficient mutants display variable, and decreased, colony size after recovery from murine models of infection (unpublished observation). Thus, it would appear that the significant compositional rearrangement within ω-less RNAP is far from random. Rather, these changes in binding of accessory factors are seemingly directed to specifically adjust promoter selectivity and preference to facilitate survival. Furthermore, these findings regarding subunit swapping within the transcription complex appear to confirm long-made contentions regarding a level of cooperativity for δ and σ factor binding within RNAP (56–58).
FIG 7.
Model for the role of ω in the Gram-positive RNA polymerase.
In spite of these changes in subunit abundance, which we suggest are utilized by the cell as a survival strategy, rpoZ deletion itself results in wide-reaching cellular stress that is associated with an impaired transcriptional process. This contention is supported by our RNA sequencing data, which demonstrate the upregulation of various genes coding for transcriptional and translational factors, putatively in an attempt to overcome diminished or inefficient RNAP function. Furthermore, we identified increased expression of genes involved in the general stress response, sugar uptake pathways, and energy metabolism, all of which may be designed to overcome unfavorable conditions (59). The latter group emphasizes that intracellular stress resulting from ω deletion, and the subsequent countermeasures it elicits (e.g., production of chaperones, RNAP subunits, and ribosomal subunits), consumes energy and therefore has the potential to deplete cellular energy pools. This in turn leads to the upregulation of sugar uptake and energy metabolism, which in our study resulted in a minor increase in ATP pools (data not shown). In accordance with this, studies investigating S. aureus heat stress demonstrate that following exposure to elevated temperatures, cellular energy levels are in fact not depleted, but rather a steady (60) or transient (61) increase in ATP pools is observed, presumably due to the upregulation of energy-generating processes. It is perhaps not surprising that the transcriptional and physiological changes observed from our in vitro experiments also translate into the impairment of virulence-related processes, as shown by the impaired biofilm formation in the rpoZ mutant.
We suggest that, collectively, our findings extend the existing knowledge regarding ω and its role within RNAP, provide a unique insight into structural rearrangements within the transcription complex in response to stress, and demonstrate the necessity of this important subunit for the cellular homeostasis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains used in this study are listed in Table 1. Unless otherwise indicated, cultures were grown as described by Shaw et al. (62). Briefly, S. aureus was grown at 37°C with shaking (250 rpm) in tryptic soy broth (TSB) or chemically defined medium (CDM) (45), while E. coli was cultivated in lysogeny broth (LB) with agitation (250 rpm) at 37°C. Overnight cultures of S. aureus were seeded at a 1:100 dilution into fresh medium. After 3 h of growth, cultures were used to inoculate fresh TSB to an optical density at 600 nm (OD600) of 0.05 and grown to the desired density in a Synergy2 plate reader (BioTek) in 96-well plates.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Description or sequencea | Reference or source |
|---|---|---|
| Species and strains | ||
| E. coli DH5α | Cloning strain | 79 |
| S. aureus | ||
| RN4220 | Restriction-deficient transformation recipient | Lab Stocks |
| USA300 HOU | MRSA isolate cured of pUSA300-HOU-MRSA | 72 |
| HKM850 | USA300 HOU codY mutant; codY::tet | 75 |
| SH1000 | 8325-4 derivate with functional rsbU; rsbU+ | 80 |
| MJH502 | SH1000 sigB mutant; sigB::tet | 80 |
| BM2393 | USA300 HOU ΔrpoZ mutant | This study |
| AW2394 | USA300 HOU ΔrpoZ/pMK4::rpoZ rpoZ+ | This study |
| Plasmids | ||
| pMK4 | Gram-positive shuttle vector | 81 |
| pJB38 | Counterselectable plasmid to create silent mutations in S. aureus | 63 |
| Primers | ||
| OL2889 | ATGGAGCTCGCCATAATTTATCTTCCACCTTC | This study |
| OL2890 | ATGACGCGTGCAGTTGTTGCAATTAAATAC | This study |
| OL2891 | ATGACGCGTGTTTCGACCATTAAAAATATGTG | This study |
| OL2892 | ATGGGTACCCATTTCTTCAGCACTTTGAAC | This study |
| OL2991 | GCGTGAAATTGATGAACAAC | This study |
| OL2992 | CAGCAATTTCTTCTAACGCTC | This study |
| OL3008 | AGAAGATTAGCTTAGAGAGGTC | This study |
| OL3009 | GAAATGCTAATGGTGTCACA | This study |
| OL3377 | CGTCAGCAATTTCTTCTAACGCTCTACCAAC | This study |
| OL3753 | ATGGGATCCATCGCCAATTTCATCTTT | This study |
| OL3807 | ATGGGATCCATCTCCCTTAAATATCACTATG | This study |
| OL2393 | TCGTATGTTGTGTGGAATTG | This study |
| OL2394 | GTGCTGCAAGGCGATTAAG | This study |
| OL1297 | GTGGTGGCGTTTGTGC | This study |
| OL1298 | CCGACAGCAAACACACCCAT | This study |
| OL2129 | TAACATTCTCTGATGAAGTTG | This study |
| OL2130 | TTAAGTCTACAGCAGCTTG | This study |
| OL3136 | TCAATATTATCGGTGGATTTA | This study |
| OL3137 | GCAAATTATGATGTTGAAATAG | This study |
| OL3747 | CGTAGATGCAAATTATTACG | This study |
| OL3748 | CGTTAATGAAACAATTGGAC | This study |
| OL4099 | GGTTCACGTTCCTTTATC | This study |
| OL4100 | CAGGTTTACCATCTTTAGG | This study |
| OL4109 | GTTAAATGGTTTAATGCAGAA | This study |
| OL4110 | GTATCCATCTTCAGCGA | This study |
MRSA, methicillin-resistant S. aureus. Underlining denotes restriction sites.
Mutant construction.
In order to investigate the role of the ω subunit, a strain carrying a markerless deletion of the rpoZ gene was created, using a method based on the plasmid pJB38 as outlined by Bose and coworkers (63). Briefly, the majority of the rpoZ gene was removed via allelic replacement, as initially described elsewhere (64). To do this, both the 5′ and 3′ ends of the rpoZ gene, together with ∼1 kb of up- and downstream DNA, were PCR amplified using primers OL2889 to OL2892 and fused together using MluI sites contained in the amplification primers. This was then cloned into pJB38 using SacI and KpnI sites in primers OL2889 and OL2892. Following several selection and counterselection steps in S. aureus as described previously (63), rpoZ deletion was confirmed using internal (OL2991 and OL2992) and external (OL3008 and OL3009) primer pairs relative to the deleted region within the gene.
Complement construction.
In order to exclude that secondary mutations were causative for phenotypic changes observed in the ΔrpoZ strain, a plasmid was constructed to complement the rpoZ gene in trans. Here, the complete rpoZ operon, including its native promoter, was PCR amplified using primers OL3753 and OL3807. After BamHI digestion of the amplicon and the pMK4 shuttle vector, the PCR fragment and plasmid were ligated together and transformed into chemically competent E. coli DH5α. The plasmid isolated from ampicillin-resistant clones was confirmed via Sanger sequencing (using standard M13 primers). Correct plasmids were transformed into electrocompetent S. aureus RN4220 (65) and finally transduced into the S. aureus USA300 rpoZ mutant using ϕ11 (66).
RNA isolation and Northern blots.
The detection of RNA transcripts was performed using a Northern blotting protocol outlined by Caswell et al. (67). Briefly, RNA from cultures grown for 3 h was isolated using an RNeasy kit (Qiagen), with DNA removed using a Turbo DNA-free kit (Ambion). RNA concentrations were determined using an Agilent 2100 Bioanalyzer and an RNA 6000 Nano kit (Agilent). RNA (15 μg) was separated by gel electrophoresis (10% polyacrylamide containing 7 M urea and 1× Tris-borate-EDTA [TBE]) before being transferred to an Amersham Hybond N+ membrane (GE Healthcare) by electroblotting. In order to cross-link samples, membranes were exposed to UV radiation. Following this, membranes were prehybridized (1 h, 43°C) in ULTRAhyb-Oligo buffer (Ambion) and incubated (16 h, 43°C) with a [γ-32]ATP-end-labeled oligonucleotide (OL3377) specific to the rpoZ target sequence. Labeling was carried out using T4 polynucleotide kinase (Thermo Scientific) according to the manufacturer's protocol. After overnight incubation, each membrane was washed (30 min, 43°C) with decreasing (2×, 1×, and 0.5×) concentrations of SSC buffer (300 mM sodium chloride, 30 mM sodium citrate). Finally, membranes were exposed to X-ray film in order to detect the size and abundance of the rpoZ transcript.
RNA sequencing and data analyses.
RNA sequencing was performed for rpoZ and sigB mutants and their respective parental strains as previously outlined by our group (8, 68–70). Briefly, RNA was isolated and the quality determined as described for Northern blot experiments. Following this, RNA from three biological replicates was pooled in equimolar amounts and rRNA removed by consecutive treatment with the MICROBExpress (Ambion) and RiboZero (Epicentre) kits. Removal of rRNA was confirmed using an Agilent 2100 Bioanalyzer (RNA 6000 Nano kit; Agilent). The rRNA-depleted RNA samples were prepared for sequencing with an Ion Personal Genome Machine (PGM) system as described previously (68). First, cDNA libraries were constructed with an Ion Total RNA-seq kit, v2 (Ion Torrent). The prepared libraries were then used to generate template-positive ion sphere particles (ISPs) using an Ion PGM Template OT2 200 kit (Ion Torrent) in combination with an Ion OneTouch 2 system (Ion Torrent). The template-positive ISPs were loaded onto Ion 318 v2 chips (Ion Torrent) and sequencing runs performed with an Ion PGM Sequencing 200 kit, v2 (Ion Torrent). Raw data files in fastq format were exported and analyzed using the CLC Genomics Workbench software (Qiagen) and the USA300_FPR3757 and NCTC 8325 reference genomes (GenBank accession numbers CP000255 and NC_007795, respectively). Expression values for each gene were calculated as reads per kilobase per million mapped reads (RPKM), and a quantile normalization approach was applied (71) with a lower limit of 10 RPKM. Genes that displayed fold changes of ≥2 when comparing expression in the mutant to that in the wild-type strain were included in further analyses.
RT-qPCR.
In order to confirm expression values that were generated by RNA sequencing, a subset of genes were chosen to confirm transcriptional changes via quantitative reverse transcription-PCR (RT-qPCR), as described by us previously (8, 69, 72, 73). PCR amplification was performed using primers for each gene (Table 1) (glpF, OL1297/OL1298; asp23, OL2129/OL2130; SAUSA300_0174, OL3136/OL3137; aur, OL3747/OL3748; rpoC, OL4099/OL4100; and SAUSA300_0777, OL4109/OL4110), alongside 16S rRNA-specific primers as standard controls (OL1184/OL1185) (74).
Investigation of RNA polymerase composition.
In order to determine the effects of ω loss on the stability and composition of RNAP, we adopted an approach first outlined by Gunnelius et al. (an overview of this procedure can be found in Fig. S1 in the supplemental material) (25). Wild-type and ΔrpoZ strains were grown to exponential phase, and their cytoplasmic protein fractions were isolated as outlined previously (75). In order to differentiate between RNAP subunits unbound or within the RNAP complex, we performed two size selection steps. To do so, we first added 10 ml of phosphate-buffered saline (PBS) to the isolated protein fraction and loaded the complete mixture onto an Amicon Ultra-15 centrifugal filter unit (EMD Millipore) with a 100-kDa cutoff. After centrifugation (4,000 × g, 45 min), the approximately 300 μl of remaining protein fraction was washed with 10 ml of PBS and the centrifugation step repeated. The fraction on top of the filter was then recovered and stored (−80°C) for further analysis. The complete flowthrough of the first filter step was then applied to a second Amicon Ultra-15 centrifugal filter unit (EMD Millipore) with a 30-kDa cutoff. The flowthrough was collected and trichloroacetic acid (TCA) precipitated (overnight, 4°C). The pelleted proteins were washed three times in ice-cold absolute ethanol and subsequently resuspended in PBS. The different fractions were then subjected to either mass spectrometry or Western blot analysis.
Characterization of protein content using mass spectrometry.
Protein extracts isolated via the size selection protocol outlined above were processed by filter-aided sample preparation (FASP), as described by us previously (73, 75). Proteins were reduced with dithiothreitol (DTT), alkylated with iodoacetamide (IAA), and digested with trypsin–Lys-C (Promega) overnight at 37°C. Peptides were collected by centrifugation, desalted using Pierce SPE C18 columns with a Supelco vacuum manifold, and dried in a vacuum concentrator (Labconco). Peptides were resuspended in H2O–0.1% formic acid and separated using a 75-μm by 10-cm C18 reversed-phase high-pressure liquid chromatography (HPLC) column (New Objective) on a NanoLC Ultra (Eksigent) with a 120-min gradient (4 to 40% acetonitrile [ACN] with 0.1% formic acid). Fractions were analyzed on a linear ion trap Orbitrap instrument (Orbitrap XL; Thermo Fisher Scientific), with full MS survey scans acquired at 60,000 resolution. The top 10 most abundant ions were selected for tandem MS (MS/MS) analysis in the linear ion trap. Raw data files were processed in MaxQuant (www.maxquant.org) and searched against the UniProtKB S. aureus USA300 protein sequence database. Search parameters included constant modification of cysteine by carbamidomethylation and the variable modification methionine oxidation. Proteins were identified using filtering criteria of 1% protein and peptide false-discovery rates. Protein levels were normalized to the overall protein content in each of the investigated fractions.
Western blotting.
The assessment of β subunit abundance was performed as described by us previously (66). Size-selected protein fractions containing proteins and protein complexes of >100 kDa were prepared as for experiments determining RNAP composition. Samples were separated using 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), as described by us previously (76). Separated proteins were then transferred to a polyvinylidene difluoride (PVDF) membrane and detected using a monoclonal mouse RNAP β antibody (8RB13; Santa Cruz Biotechnology) and a horseradish peroxidase (HRP)-conjugated secondary antibody. HRP activity was assessed using the SuperSignal West Pico substrate (Thermo Fisher Scientific) and visualized on X-ray film.
MIC assessment assay.
To determine whether transcriptional changes observed within the mutant strain resulted in detectable physiological sensitivities, we assessed MICs for various antibacterial agents, as described by our group previously (77). Briefly, overnight cultures were diluted 1:1,000 into fresh medium in 96-well plates, and antibiotics at various concentrations were added at amounts no greater than 2% of the final volume. Plates were incubated for 12 h (static, 37°C), and turbidity, as a sign of bacterial growth, was manually assessed. MICs were defined as the minimum concentration of a given agent to result in no detectable turbidity in individual wells.
Secretome analysis.
To evaluate alterations in secreted proteins, bacteria were grown for 24 h before being centrifuged (6,000 × g, 10 min). Supernatants were removed and subjected to 12% SDS-PAGE as described by us previously (76). Gels were assessed either by silver staining using a Pierce silver stain kit (Thermo Fisher Scientific) according to the manufacturer's instructions or by Coomassie brilliant blue staining, in-gel trypsin digestion, and mass spectrometric analysis, as described by us previously (76). Briefly, gel pieces were minced and destained before reduction and alkylation with dithiothreitol (DTT) and iodoacetamide (IAA), respectively. Proteins were digested with trypsin–Lys-C overnight at 37°C, and peptides were extracted using 50:50 ACN and H2O–0.1% formic acid and dried in a vacuum concentrator (Labconco). Peptides were resuspended in H2O–0.1% formic acid for LC-MS/MS analysis, which was performed as described above.
Biofilm assay.
The ability of strains to form biofilms was assessed as described by us previously (78). The wells of non-tissue-culture treated 12-well polystyrene plates were incubated for 48 h (static, 4°C) with 1 ml of human serum in order to facilitate attachment of cells. Concurrently, bacteria were cultured overnight in biofilm medium (BFM) (TSB supplemented with 0.5% [wt/vol] dextrose and 3% [wt/vol] NaCl) and used to seed fresh BFM to an OD600 of 0.05. After removal of the human serum from the wells of plates, 1 ml of these fresh cultures was transferred into each well and incubated for 48 h (static, 37°C). After incubation, supernatants were gently removed from each well and the biofilm washed twice with PBS before fixation with absolute ethanol. Biofilms were stained with crystal violet solution (2% [wt/vol]) for 10 min, followed by two more rounds of washing with PBS. Residual supernatants were aspirated, and plates were dried overnight. After this time, 300 μl of absolute ethanol was added to the wells and incubated for 10 min before being removed, and the absorbance was measured at 570 nm using a Synergy2 plate reader (BioTek).
Accession number(s).
All data sets have been deposited to the NCBI Gene Expression Omnibus (GEO) (accession numbers GSE87033 and GSE87036).
Supplementary Material
ACKNOWLEDGMENT
This study was supported in part by grant AI080626 (to L.N.S.) from the National Institute of Allergy and Infectious Diseases.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00459-16.
REFERENCES
- 1.Sekine S, Tagami S, Yokoyama S. 2012. Structural basis of transcription by bacterial and eukaryotic RNA polymerases. Curr Opin Struct Biol 22:110–118. doi: 10.1016/j.sbi.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Ebright RH. 2000. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol 304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 3.Burgess RR. 1969. Separation and characterization of the subunits of ribonucleic acid polymerase. J Biol Chem 244:6168–6176. [PubMed] [Google Scholar]
- 4.Pero J, Nelson J, Fox TD. 1975. Highly asymmetric transcription by RNA polymerase containing phage-SP01-induced polypeptides and a new host protein. Proc Natl Acad Sci U S A 72:1589–1593. doi: 10.1073/pnas.72.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller AN, Yang X, Wiedermannová J, Delumeau O, Krásný L, Lewis PJ. 2014. ε, a new subunit of RNA polymerase found in Gram-positive bacteria. J Bacteriol 196:3622–3632. doi: 10.1128/JB.02020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss A, Shaw LN. 2015. Small things considered: the small accessory subunits of RNA polymerase in Gram-positive bacteria. FEMS Microbiol Rev 39:541–554. doi: 10.1093/femsre/fuv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue X, Tomasch J, Sztajer H, Wagner-Dobler I. 2010. The delta subunit of RNA polymerase, RpoE, is a global modulator of Streptococcus mutans environmental adaptation. J Bacteriol 192:5081–5092. doi: 10.1128/JB.00653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss A, Ibarra JA, Paoletti J, Carroll RK, Shaw LN. 2014. The delta subunit of RNA polymerase guides promoter selectivity and virulence in Staphylococcus aureus. Infect Immun 82:1424–1435. doi: 10.1128/IAI.01508-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. 2001. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc Natl Acad Sci U S A 98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew R, Chatterji D. 2006. The evolving story of the omega subunit of bacterial RNA polymerase. Trends Microbiol 14:450–455. doi: 10.1016/j.tim.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Nouraini S, Archambault J, Friesen JD. 1996. Rpo26p, a subunit common to yeast RNA polymerases, is essential for the assembly of RNA polymerases I and II and for the stability of the largest subunits of these enzymes. Mol Cell Biol 16:5985–5996. doi: 10.1128/MCB.16.11.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gentry DR, Burgess RR. 1993. Cross-linking of Escherichia coli RNA polymerase subunits: identification of beta′ as the binding site of omega. Biochemistry 32:11224–11227. doi: 10.1021/bi00092a036. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh P, Ishihama A, Chatterji D. 2001. Escherichia coli RNA polymerase subunit omega and its N-terminal domain bind full-length beta′ to facilitate incorporation into the alpha2beta subassembly. Eur J Biochem 268:4621–4627. doi: 10.1046/j.1432-1327.2001.02381.x. [DOI] [PubMed] [Google Scholar]
- 14.Ganguly A, Chatterji D. 2011. Sequential assembly of an active RNA polymerase molecule at the air-water interface. Langmuir 27:3808–3814. doi: 10.1021/la200225t. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh P, Ramakrishnan C, Chatterji D. 2003. Inter-subunit recognition and manifestation of segmental mobility in Escherichia coli RNA polymerase: a case study with omega-beta′ interaction. Biophys Chem 103:223–237. doi: 10.1016/S0301-4622(02)00271-5. [DOI] [PubMed] [Google Scholar]
- 16.Mathew R, Ramakanth M, Chatterji D. 2005. Deletion of the gene rpoZ, encoding the omega subunit of RNA polymerase, in Mycobacterium smegmatis results in fragmentation of the beta′ subunit in the enzyme assembly. J Bacteriol 187:6565–6570. doi: 10.1128/JB.187.18.6565-6570.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami KS. 2013. X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme. J Biol Chem 288:9126–9134. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. 1999. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98:811–824. doi: 10.1016/S0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 19.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. 2013. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res 41:6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. 2013. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo Y, Wang Y, Steitz TA. 2013. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell 50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krasny L, Gourse RL. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J 23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. 2012. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol cell 48:231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geertz M, Travers A, Mehandziska S, Sobetzko P, Chandra-Janga S, Shimamoto N, Muskhelishvili G. 2011. Structural coupling between RNA polymerase composition and DNA supercoiling in coordinating transcription: a global role for the omega subunit? mBio 2:e00034-11. doi: 10.1128/mBio.00034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunnelius L, Hakkila K, Kurkela J, Wada H, Tyystjarvi E, Tyystjarvi T. 2014. The omega subunit of the RNA polymerase core directs transcription efficiency in cyanobacteria. Nucleic Acids Res 42:4606–4614. doi: 10.1093/nar/gku084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos-Beneit F, Barriuso-Iglesias M, Fernandez-Martinez LT, Martinez-Castro M, Sola-Landa A, Rodriguez-Garcia A, Martin JF. 2011. The RNA polymerase omega factor RpoZ is regulated by PhoP and has an important role in antibiotic biosynthesis and morphological differentiation in Streptomyces coelicolor. Appl Environ Microbiol 77:7586–7594. doi: 10.1128/AEM.00465-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojima I, Kasuga K, Kobayashi M, Fukasawa A, Mizuno S, Arisawa A, Akagawa H. 2002. The rpoZ gene, encoding the RNA polymerase omega subunit, is required for antibiotic production and morphological differentiation in Streptomyces kasugaensis. J Bacteriol 184:6417–6423. doi: 10.1128/JB.184.23.6417-6423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kojima I, Kasuga K, Kobayashi M, Tsuchiya KS, Ikeno S. 2006. The biosynthesis of kasugamycin, an antibiotic against rice blast disease, with particular reference to the involvement of rpoZ, a gene encoding RNA polymerase omega subunit. Int J Soc Mat Eng Res 14:28–32. doi: 10.5188/ijsmer.14.28. [DOI] [Google Scholar]
- 29.Mathew R, Mukherjee R, Balachandar R, Chatterji D. 2006. Deletion of the rpoZ gene, encoding the omega subunit of RNA polymerase, results in pleiotropic surface-related phenotypes in Mycobacterium smegmatis. Microbiology 152:1741–1750. doi: 10.1099/mic.0.28879-0. [DOI] [PubMed] [Google Scholar]
- 30.Reference deleted.
- 31.Weiss A, Broach WH, Shaw LN. 2016. Characterizing the transcriptional adaptation of Staphylococcus aureus to stationary phase growth. Pathog Dis 74:ftw046. doi: 10.1093/femspd/ftw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geiger T, Francois P, Liebeke M, Fraunholz M, Goerke C, Krismer B, Schrenzel J, Lalk M, Wolz C. 2012. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog 8:e1003016. doi: 10.1371/journal.ppat.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geiger T, Goerke C, Fritz M, Schafer T, Ohlsen K, Liebeke M, Lalk M, Wolz C. 2010. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect Immun 78:1873–1883. doi: 10.1128/IAI.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. 2011. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lister JL, Horswill AR. 2014. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol 4:178. doi: 10.3389/fcimb.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu C, Xiong N, Zhang Y, Rayner S, Chen S. 2012. Functional characterization of lipase in the pathogenesis of Staphylococcus aureus. Biochem Biophys Res Commun 419:617–620. doi: 10.1016/j.bbrc.2012.02.057. [DOI] [PubMed] [Google Scholar]
- 38.Nanamiya H, Kasai K, Nozawa A, Yun CS, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. 2008. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol 67:291–304. [DOI] [PubMed] [Google Scholar]
- 39.Lopez JM, Dromerick A, Freese E. 1981. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J Bacteriol 146:605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochi K, Kandala J, Freese E. 1982. Evidence that Bacillus subtilis sporulation induced by the stringent response is caused by the decrease in GTP or GDP. J Bacteriol 151:1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sojka L, Kouba T, Barvik I, Sanderova H, Maderova Z, Jonak J, Krasny L. 2011. Rapid changes in gene expression: DNA determinants of promoter regulation by the concentration of the transcription initiating NTP in Bacillus subtilis. Nucleic Acids Res 39:4598–4611. doi: 10.1093/nar/gkr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krasny L, Tiserova H, Jonak J, Rejman D, Sanderova H. 2008. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol Microbiol 69:42–54. doi: 10.1111/j.1365-2958.2008.06256.x. [DOI] [PubMed] [Google Scholar]
- 43.Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 15:1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan PF, Foster SJ, Ingham E, Clements MO. 1998. The Staphylococcus aureus alternative sigma factor sigmaB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol 180:6082–6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollari M, Gunnelius L, Tuominen I, Ruotsalainen V, Tyystjarvi E, Salminen T, Tyystjarvi T. 2008. Characterization of single and double inactivation strains reveals new physiological roles for group 2 sigma factors in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 147:1994–2005. doi: 10.1104/pp.108.122713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huckauf J, Nomura C, Forchhammer K, Hagemann M. 2000. Stress responses of Synechocystis sp. strain PCC 6803 mutants impaired in genes encoding putative alternative sigma factors. Microbiology 146:2877–2889. doi: 10.1099/00221287-146-11-2877. [DOI] [PubMed] [Google Scholar]
- 49.Kullik II, Giachino P. 1997. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol 167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 50.Prajapati RK, Sengupta S, Rudra P, Mukhopadhyay J. 2016. Bacillus subtilis delta factor functions as a transcriptional regulator by facilitating the open complex formation. J Biol Chem 291:1064–1075. doi: 10.1074/jbc.M115.686170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez de Saro FJ, Yoshikawa N, Helmann JD. 1999. Expression, abundance, and RNA polymerase binding properties of the delta factor of Bacillus subtilis. J Biol Chem 274:15953–15958. doi: 10.1074/jbc.274.22.15953. [DOI] [PubMed] [Google Scholar]
- 52.Jones AL, Needham RH, Rubens CE. 2003. The delta subunit of RNA polymerase is required for virulence of Streptococcus agalactiae. Infect Immun 71:4011–4017. doi: 10.1128/IAI.71.7.4011-4017.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Churton NW, Misra RV, Howlin RP, Allan RN, Jefferies J, Faust SN, Gharbia SE, Edwards RJ, Clarke SC, Webb JS. 2016. Parallel evolution in Streptococcus pneumoniae biofilms. Genome Biol Evol 8:1316–1326. doi: 10.1093/gbe/evw072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mirani ZA, Aziz M, Khan SI. 2015. Small colony variants have a major role in stability and persistence of Staphylococcus aureus biofilms. J Antibiot 68:98–105. doi: 10.1038/ja.2014.115. [DOI] [PubMed] [Google Scholar]
- 55.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 56.Achberger EC, Whiteley HR. 1981. The role of the delta peptide of the Bacillus subtilis RNA polymerase in promoter selection. J Biol Chem 256:7424–7432. [PubMed] [Google Scholar]
- 57.Williamson VM, Doi RH. 1978. Delta factor can displace sigma factor from Bacillus subtilis RNA polymerase holoenzyme and regulate its initiation activity. Mol Gen Genet 161:135–141. [DOI] [PubMed] [Google Scholar]
- 58.Hyde EI, Hilton MD, Whiteley HR. 1986. Interactions of Bacillus subtilis RNA polymerase with subunits determining the specificity of initiation. Sigma and delta peptides can bind simultaneously to core. J Biol Chem 261:16565–16570. [PubMed] [Google Scholar]
- 59.Kultz D. 2005. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol 67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- 60.Fleury B, Kelley WL, Lew D, Gotz F, Proctor RA, Vaudaux P. 2009. Transcriptomic and metabolic responses of Staphylococcus aureus exposed to supra-physiological temperatures. BMC Microbiol 9:76. doi: 10.1186/1471-2180-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pane-Farre J, Jonas B, Forstner K, Engelmann S, Hecker M. 2006. The sigmaB regulon in Staphylococcus aureus and its regulation. Int J Med Microbiol 296:237–258. doi: 10.1016/j.ijmm.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 62.Shaw LN, Lindholm C, Prajsnar TK, Miller HK, Brown MC, Golonka E, Stewart GC, Tarkowski A, Potempa J. 2008. Identification and characterization of sigmaS, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS One 3:e3844. doi: 10.1371/journal.pone.0003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 65.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carroll RK, Robison TM, Rivera FE, Davenport JE, Jonsson IM, Florczyk D, Tarkowski A, Potempa J, Koziel J, Shaw LN. 2012. Identification of an intracellular M17 family leucine aminopeptidase that is required for virulence in Staphylococcus aureus. Microbes Infect 14:989–999. doi: 10.1016/j.micinf.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, Sayood K, Dunman PM, Roop Ii RM. 2012. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol 85:345–360. doi: 10.1111/j.1365-2958.2012.08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carroll RK, Weiss A, Shaw LN. 2016. RNA-sequencing of Staphylococcus aureus messenger RNA. Methods Mol Biol 1373:131–141. doi: 10.1007/7651_2014_192. [DOI] [PubMed] [Google Scholar]
- 69.Krute CN, Carroll RK, Rivera FE, Weiss A, Young RM, Shilling A, Botlani M, Varma S, Baker BJ, Shaw LN. 2015. The disruption of prenylation leads to pleiotropic rearrangements in cellular behavior in Staphylococcus aureus. Mol Microbiol 95:819–832. doi: 10.1111/mmi.12900. [DOI] [PubMed] [Google Scholar]
- 70.Weiss A, Broach WH, Lee MC, Shaw LN. 2016. Towards the complete small RNome of Acinetobacter baumannii. Microb Genomics doi: 10.1099/mgen.0.000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kolar SL, Nagarajan V, Oszmiana A, Rivera FE, Miller HK, Davenport JE, Riordan JT, Potempa J, Barber DS, Koziel J, Elasri MO, Shaw LN. 2011. NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology 157:2206–2219. doi: 10.1099/mic.0.049692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krute CN, Bell-Temin H, Miller HK, Rivera FE, Weiss A, Stevens SM, Shaw LN. 2015. The membrane protein PrsS mimics sigmaS in protecting Staphylococcus aureus against cell wall-targeting antibiotics and DNA-damaging agents. Microbiology 16:1136–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koprivnjak T, Mlakar V, Swanson L, Fournier B, Peschel A, Weiss JP. 2006. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J Bacteriol 188:3622–3630. doi: 10.1128/JB.188.10.3622-3630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rivera FE, Miller HK, Kolar SL, Stevens SM Jr, Shaw LN. 2012. The impact of CodY on virulence determinant production in community-associated methicillin-resistant Staphylococcus aureus. Proteomics 12:263–268. doi: 10.1002/pmic.201100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolar SL, Ibarra JA, Rivera FE, Mootz JM, Davenport JE, Stevens SM, Horswill AR, Shaw LN. 2013. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiol Open 2:18–34. doi: 10.1002/mbo3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fleeman R, LaVoi TM, Santos RG, Morales A, Nefzi A, Welmaker GS, Medina-Franco JL, Giulianotti MA, Houghten RA, Shaw LN. 2015. Combinatorial libraries as a tool for the discovery of novel, broad-spectrum antibacterial agents targeting the ESKAPE pathogens. J Med Chem 58:3340–3355. doi: 10.1021/jm501628s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shaw LN, Golonka E, Szmyd G, Foster SJ, Travis J, Potempa J. 2005. Cytoplasmic control of premature activation of a secreted protease zymogen: deletion of staphostatin B (SspC) in Staphylococcus aureus 8325-4 yields a profound pleiotropic phenotype. J Bacteriol 187:1751–1762. doi: 10.1128/JB.187.5.1751-1762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salisbury V, Hedges RW, Datta N. 1972. Two modes of “curing” transmissible bacterial plasmids. J Gen Microbiol 70:443–452. doi: 10.1099/00221287-70-3-443. [DOI] [PubMed] [Google Scholar]
- 80.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sullivan MA, Yasbin RE, Young FE. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.