Abstract
Iron is an essential element in biology, required for numerous cellular processes. Either too much or too little iron can be detrimental, and organisms have developed mechanisms for balancing iron within safe limits. In mammals there are no controlled mechanisms for the excretion of excess iron, hence body iron homeostasis is regulated at the sites of absorption, utilisation and recycling. This review will discuss the discoveries that have been made in the past 20 years into advancing our understanding of iron homeostasis and its regulation. The study of iron-associated disorders, such as the iron overload condition hereditary haemochromatosis and various forms of anaemia have been instrumental in increasing our knowledge in this area, as have cellular and animal model studies. The liver has emerged as the major site of systemic iron regulation, being the location where the iron regulatory hormone hepcidin is produced. Hepcidin is a negative regulator of iron absorption and recycling, achieving this by binding to the only known cellular iron exporter ferroportin and causing its internalisation and degradation, thereby reducing iron efflux from target cells and reducing serum iron levels. Much of the research in the iron metabolism field has focussed on the regulation of hepcidin and its interaction with ferroportin. The advances in this area have greatly increased our knowledge of iron metabolism and its regulation and have led to the development of novel diagnostics and therapeutics for iron-associated disorders.
Iron in Biology
Iron is an essential element in biology. Its ability to readily undergo redox cycling between its two predominant oxidation states, Fe3+ (ferric) and Fe2+ (ferrous), underlies its functional importance as a cofactor required for the activity of many essential enzymes and other molecules. In particular, iron is contained within the functional haem group, a component in the electron transport chain as well as the oxygen carrying molecule haemoglobin. Indeed, most of the iron in the human body (approximately 65%) is contained within the haemoglobin carrying red blood cells. Even though iron is plentiful on Earth, most of it is in the largely insoluble and biologically unavailable Fe3+ state, hence organisms have evolved intricate mechanisms of acquiring iron from their environment. In this review the mechanisms by which mammals absorb iron from their diet as well as how iron homeostasis is regulated to maintain body iron levels within safe limits will be discussed.
The study of diseases associated with iron deficiency or iron overload have been key to increasing our understanding of iron homeostasis and its regulation. Iron deficiency is the most common nutritional deficiency world-wide. According to the World Health Organisation, an estimated 25% of the world’s population suffer from iron deficiency anaemia.1 Much of this anaemia is due to poor dietary intake of iron but infectious disease and other causes of chronic inflammation can also reduce iron absorption and availability by mechanisms that will be discussed later. Reduced iron availability causes iron-restricted erythropoiesis in the bone marrow leading to anaemia, characterised by smaller red blood cells containing less haemoglobin. The reduction in oxygen supply to the tissues caused by anaemia can lead to weakness, fatigue and cognitive impairment.1
In addition to the detrimental effects of iron deficiency, iron overload can also be detrimental to health. Iron overload is usually genetically inherited and caused by primary defects in molecules regulating iron homeostasis and termed hereditary haemochromatosis (HH). Other forms of iron overload, termed secondary iron overload, are not due to primary defects in iron homeostatic mechanisms and can have a variety of acquired causes that will be discussed later. Excess iron accumulation in tissues can lead to tissue damage and disease, including liver fibrosis, diabetes mellitus, arthropathy, endocrine dysfunction and cardiomyopathy.2 Mutations in the HFE gene are the most common cause of HH, with homozygosity for the p.C282Y mutation affecting around 1 in 200 individuals of northern European descent.3–7 While the p.C282Y mutation is relatively common in northern European populations, it is less common in southern Europeans8, 9 and rare in non-Europeans.10 Biochemical indicators of iron overload include elevated serum ferritin levels, serum iron and transferrin saturation.11, 12 Serum ferritin is a marker of tissue iron loading, ferritin being a cellular iron storage protein. Transferrin is an iron transport protein that is often saturated with iron in conditions of iron overload. When the HFE gene was discovered in 1996, very little was known about the molecular mechanisms responsible for the absorption of iron and maintenance of iron homeostasis.3 Since this time, the identification of other forms of iron overload and anaemia and elucidation of their causes have been instrumental in determining the mechanisms regulating iron absorption and homeostasis.
Iron Absorption
Approximately 2 mg of iron is absorbed daily in the duodenum and proximal jejunum. This is balanced by losses resulting from the desquamation of skin, sloughing of intestinal epithelial cells and blood loss. The human body has no controlled mechanisms for the excretion of iron and the levels are balanced by regulating iron absorption. Iron in the diet can be in the form of haem or non-haem iron. As most non-haem iron in the diet is in the ferric form, it first needs to be reduced to Fe2+ before it can be absorbed; this can be achieved by the actions of the membrane bound ferric reductase duodenal cytochrome B (DCYTB or CYBRD1), which is expressed on the apical brush border membrane of intestinal epithelial cells.13 Ferrous iron is then transported across the apical membrane of enterocytes by the divalent metal transporter 1 (DMT1), an integral 12 transmembrane domain protein that has the ability to transport a number of divalent cations including Fe2+.14, 15 To enter the systemic circulation iron must cross the basolateral membrane of intestinal enterocytes. This is achieved by the only known iron exporter, ferroportin, a 12 transmembrane domain protein, encoded by the SLC40A1 gene.16–18. Ferroportin is also required for the release of iron from other cell types, in particular macrophages and hepatocytes where it is also highly expressed.18 The release of ferrous iron from stores by ferroportin is assisted by the copper-containing ferroxidase enzyme caeruloplasmin19 or, in the intestine, by its membrane-bound counterpart hephaestin.20 These enzymes oxidise Fe2+ to Fe3+ before the iron binds to the iron-transport protein transferrin.
Iron Uptake by Tissues
Iron is transported in the circulation bound to transferrin, although circulating iron can also exist in a non-transferrin bound form, especially when serum iron levels are high and transferrin is saturated, as is the case in HH and other iron loading conditions.21 Transferrin receptor 1 (TFR1) is expressed ubiquitously on the cell surface and is responsible for taking up transferrin-bound iron through well studied mechanisms that involve receptor mediated endocytosis.22 Once internalised the endocytic vesicles are acidified, allowing iron to be released from transferrin, and the apotransferrin, still bound to TFR1, recycled back to the cell surface where it is released.22 Iron exits the endosome and enters the cytoplasm of cells via DMT1. The ferrireductase six-transmembrane epithelial antigen of the prostate 3 (STEAP3) also facilitates this process by reducing Fe3+ to Fe2+ prior to transport through DMT1.23 The levels of many iron metabolism related proteins, including TFR1 and DMT1, are regulated at the post-transcriptional level via the iron responsive element/iron responsive protein (IRE/IRP) system. IRE-stem loop structures in the 3′ untranslated regions (UTRs) of these mRNAs bind to IRPs 1 or 2 under conditions of iron deficiency and stabilise the mRNA, enhancing translation of the proteins and increasing iron uptake. IREs are also present in the 5′UTRs of ferritin and ferroportin mRNAs among others and in contrast to TFR1 and DMT1, the translation of these proteins are repressed under iron deficient conditions. More details of this important and elegant system for balancing cellular iron homeostasis can be found elsewhere.24
The erythroid bone marrow has a high demand for iron, hence mutation or deletion of genes involved in the uptake of transferrin-bound iron in humans and mice lead to varying degrees of anaemia due to a decrease in the transport of iron into cells. For example, deletion of TFR1 in mice results in embryonic lethality due to severe anaemia.25 Mutations in DMT1 lead to microcytic anaemia in mice and rats.15, 26 Humans with DMT1 mutations also develop microcytic anaemia, however, unlike in mice and rats, this is also accompanied by liver iron overload.27 STEAP3 mutations also cause a form of microcytic anaemia in humans and mice.28
Recently, a mechanism for the uptake of non-transferrin-bound iron (NTBI) was proposed that involves the zinc transporter, ZIP14 (SLC39A14).29 Mice with ablation of SLC39A14 had markedly reduced uptake of NTBI in the liver and pancreas and ablation of SLC39A14 also prevented iron loading of the parenchymal cells in the liver and pancreas of mouse models of HH.29
Iron Recycling
As mentioned earlier, most of the iron in the human body is contained within red blood cells. The iron released from these cells, when they reach the end of their lifespan, is a major source of systemically available iron that can be reused for the production of new erythrocytes in the bone marrow. Reticuloendothelial macrophages are responsible for engulfing senescent erythrocytes in a process termed erythrophagocytosis.30 During this process red blood cells are digested in the phagolysosome. From here, iron released from haem, by as yet unknown mechanisms, is transported into the cytosol via natural resistance associated macrophage protein 1 (NRAMP1), a divalent metal transporter and paralogue of DMT1.31 Haem itself can also be transported across the phagolysosomal membrane via a recently described haem transporter, a homologue of the Caenorhabditis elegans haem-responsive gene 1 (HRG1).32 Once in the cytosol, iron can be released from haem by the actions of haem oxygenase 1.33 The resultant iron liberated from the breakdown of red blood cells can then either be stored in ferritin or released back to the systemic circulation by ferroportin-mediated transport across the plasma membrane.
Systemic Regulation of Iron
Maintaining the optimal levels of iron in the circulation is critical for the functioning of cells and tissues. For example, too little iron can lead to iron restricted erythropoiesis and consequent anaemia, whereas too much can lead to tissue iron overload and related diseases.
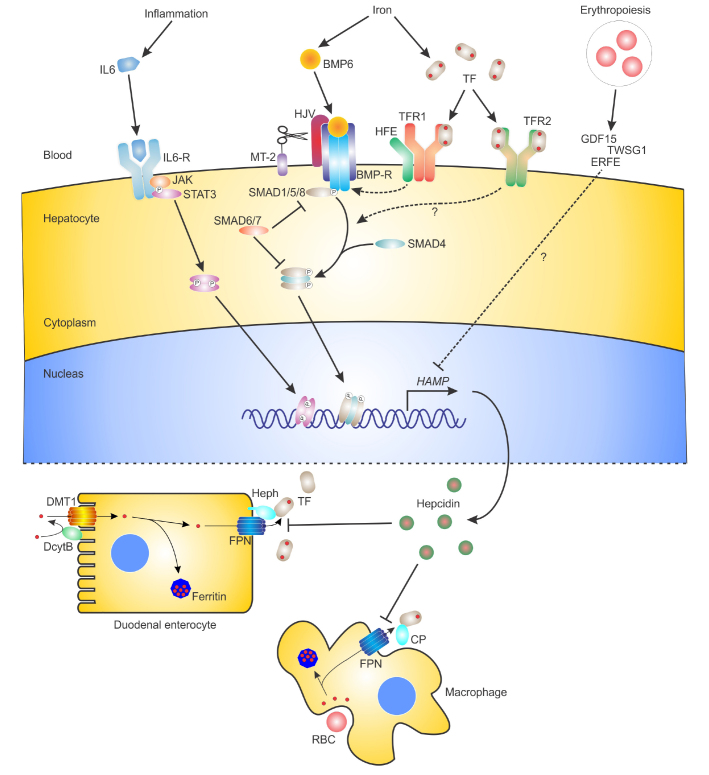
Regulating iron levels during infections is also important in the innate immune response to pathogens. To meet the body’s requirements for iron, elaborate mechanisms have evolved to sense iron levels and adjust iron absorption and recycling accordingly to maintain iron homeostasis. These mechanisms also respond to inflammatory/infectious stimuli, as well as hypoxia and erythropoietic signals, to either decrease or increase iron availability. The mechanisms regulating systemic iron homeostasis are largely centred on the liver and involve two molecules, hepcidin and ferroportin, that work together to regulate the flow of iron from cells into the systemic circulation. The diagram in figure 1 summarises the most important players involved in the systemic regulation of iron homeostasis.
Figure 1.
The regulation of hepcidin and iron homeostasis.
Schematic showing the major molecules and pathways involved in the regulation of hepcidin (HAMP) gene expression in hepatocytes and the functions of hepcidin in regulating the surface expression of the iron export protein ferroportin (FPN) in other cell types. The major molecules and pathways responsible for the iron, inflammation and erythropoietic regulation of HAMP in hepatocytes are depicted at the top of the figure. The role of hepatocyte-derived hepcidin in regulating iron absorption in duodenal enterocytes and iron recycling in macrophages via its interaction with FPN is depicted at the bottom of the figure. Small red circles represent iron. IL-6, interleukin 6; IL-6-R, IL-6 receptor; JAK, Janus kinase; STAT3, signal transducer and activator of transcription 3; BMP6, bone morphogenetic protein 6; BMP-R, BMP receptor; HJV, hemojuvelin; MT-2, matriptase-2; HFE, haemochromatosis protein; TF, transferrin; TFR1, TF receptor 1; TFR2, TF receptor 2; GDF15, growth differentiation factor 15; TWSG1, twisted gastrulation; ERFE, erythroferrone; SMAD, mothers against decapentaplegic homologue; DMT1, divalent metal transporter 1; DcytB, duodenal cytochrome B; Heph, hephaestin; CP, caeruloplasmin; RBC, red blood cell.
As mentioned previously, ferroportin is the only known cellular iron export protein. It is expressed most highly in macrophages, duodenal enterocytes and hepatocytes, important cell types involved in iron recycling, absorption, storage and regulation.18 The expression of ferroportin can be controlled at the transcriptional, translational and post-translational levels. The ferroportin mRNA contains a functional IRE in its 5′UTR and, similar to H and L ferritin, its translation is repressed under iron deficient conditions, with resultant reduction in cellular iron export.18 Transcription of the macrophage ferroportin gene can be promoted by haem34 and inhibited by inflammatory stimuli.35 At a systemic level the most important mechanism regulating ferroportin involves the liver-expressed iron regulatory hormone hepcidin. Hepcidin was originally identified in plasma and urine as a small, 25 amino acid, highly disulphide bonded, liver-expressed antimicrobial peptide.36, 37 It soon became apparent that it also played a major role in the regulation of systemic iron homeostasis.38–40 In 2004 it was shown that hepcidin functions to reduce cellular iron export by binding to ferroportin and causing its internalisation and degradation.41 Hepcidin binding to ferroportin induces the rapid ubiquitination and internalisation of the hepcidin-ferroportin complex, thereby reducing cell surface expression and iron export.42, 43 Since the discovery of hepcidin in 2000, it has become apparent that its interaction with ferroportin is critical for systemic iron regulation and that perturbations in this hepcidin-ferroportin axis are the basis for many iron-associated disorders.44
Iron Disorders
Primary iron overload
HFE-associated HH (HFE-HH or type 1 HH), the most common form of HH, has autosomal recessive inheritance.3 It wasn’t until 2003 that hepcidin deficiency as a cause of the iron overload was first uncovered.45 It was shown that patients and animal models of HFE-HH had lower expression of hepcidin in the liver compared to controls.45, 46 These observations also indicated that the liver plays a major role in the pathophysiology of HH and the systemic regulation of iron homeostasis.
After the HFE gene was identified in 1996, it became immediately apparent that there were forms of primary iron overload that were not caused by mutations in the HFE gene. Among these non-HFE forms were cases of severe, early onset, juvenile haemochromatosis (JH), that had previously been recognised as distinct from the typical HFE form of disease.47 The gene for a subset of JH (also known as type 2A HH) mapped to chromosome 1,48 and the gene (HFE2) was eventually found to encode hemojuvelin (HJV) or RGMc, a glycosylphosphatidylinositol (GPI) anchored protein in the repulsive guidance molecule (RGM) family of proteins.49 Patients with HFE2 mutations also had low levels of serum hepcidin, indicating that hepcidin deficiency also underlies this more severe form of iron overload.49 Another less common form of JH (type 2B) is caused by mutations in the gene encoding the iron-regulatory hormone hepcidin itself (HAMP).50 Patients homozygous or compound heterozygous for mutations in HAMP cannot regulate the surface expression of ferroportin systemically and hence develop severe, early onset iron overload due to constitutively active ferroportin. The importance of HJV and hepcidin as a cause of JH and their roles in regulating iron homeostasis have been confirmed by the analysis of mouse models of these diseases.51, 52
The first form of non-HFE HH to be genetically characterised was TFR2-HH or type 3 HH.53 Mutations in the gene encoding transferrin receptor 2 (TFR2) have been shown to cause disease phenotypically very similar to HFE-HH but with slightly earlier onset.54, 55 Studies in patients and animal models of TFR2-HH have also shown that, similar to HFE-HH, reduced hepcidin relative to iron stores underlies the iron overload.56–58 TFR2, a paralogue of TFR1, is expressed predominantly in hepatocytes and rather than being responsible for the uptake of transferrin-bound iron, is involved in the regulation of hepcidin in response to iron, although the mechanisms involved are not fully understood. Recent research has also suggested that TFR2 has another functional role in erythroid cells, where it is also expressed, being involved in the differentiation of red blood cells.59, 60
Patients have also been described with homozygous or compound heterozygous mutations in both the HFE and TFR2 genes, with earlier onset severe iron overload, similar to JH.61 Mouse models with deletion of both Hfe and Tfr2 recapitulate this severe iron overload and show that it is due to markedly reduced hepcidin, suggesting that HFE and TFR2 can function independently to regulate hepcidin.62, 63
Autosomal dominant forms of iron overload have also been described.64 Mutations in ferroportin (SLC40A1) have been linked to an autosomal dominant form of iron overload, termed ferroportin disease or type 4 HH.65–67 Interestingly, mutations which affect different aspects of ferroportin function lead to two distinct subtypes of ferroportin disease, which have differing patterns of iron overload.68 Mutations which cause defects in the iron transport ability of ferroportin lead to classical ferroportin disease, which is characterised by normal or only mildly elevated transferrin saturation and iron loading predominantly in reticuloendothelial cells.69, 70 An atypical form of ferroportin disease is caused by mutations that lead to hepcidin insensitivity of ferroportin and has a phenotype more similar to the autosomal recessive forms of HH, with elevated transferrin saturation and iron loading predominantly in hepatocytes.70, 71
Secondary iron overload
Iron overload that is not caused by primary genetic defects in the regulation of iron homeostasis is referred to as secondary iron overload. Secondary iron overload may occur due to a variety of causes. These include the ingestion of large amounts of dietary iron, repeated blood transfusions and various haematological conditions that result in the increased absorption and storage of excess iron. Clinically, an important cause of secondary iron overload is repeated blood transfusion due to inherited haemoglobinopathies, such as the thalassaemias.72 The buildup of excess iron over time is a major cause of morbidity in thalassaemia patients and treatment with iron chelators is a mainstay treatment to combat iron overload related disease.73 In addition to iron overload caused by blood transfusions, non-transfusion dependent iron overload can occur in some haemoglobinopathies due to increased iron absorption. In β-thalassaemia, ineffective erythropoiesis and an expanded erythroid compartment can influence systemic iron homeostasis by suppressing liver hepcidin, with resultant increased iron absorption.74, 75
Iron deficiency
Genetic forms of iron deficiency can be caused by mutations in genes involved in the regulation of iron transport and metabolism. As mentioned previously, mutations in STEAP3 and DMT1 lead to microcytic anaemia,27, 28 Iron refractory iron deficiency anaemia (IRIDA), a form of anaemia that is resistant to oral iron therapy, is caused by mutations in the transmembrane protease, serine 6 (TMPRSS6) gene, which encodes matriptase-2, a membrane bound serine protease expressed predominantly in hepatocytes that has been shown to regulate hepcidin expression.76–78 In contrast to patients with HH, patients with IRIDA have abnormally elevated levels of serum hepcidin, resulting in suppression of iron absorption and recycling.76
BMP Signalling in Iron Regulation
The bone morphogenetic protein-mothers against decapentaplegic homologue (BMP-SMAD) signalling pathway has emerged as the major signalling pathway responsible for the iron-regulated expression of hepcidin.79 BMPs are part of the transforming growth factor beta (TGF-β) superfamily of ligands and similar to TGF-β, signal through cell-associated receptors and downstream SMAD proteins to regulate gene expression. Type I and II BMP receptors are serine-threonine kinase receptors; upon BMP binding, the type II receptors activate the type I receptors by phosphorylation. Different combinations of 3 type I and 3 type II BMP receptors are involved in BMP signalling. Once activated the type I receptors then phosphorylate receptor regulated SMADs (R-SMADs), which bind to the common mediator SMAD (SMAD4) and the complex then translocates to the nucleus to regulate the expression of target genes (Figure 1).80 The functionally related R-SMADs 1, 5 and 8 are utilised by the BMP signalling pathway, whereas the R-SMADs 2 and 3 are utilised by the TGF-β pathway. The inhibitory SMADs (SMAD 6 and 7) are paralogous to other SMAD proteins and function to reduce signalling through the BMP and TGF-β pathways by competitively binding with either the BMP receptors or other SMAD proteins.80
The first clue to the role of the BMP-SMAD pathway in hepcidin regulation came with the observation that hepatocyte-specific ablation of SMAD4 in mice led to severe iron overload.81 Further research showed that HJV is a BMP co-receptor that enhances signalling through the SMAD pathway.82 Loss of HJV, as in patients with JH, results in greatly reduced BMP signalling and low hepcidin expression. HJV can also be cleaved by proprotein convertases and exist as a secreted soluble form (sHJV) that can compete with cell associated HJV and inhibit BMP-SMAD signalling to reduce hepcidin expression.83, 84 In terms of iron-regulated signalling it has been shown that HJV can utilise type II receptors BMPRII and ACTRIIA and type I receptors ALK2 and ALK3.85 Hepatocyte-specific deletion of Alk2 or Alk3 in mice results in iron overload, confirming the importance of these type I receptors in the regulation of iron homeostasis.86 Similarly, simultaneous deletion of BmprII and ActrIIa results in iron overload, whereas individual deletion of the 2 genes does not, suggesting that these type II receptors have redundant functions in the regulation of hepcidin and iron homeostasis.87 Other molecules involved in iron homeostasis are also important components in BMP-SMAD signalling. Matriptase-2 was shown to be a negative regulator of hepcidin expression by reducing BMP-SMAD signalling via cleavage of HJV.78 It has been suggested that HFE may enhance BMP-SMAD signalling via an interaction with the BMP type 1 receptor ALK3.88 It is likely that TFR2 also alters SMAD signalling through mechanisms that are not entirely understood but may involve BMP6.89
Many BMPs can upregulate hepcidin expression in vitro, including BMPs 2, 4, 5, 6, 7 and 9.90 The BMP that appears to be most physiologically relevant to the regulation of iron homeostasis is BMP6. It was shown that iron regulates the expression of BMP6 in the liver91 and that knockout of Bmp6 in mice results in severe iron overload due to hepcidin deficiency.92, 93 Which cell types contribute to BMP6 expression and how it is upregulated by iron are not yet fully understood, although it has been suggested that BMP6 expressed in nonparenchymal cells of the liver can be regulated by iron and may function in a paracrine manner to regulate hepcidin in hepatocytes.94
Other evidence suggests that the BMP-SMAD pathway is central to hepcidin and iron regulation. For example the inhibitory SMAD, SMAD7 has been shown to be a potent inhibitor of hepcidin expression.95 Another potential negative regulator of hepcidin is the known BMP inhibitor BMP-binding endothelial cell precursor-derived regulator (BMPER).96 Recently the SMAD adapter protein endofin was shown to be important for enhancing signalling through the BMP-SMAD pathway to regulate hepcidin.97 There is now a large body of evidence to implicate the BMP-SMAD signalling pathway in the iron-dependent regulation of hepcidin and iron homeostasis.
Inflammatory Regulation of Iron Homeostasis
The regulation of iron homeostasis by inflammatory stimuli is important in the innate immune response to infections and cancers. Increasing hepcidin levels during infection has the effect of sequestering iron in tissues and reducing serum iron levels, effectively withholding iron from invading pathogens. Over prolonged periods of time, such as during chronic inflammation, this can result in reduced iron availability for the production of red blood cells and resultant anaemia. It is thought that elevated hepcidin levels are a major contributor to the anaemia of chronic disease, the most common form of anaemia in hospitalised patients.98, 99
It has been shown that liver hepcidin is upregulated by inflammatory stimuli.98, 100 The inflammatory cytokine interleukin 6 (IL-6) is largely responsible for this process, as the characteristic hypoferraemic response to an acute inflammatory stimuli was completely ablated in mice lacking IL-6.101 Elegant transgenic mouse experiments showed that the signal transducer and activator of transcription 3 (STAT3) signalling pathway is responsible for the IL-6-mediated regulation of hepcidin in the liver.102 Other inflammatory cytokines have also been proposed as hepatic hepcidin regulators, such as IL-1103 and IL-22,104, 105 however, the role of these cytokines in inducing the hypoferraemic response to inflammation in vivo is less clear.106
Erythropoietic and Hypoxic Regulation of Iron Homeostasis
As the majority of iron in the human body is required for the production of red blood cells, it is not surprising that erythropoiesis and the regulation of iron homeostasis are intrinsically linked. It has been known for a long time that erythropoietic demand increases iron absorption,107 although the mechanisms underlying this association have only recently been uncovered and are still not fully understood. Erythropoiesis and hypoxia have the effect of suppressing liver hepcidin expression to increase iron absorption.98
Tissue hypoxia can result from anaemia and can lead to the production of the major erythropoiesis stimulating hormone erythropoietin (EPO). It has been suggested that hypoxia itself can directly suppress hepcidin transcription in the liver via the induction of hypoxia inducible factors108 and via effects on matriptase-2 and BMP-SMAD signalling.109 It has also been suggested that EPO can directly suppress hepcidin in hepatocytes,110 although others have suggested a more indirect effect of EPO via the stimulation of erythropoiesis and release of a hepcidin-suppressive humoral factor.111 Several erythroid-derived factors have been suggested, including growth differentiation factor 15 (GDF15) and twisted gastrulation (TWSG1), both of which are released by erythroid precursors and can suppress hepatic hepcidin gene expression.74, 112 More recently another candidate erythroid-derived regulator of hepcidin has been proposed, erythroferrone (ERFE), which is expressed by erythroid precursors and represses hepcidin during stress erythropoiesis.113 Following bleeding, it was shown that Erfe deficient mice fail to suppress hepcidin and recover from anaemia more slowly than wild-type mice.113
Diagnostics and Therapeutics for Iron Disorders
The increase in our knowledge about the mechanisms regulating iron absorption and homeostasis has led to new diagnostic and therapeutic possibilities for iron-related disorders. Serum iron, transferrin saturation and serum ferritin measurements have been the mainstay blood tests for assessing iron levels. With the discovery of hepcidin, new diagnostic tests have been developed for measuring serum levels that may soon enter routine clinical practice for the differential diagnosis of various forms of iron overload and anaemia.114
While the HFE gene test has been around for nearly 20 years,3 the discovery of genes for non-HFE forms of HH and anaemia and recent advances in DNA sequencing means that it is now becoming increasingly possible to genetically diagnose these more unusual iron-associated disorders.115
The hepcidin-ferroportin axis is a very attractive target for the development of novel therapeutics for treating iron disorders. For example, hepcidin agonists can be used to treat iron overload caused by hepcidin deficiency, such as the various forms of HH or iron loading anaemias such as thalassaemia. Smaller hepcidin-based peptides (minihepcidins) have been developed, that have shown some success in preventing iron overload in mouse models of HH.116 Targeting the upstream regulator of hepcidin, TMPRSS6, using either lipid encapsulated small interfering RNAs (siRNAs)117 or antisense oligonucleotides,118 has proven successful in treating iron overload in mouse models of HH and β-thalassaemia.
Hepcidin antagonists may be equally beneficial in treating iron deficiency anaemia that is related to abnormally elevated hepcidin, such as IRIDA or the anaemia of chronic disease. The Speigelmer lexaptepid, an RNA-like molecule, binds to and inhibits hepcidin and has shown promise in clinical trials for the treatment of the anaemia of chronic disease.119
Conclusions
In summary, this review has highlighted the major breakthroughs that have occurred in the past 20 years or so, that have greatly enhanced our understanding of how iron absorption and homeostasis are regulated. A combination of clinical medicine, basic cellular and animal model based research has been largely responsible for these discoveries. While our understanding of iron regulation is now more complete, there are still many aspects that are unknown and new players to be discovered that will form the basis of future research in this area.
Footnotes
Competing Interests: None declared.
References
- 1.McLean E, Cogswell M, Egli I, Wojdyla D, de Benoist B. Worldwide prevalence of anaemia, WHO Vitamin and Mineral Nutrition Information System, 1993–2005. Public Health Nutr. 2009;12:444–54. doi: 10.1017/S1368980008002401. [DOI] [PubMed] [Google Scholar]
- 2.Niederau C, Fischer R, Pürschel A, Stremmel W, Häussinger D, Strohmeyer G. Long-term survival in patients with hereditary hemochromatosis. Gastroenterology. 1996;110:1107–19. doi: 10.1053/gast.1996.v110.pm8613000. [DOI] [PubMed] [Google Scholar]
- 3.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 4.Bacon BR, Powell LW, Adams PC, Kresina TF, Hoofnagle JH. Molecular medicine and hemochromatosis: at the crossroads. Gastroenterology. 1999;116:193–207. doi: 10.1016/s0016-5085(99)70244-1. [DOI] [PubMed] [Google Scholar]
- 5.Olynyk JK, Cullen DJ, Aquilia S, Rossi E, Summerville L, Powell LW. A population-based study of the clinical expression of the hemochromatosis gene. N Engl J Med. 1999;341:718–24. doi: 10.1056/NEJM199909023411002. [DOI] [PubMed] [Google Scholar]
- 6.Adams PC, Reboussin DM, Barton JC, McLaren CE, Eckfeldt JH, McLaren GD, et al. Hemochromatosis and Iron Overload Screening (HEIRS) Study Research Investigators. Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med. 2005;352:1769–78. doi: 10.1056/NEJMoa041534. [DOI] [PubMed] [Google Scholar]
- 7.Allen KJ, Gurrin LC, Constantine CC, Osborne NJ, Delatycki MB, Nicoll AJ, et al. Iron-overload-related disease in HFE hereditary hemochromatosis. N Engl J Med. 2008;358:221–30. doi: 10.1056/NEJMoa073286. [DOI] [PubMed] [Google Scholar]
- 8.Piperno A, Sampietro M, Pietrangelo A, Arosio C, Lupica L, Montosi G, et al. Heterogeneity of hemochromatosis in Italy. Gastroenterology. 1998;114:996–1002. doi: 10.1016/s0016-5085(98)70319-1. [DOI] [PubMed] [Google Scholar]
- 9.Sánchez M, Villa M, Ingelmo M, Sanz C, Bruguera M, Ascaso C, et al. Population screening for hemochromatosis: a study in 5370 Spanish blood donors. J Hepatol. 2003;38:745–50. doi: 10.1016/s0168-8278(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 10.Wallace DF, Subramaniam VN. The global prevalence of HFE and non-HFE hemochromatosis estimated from analysis of next-generation sequencing data. Genet Med. 2015 doi: 10.1038/gim.2015.140. [DOI] [PubMed] [Google Scholar]
- 11.Addison GM, Beamish MR, Hales CN, Hodgkins M, Jacobs A, Llewellin P. An immunoradiometric assay for ferritin in the serum of normal subjects and patients with iron deficiency and iron overload. J Clin Pathol. 1972;25:326–9. doi: 10.1136/jcp.25.4.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs A, Miller F, Worwood M, Beamish MR, Wardrop CA. Ferritin in the serum of normal subjects and patients with iron deficiency and iron overload. Br Med J. 1972;4:206–8. doi: 10.1136/bmj.4.5834.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, et al. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291:1755–9. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- 14.Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–8. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 15.Fleming MD, Trenor CC, 3rd, Su MA, Foernzler D, Beier DR, Dietrich WF, et al. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16:383–6. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 16.Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt SJ, Moynihan J, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403:776–81. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- 17.McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, et al. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000;5:299–309. doi: 10.1016/s1097-2765(00)80425-6. [DOI] [PubMed] [Google Scholar]
- 18.Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem. 2000;275:19906–12. doi: 10.1074/jbc.M000713200. [DOI] [PubMed] [Google Scholar]
- 19.Osaki S, Johnson DA, Frieden E. The possible significance of the ferrous oxidase activity of ceruloplasmin in normal human serum. J Biol Chem. 1966;241:2746–51. [PubMed] [Google Scholar]
- 20.Vulpe CD, Kuo YM, Murphy TL, Cowley L, Askwith C, Libina N, et al. Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat Genet. 1999;21:195–9. doi: 10.1038/5979. [DOI] [PubMed] [Google Scholar]
- 21.Chua AC, Olynyk JK, Leedman PJ, Trinder D. Nontransferrin-bound iron uptake by hepatocytes is increased in the Hfe knockout mouse model of hereditary hemochromatosis. Blood. 2004;104:1519–25. doi: 10.1182/blood-2003-11-3872. [DOI] [PubMed] [Google Scholar]
- 22.Qian ZM, Tang PL. Mechanisms of iron uptake by mammalian cells. Biochim Biophys Acta. 1995;1269:205–14. doi: 10.1016/0167-4889(95)00098-x. [DOI] [PubMed] [Google Scholar]
- 23.Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, et al. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37:1264–9. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang DL, Ghosh MC, Rouault TA. The physiological functions of iron regulatory proteins in iron homeostasis - an update. Front Pharmacol. 2014;5:124. doi: 10.3389/fphar.2014.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy JE, Jin O, Fujiwara Y, Kuo F, Andrews NC. Transferrin receptor is necessary for development of erythrocytes and the nervous system. Nat Genet. 1999;21:396–9. doi: 10.1038/7727. [DOI] [PubMed] [Google Scholar]
- 26.Fleming MD, Romano MA, Su MA, Garrick LM, Garrick MD, Andrews NC. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc Natl Acad Sci USA. 1998;95:1148–53. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mims MP, Guan Y, Pospisilova D, Priwitzerova M, Indrak K, Ponka P, et al. Identification of a human mutation of DMT1 in a patient with microcytic anemia and iron overload. Blood. 2005;105:1337–42. doi: 10.1182/blood-2004-07-2966. [DOI] [PubMed] [Google Scholar]
- 28.Grandchamp B, Hetet G, Kannengiesser C, Oudin C, Beaumont C, Rodrigues-Ferreira S, et al. A novel type of congenital hypochromic anemia associated with a nonsense mutation in the STEAP3/TSAP6 gene. Blood. 2011;118:6660–6. doi: 10.1182/blood-2011-01-329011. [DOI] [PubMed] [Google Scholar]
- 29.Jenkitkasemwong S, Wang CY, Coffey R, Zhang W, Chan A, Biel T, et al. SLC39A14 Is Required for the Development of Hepatocellular Iron Overload in Murine Models of Hereditary Hemochromatosis. Cell Metab. 2015;22:138–50. doi: 10.1016/j.cmet.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korolnek T, Hamza I. Macrophages and iron trafficking at the birth and death of red cells. Blood. 2015;125:2893–7. doi: 10.1182/blood-2014-12-567776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soe-Lin S, Apte SS, Andriopoulos B, Jr, Andrews MC, Schranzhofer M, Kahawita T, et al. Nramp1 promotes efficient macrophage recycling of iron following erythrophagocytosis in vivo. Proc Natl Acad Sci USA. 2009;106:5960–5. doi: 10.1073/pnas.0900808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White C, Yuan X, Schmidt PJ, Bresciani E, Samuel TK, Campagna D, et al. HRG1 is essential for heme transport from the phagolysosome of macrophages during erythrophagocytosis. Cell Metab. 2013;17:261–70. doi: 10.1016/j.cmet.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poss KD, Tonegawa S. Heme oxygenase 1 is required for mammalian iron reutilization. Proc Natl Acad Sci USA. 1997;94:10919–24. doi: 10.1073/pnas.94.20.10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marro S, Chiabrando D, Messana E, Stolte J, Turco E, Tolosano E, et al. Heme controls ferroportin1 (FPN1) transcription involving Bach1, Nrf2 and a MARE/ARE sequence motif at position -7007 of the FPN1 promoter. Haematologica. 2010;95:1261–8. doi: 10.3324/haematol.2009.020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu XB, Nguyen NB, Marquess KD, Yang F, Haile DJ. Regulation of hepcidin and ferroportin expression by lipopolysaccharide in splenic macrophages. Blood Cells Mol Dis. 2005;35:47–56. doi: 10.1016/j.bcmd.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Krause A, Neitz S, Mägert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, et al. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000;480:147–50. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 37.Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–10. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 38.Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, et al. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–9. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- 39.Nicolas G, Bennoun M, Devaux I, Beaumont C, Grandchamp B, Kahn A, et al. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc Natl Acad Sci USA. 2001;98:8780–5. doi: 10.1073/pnas.151179498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicolas G, Bennoun M, Porteu A, Mativet S, Beaumont C, Grandchamp B, et al. Severe iron deficiency anemia in transgenic mice expressing liver hepcidin. Proc Natl Acad Sci USA. 2002;99:4596–601. doi: 10.1073/pnas.072632499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–3. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 42.Ross SL, Tran L, Winters A, Lee KJ, Plewa C, Foltz I, et al. Molecular mechanism of hepcidin-mediated ferroportin internalization requires ferroportin lysines, not tyrosines or JAK-STAT. Cell Metab. 2012;15:905–17. doi: 10.1016/j.cmet.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 43.Qiao B, Sugianto P, Fung E, Del-Castillo-Rueda A, Moran-Jimenez MJ, Ganz T, et al. Hepcidin-induced endocytosis of ferroportin is dependent on ferroportin ubiquitination. Cell Metab. 2012;15:918–24. doi: 10.1016/j.cmet.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganz T. Hepcidin and iron regulation, 10 years later. Blood. 2011;117:4425–33. doi: 10.1182/blood-2011-01-258467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, et al. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361:669–73. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad KA, Ahmann JR, Migas MC, Waheed A, Britton RS, Bacon BR, et al. Decreased liver hepcidin expression in the Hfe knockout mouse. Blood Cells Mol Dis. 2002;29:361–6. doi: 10.1006/bcmd.2002.0575. [DOI] [PubMed] [Google Scholar]
- 47.Lamon JM, Marynick SP, Roseblatt R, Donnelly S. Idiopathic hemochromatosis in a young female. A case study and review of the syndrome in young people. Gastroenterology. 1979;76:178–83. [PubMed] [Google Scholar]
- 48.Roetto A, Totaro A, Cazzola M, Cicilano M, Bosio S, D’Ascola G, et al. Juvenile hemochromatosis locus maps to chromosome 1q. Am J Hum Genet. 1999;64:1388–93. doi: 10.1086/302379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dubé MP, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 50.Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–2. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- 51.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115:2180–6. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lesbordes-Brion JC, Viatte L, Bennoun M, Lou DQ, Ramey G, Houbron C, et al. Targeted disruption of the hepcidin 1 gene results in severe hemochromatosis. Blood. 2006;108:1402–5. doi: 10.1182/blood-2006-02-003376. [DOI] [PubMed] [Google Scholar]
- 53.Camaschella C, Roetto A, Calì A, De Gobbi M, Garozzo G, Carella M, et al. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25:14–5. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- 54.Le Gac G, Mons F, Jacolot S, Scotet V, Férec C, Frébourg T. Early onset hereditary hemochromatosis resulting from a novel TFR2 gene nonsense mutation (R105X) in two siblings of north French descent. Br J Haematol. 2004;125:674–8. doi: 10.1111/j.1365-2141.2004.04950.x. [DOI] [PubMed] [Google Scholar]
- 55.Majore S, Milano F, Binni F, Stuppia L, Cerrone A, Tafuri A, et al. Homozygous p.M172K mutation of the TFR2 gene in an Italian family with type 3 hereditary hemochromatosis and early onset iron overload. Haematologica. 2006;91(Suppl):ECR33. [Google Scholar]
- 56.Nemeth E, Roetto A, Garozzo G, Ganz T, Camaschella C. Hepcidin is decreased in TFR2 hemochromatosis. Blood. 2005;105:1803–6. doi: 10.1182/blood-2004-08-3042. [DOI] [PubMed] [Google Scholar]
- 57.Wallace DF, Summerville L, Lusby PE, Subramaniam VN. First phenotypic description of transferrin receptor 2 knockout mouse, and the role of hepcidin. Gut. 2005;54:980–6. doi: 10.1136/gut.2004.062018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawabata H, Fleming RE, Gui D, Moon SY, Saitoh T, O’Kelly J, et al. Expression of hepcidin is down-regulated in TfR2 mutant mice manifesting a phenotype of hereditary hemochromatosis. Blood. 2005;105:376–81. doi: 10.1182/blood-2004-04-1416. [DOI] [PubMed] [Google Scholar]
- 59.Nai A, Pellegrino RM, Rausa M, Pagani A, Boero M, Silvestri L, et al. The erythroid function of transferrin receptor 2 revealed by Tmprss6 inactivation in different models of transferrin receptor 2 knockout mice. Haematologica. 2014;99:1016–21. doi: 10.3324/haematol.2013.103143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallace DF, Secondes ES, Rishi G, Ostini L, McDonald CJ, Lane SW, et al. A critical role for murine transferrin receptor 2 in erythropoiesis during iron restriction. Br J Haematol. 2015;168:891–901. doi: 10.1111/bjh.13225. [DOI] [PubMed] [Google Scholar]
- 61.Pietrangelo A, Caleffi A, Henrion J, Ferrara F, Corradini E, Kulaksiz H, et al. Juvenile hemochromatosis associated with pathogenic mutations of adult hemochromatosis genes. Gastroenterology. 2005;128:470–9. doi: 10.1053/j.gastro.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 62.Wallace DF, Summerville L, Crampton EM, Frazer DM, Anderson GJ, Subramaniam VN. Combined deletion of Hfe and transferrin receptor 2 in mice leads to marked dysregulation of hepcidin and iron overload. Hepatology. 2009;50:1992–2000. doi: 10.1002/hep.23198. [DOI] [PubMed] [Google Scholar]
- 63.Delima RD, Chua AC, Tirnitz-Parker JE, Gan EK, Croft KD, Graham RM, et al. Disruption of hemochromatosis protein and transferrin receptor 2 causes iron-induced liver injury in mice. Hepatology. 2012;56:585–93. doi: 10.1002/hep.25689. [DOI] [PubMed] [Google Scholar]
- 64.Eason RJ, Adams PC, Aston CE, Searle J. Familial iron overload with possible autosomal dominant inheritance. Aust N Z J Med. 1990;20:226–30. doi: 10.1111/j.1445-5994.1990.tb01024.x. [DOI] [PubMed] [Google Scholar]
- 65.Njajou OT, Vaessen N, Joosse M, Berghuis B, van Dongen JW, Breuning MH, et al. A mutation in SLC11A3 is associated with autosomal dominant hemochromatosis. Nat Genet. 2001;28:213–4. doi: 10.1038/90038. [DOI] [PubMed] [Google Scholar]
- 66.Montosi G, Donovan A, Totaro A, Garuti C, Pignatti E, Cassanelli S, et al. Autosomal-dominant hemochromatosis is associated with a mutation in the ferroportin (SLC11A3) gene. J Clin Invest. 2001;108:619–23. doi: 10.1172/JCI13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pietrangelo A. The ferroportin disease. Blood Cells Mol Dis. 2004;32:131–8. doi: 10.1016/j.bcmd.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 68.Wallace DF, Subramaniam VN. Non-HFE haemochromatosis. World J Gastroenterol. 2007;13:4690–8. doi: 10.3748/wjg.v13.i35.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schimanski LM, Drakesmith H, Merryweather-Clarke AT, Viprakasit V, Edwards JP, Sweetland E, et al. In vitro functional analysis of human ferroportin (FPN) and hemochromatosis-associated FPN mutations. Blood. 2005;105:4096–102. doi: 10.1182/blood-2004-11-4502. [DOI] [PubMed] [Google Scholar]
- 70.De Domenico I, Ward DM, Nemeth E, Vaughn MB, Musci G, Ganz T, et al. The molecular basis of ferroportin-linked hemochromatosis. Proc Natl Acad Sci USA. 2005;102:89:55–60. doi: 10.1073/pnas.0503804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drakesmith H, Schimanski LM, Ormerod E, Merryweather-Clarke AT, Viprakasit V, Edwards JP, et al. Resistance to hepcidin is conferred by hemochromatosis-associated mutations of ferroportin. Blood. 2005;106:1092–7. doi: 10.1182/blood-2005-02-0561. [DOI] [PubMed] [Google Scholar]
- 72.Porter JB. Pathophysiology of transfusional iron overload: contrasting patterns in thalassemia major and sickle cell disease. Hemoglobin. 2009;33(Suppl 1):S37–45. doi: 10.3109/03630260903346627. [DOI] [PubMed] [Google Scholar]
- 73.Olivieri NF, Brittenham GM. Management of the thalassemias. Cold Spring Harb Perspect Med. 2013;3:a011767. doi: 10.1101/cshperspect.a011767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanno T, Bhanu NV, Oneal PA, Goh SH, Staker P, Lee YT, et al. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med. 2007;13:1096–101. doi: 10.1038/nm1629. [DOI] [PubMed] [Google Scholar]
- 75.Kautz L, Jung G, Du X, Gabayan V, Chapman J, Nasoff M, et al. Erythroferrone contributes to hepcidin suppression and iron overload in a mouse model of β-thalassemia. Blood. 2015;126:2031–7. doi: 10.1182/blood-2015-07-658419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA) Nat Genet. 2008;40:569–71. doi: 10.1038/ng.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hooper JD, Campagnolo L, Goodarzi G, Truong TN, Stuhlmann H, Quigley JP. Mouse matriptase-2: identification, characterization and comparative mRNA expression analysis with mouse hepsin in adult and embryonic tissues. Biochem J. 2003;373:689–702. doi: 10.1042/BJ20030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8:502–11. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parrow NL, Fleming RE. Bone morphogenetic proteins as regulators of iron metabolism. Annu Rev Nutr. 2014;34:77–94. doi: 10.1146/annurev-nutr-071813-105646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 81.Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 82.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–9. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 83.Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106:2884–9. doi: 10.1182/blood-2005-05-1845. [DOI] [PubMed] [Google Scholar]
- 84.Lin L, Nemeth E, Goodnough JB, Thapa DR, Gabayan V, Ganz T. Soluble hemojuvelin is released by proprotein convertase-mediated cleavage at a conserved polybasic RNRR site. Blood Cells Mol Dis. 2008;40:122–31. doi: 10.1016/j.bcmd.2007.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xia Y, Babitt JL, Sidis Y, Chung RT, Lin HY. Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood. 2008;111:5195–204. doi: 10.1182/blood-2007-09-111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Steinbicker AU, Bartnikas TB, Lohmeyer LK, Leyton P, Mayeur C, Kao SM, et al. Perturbation of hepcidin expression by BMP type I receptor deletion induces iron overload in mice. Blood. 2011;118:4224–30. doi: 10.1182/blood-2011-03-339952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mayeur C, Leyton PA, Kolodziej SA, Yu B, Bloch KD. BMP type II receptors have redundant roles in the regulation of hepatic hepcidin gene expression and iron metabolism. Blood. 2014;124:2116–23. doi: 10.1182/blood-2014-04-572644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu XG, Wang Y, Wu Q, Cheng WH, Liu W, Zhao Y, et al. HFE interacts with the BMP type I receptor ALK3 to regulate hepcidin expression. Blood. 2014;124:1335–43. doi: 10.1182/blood-2014-01-552281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McDonald CJ, Wallace DF, Ostini L, Subramaniam VN. Parenteral vs. oral iron: influence on hepcidin signaling pathways through analysis of Hfe/Tfr2-null mice. Am J Physiol Gastrointest Liver Physiol. 2014;306:G132–9. doi: 10.1152/ajpgi.00256.2013. [DOI] [PubMed] [Google Scholar]
- 90.Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117:1933–9. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang RH, et al. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112:1503–9. doi: 10.1182/blood-2008-03-143354. [DOI] [PubMed] [Google Scholar]
- 92.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41:478–81. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 93.Andriopoulos B, Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41:482–7. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Enns CA, Ahmed R, Wang J, Ueno A, Worthen C, Tsukamoto H, et al. Increased iron loading induces Bmp6 expression in the non-parenchymal cells of the liver independent of the BMP-signaling pathway. PLoS One. 2013;8:e60534. doi: 10.1371/journal.pone.0060534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, Müller A, Boutros M, et al. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood. 2010;115:2657–65. doi: 10.1182/blood-2009-09-238105. [DOI] [PubMed] [Google Scholar]
- 96.Patel N, Masaratana P, Diaz-Castro J, Latunde-Dada GO, Qureshi A, Lockyer P, et al. BMPER protein is a negative regulator of hepcidin and is up-regulated in hypotransferrinemic mice. J Biol Chem. 2012;287:4099–106. doi: 10.1074/jbc.M111.310789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goh JB, Wallace DF, Hong W, Subramaniam VN. Endofin, a novel BMP-SMAD regulator of the iron-regulatory hormone, hepcidin. Sci Rep. 2015;5:13986. doi: 10.1038/srep13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, Devaux I, et al. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110:1037–44. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ganz T. Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood. 2003;102:783–8. doi: 10.1182/blood-2003-03-0672. [DOI] [PubMed] [Google Scholar]
- 100.Nemeth E, Valore EV, Territo M, Schiller G, Lichtenstein A, Ganz T. Hepcidin, a putative mediator of anemia of inflammation, is a type II acute-phase protein. Blood. 2003;101:2461–3. doi: 10.1182/blood-2002-10-3235. [DOI] [PubMed] [Google Scholar]
- 101.Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, et al. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271–6. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, et al. STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology. 2007;132:294–300. doi: 10.1053/j.gastro.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 103.Lee P, Peng H, Gelbart T, Wang L, Beutler E. Regulation of hepcidin transcription by interleukin-1 and interleukin-6. Proc Natl Acad Sci USA. 2005;102:1906–10. doi: 10.1073/pnas.0409808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Armitage AE, Eddowes LA, Gileadi U, Cole S, Spottiswoode N, Selvakumar TA, et al. Hepcidin regulation by innate immune and infectious stimuli. Blood. 2011;118:4129–39. doi: 10.1182/blood-2011-04-351957. [DOI] [PubMed] [Google Scholar]
- 105.Smith CL, Arvedson TL, Cooke KS, Dickmann LJ, Forte C, Li H, et al. IL-22 regulates iron availability in vivo through the induction of hepcidin. J Immunol. 2013;191:1845–55. doi: 10.4049/jimmunol.1202716. [DOI] [PubMed] [Google Scholar]
- 106.Wallace DF, Subramaniam VN. Analysis of IL-22 contribution to hepcidin induction and hypoferremia during the response to LPS in vivo. Int Immunol. 2015;27:281–7. doi: 10.1093/intimm/dxu144. [DOI] [PubMed] [Google Scholar]
- 107.Mendel GA. Studies on iron absorption. I. The relationships between the rate of erythropoiesis, hypoxia and iron absorption. Blood. 1961;18:727–36. [PubMed] [Google Scholar]
- 108.Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117:1926–32. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lakhal S, Schödel J, Townsend AR, Pugh CW, Ratcliffe PJ, Mole DR. Regulation of type II transmembrane serine proteinase TMPRSS6 by hypoxia-inducible factors: new link between hypoxia signaling and iron homeostasis. J Biol Chem. 2011;286:4090–7. doi: 10.1074/jbc.M110.173096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pinto JP, Ribeiro S, Pontes H, Thowfeequ S, Tosh D, Carvalho F, et al. Erythropoietin mediates hepcidin expression in hepatocytes through EPOR signaling and regulation of C/EBPalpha. Blood. 2008;111:5727–33. doi: 10.1182/blood-2007-08-106195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Q, Davidoff O, Niss K, Haase VH. Hypoxia-inducible factor regulates hepcidin via erythropoietin-induced erythropoiesis. J Clin Invest. 2012;122:4635–44. doi: 10.1172/JCI63924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tanno T, Porayette P, Sripichai O, Noh SJ, Byrnes C, Bhupatiraju A, et al. Identification of TWSG1 as a second novel erythroid regulator of hepcidin expression in murine and human cells. Blood. 2009;114:181–6. doi: 10.1182/blood-2008-12-195503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kautz L, Jung G, Valore EV, Rivella S, Nemeth E, Ganz T. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat Genet. 2014;46:678–84. doi: 10.1038/ng.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kroot JJ, van Herwaarden AE, Tjalsma H, Jansen RT, Hendriks JC, Swinkels DW. Second round robin for plasma hepcidin methods: first steps toward harmonization. Am J Hematol. 2012;87:977–83. doi: 10.1002/ajh.23289. [DOI] [PubMed] [Google Scholar]
- 115.McDonald CJ, Ostini L, Wallace DF, Lyons A, Crawford DH, Subramaniam VN. Next-generation sequencing: Application of a novel platform to analyze atypical iron disorders. J Hepatol. 2015;63:1288–93. doi: 10.1016/j.jhep.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 116.Ramos E, Ruchala P, Goodnough JB, Kautz L, Preza GC, Nemeth E, et al. Minihepcidins prevent iron overload in a hepcidin-deficient mouse model of severe hemochromatosis. Blood. 2012;120:3829–36. doi: 10.1182/blood-2012-07-440743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schmidt PJ, Toudjarska I, Sendamarai AK, Racie T, Milstein S, Bettencourt BR, et al. An RNAi therapeutic targeting Tmprss6 decreases iron overload in Hfe(−/−) mice and ameliorates anemia and iron overload in murine β-thalassemia intermedia. Blood. 2013;121:1200–8. doi: 10.1182/blood-2012-09-453977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guo S, Casu C, Gardenghi S, Booten S, Aghajan M, Peralta R, et al. Reducing TMPRSS6 ameliorates hemochromatosis and β-thalassemia in mice. J Clin Invest. 2013;123:1531–41. doi: 10.1172/JCI66969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van Eijk LT, John AS, Schwoebel F, Summo L, Vauléon S, Zöllner S, et al. Effect of the antihepcidin Spiegelmer lexaptepid on inflammation-induced decrease in serum iron in humans. Blood. 2014;124:2643–6. doi: 10.1182/blood-2014-03-559484. [DOI] [PMC free article] [PubMed] [Google Scholar]