Abstract
Split spinach aptamer (SSA) probes for fluorescent analysis of nucleic acids were designed and tested. In SSA design, two RNA or RNA/DNA strands hybridized to a specific nucleic acid analyte and formed a binding site for DFHBI dye, which was accompanied by up to 270-fold increase in fluorescence. The major advantage of the SSA probe over state-of-the art fluorescent probes is high selectivity: it produces only the background fluorescence in the presence of single base mismatched analyte even at room temperature. SSA is a promising tool for label-free analysis of nucleic acids at ambient temperatures.
Keywords: Label-free probe, high selectivity, RNA analysis, split probe, aptameric sensor
Table of Contents
Two strands (SSA_m and SSA_f) hybridize to DNA or RNA analyte and form a binding site for organic dye DFHBI. Binding of DFHBI results in up to 270-fold fluorescence increase. The probe is highly selective to a single base mismatch at ambient temperatures.
Hybridization probes that fluoresce upon binding to specific nucleic acid sequences (instantaneous probes) have attracted significant attention due to the possibility of immediate detection of specific nucleic acids in mix-and-read format i. e. without the need for time-consuming and effort-intensive downstream analysis (e.g. by electrophoresis).[1] Practically significant representatives of such probes include adjacent hybridization probes,[2] molecular beacon (MB) probes,[3] and their variations.[3c, 4] Special emphasis is given to the development of the probes that enable RNA detection in live cells.[5] Such probes should operate under physiological conditions (pH, salt concentration, temperature) and should be selective enough to fluoresce only in the presence of specific RNA. Here, we report on new fluorescent probes that operate in the mix-and-read format. The major advantages of the probes are (i) label-free design and (ii) high selectivity of DNA and RNA recognition under physiological conditions.
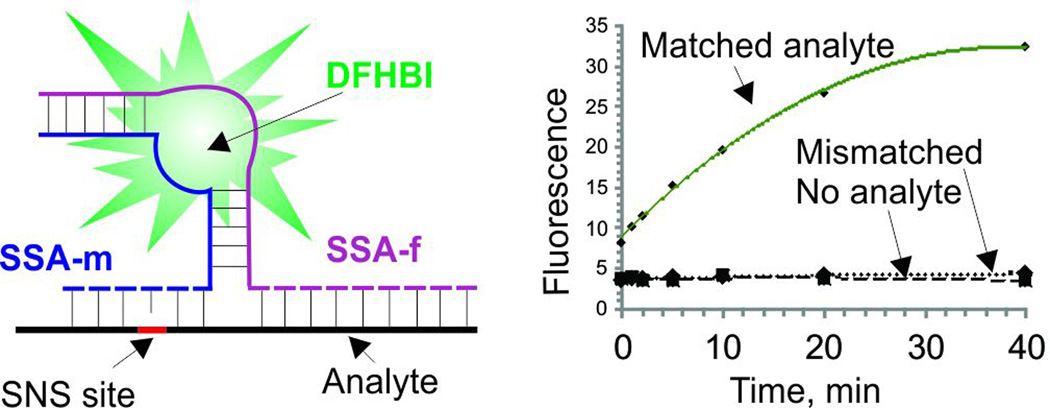
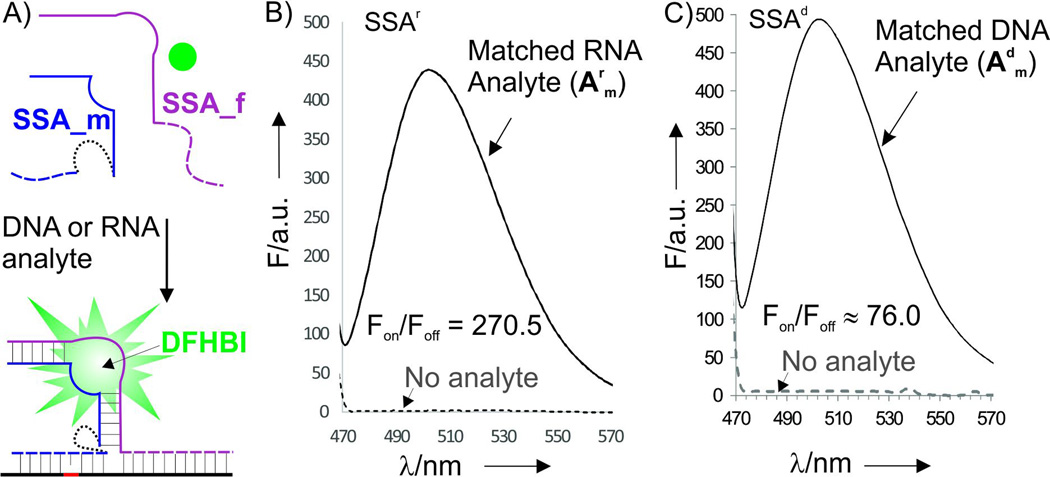
The probes were designed based on the recently isolated spinach aptamer,[6] an RNA molecule with the affinity to a low-fluorescent dye 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI, Fig. 1). Binding of DFHBI to the aptamer increases its fluorescence.[6] To design the split spinach aptamer (SSA) probes, we divided the structure of spinach aptamer into two parts (SSA_f and SSA_m) and linked each part with an analyte-binding arm, a fragment complementary to DNA or RNA analyte (dashed lines in Fig. 1A). Splitting of the dye-binding core of the aptamer prevented DFHBI from binding, and the fluorescence of the dye remained low, when no nucleic acid analyte was present. Hybridization of strands SSA_f and SSA_m to the adjacent fragments of the analyte stabilized the DFHBI-binding site, thus resulting in tighter binding of the dye to the aptameric core, which was accompanied by the increase in fluoresce (Fig. 1A bottom).
Figure 1.
General design and fluorescent response of the split spinach aptamer (SSA) probe and the fluorescent reposed of SSAr and SSAd probes. A) Two strands, SSA_m and SSA_f hybridize to a specific DNA or RNA analyte and re-form a binding site for DFHBI organic dye. Binding of the dye by the aptamer results in fluorescent increase. Dashed lines represent analyte-binding arms, which were DNA in SSAd or RNA in SSAr. Dotted line is either diuridylate (UU) linker for SSAr or triethylene glycol for SSAd (see Fig. S1 for detailed design). B) Fluorescent response of SSAr_m (2.6 µM), SSAr_f (3.6 µM) and DFHBI dye (1 µM) in the absence or presence of fully matched analyte Amr (1.38 µM). Emission spectrum (λex = 450 nm) were recorded after 90 min of incubation. C) Fluorescent response of SSAd_m (2 µM), SSAd_f (3.6 µM) and DFHBI dye (2 µM) in the absence or presence of fully matched DNA analyte Amd (5.5 µM). Emission spectrum (λex = 450 nm) were recorded after 30 min of incubation.
Two types of probes were designed: SSAr and SSDd. SSAr consisted entirely of ribonucleotides. To enable greater conformational flexibility, diuridylate (UU) linker was used to attach the analyte-binding arm of strand m to the aptamer half-core (Fig. 1A). The linker was crucial to prevent the interference between the hybridization of the strand to the analyte and the correct formation of the aptamer’s DFHBI binding pocket (see examples of less successful SSA designs in supporting materials, Fig. S1-S3). SSAr can potentially be expressed in live cells and used for imaging of specific mRNAs.[5] On the other hand, for in vitro RNA/DNA analysis, DNA probes are preferable due to greater chemical stability, lower synthetic cost and reduced ability to form intramolecular structures. Therefore, in our SSAd design the analyte-binding arms were made of deoxyribonucleotides, and connected to the aptameric portion of SSAd_m via triethylene glycol linker. In this proof-of-concept study, we targeted a fragment of inhA gene from Mycobacterium tuberculosis (Mtb), which contains point mutation associated with Mtb resistance to one of the key drug of tuberculosis treatment – isoniazid. In this work, a C->T mutation was targeted using the following DNA and RNA analytes: matched DNA analyte Adm (5’-GCG GCA TGG GTA TGG GCC ACT GAC A C A ACA CAA GGA C) and a single-base mismatched Admm (5’-GCG GCA TGG GTA TGG GCC ACT GAC A T A ACA CAA GGA C), as well as their RNA counterparts Arm and Armm.
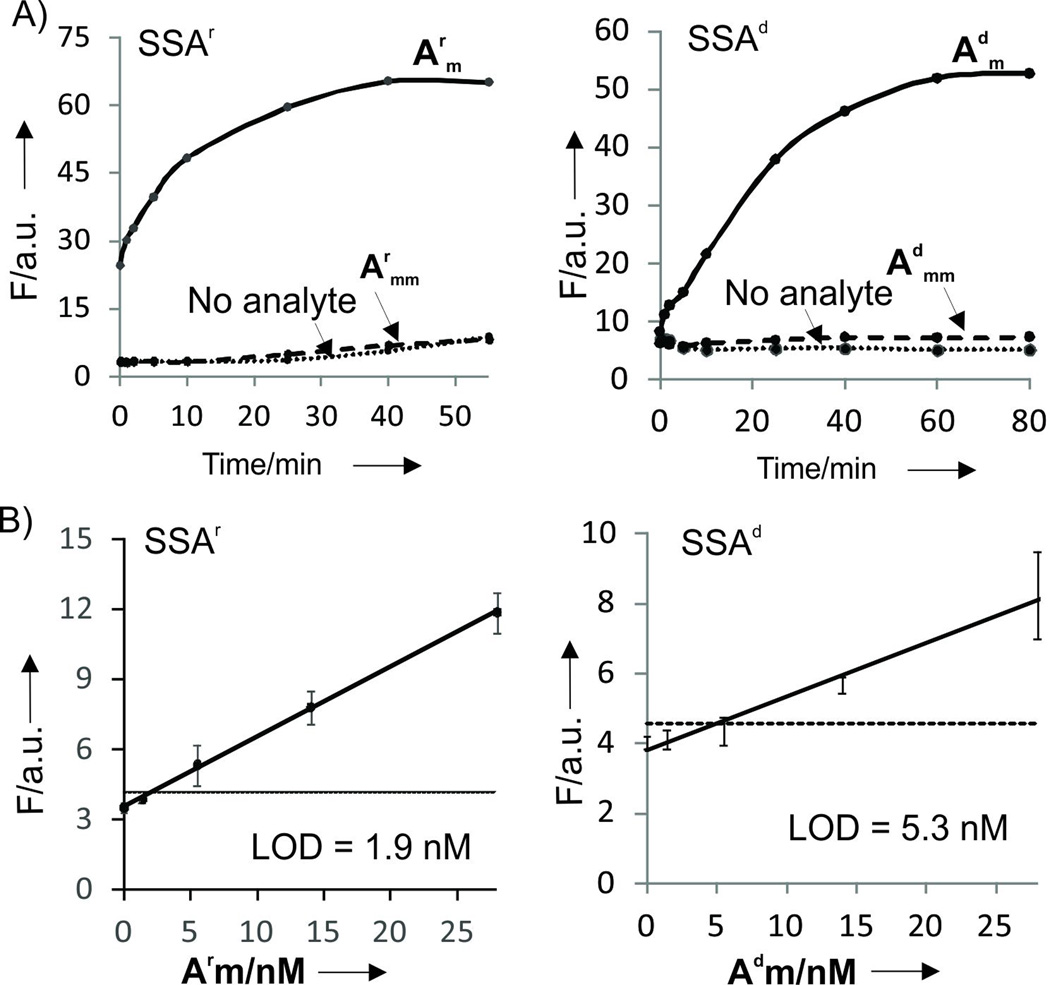
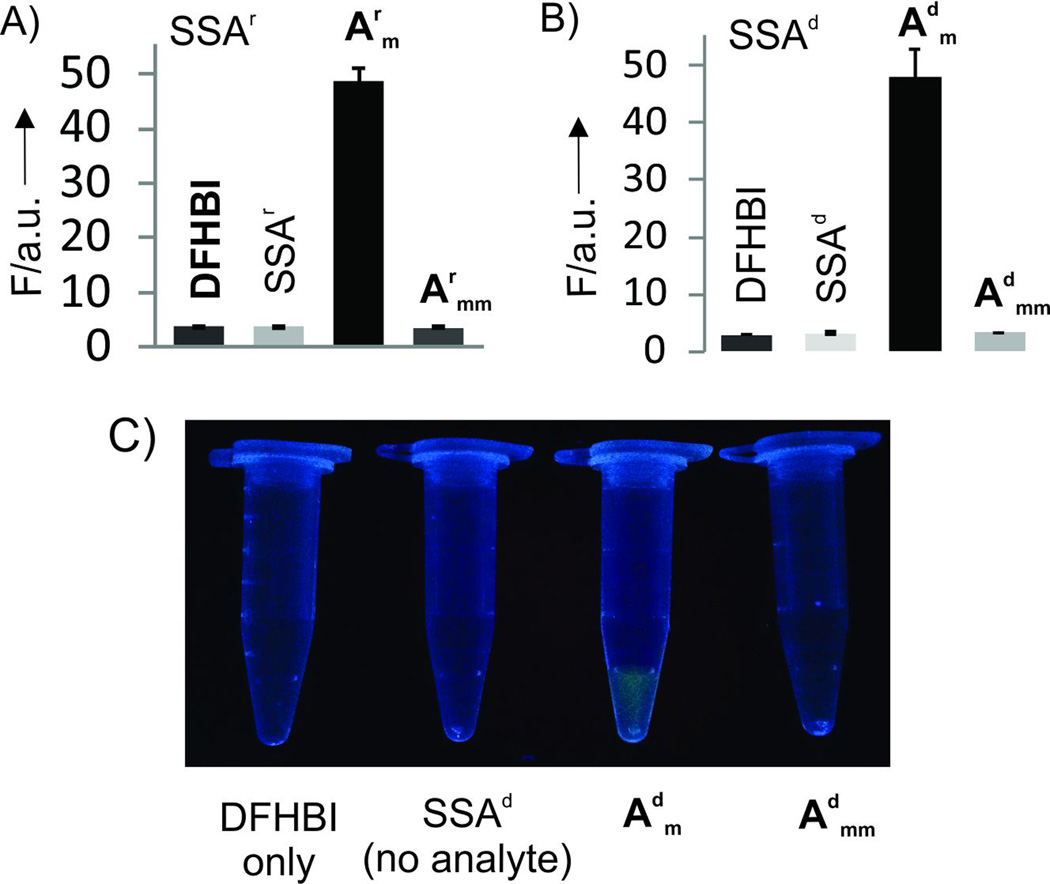
Emission spectra of SSA probes recorded in the absence or presence of the fully matched analytes demonstrated turn-on ratio (Fon/Foff) up to 270 and 76 for SSAr and SSAd probes, respectively (Fig. 1B,C), which exceeds that of the instantaneous probes currently used for nucleic acid analysis.[2, 3c, 7] Importantly, the increase in fluorescence was observed immediately after addition of analytes (Fig. 2A). Both SSAd and SSAr demonstrated limits of detection (LODs) in the low nanomolar range: 1.8 nM for SSAr/Arm, 5.3 nM for SSAd/Arm or 1.5 nM SSAd/Adm (Figs. 2B and S4). These LODs fall in the range of that reported for a typical MB probe.[3c] Further we demonstrated that SSA probes are highly selective and can differentiate single base substituted analytes even at room temperature. Indeed, no fluorescence above the background was observed when fully matched analyte was replaced with a single based mismatched one (Fig. 3 A and B). The high selectivity of the analyte recognition can be visually monitored upon light irradiation of the samples (Fig. 3C). This high selectivity of SSAr was preserved at 37°C (Fig. S5), which might be important for future application of the probe for RNA monitoring in live cells. Overall, the performance of SSA probes is comparable with currently used state-of-the-art practically useful MB probes in terms of LOD, but SSA probes have better turn-on ratios and selectivity at ambient temperatures.[3c]
Figure 2.
Kinetics and limit of detection (LOD) of the SSA probes. A) Time dependence of fluorescent response of SSAr and RNA analytes (left) and SSAd and DNA analytes (right). The apparent difference in signal-to-noise ratio in comparison with Figure 1 B and C is due to the different analyte concentrations. Reaction mixtures contained: 1 µM DFHBI, 2.6 µM SSAr_m and 3.6 µM SSAr_f, 275 nM RNA analytes; or 2 µM DFHBI, 2 µM SSAd_m and 3.6 µM SSAd_f, 275 nM DNA analytes. B) Limits of detection (LOD) for SSAr and SSAd after 30 min of incubation. Averaged data from three independent experiments with standard deviations are presented.
Figure 3.
Selectivity of the SSA probes. A) Fluorescence response of SSAr in the presence of 100 nM of either matched (Arm) or single base mismatched (Armm) RNA analytes. B) Fluorescence response of SSAd in the presence of 100 nM of either matched (Adm) or single base mismatched (Admm) DNA analytes. The data are average values of 3 independent experiments with standard deviations. C) Photograph of the SSAd samples from panel B upon excitation with transilluminator. The controls samples were as follows: DFHBI, DFHBI dye only; SAA, DFHBI dye, SSA_m, and SSA_f (no analyte). The concentrations were as specified in Figure 2 legend.
Split (binary) hybridization probes have attracted significant attention due to their high selectivity in recognition of nucleic acids at ambient temperatures,[2, 7, 8] which otherwise is difficult to achieve. In split probe design, one of the analyte-binding arms can be short (7–9 nucleotides in this study, see SI for more details) to form stable hybrid only with a perfectly matched sequence, while the entire recognition site remains long (e.g. 28 nucleotides in this study). Earlier, we introduced the strategy of split aptameric probes for nucleic acid recognition by designing split malachite green (MGA) aptamer probe.[8a] The probe was proven to be a versatile tool of RNA nanotechnology and synthetic biology.[9] The disadvantages of split MGA probe was low fluorescence intensity and strong photobleaching, which limited its practical applications. Spinach aptamer isolated by Paige et al.[6] has attracted significant attention both as a tool for fluorescent monitoring of endogenous RNA in live cells[10] and as a sensor platform for detection of biological molecules and metal ions in vitro.[11] Self-reconstituting split spinach aptamer constructs were designed recently for fluorescent monitoring of RNA assembly, functional imaging of viral genome trafficking and for monitoring of ribozyme activity.[12] However, these constracts did not operate as missmatch-selective sensors for nucleic acid analysis. Additionally, structural swithching spinach aptamer-based monolith sensors were developed for the detection of RNA and DNA.[13] However, long RNA probes have a disadvntage of misfolding in non-fuctional structure, which can be as much as 80% in case of spinach apatamer,[14] and lead to low (ca. 5) turn-on ratio.[13a] They may also respond slowly[13a] due to the time required for structural switch between the two energetically close RNA conformations. Even though spinach molecular beacon has been shown to diffrentiate several specific single nucleotide substitutions,[13a] in general, structural switching constructs are shown to poorly differentiate single base mismatches.[15] Moreover, long RNA structure-switching sensors requier tedious optimization and are expesive commersial products. Finally, structural swithcing spinach construct was able to detect only about 25 nM analytes even when sophisticated amplification stratages are implemented.[13c]
In this work, we took advantage of the recently published X-ray structure of spinach aptamer,[16] which reviled actual folding of spinach aptamer and localized the G- quadruplex -based binding site for DHFBI. Our prior attempts to design SSA based on predicted secondary structure[6] were unsuccessful. Practically important features of SSA probes are the following. (i) High selectivity at ambient temperatures. (ii) Mix-and-read reporting format with up to 270-fold turn-on ratios, which is better than that of other mix-and-read probes including adjacent[2] and MB probes.[3] (iii) LOD in low nanamolar range, which is about one order of magnitude better than that for structure switching spinach sensors.[13b] (iv) Label-free design: there is no need for conjugation of oligonucleotides with a fluorophore or quencher dye, which eliminates the need for purification of SSA strands prior to usage (note that all SSA_f and SSA_m stands used in this study were only desaulted after solid-phase synthesis, see Table S1). (v) It is easy to tailor SSA probes for recognition of each new analyte by simple change of the analyte-binding arms. (vi) Finally, being short, RNA or RNA/DNA strands, SSA components can be convinently obtained from industrial suppliers of custom-made nucleic acids.
In conclusion, we have designed two label-free fuorescent probes for nucleic acid analysis. The probes demmostrate supperior performance in comparison with relevant atate-of–the art probes. The probes can be custom-designed and purchased as synthetic products, which makes them affordable by any laboratory. The applicability of SSA for in vivo detection of single nucleotide differences in RNA or in PCR products are the subjects of the follow up studies.
Experimental Section
All oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). DNAse/RNAse free water was purchased from Fisher Scientific and used for all assays. DFHBI was purchased from Lucerna, Inc. (New York, NY). All experiments were done at room temperature (22.5°C) or at 37°C in a buffer containing 10 mM Tris-HCl (pH 7.4) 100 mM KCl, 5 mM MgCl2. The fluorescent spectra were taken on a Fluorescence Spectrometer LS55 (PerkinElmer). The emission was registered at 500 nm upon excitation at 450 nm. The data were processed using MS Excel. The data of at least 3 independent experiments are included in the calculation of average values and standard deviations.
Supplementary Material
Acknowledgments
We greatful to Dr. Gerasimova for discussion and cariful reading of the manuscript. Funding from NSF CCF (1423219) and NIH NIAID R15AI10388001A1 is greatly appreciated.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- 1.a) Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Khakshoor O, Kool ET. Chem. Commun. 2011;47:7018–7024. doi: 10.1039/c1cc11021g. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Marras SA, Tyagi S, Kramer FR. Clin. Chim. Acta. 2006;363:48–60. doi: 10.1016/j.cccn.2005.04.037. [DOI] [PubMed] [Google Scholar]; (d) Juskowiak B. Anal. Bioanal. Chem. 2011;399:3157–3176. doi: 10.1007/s00216-010-4304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Obliosca JM, Liu C, Batson RA, Babin MC, Werner JH, Yeh HC. Biosensors (Basel) 2013;3:185–200. doi: 10.3390/bios3020185. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Suzuki Y, Yokoyama K. Biosensors. 2015;5:337–363. doi: 10.3390/bios5020337. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Kato T, Shimada I, Kimura R, Hyuga M. Chem Commun. 2016;52:4041–4044. doi: 10.1039/c5cc08816j. [DOI] [PubMed] [Google Scholar]; h) Knez K, Spasic D, Janssen KP, Lammertyn J. Analyst. 2014;139:353–370. doi: 10.1039/c3an01436c. [DOI] [PubMed] [Google Scholar]; i) Lai YH, Sun SC, Chuang MC. Biosensors (Basel) 2014;4:273–300. doi: 10.3390/bios4030273. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Østergaard ME, Hrdlicka PJ. Chem. Soc. Rev. 2011;40:5771–5788. doi: 10.1039/c1cs15014f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Cardullo RA, Agrawal S, Flores C, Zamecnik PC, Wolf DE. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Oser A, Valet G. Angew. Chem., Int. Ed. Engl. 1990;29:1167–1168. [Google Scholar]; d) Silverman AP, Kool ET. Chem. Rev. 2006;106:3775–3789. doi: 10.1021/cr050057+. [DOI] [PubMed] [Google Scholar]; c) Guo J, Ju J, Turro NJ. Anal Bioanal Chem. 2012;402:3115–3125. doi: 10.1007/s00216-011-5526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]; b) Wang K, Tang Z, Yang CJ, Kim Y, Fang X, Li W, Wu Y, Medley CD, Cao Z, Li J, Colon P, Lin H, Tan W. Angew Chem Int Ed Engl. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kolpashchikov DM. Scientifica (Cairo) 2012;2012:928783. doi: 10.6064/2012/928783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Venkatesan N, Seo YJ, Kim BH. Chem. Soc. Rev. 2008;37:648–663. doi: 10.1039/b705468h. [DOI] [PubMed] [Google Scholar]; b) Satterfield BC, West JA, Caplan MR. Nucleic Acids Res. 2007;35:e76. doi: 10.1093/nar/gkm113. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tan X, Wang Y, Armitage BA, Bruchez MP. Anal Chem. Anal Chem. 2014;86:10864–10869. doi: 10.1021/ac502986g. [DOI] [PubMed] [Google Scholar]; d) Juul S, Obliosca JM, Liu C, Liu YL, Chen YA, Imphean DM, Knudsen BR, Ho YP, Leong KW, Yeh HC. Nanoscale. 2015;7:8332–8337. doi: 10.1039/c5nr01705j. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Obliosca JM, Babin MC, Liu C, Liu YL, Chen YA, Batson RA, Ganguly M, Petty JT, Yeh HC. ACS Nano. 2014;8:10150–10160. doi: 10.1021/nn505338e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Tyagi S. Nat. Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]; b) Weil TT, Parton RM, Davis I. Trends Cell Biol. 2010;7:380–390. doi: 10.1016/j.tcb.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Broude NE. Mol Microbiol. 2011;5:1137–1147. doi: 10.1111/j.1365-2958.2011.07652.x. [DOI] [PubMed] [Google Scholar]; d) Santangelo PJ, Alonas E, Jung J, Lifland AW, Zurla C. Methods Enzymol. 2012;505:383–399. doi: 10.1016/B978-0-12-388448-0.00028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Nakamura Y, Ishiguro A, Miyakawa S. Genes Cells. 2012;17:344–364. doi: 10.1111/j.1365-2443.2012.01596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Wang T, Ray J. Protein Cell. 2012;3:739–754. doi: 10.1007/s13238-012-2072-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Boutorine AS, Novopashina DS, Krasheninina OA, Nozeret K, Venyaminova AG. Molecules. 2013;18:15357–15397. doi: 10.3390/molecules181215357. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Urbanek MO, Galka-Marciniak P, Olejniczak M, Krzyzosiak WJ. RNA Biol. 2014;11:1083–1095. doi: 10.4161/rna.35506. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Urbanek MO, Galka-Marciniak P, Olejniczak M, Krzyzosiak WJ. RNA Biol. 2014;8:1083–1095. doi: 10.4161/rna.35506. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Holstein JM, Rentmeister A. Methods. 2016;98:18–25. doi: 10.1016/j.ymeth.2015.11.016. [DOI] [PubMed] [Google Scholar]; k) Wang Z, Zhang K, Wooley KL, Taylor JS. J. Nucleic Acids. 2012;2012:962652. doi: 10.1155/2012/962652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Paige JS, Wu KY, Jaffrey SR. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Strack RL, Disney MD, Jaffrey SR. Nature Methods. 2013;10:1219–1224. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Kolpashchikov DM. Chem. Rev. 2010;110:4709–4723. doi: 10.1021/cr900323b. [DOI] [PubMed] [Google Scholar]; b) Karhunena U, Malmi E, Brunet E, Rodríguez-Ubis JC, Soukka T. Sensors and Actuators B. 2015;211:297–302. [Google Scholar]; c) Ma JL, Yin BC, Ye BC. Analyst. 2016;141:1301–1306. doi: 10.1039/c5an02446c. [DOI] [PubMed] [Google Scholar]
- 8.a) Kolpashchikov DM. J. Am. Chem. Soc. 2005;127:12442–12443. doi: 10.1021/ja0529788. [DOI] [PubMed] [Google Scholar]; b) Sando S, Narita A, Aoyama Y. Chembiochem. 2007;8:1795–1803. doi: 10.1002/cbic.200700325. [DOI] [PubMed] [Google Scholar]
- 9.a) Afonin KA, Danilov EO, Novikova IV, Leontis NB. Chembiochem. 2008;9:1902–1905. doi: 10.1002/cbic.200800183. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, Desai R, Santhanam A, Grabow WW, Jaeger L, Heldman E, Reiser J, Chiu W, Freed EO, Shapiro BA. Nano Lett. 2014;14:5662–5671. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Afonin KA, Desai R, Viard M, Kireeva ML, Bindewald E, Case CL, Maciag AE, Kasprzak WK, Kim T, Sappe A, Stepler M, Kewalramani VN, Kashlev M, Blumenthal R, Shapiro BA. Nucleic Acids Res. 2014;42:2085–2097. doi: 10.1093/nar/gkt1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Kellenberger CA, Hallberg ZF, Hammond MC. Methods Mol. Biol. 2015;1316:87. doi: 10.1007/978-1-4939-2730-2_8. [DOI] [PubMed] [Google Scholar]; b) Guet D, Burns LT, Maji S, Boulanger J, Hersen P, Wente SR, Salamero J, Dargemont C. Nat. Commun. 2015;6:8882. doi: 10.1038/ncomms9882. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Han KY, Leslie BJ, Fei J, Zhang J, Ha T. J. Am. Chem. Soc. 2013;135:19033–19038. doi: 10.1021/ja411060p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) DasGupta S, Shelke SA, Li NS, Piccirilli JA. Chem. Commun. 2015;51:9034–9037. doi: 10.1039/c5cc01526j. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Strack RL, Song W, Jaffrey SR. Nat. Protoc. 2014;9:146–155. doi: 10.1038/nprot.2014.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Nakayama S, Luo Y, Zhou J, Dayie TK, Sintim HO. Chem Commun. 2012;48:9059–9061. doi: 10.1039/c2cc34379g. [DOI] [PubMed] [Google Scholar]
- 12.a) Rogers TA, Andrews GE, Jaeger L, Grabow WW. ACS Synth Biol. 2015;4:162–166. doi: 10.1021/sb5000725. [DOI] [PubMed] [Google Scholar]; b) Tsvetkova IB, Yi G, Yi Y, Kao CC, Dragnea BG. Virus Res. 2015;210:291–297. doi: 10.1016/j.virusres.2015.08.023. [DOI] [PubMed] [Google Scholar]; c) Ausländer S, Fuchs D, Hürlemann S, Ausländer D, Fussenegger M. Nucleic Acids Res. 2016;44:e94. doi: 10.1093/nar/gkw117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Bhadra S, Ellington AD. RNA. 2014;20:1183–1194. doi: 10.1261/rna.045047.114. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bhadra S, Ellington AD. Methods Enzymol. 2015;550:215–249. doi: 10.1016/bs.mie.2014.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Akter F, Yokobayashi Y. ACS Synth Biol. 2015;5:655–658. doi: 10.1021/sb500314r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Autour A, Westhof E, Ryckelynck M. Nucleic Acids Res. 2016;44:2491–2500. doi: 10.1093/nar/gkw083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Stojanovic MN, de Prada P, Landry DW. Chembiochem. 2001;2:411–415. doi: 10.1002/1439-7633(20010601)2:6<411::AID-CBIC411>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]; b) Sando S, Sasaki T, Kanatani K, Aoyama Y. J. Am. Chem. 2003;125:15720–15721. doi: 10.1021/ja0386492. [DOI] [PubMed] [Google Scholar]
- 16.Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, Rice PA, Piccirilli JA. Nat. Chem. Biol. 2014;10:686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.