Abstract
We provide datasets from combined ex vivo diffusion tensor imaging (DTI) and Clear Lipid-exchanged, Anatomically Rigid, Imaging/immunostaining compatible, Tissue hYdrogel (CLARITY) performed on intact mouse brains. DTI-derived measures of fractional anisotropy (FA), radial diffusivity (RD), and axial diffusivity (AD) were compared to antibody-based labeling of myelin basic protein (MBP), as measured by fluorescence microscopy. We used a customized CLARITY hydrogel solution to facilitate whole brain tissue clearing and subsequent immunolabeling. We describe how CLARITY was made compatible with magnetic resonance imaging with the intention of facilitating future multimodal imaging studies that may combine noninvasive imaging with 3D immunohistochemistry. These data and methods are related to the accompanying research article entitled, ‘The role of myelination in measures of white matter integrity: Combination of diffusion tensor imaging and two-photon microscopy of CLARITY intact brains’ (E.H. Chang, M. Argyelan, M. Aggarwal, T-S. Chandon, K.H. Karlsgodt, S. Mori, A.K. Malhotra, 2016) [1].
Keywords: DTI, CLARITY, Multimodal imaging, Myelination, Radial diffusivity
Specifications Table
| Subject area | Neuroscience, Radiology |
| More specific subject area | Diffusion tensor imaging and immunohistochemistry |
| Type of data | Table, fluorescence microscopy images, summary graphs |
| How data was acquired | Diffusion MRI was acquired on Bruker 11.7T NMR spectrometer. Two-photon laser microscopy images were acquired on Olympus FV-1000-MPE microscope. |
| Data format | Images and graphs |
| Experimental factors | Mice were transcardially perfused with a CLARITY liquid hydrogel, ex vivo brains were then immersed in PBS and 0.1 mM gadopentetate dimeglumine prior to DTI. During scanning, brains were immersed in Fomblin for susceptibility matching and to limit tissue dehydration. |
| Experimental features | TrackVis was used to create white matter regions-of-interest (ROIs) from DTI-derived FA maps. CLARITY images were analyzed using Imaris 8.0 with the Surfaces module to trace individual MBP-positive ROIs. |
| Data source location | Manhasset, New York, USA |
| Data accessibility | Data is provided in this article and is related to the research article [1]. |
Value of the data
-
•
This is the first dataset of CLARITY whole-brain MBP immunolabeling and the first study, to our knowledge, combining DTI with CLARITY in the same brain samples.
-
•
CLARITY immunolabeling can be used in future studies to examine other protein targets found in brain structures with complex 3D architectures and distributions. This enables molecular analysis of brain targets across intact macroscopic circuits, while maintaining microscopic resolution.
-
•
Our dataset demonstrates that within-subject multimodal imaging studies can provide useful insight into the biological basis of noninvasive imaging methods.
1. Data
The dataset includes images, videos, and plots from experiments combining ex vivo DTI and two-photon microscopy of CLARITY mouse brains. Fig. 1, Fig. 2, Fig. 3 show both raw and analyzed data from these experiments indicating the relationship between various diffusivity measures and MBP immunofluorescence. Table 1 shows DTI-derived measures and MBP immunofluorescence values for major myelinated white matter tracts of the mouse brain.
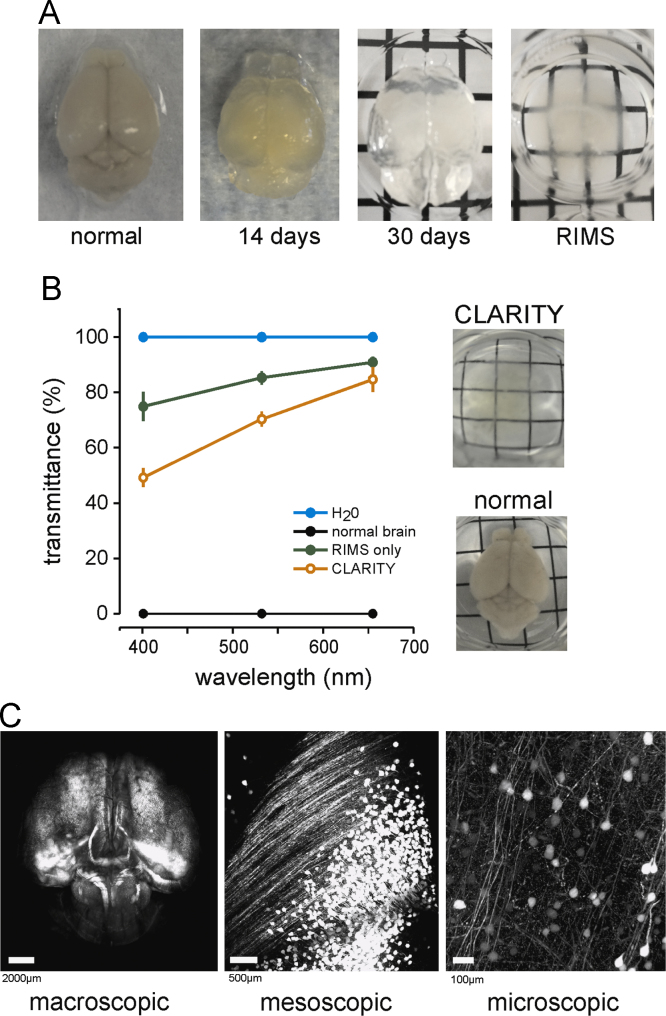
Fig. 1.
CLARITY clearing and Thy1-YFP-H images across multiple scales (A) Example images of normal PFA-fixed brain and CLARITY brains during the clearing process. (B) Plot shows mean±SEM tissue transparency of CLARITY samples at three laser wavelengths. Transmittance values were normalized to H20. Images on the right show an example whole brain prior to clearing and a CLARITY-cleared brain. (C) Example images of a CLARITY Thy1-eYFP-H intact brain demonstrating the potential for examination of fluorescence on multiple biological scales.
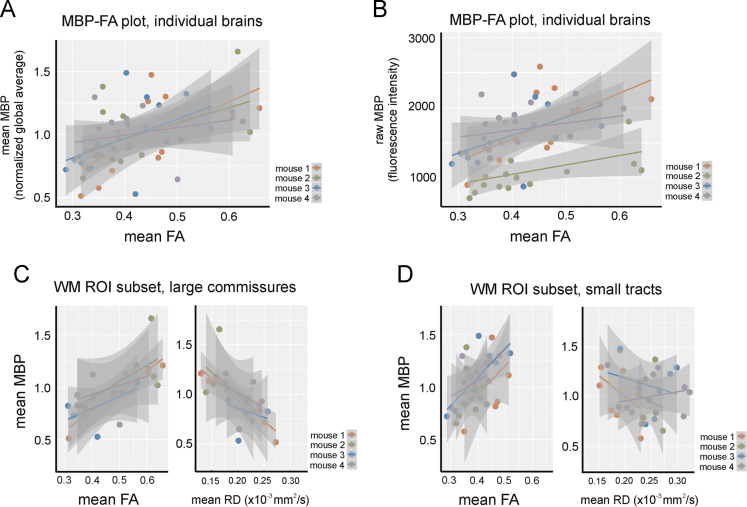
Fig. 2.
Raw MBP immunofluorescence and individual brain data from FA, AD, and RD correlations. (A) Scatter plots showing MBP-FA correlations for each individual mouse brain across the entire set of WM ROIs. MBP immunofluorescence was normalized to a global average. (B) Scatter plots showing MBP-FA correlations for each individual mouse brain, with raw MBP fluorescence intensity values. (C) Scatter plots of MBP-FA and MBP-RD correlations for a subset of WM tracts (corpus callosum, fimbria, and anterior commissure) showing data from each individual brain, with MBP normalization to a global average. (D) Scatter plots of MBP-FA and MBP-RD correlations for a different subset of WM tracts (fornix, stria medullaris, fasciculus retroflexus, and mammillothalamic tract) showing data from each individual brain, with MBP normalization to a global average. For all plots, linear regressions are shown with 95% confidence intervals (grey).
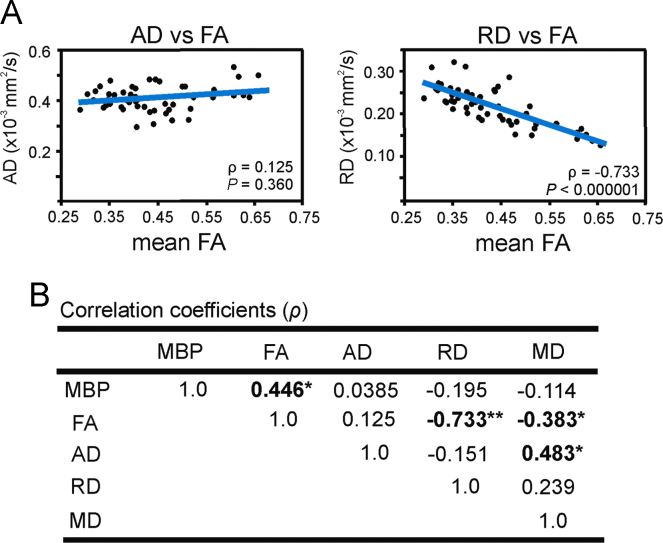
Fig. 3.
Additional FA and directional diffusivity correlation plots. (A) Correlation plots of directional diffusivity and FA measures. Left, FA did not correlate significantly with AD. Right, RD had a significant negative correlation with FA. (B) Correlation matrix for MBP, FA, AD, RD, and MD. Statistically significant correlations are indicated in bold, *P<0.005, **P<0.00001.
Table 1.
Mean values of DTI-derived metrics and MBP immunofluorescence Table displaying mean±SEM values of DTI measures and mean MBP immunofluorescence for each WM ROI identified in this dataset. Mean values are averaged from the four mouse brains analyzed in this study. AD, RD, and MD values are in units of ×10−3 mm2/s. Raw MBP fluorescence intensities are unit-less with a range of 0–4095.
| White matter ROI | FA | AD | RD | MD | MBP |
|---|---|---|---|---|---|
| Corpus collosum, genu | 0.41±0.0042 | 0.39±0.0086 | 0.22±0.0085 | 0.28±0.0081 | 1333±239 |
| Corpus collosum, body | 0.34±0.012 | 0.39±0.010 | 0.24±0.012 | 0.29±0.011 | 1139±162 |
| Corpus collosum, splenium | 0.35±0.013 | 0.41±0.0087 | 0.25±0.0060 | 0.30±0.0061 | 1231±115 |
| Anterior commissure, posterior | 0.53±0.030 | 0.44±0.025 | 0.18±0.0079 | 0.27±0.0090 | 1609±193 |
| Fimbria, left | 0.56±0.024 | 0.46±0.027 | 0.17±0.016 | 0.29±0.0031 | 1707±196 |
| Fimbria, right | 0.58±0.025 | 0.45±0.019 | 0.15±0.011 | 0.27±0.027 | 1739±225 |
| Fornix, left | 0.40±0.025 | 0.41±0.016 | 0.21±0.019 | 0.27±0.015 | 1738±367 |
| Fornix, right | 0.46±0.020 | 0.40±0.031 | 0.21±0.025 | 0.24±0.015 | 1674±295 |
| Stria medullaris, left | 0.41±0.23 | 0.43±0.024 | 0.24±0.025 | 0.29±0.020 | 1935±266 |
| Stria medullaris, right | 0.41±0.026 | 0.43±0.028 | 0.23±0.019 | 0.28±0.018 | 1734±236 |
| Fasciculus retroflexus, left | 0.42±0.026 | 0.36±0.027 | 0.21±0.025 | 0.27±0.032 | 1709±214 |
| Fasciculus retroflexus, right | 0.42±0.031 | 0.35±0.022 | 0.23±0.033 | 0.25±0.021 | 1664±206 |
| Mammillothalamic tract, left | 0.33±0.015 | 0.42±0.026 | 0.27±0.021 | 0.32±0.023 | 1172±222 |
| Mammillothalamic tract, right | 0.33±0.018 | 0.42±0.014 | 0.26±0.020 | 0.32±0.014 | 1306±205 |
Supplementary material related to this article can be found online at doi:10.1016/j.dib.2016.12.018.
The following is Supplementary material related to this article Video 1, Video 2.
3D visualization of CLARITY MBP-labeled whole mouse brain. Imaris animation shows a MBP immunolabeled whole brain. Major myelinated white matter structures such as the hippocampal commissure, corpus callosum, and stria medullaris are plainly visible.
Optical section through CLARITY MBP-labeled mouse brain. 1500 µm thick sagittal optical section near the midline showing many of the major myelinated white matter tracts used for comparison in this study. White matter in the cerebellum and brainstem (not quantified in this study) were also visibly labeled by MBP antibody.
.
2. Experimental design, materials and methods
2.1. Experimental design and animal subjects
For all experiments, we used C57BL/6J mice from Jackson Laboratories (Bar Harbor, ME). All animal procedures were approved by the Feinstein Institute Medical Research Institutional Animal Care and Use Committee and maintained according to National Institutes of Health guidelines. We first piloted our combined DTI and CLARITY [2] approach using Thy1-eYFP-H mice (n=3) that endogenously produce fluorescence signal without immunolabeling (Fig. 1C). Once we finalized a working protocol that enabled DTI followed by CLARITY, we used four C57BL/6J mice for the main experiments. The mice were first perfused with a CLARITY liquid hydrogel solution, then brains were removed and underwent ex vivo DTI scanning. Subsequently the CLARITY hydrogels were polymerized, tissue cleared, MBP immunolabeled, and finally imaged intact using a two-photon laser scanning microscope. Details of each step can be found below and also in the research article [1].
2.2. ex vivo DTI
Mouse brains were scanned in a CLARITY liquid hydrogel containing 4% paraformaldehyde and 0.1 mM gadopentetate dimeglumine (Gd-DTPA). The Gd-DTPA was used as a T1 shortening contrast agent to achieve shorter repetition times (TR), thereby allowed faster DTI acquisitions while maintaining good signal-to-noise [3]. Diffusion-weighted magnetic resonance (MR) images were acquired on an 11.7 T NMR spectrometer (Bruker BioSpin, Billerica, MA) using a three-dimensional gradient-and-spin-echo (3D DW-GRASE) sequence with twin navigator-echo phase correction [3], along 15 independent directions. A b-value of 1500 s/mm2 was used and scanning time was ~16.5 h for each mouse brain.
2.3. CLARITY clearing and MBP immunolabeling
CLARITY tissue clearing and immunolabeling were performed as previously described [2] with important modifications made to the hydrogel solution in order to obtain sufficient whole brain immunolabeling, while also maintaining tissue rigidity. We experimented with different concentrations of paraformaldehyde (PFA) and bis-acrylamide before arriving at an optimal hydrogel mixture containing 4% paraformaldehyde, 1.75% acrylamide, 0.01875% bis-acrylamide, 0.25% VA-044 initiator (Wako Chemicals USA) and 1X PBS. This resulted in a hydrogel-tissue matrix that facilitated lipid clearance and allowed for deep immunolabeling of whole mouse brains. Instead of electrophoretic tissue clearing (ETC) at high temperature, we used passive clearing at 37 °C (without agitation) in order to minimize changes in overall brain volume that commonly occur with ETC. While this passive technique is markedly slower, we found that it achieved excellent tissue transparency and structural preservation without tissue inflation (Fig. 1A and B). Following clearing, intact brains were immunlabeled with anti-MBP (1:50, EMD Millipore), followed by secondary antibody labeling with AlexaFluor 633 (1:50, Life technologies). The entire clearing and immunolabeling procedure took approximately 80 days to complete for each whole brain. Once MBP labeled, brains were immersed in refractive index matching solution (RIMS; [4]) and imaged using an Olympus FV1000-MPE with Mai Tai DeepSee Ti:Sapphire laser (SpectraPhysics).
2.4. DTI-CLARITY analysis
From the diffusion-weighted MR images, maps of FA, AD, RD, and MD (Table 1; Fig. 3) were calculated using FSL (http://www.fmrib.ox.ac.uk/fsl) software. TrackVis (http://www.trackvis.org) was used for post hoc calculations and visualization. White matter regions-of-interest (ROIs) were drawn in TrackVis from the FA map. For visualization and analysis of two-photon image stacks, we used Imaris 8.0 software (Bitplane). ROI-based analyses (Fig. 2) were conducted in the DTI space and CLARITY space with statistical comparisons using Spearman׳s rank correlations or mixed effect models performed in R software (version 3.0). Statistical significance for all tests was defined as P<0.05.
Acknowledgements
The authors would like to thank Dr. Amanda Chan for valuable technical assistance. This work was supported by Center for Intervention Development and Applied Research (P50MH080173). Funding agencies had no role in the study design, acquisition or interpretation of data, or in writing the manuscript.
Footnotes
Transparency data associated with this article can be found in the online version at doi:10.1016/j.dib.2016.12.018.
Transparency document. Supporting Material
Supplementary material.
.
References
- 1.Chang E.H., Argyelan M., Aggarwal M., Chandon T.-S., Karlsgodt K.H., Mori S., Malhotra A.K. The role of myelination in measures of white matter integrity: combination of diffusion tensor imaging and two-photon microscopy of CLARITY intact brains. Neuroimage. 2016 doi: 10.1016/j.neuroimage.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung K., Wallace J., Kim S.Y., Kalyanasundaram S., Andalman A.S., Davidson T.J., Mirzabekov J.J., Zalocusky K.A., Mattis J., Denisin A.K., Pak S., Bernstein H., Ramakrishnan C., Grosenick L., Gradinaru V., Deisseroth K. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aggarwal M., Mori S., Shimogori T., Blackshaw S., Zhang J. Three-dimensional diffusion tensor microimaging for anatomical characterization of the mouse brain. Magn. Reson. Med. 2010;64:249–261. doi: 10.1002/mrm.22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang B., Treweek J.B., Kulkarni R.P., Deverman B.E., Chen C.K., Lubeck E., Shah S., Cai L., Gradinaru V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D visualization of CLARITY MBP-labeled whole mouse brain. Imaris animation shows a MBP immunolabeled whole brain. Major myelinated white matter structures such as the hippocampal commissure, corpus callosum, and stria medullaris are plainly visible.
Optical section through CLARITY MBP-labeled mouse brain. 1500 µm thick sagittal optical section near the midline showing many of the major myelinated white matter tracts used for comparison in this study. White matter in the cerebellum and brainstem (not quantified in this study) were also visibly labeled by MBP antibody.
Supplementary material.