Abstract
The pathway of CRISPR-Cas immunity redefines the roles of RNA in the flow of genetic information and ignites excitement for next-generation gene therapy tools. CRISPR-Cas machineries offer a fascinating set of new enzyme assemblies from which one can learn principles of molecular interactions and chemical activities. The interference step of the CRISPR-Cas immunity pathway congregates proteins, RNA, and DNA into a single molecular entity that selectively destroys invading nucleic acids. Although much remains to be discovered, a picture of how the interference process takes place is emerging. This review focuses on the current structural data for the three known types of RNA-guided nucleic acid interference mechanisms. In it, we describe key features of individual complexes and we emphasize comparisons across types and along functional stages. We aim to provide readers with a set of core principles learned from the three types of interference complexes and a deep appreciation of the diversity among them.
Keywords: DNA interference, RNA silencing, ribonucleoprotein particles, prokaryote immunity
INTRODUCTION
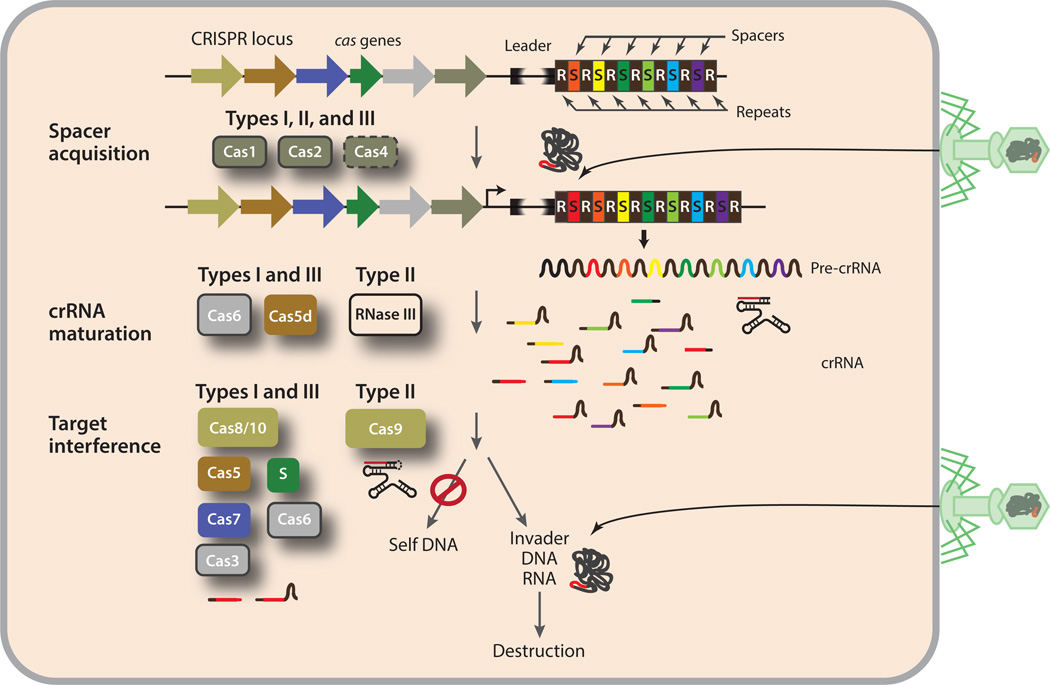
The recent discovery of adaptive immunity in prokaryotes conferred by clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins (3, 8, 18) has excited several scientific communities, including but not limited to those focused on microbiology, RNA biology, and biotechnology. The CRISPR-Cas loci are widespread in sequenced genomes of bacteria (~48%) and archaea (~84%). These loci provide microbes with defense against invading genetic elements in a sequence-specific manner; at a conceptual level, this defense mechanism is reminiscent of RNA interference in eukaryotes. The immunity mediated by CRISPR-Cas includes three functional processes: spacer acquisition, CRISPR RNA (crRNA) maturation, and target interference (Figure 1). The spacer acquisition step incorporates new spacers from the invading virus or plasmids into the CRISPR locus, the crRNA maturation step prepares functional crRNAs from the transcript of the repeat–spacer array, and the target interference step directs crRNA-guided cleavage of invading genetic elements. For extensive reviews on the CRISPR-Cas pathway, the reader is referred to References 5, 21, 35, 53, 77, 84, 86, 88, and 91.
Figure 1.
Overview of the CRISPR-Cas immunity pathways and their associated Cas proteins. In the spacer acquisition step, DNA segments are incorporated into the repeat–spacer array near the leader sequence. Cas1 and Cas2, and Cas4 in some organisms, are believed to be responsible for this process. Cas6, and in some cases Cas5d or endogenous RNase III, is the primary enzyme that processes precursor crRNA transcripts to generate mature crRNA. Additional maturation steps may take place in some systems, but these steps have not yet been characterized. During the target interference step, crRNA and the remaining Cas proteins assemble into three different types of ribonucleoprotein particles (crRNPs) that destroy invader nucleic acids and spare self nucleic acids. Abbreviations: Cas, CRISPR-associated; CRISPR, clustered regularly interspaced short palindromic repeats; crRNA, CRISPR RNA.
All three functional processes are essential for CRISPR-Cas-mediated immunity and are of great interest to biotechnology and for industry applications. Both scholarly and practical interests in CRISPR-Cas biology demand a thorough understanding of its inner workings, especially the mechanisms that underlie molecular assembly, enzyme specificity, and regulatory interactions. Over the past several years, functional and structural data on CRISPR-Cas machineries have rapidly become available, providing unprecedented access to information about the mechanism(s) of multicomponent protein–nucleic acid assembly and nucleic acid interference. This review summarizes the biochemical and structural discoveries related to the machineries responsible for nucleic acid interference mediated by CRISPR-Cas. Other excellent reviews of the similar and related processes can be found in References 2, 31, 32, 39, 45, 66, 86. In the following sections, we describe the nomenclature, function, organization, and nucleic acid interference models of all known interference complexes, and we aim to make mechanistic connections by comparing the available functional and structural data. We conclude with a discussion of the structural basis for genome editing applications.
CRISPR-Cas SYSTEMS
Functional CRISPR-Cas loci comprise a DNA repeat–spacer array, a set of cas genes, and a leader sequence. The locus-specific repeat sequences range from 23 to 48 nucleotides (nt) in length, and each bears the identity of a particular CRISPR-Cas system and dictates CRISPR-specific interactions (44). The repeats are interspersed with distinct spacer sequences that are roughly 24–70 nt long, and some of these sequences match regions of bacteriophage or plasmid DNA (7, 34, 55, 62). The complementarity between the spacers and the invader genetic elements is the basis upon which the CRISPR-Cas systems target invaders for destruction. The leader sequence is believed to direct transcription of the repeat–spacer array and acquisition of new spacers (63, 94). With only one exception (12), Cas proteins are responsible for all three functional steps of the CRISPR-Cas immunity (21, 86).
Although the Cas proteins are overwhelmingly diverse (25, 48), bioinformatics and functional testing have helped to categorize most of them into ten broad superfamilies, Cas1–Cas10 (49, 50). Cas1 and Cas2 (11, 94), and possibly Cas4 (47), are involved in the spacer acquisition step, but their mechanism(s) of action remains uncharacterized. The crRNA maturation step is carried out by the Cas6 superfamily of endoribonucleases in most CRISPR-Cas systems (8, 10, 30, 46, 68), but, in systems that lack Cas6, this step is carried out either by a distinct Cas5 protein, Cas5d (58), or by the host RNase III in a trans-activating RNA-dependent manner (12) (Figure 1). During the interference stage, the crRNA is assembled with specific Cas proteins into CRISPR ribonucleoprotein (crRNP) complexes (Figure 1). crRNPs that search for but do not cleave target nucleic acids, termed surveillance crRNPs, recruit additional effector proteins to cleave target nucleic acids. Those that do cleave target nucleic acids are termed effector crRNPs.
TYPES AND SUBTYPES
CRISPR-Cas systems are categorized into three major types (I, II and III) primarily on the basis of the phylogeny of the best-conserved cas1 gene and the combined presence of other cas genes, and each type is further divided into subtypes (49). All three types share the same functional paths (Figure 1), but they differ in the methods and machineries used to carry out these functions (Figure 2). Although types I and type III crRNPs are prevalent in both bacteria and archaea, type II crRNPs are found only in bacterial organisms. A given organism may possess one, two, or all of the CRISPR-Cas types; however, in organisms containing more than one CRISPR-Cas type, neither the interaction among different types nor the distribution of functional roles is known (9).
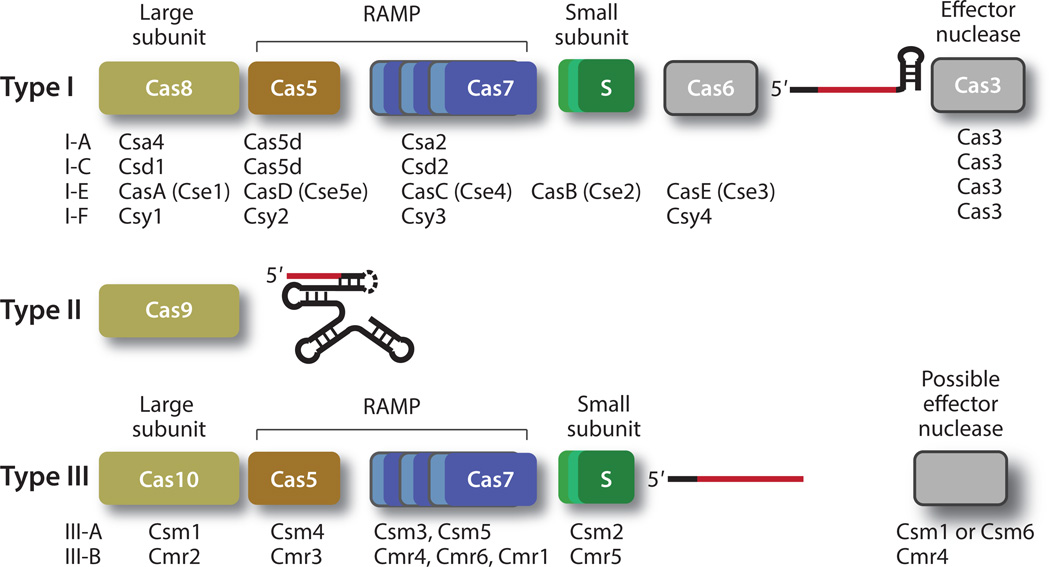
Figure 2.
The principal components of the three types of interference complexes. Proteins are represented by colored bars, and crRNA are represented by lines. Type I and type III crRNPs share the same four fundamental protein classes (large subunit, small subunit, Cas5, and Cas7). The large subunit is Cas8 for type I crRNPs and Cas10 for type III crRNPs, and Cas5 and Cas7 are distinct classes of the repeat associated mysterious protein (RAMP) superfamily. Type I crRNPs recruit an effector protein, Cas3, for DNA cleavage. The effector protein for the DNA-targeting type III crRNPs is not yet known, although Csm1 and Csm6 are the speculated effector proteins for type III-A, and Cmr4 is believed to be the effector protein of the type III-B crRNP. The guide region of the crRNA is shown in red, and the region derived from the repeat sequences is shown in black. The type II crRNPs comprise Cas9, a crRNA, and a trans-activating RNA (tracrRNA). The crRNA and the tracrRNA maybe covalently linked by a stem loop (dashed line), resulting in a single guide RNA (sgRNA). The names of other individual Cas proteins used in specific crRNPs that have been identified so far are listed under the cartoon for each type. Abbreviations: Cmr, Cas module RAMP antiviral complex; Csm, Cas subtype Mycobacterium tuberculosis antiviral complex (Csm).
The three types of CRISPR-Cas systems produce functional crRNA using different methods. In the type I and type III systems, the crRNA is processed primarily by the Cas6 endoribonuclease. The mature crRNA bears a 7- or 8-nt 5′ tag derived from the 3′ portion of the repeat, the guide, and, in the cases where the 3′ region is not processed further, a stem loop derived from the 5′ portion of the repeat (10, 22, 30, 67, 68, 72, 74, 81) (Figure 2). In the type I-C system, in which Cas6 is absent, this process is carried out by the Cas5d endoribonuclease, and the crRNA bears a similar overall structure but has an 11-nt 5′ tag (58). Both the Cas6 and Cas5d associated with the type I system are typically single-turnover enzymes and are part of the interference complex (22, 30, 58, 72, 81). The Cas6 associated with the type III system often dissociates from the product after processing, however, and remains independent from the interference complexes (74, 76, 87) (Figure 2). The type II system employs the host RNase III endoribonuclease in a trans-activating crRNA (tracrRNA)-dependent manner and produces crRNAs bearing a 3′ tag that remains paired with the tracrRNA throughout the interference function (Figure 2) (12).
The three CRISPR-Cas types also differ in interference target and in the composition of their interference complexes. The type I CRISPR-Cas system targets DNA exclusively, whereas some subtypes of the type II and type III systems target DNA and others target RNA. Both type I and type III interference complexes are multi–Cas protein crRNPs that have a single RNA subunit (Figure 2). The genes encoding these crRNP proteins are typically arranged as a cassette or as an operon, but not all proteins encoded within the cassette are part of the interference complexes. Type II interference complexes comprise a single Cas9 protein and two different RNA subunits (Figure 2).
PRINCIPAL BUILDING BLOCKS
The need to define a common set of principal components of crRNPs arises from the multiple naming systems used for these complexes and the observed similarity in their three-dimensional organizations. The systematic naming system for the Cas proteins first proposed by Makarova & Koonin in 2011 (49), and refined in 2013 (50), best reflects the structural and functional similarities among the different types and subtypes of crRNPs. In their unified system, type II crRNPs comprise a single class of protein, Cas9, whereas both type I and type III crRNPs share four fundamental classes of proteins: the large subunit, the small subunit, Cas5, and Cas7 (Figure 2). The large subunit comprises the Cas8 class of proteins for type I crRNPs and the Cas10 class for the type III crRNPs. These two protein classes contain polymerase-like features, and proteins belonging to the Cas10 class are often fused with a histidine–aspartate (HD) nuclease domain. The small subunit in type I and type III crRNPs is a small α-helical protein that plays essential roles in complex assembly. Cas5 and Cas7 are distinct classes of the repeat associated mysterious protein (RAMP) superfamily that are characterized by the presence of the ferredoxin-like fold (47). The RAMP superfamily also includes other Cas protein classes such as Cas6 proteins, which process crRNA (47) and form part of the mature complexes for the subtype I-E, I-B, and I-F crRNPs because of their tight association with the processed crRNA.
In addition to the stably bound protein factors, type I, and possibly type III-A, crRNPs recruit additional effector proteins to cleave their DNA targets. Type I crRNPs recruit the Cas3 nucleasehelicase for cleavage of target DNA after substrate binding, and this protein is thus a key component of the holo-complexes (Figure 2). Note that Cas3 also contains the same HD nuclease domain as that in the Cas10 protein, suggesting a frequent exchange of domains among Cas proteins. The identity of the effector protein for the type III-A crRNP remains elusive owing to limited biochemical data. A recent study showed that Staphylococcus epidermidis type III-A crRNP achieves DNA targeting in a transcription-dependent manner (23). In this case, the HD domain associated with the Cas10 subunit may be responsible for cleaving DNA (23, 65). Alternatively, type III-A crRNP recruits an unidentified effector protein to cleave DNA. The stable assemblies for both type III-B (RNA-cleaving) and type II-A (DNA-cleaving) crRNPs are associated with catalytic activities. Cas7 is believed to be the catalytic subunit of the III-B crRNPs, and Cas9 bears a RuvC domain and an HNH-like nuclease domains that are responsible for DNA cleavage.
DISCOVERY
Works from laboratories across the globe have resulted in isolation and biochemical characterization of representatives of type I, type II, and type III crRNPs. In all cases, bioinformatics analysis provided the initial guidance in the search for interference activities, and such analysis has made selective purification and reconstitution possible. In combination, traditional biochemical purification methods and modern sequencing and mass spectrometry techniques ultimately led to the isolation and characterization of nucleic acid–targeting crRNPs.
Among all types of crRNPs, the first to be isolated was a type I-E surveillance complex termed Cascade (CRISPR-associated complex for antiviral defense) isolated from Escherichia coli by Brouns et al. in 2008 (8). This complex was purified from cells overexpressing the CRISPR locus with a single affinity tag incorporated in various cas genes, and it was shown to contain five Cas proteins and a ~57-nt crRNA (8). The five proteins, CasA (also called Cse1 or Cas8), CasB (also called Cse2 or small subunit), CasC (also called Cse4 or Cas7), CasD (also called Cas5e), and CasE (also called Cse3 or Cas6e) (Figure 2) are encoded by an operon containing casA–casE. CasE is the endoribonuclease responsible for processing crRNA by the Cascade (8). Specific gene knockout followed by phage resistance assay demonstrated the essential function of each protein against viral infection in vivo (8). The Cascades of type I-A (46), I-C (58), and I-F (92) were later isolated in a similar manner and found to contain analogous protein subunits, with two exceptions. The type I-C and type I-F Cascade contain only three (Csd1, Csd2, and Cas5d) and four (Csy1, Csy2, Csy3, and Csy4) subunits, respectively, although both complexes also contain a crRNA-processing endoribonuclease. The type I-C endoribonuclease Cas5d shares RNA recognition properties with both CasE and CasD of type I-E, and these shared properties led to a provocative model in which Cas5d plays the roles of both CasE and CasD (58). The type I-F Cascade lacks the CasB-equivalent subunit but its Csy1 subunit appears to contain a CasB-equivalent domain (92), suggesting a conserved Cascade organization.
The type II effector crRNPs were first demonstrated to have an in vivo DNA interference function in 2007 and 2010 (3, 18). In 2012, two different groups (19, 40) subsequently characterized the subunit composition and in vitro DNA cleavage activity of the Streptococcus pyogenes and Streptococcus thermophilus type II-A crRNPs. Type II-A crRNPs contain a single protein subunit, Cas9 (also Csn1), a crRNA of 39–42 nt in length, and a tracrRNA of ~75 nt. The tracrRNA base pairs with the repeat region of the crRNA and is required for crRNA processing by the host RNase III in the presence of Cas9 (12, 24). Jinek et al. (40) further demonstrated that for the type II-A system, the crRNA and tracrRNA may be linked into a single guide RNA (sgRNA) by a tetraloop without affecting its DNA interference function, making this complex the simplest DNA interference complex known. A type II-B crRNP from Francisella novicida was reported around the same time, and this complex regulates endogenous gene expression in order to promote bacterial virulence without requiring catalytic centers for DNA cleavage (42, 70). The type II-B crRNP is believed to target mRNA and to contain Cas9, a tracrRNA of ~91 nt, and a small CRISPR/Cas-associated RNA (scaRNA) of ~48 nt (70), although the F. novicida Cas9 is also able to cleave DNA in vitro using its tracrRNA and crRNA (16). Several other type II systems similar to the F. novicida crRNP also contribute to pathogenesis of their host bacteria (70). The type II effector crRNPs are considered one of the most important discoveries in CRISPR-Cas studies, and these discoveries have resulted in an explosion of applications in many areas of biology.
The first antiplasmid immunity function by a type III crRNP was reported in 2008, when it was described for the type III-A Csm (Cas subtype Mycobacterium tuberculosis antiviral complex) from S. epidermidis (52). Affinity purification of the crRNA-containing Csm later resulted in identification of five associated Csm proteins (Csm1–Csm5) all encoded from the cas10-csm operon and a 31–67-nt crRNA (28, 29). The native Csm from Sulfolobus solfataricus (Sso) was also purified by a combination of affinity and ion exchange methods, but this complex was shown to contain eight Csm proteins and a crRNA (69). Surprisingly, one recent study showed that a type III-A Csm cleaves RNA rather than DNA in vitro (80). In 2009, Terns and colleagues (27) isolated another type III crRNP, the type III-B Cmr (Cas module RAMP antiviral complex), from the cell extract of the archaeon Pyrococcus furiosus (Pf). Interestingly, the Cmr was also shown to have RNA cleavage (27). The PfCmr contains six proteins, Cmr1–Cmr6, all of which are encoded from a tightly linked RAMP module (named after the predominant presence of RAMP proteins), and one of two distinct crRNAs, a 45-mer and a 39-mer species (27). Two other Cmrs were later isolated from Thermus thermophilus (Tt) (79) and S. solfataricus (95) and were found to have the same core composition, although the two complexes varied somewhat in the size of the crRNA and the number of Cmr proteins. The TtCmr has the same Cmr1–Cmr6 subunits as in the PfCmr, along with a 40- or 46-mer crRNA (79). In addition to containing Cmr1–Cmr6, the SsoCmr also has an Sso-specific Cmr7 protein (95). The precise size of the SsoCmr-associated crRNA has not been characterized, but it was estimated to be ~46 nucleotides (95). Similar to the PfCmr, both TtCmr and SsoCmr have RNA cleavage activities in vitro.
FUNCTIONS
The process of RNA-guided nucleic acid destruction begins with assembly of functional crRNPs and ends with the capture and cleavage of the target nucleic acids. Similar to other ribonucleoprotein particles, this process involves intricate protein–protein, protein–nucleic acid, and nucleic acid–nucleic acid interactions, and it requires enzymatic activities that break the phosphodiester bond. In addition to these shared processes, the three types of crRNPs also exhibit type-specific functions, which we describe below.
Most crRNPs target DNA for destruction; type II-B and type III-B crRNPs are believed to also target RNA. The in vitro RNA cleavage activity of the type III-B complex has allowed us to learn its functional properties. Both a region of ~31–38 nt (depending on the size of crRNA) that is complementary to the guide region of the crRNA and a correctly assembled multisubunit Cmr were shown to be necessary and sufficient for RNA cleavage. Importantly, two of the three reported Cmrs exhibit the ability to make regularly spaced excisions, and this ability may be explained by two possible models of assembly (26, 64, 79). The first model is a single Cmr assembly bearing regularly spaced RNA cleavage centers. The second entails multiple Cmr assemblies of a regular size, each of which contains a single RNA cleavage center. Current structural data are consistent with the former model, but they do not exclude the latter. RNA cleavage by the Cmrs requires divalent ions, but the cleavage products are consistent with a metal-independent mechanism, suggesting that the metal ions play a structural rather than catalytic role.
In contrast to the type III-B crRNPs, the type I, type II-A, and type III-A crRNPs target DNA in vivo. Both type II-A and type III-A crRNPs were recently shown to also have RNA cleavage activity in vitro (60, 80), however, suggesting an RNA binding and cleavage mechanism that may be related to that for DNA. The DNA targeted by these crRNPs contains a sequence segment (protospacer) that is complementary to the guide region of the crRNA (spacer). During interference, the guide region of the crRNA pairs with the complementary strand of the protospacer to form a structure called an R-loop while a protein or proteins cleave both the complementary and the noncomplementary strands. Importantly, because the crRNA is also complementary to its encoding DNA, the crRNPs must be able to distinguish the self DNA from the invader DNA. The type III-A crRNP is believed to discriminate its own DNA on the basis of the repeat sequences flanking the complementary spacers (52). For type I and type II crRNPs, a 2–5-nt segment adjacent to the protospacer [protospacer-adjacent motif (PAM)], as well as the paired crRNA, is sufficient for crRNPs to control this specificity (13, 54). Protospacers lacking a PAM remain intact when challenged by crRNPs. Finally, the type I and type III-A DNA-targeting crRNPs must recruit effector proteins that perform the actual DNA cleavage.
Recent structural and biochemical data have provided important insights about each of the functions described above. Intricate molecular interaction networks and large molecular motions have been observed. Successful studies of distinct crRNPs have revealed unique mechanisms that are pertinent to individual complexes. However, comparisons of the overall and subunit structures have revealed surprising mechanistic links among remotely related crRNPs, including links between those that target DNA versus RNA.
ASSEMBLY
Studies of crRNPs by electron microscopy, X-ray crystallography, native mass spectroscopy, and biochemical techniques have revealed atomic or near atomic resolution models for several crRNPs belonging to each of the three main types. These structural models have provided important insights into crRNP assembly and function. Owing to the structural relatedness between type I and type III crRNPs, we discuss their assembly principles together, and we discuss those for the type II crRNP separately.
The preparation of homogeneous specimens in large quantities has been crucial to the success of the structural studies cited here. In all reported cases, recombinant proteins from bacterial expression systems and a combination of synthetic and in vitro transcribed RNA were used to assemble the complexes. However, assembling these complexes, including the crRNA, within the expression hosts using coexpression strategies has proven to be much more helpful and, in some cases, necessary. For the six-component type I-E Cascade complex, a plasmid bearing the gene encoding Cas8 and a tandem repeat–spacer array was cotransformed into expression E. coli cells with another plasmid bearing the operon encoding Cas5, Cas6, Cas7, and the small subunit (38, 57, 96). For the multicomponent type III complexes, either native purification (80) or subcomplex purification followed by assembly with synthetic RNA on gel filtration columns proved to be effective (6, 64, 78, 79). For the type II Cas9 protein, bacterial expression worked sufficiently well for protein production despite its large size, but the challenge of constructing the appropriate RNA and DNA molecules had to be overcome via nucleic acid engineering (41). A minimal chimera RNA between the crRNA and tracrRNA was found to be fully functional and was employed in structural studies (1, 59).
Type I and Type III: Helical Assembly
Single-particle cryo-electron microscopy (cryoEM) and X-ray crystallographic studies have revealed detailed subunit arrangement and interaction information (33, 38, 43, 57, 90, 96). Combined EM, X-ray crystallographic, and mass spectrometry data for the type III-B Cmr (6, 64, 78, 79) are now available, as are negative-stained EM data for the type III-A Csm (69). In addition, crystal structures are available for many individual subunits of both type I and type III crRNPs (Table 1). These studies show that, despite having distant phylogeny, the type I and type III crRNPs share a surprisingly similar architecture. Analysis of the available crystal structures of individual subunits across type I and type III crRNPs suggests a propensity for a common helical assembly and for binding single-stranded crRNA. Type-specific differences have also been observed that may explain functional variations among crRNPs.
Table 1.
List of crystal and electron microscopy (EM) structures of CRISPR ribonucleoproteins (crRNPs) and subunits*
| crRNP or subunit name |
Subtype and species | PDB ID (resolution) |
Components | Notes |
|---|---|---|---|---|
| Crystal structures | ||||
| Cascade | I-E Escherichia coli K12 (Ec) |
4U7U (3.0 Å) | Cas8, Cas5, Cas6, Cas7, small subunit, crRNA |
First crystal structure of multisubunit CRISPR-Cas crRNP |
| Cascade | I-E Escherichia coli K12 (Ec) |
1VY8 (3.2 Å) | Cas8, Cas5, Cas6, Cas7, small subunit, crRNA |
First crystal structure of multisubunit CRISPR-Cas crRNP |
| Cascade | I-E Escherichia coli K12 (Ec) |
4QYZ (3.0 Å) | Cas8, Cas5, Cas6, Cas7, small subunit, crRNA, ssDNA |
First crystal structure of multisubunit CRISPR-Cas crRNP bound with an ssDNA substrate |
| Cas3 | I-C Thermobifida fusca YX (Tf) |
4QQW (2.7 Å) | Cas3, Fe3+ DNA, | First Cas3 structure |
| 4QQX (3.3 Å) | Cas3, Fe3+, ATP | First Cas3 structure | ||
| 4QQY (3.1 Å) | Cas3, Fe3+ ADP | First Cas3 structure | ||
| 4QQZ (2.9 Å) | Cas3, Fe3+, DNA, ANP | First Cas3 structure | ||
| Cas7 (Csa2) | I-A Sulfolobus solfataricus P2 (Sso) |
3PS0 (2.0 Å) | Cas7 | First type I-A Cas7 structure |
| Cas7 (Cmr4) | III-B Pyrococcus furiosus DSM 3638 (Pf) |
4RDP (2.8 Å) | Cas7 | First type III-B Cas7 structure |
| Cas7 (Csc2) | I-D Thermofilum pendens (Tp) |
4TXD (1.8 Å) | Cas7 | First type I-D Cas7 structure |
| Cas7 (Csm3) | III-A Methanopyrus kandleri (Mk) |
4N0L (2.4 Å) | Cas7 | First type III-A Cas7 structure |
| Cas5 (Cas5d) | I-C Bacillus halodurans (Bh) |
4F3M (1.7 Å) | Cas5 | First type I-C Cas5 structure; first Cas5 protein to show RNA processing activity |
| Cas5 (Cmr3) | III-B Pyrococcus furiosus DSM 3638 (Pf) |
4H4K (2.8 Å) | Cas10, Cas5 | Second type III-B Cas5 structure; α1 helix observed; thumb disordered |
| Cas5 (Cmr3) | II-B Pyrococcus furiosus DSM 3638 (Pf) |
3W2W (2.5 Å) | Cas10, Cas5 | Second type III-B Cas5 structure; thumb β-hairpin observed; α1 helix disordered |
| Cas8 (Cse1, CasA) |
I-E Acidimicrobium ferrooxidans DSM 10331 (Acf) |
4H3T (2.0 Å) | Cas8 | |
| Cas10 (Cmr2) | III-B Pyrococcus furiosus DSM 3638 (Pf) |
3UNG (2.3 Å) | Cas10 | First type III-B Cas10 structure; ATP and metal binding observed |
| Small subunit (Cmr5) |
III-B Pyrococcus furiosus DSM 3638 (Pf) |
4GKF (2.1 Å) | Cmr5 | |
| Small subunit (Cmr5) |
III-B Thermus thermophilus HB8 (Tt) |
2ZOP (2.1 Å) | Cmr5 | |
| Small subunit (Cmr5) |
III-B Archaeoglobus fulgidus (Af) |
2OEB (1.6 Å) | Cmr5 | |
| Small subunit (CasB) |
I-E Thermobifida fusca (Tf) |
4H79 (1.9 Å) | CasB | |
| Small subunit (CasB) |
I-A Thermus thermophilus (Tt) |
4H7A (2.6 Å) | CasB | |
| Cas9 | II-A Streptococcus pyogenes SF370 (Sp) |
4UN3 (2.6 Å) | Cas9, sgRNA, PAM- containing dsDNA |
First Cas9 bound with a PAM-containing dsDNA |
| 4UN4 (2.4 Å) | Cas9, sgRNA, PAM- containing dsDNA with 2-nt mismatch |
|||
| 4UN5 (2.4 Å) | Cas9, sgRNA, PAM- containing dsDNA with 3-nt mismatch |
|||
| Cas9 | II-A Streptococcus pyogenes (Sp) |
4OO8 (2.5 Å) | Cas9, sgRNA, ssDNA | First Cas9 structure bound with sgRNA and ssDNA |
| Cas9 | II-A Streptococcus pyogenes SF370 (Sp) |
4CMP (2.6 Å) | Cas9 | First apo Cas9 structures (II-A and II-C) |
| 4CMQ (3.1 Å) | Cas9, Mn2+ | |||
| Cas9 | II-C Actinomyces naeslundii (An) |
4OGE (2.2 Å) | Cas9 | |
| 4OGC (2.8 Å) | Cas9, Mn2+ | |||
| EM structures | ||||
| Cmr | III-B Thermus thermophilus (Tt) |
EMD-2418 (22 Å) |
Cas10, Cas5, three different Cas7s (Cmr1, Cmr4, and Cmr6), small subunit, crRNA |
First EM structure revealing a helical feature, six Cas7 repeats, and the base structure |
| Cmr | III-B Pyrococcus furiosus DSM 3638 (Pf) |
EMD-6165 EMD-6166 (15 Å) |
Cas10, Cas5, three different Cas7s (Cmr1, Cmr4, and Cmr6), small subunit, crRNA, ssRNA |
First EM structure revealing the location of the HD domain and the α1 hooks |
| EMD-6167 (12 Å) |
Cas7 (Cmr4), small subunit |
|||
| Csm | III-A Sulfolobus solfataricus (Sso) |
EMD-2420 (30 Å) |
Cas10, Cas5, five different Cas7s, small subunit, crRNA |
First EM structure of a type III-A complex |
| Csm | III-A Thermus thermophilus (Tt) |
EMD-6122 (21 Å) |
Cas10, Cas5, two different Cas7s (Csm3 and Csm5), small subunit, crRNA |
Second EM structure of a type III-A complex |
| Cascade | I-E Escherichia coli K12 (Ec) |
EMD-5314 (8.8 Å) |
Cas8, Cas5, Cas6, Cas7, small subunit, crRNA |
First high-resolution EM structure. Six Cas7 repeats observed |
| Cascade | I-E Escherichia coli K12 (Ec) |
EMD-5315 (9.2 Å) |
Cas8, Cas5, Cas6, Cas7, small subunit, crRNA, ssRNA |
Discontinuous RNA duplex was observed |
| Cascade | I-E Escherichia coli K12 (Ec) |
EMD-5929 (9 Å) |
Cas8, Cas5, Cas6, Cas7, small subunit, crRNA, dsDNA |
Conformational changes in the small and large subunits observed |
| Cascade | I-E Escherichia coli K12 (Ec) |
EMD-5930 (20 Å) |
Cas8, Cas5, Cas6, Cas7, small subunit, crRNA, dsDNA, Cas3 |
First structure reporting the locations of the dsDNA and the Cas3 binding site |
Abbreviations: Cas, CRISPR-associated; Cascade, CRISPR-associated complex for antiviral defense; Cmr, Cas module RAMP antiviral complex; Csm, Cas subtype Mycobacterium tuberculosis antiviral complex; crRNA, CRISPR RNA; ds, double-stranded; HD, histidine–aspartate; PAM, protospacer-adjacent motif; PDB, Protein Data Bank; RAMP, repeat associated mysterious protein; sgRNA, single guide RNA; ss, single-stranded.
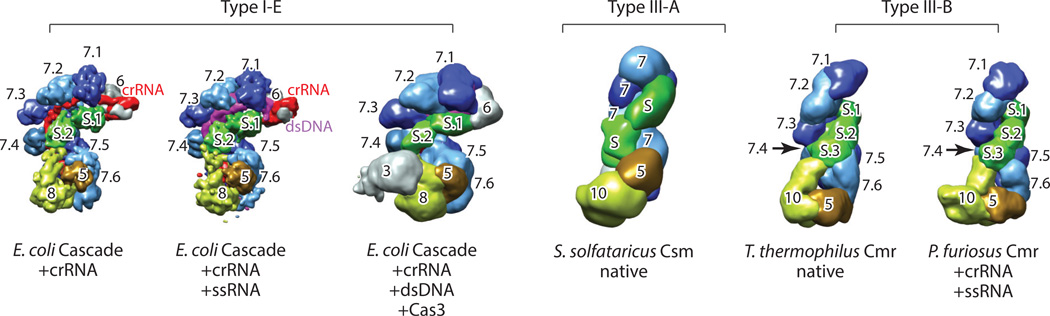
Type I and type III crRNPs are elongated in shape with a distinct head and a base flanking a helical body (Figure 3). The crRNA lies linearly along a central channel through the major groove of the helical assembly, and its 5′ and 3′ ends are anchored in the base and the head of this assembly, respectively. This architecture has been compared to the shapes of seahorses or sea worms (31). The four principal protein classes are arranged similarly in space in the crRNP structures regardless the type (Figure 3). Each crRNP contains a single copy of the large subunit, one or two Cas5 proteins, and varying numbers of copies of the small subunit and the Cas7 proteins. The most-conserved region of the crRNPs is the base, which comprises the large subunit and Cas5. The least-conserved region is the head, which comprises type-specific components. The head of the type I-E Cascade is formed by the crRNA-processing endonuclease Cas6 bound to the 3′ hairpin of the crRNA, whereas those of the type III-A and type III-B complexes are formed by heterogeneous Cas7 proteins (Figure 3).
Figure 3.
Architectural similarities between the type I and type III crRNPs. Electron microscopy densities are segmented for each complex and are colored according to the same scheme as that used for protein subunits in Figures 1 and 2. The crRNA is colored in red, and the target nucleic acids are colored in magenta. Various crRNPs belonging to different types are manually superimposed and displayed in the same orientation. The segment corresponding to a specific Cas protein is labeled by only the numerical component of the protein name (e.g., 10 indicates Cas10). Subunits that are repeated multiple times are labeled by the numerical name of the subunit followed by a number specifying the order of the repeat (e.g., 7.6 indicates the sixth Cas7). The small subunit is labeled S, S.1, S.2, or S.3. In the case of the type III-A crRNP, the exact number of repeats has not been determined for either Cas7 or the small subunit, so the corresponding densities are labeled “7” and “S,” respectively. The reader is referred to Table 1 for additional information. Abbreviations: Cas, CRISPR-associated; crRNA, CRISPR RNA; crRNP, CRISPR ribonucleoprotein; ds, double-stranded; E. coli, Escherichia coli; P. furiosus, Pyrococcus furiosus; ss, single-stranded; S. solfataricus, Sulfolubus sofataricus; T. thermophilus, Thermus theromphilus.
The most represented protein class in type I and type III crRNPs is the Cas7 class that forms the major helical backbone of the Cascade, the Csms, and the Cmrs (Figures 3 and 4, 7.1–7.6). Both the type I-E Cascade and the type III-B Cmrs contain six copies of Cas7, which may be either the same or different (Figure 4). Cascade contains six identical Cas7 proteins (CasC) (Figures 3 and 4, 7.1–7.6) (38, 57, 96), whereas the Cmr contains three different Cas7 proteins [Cmr4 (four subunits), Cmr1 (one subunit), and Cmr6 (one subunit)] (6, 64, 78, 79). The Sso type III-A Csm contains eight Cas7 proteins, which belong to five different subclasses (69), and the TtCsm contains seven Cas7 subunits, which belong to two different subclasses (80). However, the overall size and shape of the SsoCsm and the TtCsm are similar to those of the Cmr (Figure 3) (6, 69). The second multirepeat protein is the small subunit, which is represented either two or three times among the crRNPs (Figures 3 and 4, S.1–S.3). The small subunit forms the second backbone of the central body that complements the contour of the Cas7 backbone (Figures 3 and 4).
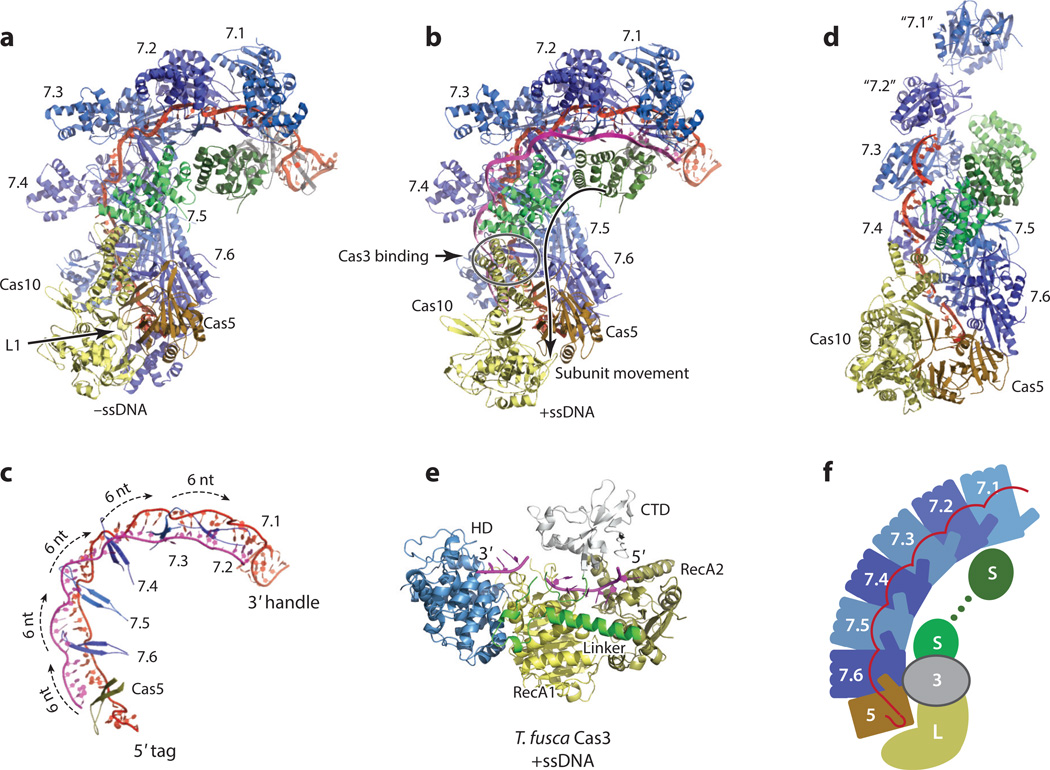
Figure 4.
Crystal or combined electron microscopy (EM) and crystal structures of the type I-E Cascade (a,b,c), the type III-B Cmr (d), and the effector protein, Cas3 (e). Cas7 subunit repeats are colored in alternating dark blue and light blue. The large subunit and the small subunit are colored in yellow and green, respectively. Cas5 is colored in orange. The crRNA is colored in red and the target is colored in magenta. (a) The crystal structures of the Escherichia coli Cascade without a bound substrate DNA (PDB IDs: 4U7U and 1VY8); (b) The crystal structure of the E. coli Cascade with a single-stranded DNA (ssDNA) bound (PDB ID: 4QYZ). The direction of movement of the small subunit proteins upon binding of the ssDNA substrate is indicated by an arrow. The Cas3 binding site on the Cascade as determined by an EM method (see Figure 3) is indicated by an open circle. (c) The arrangement of the thumb β-hairpins of Cas5 and Cas7 repeats on the crRNA:ssDNA hybrid (PDB ID: 4QYZ). The Cas5 thumb interacts with the 5′ tag in a sequence-specific manner. The Cas7 thumbs insert into the crRNA:ssDNA heteroduplex, causing every sixth base pair to unwind. (d). The EM structure of the type III-B Cmr fitted with crystal structures of the Cas10–Cas5 heterodimer (PDB ID: 4K4K), the Cas7 filament (PDB ID: 4RDP), and the small subunit (PDB ID: 4GKF) with the EM density removed. A portion of the crRNA is modeled based on crRNA–Cascade interactions that have been adjusted for the different symmetry relationships among the Cas7 repeats of the Cmr. The locations of the Cmr1 (“7.2”) and Cmr6 (“7.1”) proteins are indicated by models from Cmr4 proteins (7.3–7.6) and are labeled with quotation marks. (e) The crystal structure of Thermobifida fusca Cas3 bound with an ssDNA (PDB ID: 4QQW). Colors are arbitrarily assigned to individual domains. Cas3 first binds and cleaves the noncomplementary strand using its histidine–aspartate (HD) domain and then processes the complementary strand using an uncharacterized mechanism. (f) Cartoon representation of the key features of type I and type III crRNP assemblies. Cas3 binding applies to only type I crRNPs. Abbreviations: Cas, CRISPR-associated; Cascade, CRISPR-associated complex for antiviral defense; Cmr, Cas module RAMP antiviral complex; crRNA, CRISPR RNA; crRNP, CRISPR ribonucleoprotein; CTD, C-terminal domain; PDB, Protein Data Bank; RAMP, repeat associated mysterious protein.
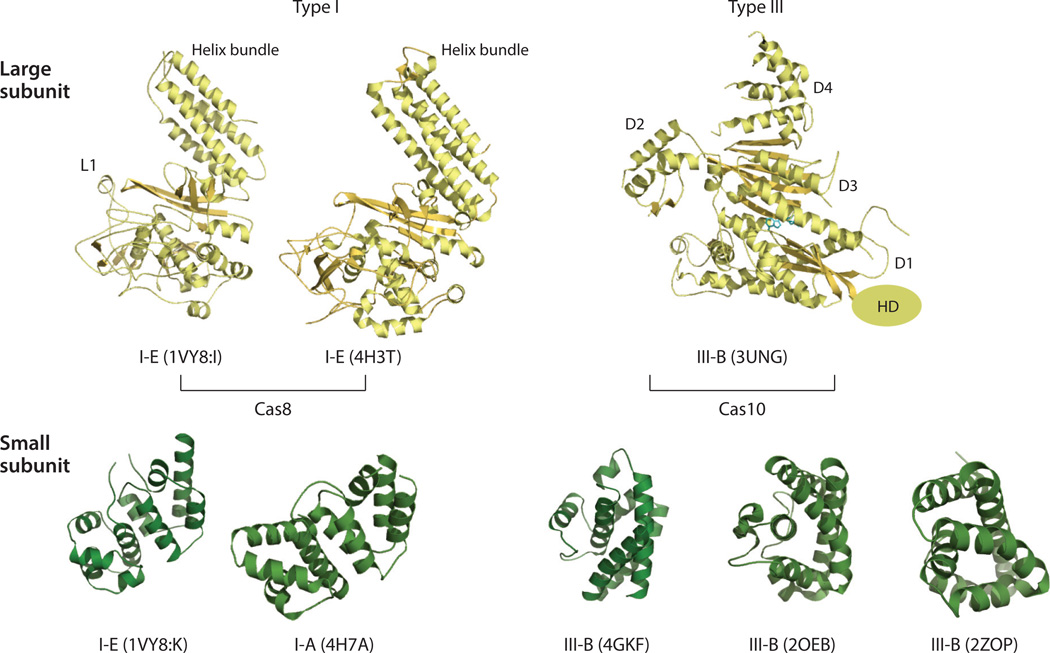
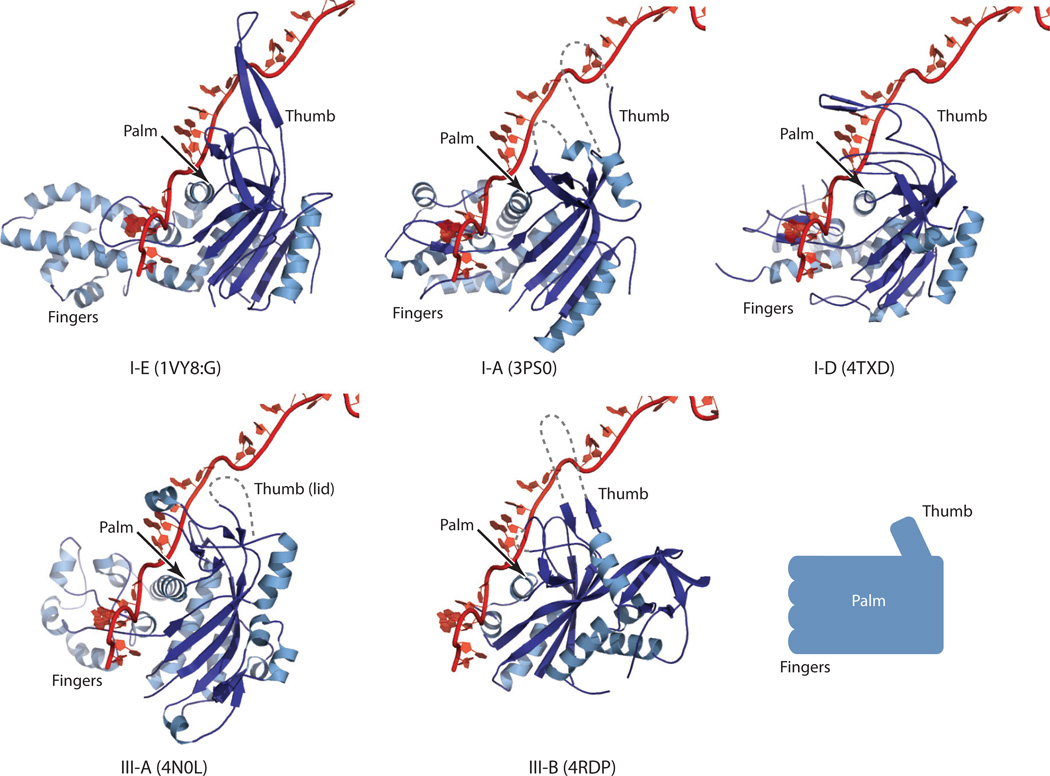
The similarity in assembly between the type I and type III crRNPs is further supported by structural similarities among the individual principal building blocks. The large subunits of both type I-E (Cas8) and type III-B (Cas10) crRNPs are multidomain and are dominated by helical secondary structures (Figure 5). Similarly, the small subunit is exclusively helical in both types (Figure 5). Both Cas7 and Cas5 belong to the RAMP superfamily, which includes proteins that are characterized by the ferredoxin-like fold and have structures that are comparable to that of a right hand (Figures 4, 6, and 7). The ferredoxin-like fold forms the core of the protein, or the palm. A long β-hairpin protruding between the second and the third core β-strands comprises the thumb, and a helical insertion between the first and the second core β-strands forms the fingers. Cas5 proteins seem to lack the fingers domain, suggesting that this domain is specific to Cas7 (Figures 4 and 6). The helical assembly of crRNPs arises from the filamentous characteristics of theCas7 family of proteins. Cas7 can self-assemble in a head-to-tail (or thumb-to-palm) fashion and can thus extend indefinitely in the absence of competing interactions (Figures 4 and 7). Structural studies of an isolated type III-B Cas7 and a Cas7–small subunit complex (Cmr4–Cmr5) support this structural property (6, 64, 78). The fact that crRNPs possess defined numbers of Cas7 subunits implies that the interactions between Cas7 and the base and head subunits must be stronger than its self-interactions.
Figure 5.
Similar structural classes of the large and small subunits between the type I and type III crRNPs. The large subunit proteins are shown in yellow, and the small subunit proteins are shown in green. The crRNP subtype and the PDB ID are indicated below the structure for each protein. Table 1 contains additional information about these proteins. The L1 loop of the Escherichia coli Cas8 protein has been shown to interact with the protospacer-adjacent motif (PAM) sequence to prevent destruction of “self” DNA. The histidine–aspartate (HD) domain of Cas10 is not included in the crystal structure, and its position is indicated only on the basis of sequence connectivity. The bound ATP molecule in the Cas10 protein is shown in the stick model. The physiological importance of the ATP binding site on Cas10 remains unclear. D1–D4 refer to domains 1–4, respectively. Abbreviations: Cas, CRISPR-associated; crRNP, CRISPR ribonucleoprotein; PDB, Protein Data Bank.
Figure 6.
Similar Cas7 structures between type I type III CRISPR-Cas systems. Currently known Cas7 structures are superimposed with that of type I-E (PDB ID: IVY8, chain G) with bound crRNA. The type and the PDB ID for each Cas7 structure are indicated below it. Table 1 contains additional information about these structures. Dashed lines indicate disordered regions in the crystal structures. The thumb hairpin loop is often disordered in RNA-free Cas7 structures, but this loop is expected to be stabilized in a manner similar to that depicted in the type I-E Cas7 structure (PDB ID: 1VY8, chain G). The right hand representation of the Cas7 structure is shown at the lower right, and key regions of this structure are labeled.
Figure 7.
Similar Cas5 structures between the type I and type III CRISPR-Cas systems. Currently known Cas5 structures are superimposed with that of type I-E (PDB ID: IVY8, chain H) with bound crRNA. The type and the PDB ID for each protein are indicated below the appropriate structure. Table 1 contains additional information about these structures. Dashed lines indicate the disordered regions in the crystal structures. The thumb hairpin loop is often disordered in RNA-free Cas5 structures, but this loop is expected to be stabilized in a manner similar to that depicted in the type I-E Cas5 structure (PDB ID: 1VY8, chain H). The right hand representation of the Cas5 structure is shown at the right, and key regions of the structure are labeled.
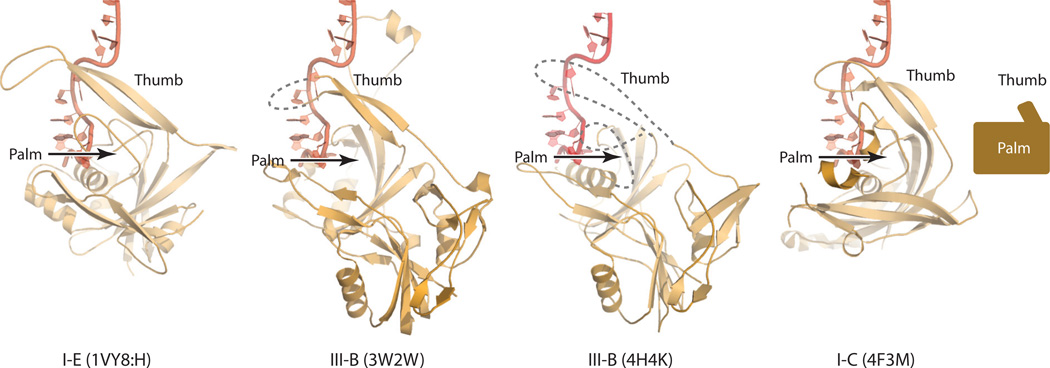
Cas3 is the effector protein of the type I crRNPs and is associated with the crRNPs upon binding of cognate double-stranded DNA (dsDNA) (61). Cas3 belongs to the SF2 family of helicases and, not surprisingly, contains structural features of this family of enzymes (39) (Figure 3). Cas3 is placed near the base of the type I-E Cascade on the back surface of the large subunit (Figure 3).
Type II: Bilobe Architecture
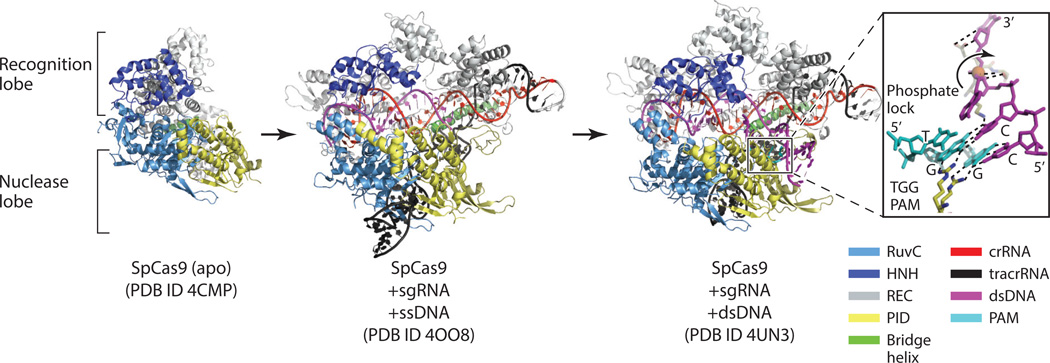
Four crystal structures of the type II crRNP principal component, Cas9, from S. pyogenes (Sp) and Actinomyces naeslundii (An) have been obtained. These structures belong to Cas9 of type II-A and type II-C subtypes and include the apo structures of SpCas9 and AnCas9 (41), a structure of SpCas9 bound with an sgRNA and a 20-nt single-stranded DNA (ssDNA) (59), and the structure of SpCas9 bound with an sgRNA and a double-stranded DNA (dsDNA) substrate (1). Cas9 is a large protein (~100–190 kDa) with multiple juxtaposed domains belonging to two major lobes: a recognition (α-helical) lobe and a nuclease lobe. The nuclease lobe is further divided into three regions of interest: a RuvC-like domain, an HNH-like domain, and a PAM-interacting domain (PID) (Figure 8). The apo II-C Cas9 structures contain most of the nuclease lobe but only a partial recognition lobe (41). Upon binding nucleic acids, the recognition lobe reorganizes to engage them by undergoing a drastic conformational change (Figure 8).
Figure 8.
Structures of Cas9 and its complexes with RNA and DNA substrates. The domain color scheme is indicated at the lower right. Cas9 has a bilobe organization comprising the recognition lobe (gray) and the nuclease lobe (dark blue, light blue, and yellow) connected by an arginine-rich bridge helix (green). Large rearrangements of the recognition lobe are observed upon binding of the single guide RNA (sgRNA) and the complementary single-stranded DNA (ssDNA). More moderate arrangements are observed when double-stranded DNA (dsDNA) bearing the protospacer-adjacent motif (PAM) sequence associates. Additional information about the structures is found in Table 1. (left) Structure of the apo Streptococcus pyogenes (Sp) Cas9. (middle) Structure of SpCas9 bound with sgRNA and a complementary ssDNA. (right) Structure of SpCas9 bound with an sgRNA and a dsDNA substrate bearing the TGG PAM sequence. The PAM–Cas9 interactions are depicted in the inset. A pair of arginine residues forms nucleobase-specific hydrogen bonds with the PAM nucleotides GG. As a result, the +1 nucleotide of the complementary DNA strand splays out of the helical stack and pairs with the guide RNA. The phosphate backbone kink is stabilized by the phosphate lock loop.
crRNA RECOGNITION
Crystal structures of the E. coli type I-E crRNP in the presence and absence of ssDNA and an atomic model of the P. furiosus type III-B crRNP have provided insights into crRNA recognition, and, more importantly, how these two distantly related crRNPs share a similar crRNA recognition mechanism. In contrast, the crystal structures of the S. pyogenes type II-A crRNP reveal a drastically different crRNA recognition mechanism.
Type I and Type III crRNPs: Thumb Grips
So far, only one crystal structure among all type I and type III crRNPs allows detailed analysis of crRNA binding to crRNP proteins. The Wiedenheft and Wang laboratories independently obtained the crystal structure of the E. coli Cascade bound with a crRNA (38, 96), and the Bailey laboratory obtained the structure of the E. coli Cascade bound with a crRNA and the cognate ssDNA (57). These important structures have advanced our understanding of crRNA recognition by type I crRNPs.
The 5′ tag of the crRNA (8 nt) is the key element of specificity. The backbone of the first eight nucleotides is bent into an S-shape; nucleotides 1–4 form the lower arc, and nucleotides 5–8 form the upper arc. The 5′ tag is completely engulfed by Cas5, Cas7.6 (the Cas7 subunit closest to Cas5), and to some degree, the L1 loop of the large subunit (Figure 4). The lower arc forms a tight loop that lies parallel to the first helix (α1) of the Cas5 palm domain. The upper arc bends less sharply and is pressed against the α1 helix of Cas7.6 by the β-hairpin thumb domain of Cas5 from below (Figure 4). The nucleobases of the first six nucleotides are engaged in close contact with the surrounding proteins. The first nucleotide interacts extensively with Cas7.6, the second and third interact with Cas5, and the fourth, fifth, and sixth interact with the L1 loop of the large subunit. These combined interactions likely impart the specificity required for crRNA association and explain the necessity of protein complex formation for specific binding of crRNA.
The guide (spacer) region of the crRNA extends along the helical contour formed by the six Cas7 repeats and is fixed on the protein surface by a set of spectacular thumb-to-palm interactions among the Cas7 proteins (Figure 4). The thumbs are spaced evenly along the RNA with a 6-nt spacing (Figure 4). Each thumb inserts into the helical stack of the bases and presses the phosphate backbone against the α1 helix of the Cas7 above, thereby splaying every sixth base, and this splay is accompanied by a sharp kink of the phosphate backbone at this position (Figure 4). The contacts established between the RNA and proteins are nonspecific and thus can accommodate any sequences.
Comparison of the Cas5 and Cas7 protein structures between the type I and type III crRNPs suggests these proteins share a similar crRNA binding mechanism (Figures 6 and 7). Superimposition of RNA-free type III-B, I-A, I-D, and III-A Cas7 structures with the structure for RNA-loaded type I-E Cas7 of Cascade (7.6) reveals striking similarities among the thumb and palm domains of these proteins, and these similarities are likely to preserve the thumb-to-palm (or thumb grip) principle observed in E. coli Cascade (Figure 6). Similarly, superimposition of the RNA-free type III-B Cas5 structures with the RNA-bound type I-E Cas5 structure shows that although the thumb and palm elements of type III-B Cas5 are often disordered in isolation, these elements are likely engaged in similar interactions with the 5′ tag of the cognate crRNA (Figure 7). Interestingly, locations of the predicted thumbs of Cas5 and Cas7 in the assembled Cmr match those of regularly spaced α1 hook densities (64), supporting a preservation of the observed interactions between the thumb and the small subunit in the type III-B Cmr.
Type II: Bilobe Clamp
The principles of RNA interaction with the type II Cas9 protein are revealed by three significant SpCas9 structures (the apo structure, the ssDNA- and sgRNA-bound structure, and the dsDNA and sgRNA-bound structure) (1, 41, 59). The key elements recognized by the Cas9 of the type II crRNP include the repeat:antirepeat hybrid formed between the crRNA, the sgRNA, and the first stem loop of the tracrRNA, although the last two stem loops of the tracrRNA also play some roles in stabilizing the crRNP (40, 59). The repeat:antirepeat hybrid contains 10 bp interrupted by a 2,4-nucleotide internal loop (Figure 8). The duplex is buried at the interface between the two lobes of Cas9 and the first helical segment of the recognition lobe. The internal loop also contacts residues from the recognition lobe. The guide region (base paired with ssDNA) is engulfed in a sequence-independent manner by the arginine-rich bridge helix that connects the recognition lobe and the nuclease lobe, the recognition lobe, and the HNH-like domain (Figure 8). The 10–12 nt near the repeat interact most extensively with the bridge helix, which provides the structural basis for the so-called seed interaction for binding substrate DNA near the PAM sequence (82) in much the same way as the seed interaction observed at the guide–repeat junction for Cascade does (92). The second and third stem loops of the tracrRNA interact with residues from the PAM-interacting domain and the RuvC-like domain (Figure 8) (59). These two stem loops are not required for Cas9 function but can dramatically increase its catalytic efficiency (40, 59).
TARGET RECOGNITION
Type I and Type III: Base Pairs with a Stretch
Most known effector and surveillance complexes target dsDNA for interference, although the type I-E Cascade and the type III-A Csm can bind complementary ssRNA (80, 90). For the crRNA to gain access to the complementary strand of a dsDNA, the dsDNA must unwind. Furthermore, the crRNPs must also specifically recognize the PAM motif to distinguish self DNA from foreign DNA. The crystal structures of E. coli Cascade bound to ssDNA (57) and the combined EM and crystal structure models of the PfCmr (6, 64) have provided insights into how the substrate DNA or RNA binds to the type I and type III crRNPs.
In the ssDNA-bound Cascade, the bound crRNA is partially exposed to solvent owing to the interdigitized thumb grips (Figure 4) and is thus unable to base pair with a target DNA. In addition, the phosphate backbone of crRNA closely follows the contour of the central filament, restricting the way in which the target can be paired with the crRNA. Interestingly, Cascade, and most likely the Cmr (with RNA substrate), overcomes these challenges via segmented base pairing. The bound crRNA–ssDNA heteroduplex deviates largely from the standard B- or A-form and does not hybridize continuously. The entire duplex is underwound and stretched longitudinally to match the entire assembly contour. Base pairing is interrupted every 6 bp by the inserted thumb β-hairpin, leading to an irregular and discontinuous RNA–DNA heteroduplex (Figure 4). The regularly disrupted base pairing is consistent with the fact that mutations occurring every 5 bp in target DNA are tolerated in vivo (73).
Each segment of 5 bp has a similar structure as a result of having similar interactions with the Cas7 repeats (Figure 4), suggesting that the protein assembly dictates the features of the target interactions. Strikingly, substrate binding is accompanied by a notable domain rotation of the large subunit and as much as 16 Å sliding of the two small subunits toward the base (Figures 3 and 4), demonstrating that DNA hybridization is a dynamic process.
In the combined EM and crystal structural models of the PfCmr, a similar irregularity in the crRNA–RNA hybrid must occur, imposed by the helical contour and by the structural similarity between its Cas7 and that of Cascade (64) (Figures 4 and 7). Thus, the basic features observed in the Cascade crRNA–DNA hybrid are likely to be conserved between type I and type III crRNPs.
Given that the 5′ end of the Cascade-bound guide region has the strongest affinity for DNA (92), hybridization likely first takes place near the PAM sequence. This hybridization leads to small subunit sliding and large subunit rotation, then to hybridization of the next segment and perhaps to more small subunit sliding and large subunit rotation (Figure 9a). The cycle continues until the last segment is hybridized. Single-molecule experiments showed that base pairing is not stable until the last base pair at the distal end is formed (22), suggesting the importance of complete base pairing in DNA cleavage. It is believed that irregular and discontinuous base pairing allows for better mismatch detection and more accurate substrate capture than a single hybridization event does (57). This principle of heteroduplex formation has been compared to that facilitated by the DNA recombination protein RecA (57, 86).
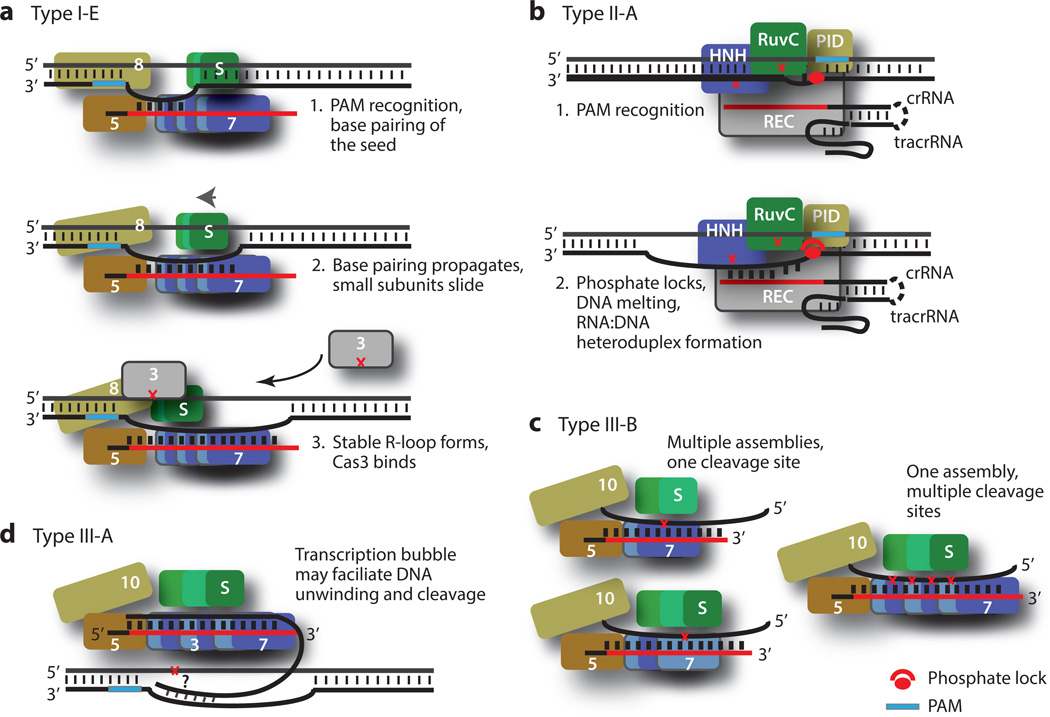
Figure 9.
Nucleic acid target binding and cleavage models for type I, type II, and type III crRNPs. Red crosses indicate DNA or RNA cleavage sites. (a) Model of type I crRNP function. Upon recognition of the protospacer-adjacent motif (PAM) sequence on the complementary strand, the surveillance complex allows the so-called seed base pairing between the crRNA and the complementary DNA to occur. This pairing triggers sliding of the small subunit toward the base of the crRNP and domain rotation in the large subunit, freeing the downstream guide RNA for additional base pairing. This process continues until the ultimate formation of the R-loop. Cas3 is then recruited to the region in which the noncomplementary strand wraps around the helix bundle domain of the Cas8 protein, where itmakes the first excision. (b) Model of type II crRNP function. The DNA duplex is inspected by the Cas9 crRNP until the PAM sequence is recognized. A PAM sequence–interacting element such as the pair of arginine residues of the SpCas9 engages the PAM nucleotides, triggering unwinding of the adjacent base pairs and splaying of the first substrate nucleotide. The DNA–crRNA base pairing forms and is made possible by a stabilizing interaction between the severely kinked phosphate backbone and the phosphate lock loop of Cas9, and the pairing propagates while the DNA substrate unwinds. The HNH-like domain cleaves the complementary strand while the RuvC-like domain cleaves the noncomplementary strand. (c) Model of type III-B crRNP function. The crRNP has a stable assembly with multiple Cas7 repeats that may facilitate cleavage of the RNA base paired with the crRNA. The alternative model of multiple assemblies with varying Cas7 repeats is not depicted. Numbered subunits indicate respective proteins (e.g., 3 indicates Cas3, 8 indicates Cas8). Abbreviations: crRNA, CRISPR-RNA; crRNP, CRISPR ribonucleoprotein; PID, PAM-interacting domain; S, small subunit; Sp, Streptococcus pyogenes; tracrRNA, trans-activating RNA.
Self Versus Foreign: The Protospacer-Adjacent Motif Meets the Phosphate Lock
DNA-targeting crRNPs must recognize and utilize the PAM sequence to prevent self-targeting. In vitro DNA binding studies of type I-E Cascade on dsDNA binding have shown that the R-loop forms only when PAM is present (75, 89). The use of a DNA substrate bearing the PAM sequence in both strands of the target DNA showed that the PAM sequence is preferentially recognized in the complementary strand (89). Similar studies with type II Cas9 complexes also demonstrated the importance of the PAM sequence in DNA interference (15, 16, 40). The type II crRNPs recognize the PAM sequence on the noncomplementary strand of the dsDNA rather than on the complementary strand (20, 40), however, suggesting a mechanistic difference between these systems at this step.
The nucleic acid structure captured by X-ray crystallography that most closely resembles an R-loop structure is that for the type II Cas9 bound with dsDNA, and this structure sheds light on how the PAM facilitates R-loop formation (1). To obtain the structure of a PAM-interacting Cas9 complex, a partially active Cas9 containing a defective HNH-like domain and an active RuvC-like domain was incubated with an 83-nt sgRNA and a dsDNA substrate bearing the TGG PAM sequence on its noncomplementary strand. In the crystal structure, the RuvC-like domain cleaved the noncomplementary strand but left its 3′ cleavage product intact, allowing examination of PAM–Cas9 interactions. The duplex containing the PAM sequence lies at an ~120° angle to the sgRNA–DNA heteroduplex, which is coaxial with the crRNA–tracrRNA duplex (Figure 8). Unlike the irregular crRNA–DNA heteroduplex observed in the type I and type III crRNPs, the sgRNA–DNA hybrid maintains a regular A-form helix, likely owing to its short length. The substrate strand is engaged in sequence-independent interactions mostly with the recognition lobe.
The PAM helix is nestled in a positively charged groove formed between the C-terminal domain (CTD) and the Topo-homology domain (collectively, the PID) (Figure 8). The PAM nucleotides TGG remain base paired (Figure 8). There is only slight tightening of the binding pocket on Cas9 compared with its PAM-free structure, suggesting a preformed PAM-binding channel. The structure identifies the DRKRY motif on a β-hairpin of the CTD in SpCas9 as the PAM-interaction unit, where the pair of arginine residues forms hydrogen bonds with the major groove edges of the two guanine nucleotides. The nucleotides on the target strand are not recognized by Cas9, providing the structural basis for the tolerance of mismatches in the PAM on this strand. The specific PAM–Cas9 interaction triggers local structural changes that destabilize the adjacent base pairing (Figure 8). In particular, a sharp kink in the target strand immediately downstream of the PAM motif is formed (Figure 8). The kink is necessary for the target strand to transition from pairing with the nontarget strand to pairing with the sgRNA. Nearby serine and lysine residues recognize the kinked phosphate group (called the phosphate lock) (Figure 8), which drives the conformational change of the target strand necessary for R-loop formation (Figure 9).
Although the protein elements interacting with the TGG PAM sequence are conserved in some type II-A Cas9 species, these elements can be different in or completely absent from others (1, 15, 16). Furthermore, the sequences and locations of the PAM vary widely (16). Thus, the principle learned from the SpCas9 studies may need to be revised once more Cas9–PAM interactions are observed.
TARGET CLEAVAGE
Various crRNPs employ different mechanisms to cleave target nucleic acids. The catalytic centers for the type I and type II crRNPs are readily identified and confirmed on the basis of both sequence homology to known nucleases and in vitro biochemical and structural studies. However, those associated with type III crRNPs are more difficult to discern because of the lack of clear homology and high-resolution structures. Thus, we summarize below the known mechanisms of how type I and type II crRNPs cleave target DNA and the predicted mechanisms of how type III-B crRNPs cleave RNA.
DNA Cleavage: Something Borrowed
The type I surveillance crRNPs do not display DNA cleavage activities; rather, they recruit Cas3-related effectors to cleave the target DNA. Many type I Cas3 proteins are large helicase–nuclease combinations, but some contain two separate interacting polypeptides [Cas3′ being the helicase and Cas3″ being an HD nuclease]. A structure of a Cas3- and dsDNA-loaded Cascade was determined using cryoEM methods (33), and an atomic model of the complex was then constructed based on this structure using the ssDNA-loaded Cascade crystal structure (57). The modeled Cas3- and dsDNA-loaded Cascade shows that the PAM-proximal dsDNA is situated between the fingers domains of Cas7.6 and Cas7.5, and the complementary strand pairs with the bound crRNA while the noncomplementary strand traverses a positive path composed of the helix bundle of the large subunit and the two small subunits. This binding model places a region near the PAM sequence of the displaced noncomplementary strand on Cas3 and therefore predicts that the noncomplementary strand is nicked and further degraded by Cas3. Consistently, in vitro DNA cleavage experiments have shown that in the absence of ATP, only the noncomplementary strand is cleaved, whereas in the presence of ATP, both strands are cleaved processively (75).
The mechanism of how Cas3 interacts with and cleaves ssDNA was learned from recent crystal structures of Thermobifida fusca (type I-C) Cas3 bound with an ssDNA substrate and various nucleotides (37). The nuclease domain of Cas3 belongs to the family of metal-dependent HD nucleases, and its structure indeed resembles that of other HD proteins (37, 56). Cas3 has four domains: the HD domain, a RecA-like domain 1, a RecA-like domain 2, and a CTD. The two RecA-like domains form the helicase core, and the HD domain and the helicase core are arranged linearly with respect to the bound ssDNA, the 3′ and 5′ ends of which are anchored in the HD domain and the helicase core, respectively (Figure 3). The 3′ terminal phosphate group of the ssDNA is engaged in coordination interactions with two bound metal ions and is believed to be cleaved via a nucleophilic attack mediated by a metal-coordinated water molecule. The fact that ATP is required for Cas3 to cleave both DNA strands processively suggests that a large rearrangement in Cas3-bound Cascade is powered by ATP hydrolysis and is necessary for Cas3 to access the complementary strand. There is also some evidence indicating that negative supercoil in DNA can increase the efficiency of Cascade cleavage, perhaps via efficient conformational change and strand separation (89).
The type II Cas9 proteins also use well-characterized nuclease domains, the HNH-like and the RuvC-like domains, to cleave DNA. The HNH-like domain contains the hallmark ββα-metal active site, which typically comprises a coordinated metal; a water molecule; and aspartate, asparagine, and histidine residues. The coordinate metal ion is not observed in the currently known Cas9 structures owing to mutation of the catalytic histidine. Studies of other HNH nucleases suggest that the histidine acts as a general base to activate the water molecule, allowing it to attack the scissile phosphate in the in-line displacement reaction. The metal ion coordinated by the aspartate and asparagine residues and the oxygen atoms of the scissile phosphate stabilizes the phosphoanion transition state and the leaving group (83). The RuvC domain is also a well-characterized nuclease domain that contains two metal ions, two aspartate residues, a glutamate residue, and a histidine residue. In general, the two metals are simultaneously coordinated with a nonbridging oxygen of the scissile phosphate and a carboxylate group, enabling an activated hydroxyl group (typically water) to attack the scissile phosphate in the in-line displacement reaction (93). The current Cas9 structures still lack the detailed interactions between the DNA substrate and the nuclease domains at the active sites needed to confirm the proposed catalytic mechanisms.
RNA Cleavage: Shaping the Scissile Phosphate
Unlike the type I, type II, or type III-A effector/surveillance complexes that target DNA, the type III-B Cmr targets complementary RNA for destruction. Both the TtCmr and the PfCmr can make three, four, or five cuts, depending on the organism(s) and guide RNA used, although four cuts seem to be the most dominate product (6, 26, 64, 79). The last cleavage site (that closest to the 3′ end) on the target RNA is always 5 nt away from the last paired target nucleotide, and each of the remaining sites is located 6 nt upstream of the preceding site. The SsoCmr also cleaves RNA, but its cleavage pattern is somewhat different from that observed for the other two Cmrs. The SsoCmr does not cleave at regularly spaced sites; rather, it cleaves preferentially within AU-rich regions. The Sso complex also requires an unpaired flap at the 3′ end of the target nucleic acid, but the sequence of this flap is not important for cleavage. In addition, the SsoCmr cleaves both its guide RNA and its target RNA, and this activity depends on the presence of a 3′ overhang of the target RNA (95).
The fact that Cmrs cleave the target RNA at sites with a 6-nt spacing and the observation that the crRNA–DNA hybrid bound within Cascade has a 6-bp periodicity (38, 57, 96) may not be a coincidence. The four cleavage sites are reasonably attributed to the four Cas7 subunits that are separated by 6 bp of stretched RNA (64) (Figure 9c), a notion supported by modeling and mutagenesis studies. Furthermore, the observed set of striking α1 hook structures is believed to act analogously to the Asp-Arg/Lys-Trp triad of the Cas7 that penetrates the cRNA–ssDNA duplex with regular 6-bp spacing (96). The distorted RNA bases at this 6-nt spacing have important implications for breakage of the phosphate backbone because the bases that are flipped outward facilitate formation of the necessary in-line conformation at the upstream scissile phosphate and can thus enhance the rate of bond breakage at these sites.
Despite the consistency between the structural data and the evenly spaced cleavage sites, the four Cmr4/four cleavage site model does not satisfactorily explain cleavage at three or five sites (26, 64, 79). Although the varying Cmr4 subunit model accounts for these cleavage sites better than the four Cmr4/four cleavage site model does (Figure 9c), the assemblies corresponding to varying Cmr4 subunits have not been captured by experiments, suggesting a high stability of the four-Cmr4 assembly. Alternatively, other Cas7 proteins may play a role in cleavage in some organisms, and the activity of a given Cmr4 may depend on its location within the helical assembly. Answers to these unresolved issues await high-resolution structures of the Cmr holo-complex.
The type III-A Csm was shown to cleave RNA in the same manner as the type III-B Cmr does (80). Furthermore, the Csm was recently shown to target DNA in a transcription-dependent manner (6). Given that these two complexes have a similar overall assembly, especially with respect to the arrangement of the Cas7 subunits (80), the Csm is believed to bind and cleave RNA according to the same principle as the Cmr. Thus, the Csm may use its ability to bind RNA in locating target DNA (Figure 9d), making it a dual-substrate cleavage crRNP.
APPLICATIONS
One of the most exciting prospects in the research on CRISPR-Cas immunity is development of new biotechnology that serves the needs of basic research and those of clinical applications. Both the CRISPR-Cas system as a whole and individual enzymes have been exploited for these purposes, which range from generating phage-resistant bacterial products (4) to altering specific genetic sequences in animal cells (85). The most powerful yet simple tool to have emerged from the CRISPR-Cas research to date is the Cas9-based genome editing system. Cloning customized sgRNA together with Cas9 in transfecting vector systems allows simultaneous delivery of the type II crRNP to cells that can then cleave almost any desired DNA target. The utility of the Cas9–sgRNA nuclease in eukaryotic cells is based on its ability to produce RNA-guided double-stranded breaks in genomes. These breaks can be repaired either by the error-prone process of nonhomologous end-joining or by the precise process of homology-directed repair. The superb adaptability and power of the programmable RNA-guided nuclease have been demonstrated in multiple organisms and cell types (for reviews on this topic, see References 36, 51, 71, 85).
Despite its promises and its rapid gain in popularity as a major genome editing tool, the Cas9–sgRNA system still has several limitations. A notable fraction of off-target nucleic acid degradation has been documented (17). A large sgRNA screening across multiple genes in different organisms suggests that the PAM sequence in use is not completely optimized (14). Although efforts using modified Cas9 resulted in fewer off-target effects, better systems with higher stringency are required for safe applications. Continued improvement in the Cas9 enzyme itself through either protein or tracrRNA engineering provides one solution to the problem. Alternatively, Cas9 enzymes from other species or multisubunit systems, such as the type I-E Cascade and the type III-A crRNP, that have tighter requirements for the PAM sequence used could be developed to eliminate the off-target activities.
Targeting DNA alone may limit our ability to achieve full knockout owing to splice variants or inaccessible chromatin structures. In cases in which the cellular contexts do impact Cas9 accessibility, RNA interference (RNAi) or CRISPR-Cas RNA-targeting complexes such as the recently characterized PAM-dependent RNA-cleaving Cas9 system (60) may be used. Moreover, the type II-B Cas9 and the type III-B Cmr may be combined with the Cas9–sgRNA nuclease to achieve the best gene silencing result. As our basic knowledge on various CRISPR-Cas interference systems grows, newer and safer biotechnology tools are expected to emerge.
CONCLUDING REMARKS
The crRNPs carry out nucleic acid silencing function by a variety of methods that were initially thought to be unrelated. Although this observation remains true for type II crRNPs, structural and mechanistic connections between type I and type III crRNPs, as well as among type III subtypes, are now evident. Despite a distant phylogeny, the protein subunits that compose both type I type III crRNPs may be categorized into four principal classes that construct a similar architecture characterized by helical repeats. More importantly, the ways in which the crRNAs, and possibly the target nucleic acids (regardless of whether they are DNA or RNA), interact with the assembled crRNPs are strikingly similar. This mode of nucleic acid interaction is believed to result in both fidelity and silencing efficiency. The segmented target–guide base pairing cycle has a low tolerance for mismatches and allows multiple cleavage of the target.
Critical interactions that allow the crRNP to distinguish self from foreign DNA and that lead to a following testable model have been observed only in the case of the type II Cas9 crRNP. A pair of arginine residues stabilize the two guanine nucleotides of the PAM sequence, resulting in a kinked phosphate backbone of the complementary strand that is stabilized by the phosphate lock loop. The phosphate lock promotes unwinding of the DNA that in turn pairs with the guide RNA. Although innovative biologists are using sophisticated screening methods to quickly identify the most efficient and broadly applicable Cas9 technology, theoretical studies that may help validate and predict experimental results should also be undertaken, and, more importantly, such studies are needed to understand the fundamental basis for the observed cellular effects. Given the available crystal structures and the growing amount of experimental data on Cas9 specificity, these studies are now within reach.
Many questions surrounding the CRISPR-Cas pathway remain unanswered. Given the fact that both RNA and DNA may be cleaved by many crRNPs, what are the true interference targets of the CRISPR-Cas immunity process in cells? How do the multisubunit crRNPs recognize the correct PAM sequence? How do type III-A crRNPs achieve transcription-dependent DNA targeting? Do the energetics of PAM–Cas9 interactions support the formation of a DNA–RNA three-way junction of the R-loop? Do the energetics of segmented pairing support its hypothesized beneficial effect to fidelity? What defines the length of the helical assemblies of crRNPs? Would multicomponent systems also be a good choice for genome editing and RNA silencing? Do all crRNPs have roles in prokaryote development and in the virulence of pathogens? Continued mechanistic studies of crRNPs may reveal surprising answers to these questions.
Acknowledgments
This work was supported by National Institutes of Health grant R01 GM099604 to H.L.
Glossary
- Clustered regularly interspaced short palindromic repeats (CRISPR)
prokaryotic DNA loci that store invader genetic elements used for defending against invaders
- Repeat
a short and repetitive DNA element within the CRISPR locus; often palindromic but can be nonpalindromic
- Spacer
a unique short DNA element flanked by identical repeats within the CRISPR locus; often match genetic elements of invaders
- crRNP
ribonucleoprotein particles formed between Cas proteins and crRNA that perform crRNA-guided surveillance or cleavage of nucleic acids
- CRISPR-Cas types
a categorization system based on the phylogeny of Cas1 and the cas gene combinations within a given CRISPR locus; an organism may contain more than one type
- Repeat associated mysterious proteins (RAMPs)
proteins containing a ferredoxin-like fold and a glycine-rich loop; these proteins are the most abundant type of Cas protein
- HD domain
a nuclease domain containing arrangements of histidine and aspartate residues, typically H—HD—D or HD—H—D
- CRISPR-associated complex for antiviral defense (Cascade)
the first reported CRISPR antiviral complex; belongs to the type I-E CRISPR-Cas system
- crRNA, tracrRNA, and sgRNA
type-specific small RNAs derived from CRISPR-related loci used by Cas proteins in guiding RNA processing or nucleic acid interference
- Cas subtype Mycobacterium tuberculosis antiviral complex (Csm)
type III-A under the unified system; first demonstrated DNA interference CRISPR-Cas system
- Cas module RAMP antiviral complex (Cmr)
type III-B under the unified system; first demonstrated RNA interference CRISPR-Cas system
- Protospacers
exogenous nucleic acid elements with sequences that are complementary to those of CRISPR spacers
- R-loop
a three-stranded structure in which an RNA is hybridized to a complementary DNA strand, resulting in displacement of the noncomplementary strand
- Protospacer-adjacent motif (PAM)
a highly conserved and system-specific 2–5 nucleotide motif flanking one side of the protospacer that triggers DNA interference
- RuvC-like domain
a nuclease domain with an RNase H fold that contains two metal ions and a combination of histidine, aspartate, and glutamate residues
- HNH-like domain
a nuclease domain containing the hallmark ββα-metal active site formed by a metal ion and a combination of histidine, asparagine, and aspartate residues
- Phosphate lock
a protein element in Cas9 that stabilizes the severely kinked phosphate backbone of the first DNA nucleotide paired with the sgRNA
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey S. The Cmr complex: an RNA-guided endoribonuclease. Biochem. Soc. Trans. 2013;41:1464–1467. doi: 10.1042/BST20130216. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R, Horvath P. CRISPR: new horizons in phage resistance and strain identification. Annu. Rev. Food Sci. Technol. 2012;3:143–162. doi: 10.1146/annurev-food-022811-101134. [DOI] [PubMed] [Google Scholar]
- 5.Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benda C, Ebert J, Scheltema RA, Schiller HB, Baumgärtner M, et al. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol. Cell. 2014;56:43–54. doi: 10.1016/j.molcel.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 8.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carte J, Christopher RT, Smith JT, Olson S, Barrangou R, et al. The three major types of CRISPR-Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Mol. Microbiol. 2014;93:98–112. doi: 10.1111/mmi.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 12.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, et al. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lecrivain AL, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2011;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 19.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. PNAS. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasiunas G, Siksnys V. RNA-dependent DNA endonuclease Cas9 of the CRISPR system: Holy Grail of genome editing? Trends Microbiol. 2013;21:562–567. doi: 10.1016/j.tim.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Gasiunas G, Sinkunas T, Siksnys V. Molecular mechanisms of CRISPR-mediated microbial immunity. Cell. Molec. Life Sci. 2014;71:449–465. doi: 10.1007/s00018-013-1438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Molec. Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottesman S. Microbiology: dicing defence in bacteria. Nature. 2011;471:588–589. doi: 10.1038/471588a. [DOI] [PubMed] [Google Scholar]
- 25.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLOS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hale CR, Cocozaki A, Li H, Terns RM, Terns MP. Target RNA capture and cleavage by the Cmr type III-B CRISPR–Cas effector complex. Genes Dev. 2014;28:2432–2443. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. PNAS. 2011;108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatoum-Aslan A, Samai P, Maniv I, Jiang W, Marraffini LA. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J. Biol. Chem. 2013;288:27888–27897. doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heidrich N, Vogel J. Same same but different: new structural insight into CRISPR-Cas complexes. Mol. Cell. 2013;52:4–7. doi: 10.1016/j.molcel.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 32.Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem. Sci. 2015;40:58–66. doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, et al. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. PNAS. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, et al. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 2005;29:435–463. doi: 10.1016/j.femsre.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 36.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huo Y, Nam KH, Ding F, Lee H, Wu L, et al. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nature Struct. Mol. Biol. 2014;21:771–777. doi: 10.1038/nsmb.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, et al. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson RN, Lavin M, Carter J, Wiedenheft B. Fitting CRISPR-associated Cas3 into the helicase family tree. Curr. Opin. Struct. Biol. 2014;24:106–114. doi: 10.1016/j.sbi.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones CL, Sampson TR, Nakaya HI, Pulendran B, Weiss DS. Repression of bacterial lipoprotein production by Francisella novicida facilitates evasion of innate immune recognition. Cell. Microbiol. 2012;14:1531–1543. doi: 10.1111/j.1462-5822.2012.01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Molec. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 44.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H. Structural principles of CRISPR RNA processing. Structure. 2015;23:13–20. doi: 10.1016/j.str.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J. Biol. Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. Aputative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makarova KS, Wolf YI, Koonin EV. The basic building blocks and evolution of CRISPR–Cas systems. Biochem. Soc. Trans. 2013;41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malina A, Mills JR, Cencic R, Yan Y, Fraser J, et al. Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev. 2013;27:2602–2614. doi: 10.1101/gad.227132.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mojica FJM, Díez-Villasñor C, Garía-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 55.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 56.Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J. Biol. Chem. 2011;286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulepati S, Héroux A, Bailey S. Structural biology. Crystal structure of a CRISPR RNA–guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nam KH, Haitjema C, Liu X, Ding F, Wang H, et al. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 63.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Molec. Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 64.Ramia NF, Spilman M, Tang L, Shao Y, Elmore J, et al. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Cell Rep. 2014;9:1610–1617. doi: 10.1016/j.celrep.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramia NF, Tang L, Cocozaki AI, Li H. Staphylococcus epidermidis Csm1 is a 3′–5′ exonuclease. Nucleic Acids Res. 2014;42:1129–1138. doi: 10.1093/nar/gkt914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reeks J, Naismith JH, White MF. CRISPR interference: a structural perspective. Biochem. J. 2013;453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reeks J, Sokolowski RD, Graham S, Liu H, Naismith JH, White MF. Structure of a dimeric crenarchaeal Cas6 enzyme with an atypical active site for CRISPR RNA processing. Biochem. J. 2013;452:223–230. doi: 10.1042/BJ20130269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richter H, Zoephel J, Schermuly J, Maticzka D, Backofen R, Randau L. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012;40:9887–9896. doi: 10.1093/nar/gks737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rouillon C, Zhou M, Zhang J, Politis A, Beilsten-Edmands V, et al. Structure of the CRISPR interference complex CSM reveals key similarities with Cascade. Mol. Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sampson TR, Saroj SD, Llewellyn AC, Tzeng Y-L, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat. Struct. Molec. Biol. 2011;18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 73.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. PNAS. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shao Y, Li H. Recognition and cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6. Structure. 2013;21:385–393. doi: 10.1016/j.str.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, et al. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sokolowski RD, Graham S, White MF. Cas6 specificity and CRISPR RNA loading in a complex CRISPR-Cas system. Nucleic Acids Res. 2014;42:6532–6541. doi: 10.1093/nar/gku308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 78.Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, et al. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol. Cell. 2013;52:146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Staals RHJ, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol. Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Staals RHJ, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, et al. RNA Targeting by the Type III-A CRISPR-Cas Csm Complex of Thermus thermophilus. Mol. Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 84.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Terns RM, Terns MP. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet. 2014;30:111–118. doi: 10.1016/j.tig.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Westra ER, Swarts DC, Staals RHJ, Jore MM, Brouns SJJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 89.Westra ER, van Erp PBG, Künne T, Wong SP, Staals RHJ, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 92.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. PNAS. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, et al. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–150. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]