Abstract
Background
The aim of this study was to investigate the diagnostic and prognostic value of microRNA (miRNA)-21, miRNA-23a, and miRNA-125b in Burkitt lymphoma (BL) in children.
Material/Methods
We recruited 41 children with BL for the case group, 56 children with lymph node inflammation for the positive control group, and 60 healthy children for the negative control group. Real-time fluorescent quantitative polymerase chain reaction (RT-qPCR) was conducted for detection of circulating miRNA-21, miRNA-23a, and miRNA-125b. A receiver operating characteristic (ROC) curve was drawn to compare the diagnostic value of miRNA-21, miRNA-23a, and miRNA-125b. Kaplan-Meier method and log-rank test were used for prognostic analyses.
Results
MiRNA-21 and miRNA-23a had significantly higher expression in cases than in positive and negative controls (all P<0.05). Overexpression of miRNA-21 and miRNA-23a were associated with staging, WBC, upregulated serum lactate dehydrogenase (LDH) level, presence of lymphoma size ≥6 cm, and cluster of differentiation 10 (CD10) expression, while miRNA-125b expression had an association with staging and upregulated serum LDH level (both P<0.05). ROC curves of miRNA-21, miRNA-23a, and miRNA-125b presented an area under curve (AUC) of 0.759, 0.853 and 0.615, respectively. MiRNA-21 and miRNA-23a in combination had an AUC of 0.869. After treatment, both miRNA-21 and miRNA-23a expression were significantly decreased (both P<0.05). Advanced clinical stage, upregulated LDH, and lymphoma size of ≥6 cm were related to low complete remission rate (all P<0.05).
Conclusions
Patients with high expression of miRNA-21 and miRNA-23a had significantly lower complete remission rates and survival rates than those with low expression. Expression of miRNA-21 and miRNA-23a may serve as useful diagnostic and prognostic biomarkers in children with BL.
MeSH Keywords: Burkitt Lymphoma, Diagnosis, Prognosis
Background
Burkitt lymphoma (BL), a member of the non-Hodgkin lymphoma family, is a high-grade mature B cell lymphoma with a cell doubling time of 24–48 h, making it the fastest-growing human tumor [1,2]. Despite efforts made in the past decades, BL remains a great challenge due to difficult diagnosis and poor prognosis after chemotherapy [3,4]. In this context, it would be of great value to understand the pathogenesis of BL at the molecular level, which would simplify the diagnosis and improve the prognosis. MicroRNAs (miRNAs) are single-stranded RNA molecules (20–23 nucleotides) that repress gene expression through interaction with the 3′ untranslated region of mRNA [5]. Accumulating studies have shown that miRNAs are important participants, acting as oncogenic or anti-oncogenic factors in the development of cancers through regulation of the cell cycle [6–8]. There is also evidence that miRNAs are useful molecular biomarkers in the classification and prognosis in aggressive B cell lymphoma [9]. We believe that expressions of miRNAs may be useful biomarkers for diagnosis and prognosis of BL.
Recently, miRNA-21 has been found to be overexpressed in diffuse large B cell lymphoma and is validated as an independent risk factor in prognosis [10–12]. miRNA-21 is also a diagnostic biomarker in primary diffuse large B cell lymphoma of the central nervous system [13]. In a mouse model of precursor B cell lymphoma, overexpression of miRNA-21 in the hematopoietic cells was identified as an oncogenic factor [14]. Thus, miRNA-21 could be considered as a potential diagnostic and prognostic biomarker for B cell malignancies. MiRNA-23a cluster is observed as a potent inhibitor of B lymphopoiesis both in vivo and in vitro, promoting myeloid development over lymphoid development [15]. The value of miRNA-23a in early detection and prognosis has been confirmed in other diseases [16–18]. In an miRNA profiling study, miRNA-23a was well established as a differentially expressed gene in BL in comparison with other B cell non-Hodgkin lymphomas [19]. In the same study, differential expression of miRNA-125b was also listed as a characteristic of BL [19]. miRNA-125b is suggested as a diagnostic and prognostic biomarker in cancers such as leukemia, breast cancer, and lung cancer [20–22]. In addition, miRNA-125b is reported to be overexpressed in pediatric lymphoma patients with high resistance to vincristine [23]. Based on these previous studies, we propose that expression of miRNA-21, miRNA-23a, and miRNA-125b may be diagnostic and prognostic biomarkers in BL.
To date, miRNA-21, miRNA-23a, and miRNA-125b are seldom studied in relation to BL in children; therefore, we conducted the present study to determine the role of miRNA-21, miRNA-23a, and miRNA-125b in the diagnosis and prognosis in children with BL.
Material and Methods
Ethics statement
The present study was performed with the approval of the Ethics Committee of the Children’s Hospital of Fudan University. All aspects of the study complied with the Declaration of Helsinki [24] and informed consent was received from all patients or their families before the examination.
Study subjects
The case group consisted of 41 children who were diagnosed with BL between September 2007 and October 2015 from our hospital. All the diagnoses conformed to the classification and criteria proposed by World Health Organization in 2008 [25] and all the patients (age range of 2–16 years; median age of 8 years) received no chemotherapy or radiotherapy before diagnosis. As observed in the histopathological examination, BLs had middle-sized lymphoma size, with round nucleus, nucleolus, moderate amount of cytoplasm and mitosis, and also had macrophagocytes interspersed like a starry sky. In addition, the BL cells went beyond the lymph node. The St Jude staging system was used for staging of BL [26]. To improve the reliability and accuracy of our study results, we excluded patients with BL that transformed from other types of lymphomas, patients with primary and/or secondary central nervous system lymphoma, patients with autoimmune diseases, and patients with other neoplastic diseases. The positive control group consisted of 56 patients with lymph node inflammation (35 boys and 21 girls; age range 3–16 years; median age of 7.6 years). All the included positive controls showed no presence of lymphoma or lymph node-related diseases in the histopathological examinations. The negative control group consisted of 60 healthy people (33 boys and 27 girls; age range 2–15 years; median age 7.4 years) who received physical examinations in our hospital during the same period. These 3 groups showed no clear differences in age or sex (all P>0.05).
Blood sampling and total RNA extraction
Peripheral blood (3–5 ml) was collected with EDTA-K2-containing tube, followed by 10-min serum centrifugation (1600×g). The upper layer of plasma was collected with a clean centrifuge tube and then centrifuged for 10 min (13300×g) to separate pure plasma from cellular debris and other pollutants. After centrifugation, pure plasma was drawn into another clean centrifuge tube and then frozen at −80°C. For RNA extraction, the pure plasma (500 μL) was placed in a clean centrifuge tube (1.5 ml) and heated at 75°C for 5 min and incubated at 42°C for 1 h. RNA extraction was conducted with a TRIzol kit (Invitrogen) in accordance with the instructions. RNA concentration was measured with NanoDrop 1000 (Thermo Scientific, USA) and the optical density ratio (1.8–2.0) at 260/280 nm was recorded. The extracted RNA was preserved at −80°C until use.
Real-time fluorescent quantitative polymerase chain reaction (RT-qPCR)
In accordance with the instructions of the RNA reverse transcription kit (TaKaRa), an RT-qPCR system (20 μL) was built for preparation of reverse transcription reagents. The reagent preparation was performed on ice. With well-mixed reagents and a qPCR machine (Applied Biosystems, USA), RT-qPCR was conducted to synthetize cDNA templates. The reaction condition was 30-min incubation at 37°C followed by 15-s incubation at 80°C. The RT-PCR products were preserved at −20°C until use. For measurement of miRNA-21, miRNA-23a, and miRNA-125b, U6 small nuclear RNA (snRNA) was selected as a reference gene and RT-qPCRs were conducted under the conditions of 40 cycles of 10-min incubation at 95°C, 15-s incubation at 95°C, and 1-min incubation at 60°C. Primers (forward and reverse) of miRNA-21, miRNA-23a, and miRNA-125b for RT-qPCR are listed in Table 1. The value of gene expression was calculated with 2−ΔΔCt method (ΔΔCt=ΔCttumor–ΔCtnormal, ΔCt=CtmiRNA–CtU6snRNA).
Table 1.
Primers of miRNA-21, miRNA-23a, miRNA-125b, and U6 snRNA for RT-qPCR.
| Gene | Primer sequence (5′-3′) | |
|---|---|---|
| miR-21 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACA |
| Forward | GCCGCTAGCTTATCAGACTGATGT | |
| Reverse | GTGCAGGGTCCGAGGT | |
| miR-23a | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGAAAT |
| Forward | GCGTAGGCAGTGTATTGCTAGC | |
| Reverse | CAGTGCAGGGTCCGAGGT | |
| miR-125b | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCATGGATACGACTCACAA |
| Forward | TGCGTCCCTGAGACCCTA | |
| Reverse | GTGCAGGGTCCGAGT | |
| U6 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAATATG |
| Forward | CTCGCTTCGGCAGCACA | |
| Reverse | AACGCTTCACGAATTTGCGT |
RT-qPCR – real-time fluorescent quantitative polymerase chain reaction, miRNA – microRNA; snRNA – small nuclear RNA; RT – reverse transcription.
Analyses after treatment
All the patients were treated with B-NHL-BFM-90 protocol [27]. For evaluation of short-term efficacy in patients with BL, the remission criteria published by National Comprehensive Cancer Network of the United States was used [28]. Therapeutic efficacy was described as complete remission, partial remission, stable disease, or recurrence/progression. The expressions of miRNA-21, miRNA-23a, and miRNA-125b were analyzed in relation to complete remission. The complete remission rate in a group was defined as the ratio between the number of patients with complete remission and the total number of patients in that group. Plasma of treated patients was collected to extract RNAs for determination of the expressions of miRNA-21, miRNA-23a, and miRNA-125b.
Follow-up
To investigate the associations between patient base-line characteristics and prognosis, we conducted telephone follow-ups and outpatient follow-ups once every 2 months. All the patients were followed up until their deaths.
Statistical analyses
SPSS 19.0 (SPSS Inc., Chicago, IL) was used for data analyses. Measurement data are expressed as mean ± standard deviation (χ̄±s). For measurement data, between-group comparisons were conducted by use of the non-paired t test. Enumeration data are expressed as frequency or percentage. Between-group comparisons were conducted by chi-square test and between-group comparisons among 3 groups were conducted by analysis of covariance model. A receiver operating characteristic (ROC) curve was drawn to present the diagnostic accuracy of miRNAs in BL. Kaplan-Meier method and log-rank test were used for prognostic analyses. P<0.05 indicated statistical difference and P<0.01 indicated a statistically significant difference.
Results
Clinical data
The base-line characteristics of included patients are presented in Table 2. Among the 41 children, 30 were boys and 11 were girls (boy/girl: 2.72/1), 27 were at stage III/IV (advanced stage, 65.85%), 23 had lymphomas in the head and neck (56.10%), 17 had lymphomas in the stomach (42.50%), 9 presented central nervous system involvement, 9 exhibited marrow involvement, 11 had bone involvement, 29 had lymph node involvement (70.73%) by imaging, 22 had B symptoms (e.g., fever, night sweats, and emaciation), 26 had anemia, 25 had increased white blood count (WBC) and 28 had increased platelet (PLT), 24 had upregulated lactate dehydrogenase (LDH) level (58.54%), 10 had higher uric acid level (24.38%), and 22 had lymphoma size of ≥6 cm. Immunohistochemical analyses showed 27 had positive expression of cluster of differentiation 10 (CD10) (65.85%), 9 had Bcl-2 (21.96%), and 31 had Bcl-6 expression (75.61%). A Ki-67-positive rate of >75% was found in 35 patients. All the included patients were free of human immunodeficiency virus (HIV) infection. Epstein-Barr virus (EBV) infection (n=8, 19.51%) and hepatitis B virus (HBV) infection (n=7, 17.07%) were found.
Table 2.
Clinical data of the 41 included Burkitt lymphoma patients.
| Base-line characteristic | Number (percentage) |
|---|---|
| Sex | |
| Male | 30 (73.17%) |
| Female | 11 (26.83%) |
| Staging | |
| I/II | 27 (65.85%) |
| III/IV | 14 (35.15%) |
| Location | |
| Head and neck | 23 (56.10%) |
| Stomach | 17 (42.50%) |
| Central nervous system involvement | 9 (21.95%) |
| Marrow involvement | 9 (21.95%) |
| Bone involvement | 11 (26.83%) |
| Lymph node | 29 (70.73%) |
| B symptom | 22 (53.66%) |
| Anemia | 26 (63.41%) |
| Increased WBC | 25 (60.98%) |
| Upregulated PLT | 28 (68.29%) |
| Upregulated LDH level | 24 (58.54%) |
| Higher uric acid level | 10 (24.38%) |
| Lymphoma size of ≥6 cm | 22 (53.66%) |
| CD10 (+) | 27 (65.85%) |
| Bcl-2 (+) | 9 (21.96%) |
| Bcl-6 (+) | 31 (75.61%) |
| Ki-67 (> 75%) | 35 (85.37%) |
| Virus infection | |
| HIV | 0 (00.00%) |
| EBV | 8 (19.51%) |
| HBV | 7 (17.07%) |
LDH – lactate dehydrogenase; HIV – human immunodeficiency virus; EBV – Epstein-Barr virus; HBV – hepatitis B virus; CD10 – cluster of differentiation 10; WBC – white blood cell; PLT – platelet.
Expressions of miRNA-21, miRNA 23a, and miRNA-125b
RT-qPCR results are presented in Figure 1. Cases presented significantly higher mean expressions of miRNA-21 and miRNA-23a (both P<0.05), but has mean expression of miRNA-125b (P>0.05) similar to that found in positive and negative controls. No significant difference was observed between positive controls and negative controls in the mean expressions of miRNA-21, miRNA-23a, or miRNA-125b (all P>0.05).
Figure 1.
RT-qPCR was used to detect the expressions of miRNA-21, miRNA-23a, and miRNA-125b in children with BL, LI, and HP. RT-qPCR – real-time fluorescent quantitative polymerase chain reaction; BL – Burkitt lymphoma; LI – lymph node inflammation; HP – healthy people; miRNA – microRNA; * P<0.05, between-group comparison among the 3 group after adjustment of such co-variants as age and sex.
Associations of miRNA-21, miRNA-23a, and miRNA-125b expressions with clinicopathologic features
The associations of miRNA-21, miRNA-23a, and miRNA-125b expressions with clinicopathologic features are presented in Table 3. MiRNA-21, miRNA-23a, and miRNA-125b expressions presented no obvious association with sex, B symptoms, uric acid level, Bcl-2 expression, Bcl-6 expression, EBV infection, or HBV infection (all P>0.05). Expressions of miRNA-21 and miRNA-23a were closely correlated with staging, increased WBC, upregulated serum lactate dehydrogenase (LDH) level, presence of lymphoma size ≥6 cm, and CD10 expression (all P<0.05), while miRNA-125b expression had a strong association with staging, anemia, increased WBC and PLT, upregulated serum LDH level, and CD10 expression (all P<0.05).
Table 3.
Associations of miRNA-2, miRNA-3a, and miRNA-125b expressions with clinicopathologic features of Burkitt lymphoma.
| Cases (n) | miRNA-21 expression | P | miRNA-23a expression | P | miRNA-125b expression | P | |
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Male | 30 | 1.23±0.50 | 0.615 | 2.19±0.47 | 0.761 | 2.50±0.36 | 0.066 |
| Female | 11 | 1.33±0.63 | 2.14±0.56 | 2.28±0.25 | |||
| Staging | |||||||
| I/II | 14 | 0.69±0.16 | <0.001 | 1.79±1.37 | <0.001 | 2.28±0.42 | 0.049 |
| III/IV | 27 | 1.554±0.35 | 2.38±0.41 | 2.53±0.27 | |||
| B symptom | |||||||
| Yes | 22 | 1.27±0.74 | 0.888 | 2.25±0.51 | 0.323 | 2.50±0.34 | 0.247 |
| No | 19 | 1.25±0.73 | 2.09±0.46 | 2.38±0.35 | |||
| Anemia | |||||||
| Yes | 26 | 1.35±0.51 | 0.151 | 2.23±0.51 | 0.328 | 2.46±0.34 | 0.707 |
| No | 15 | 1.11±0.49 | 2.08±0.43 | 2.42±0.36 | |||
| WBC | |||||||
| Increased | 25 | 1.41±0.52 | 0.016 | 2.30+0.50 | 0.043 | 2.48±0.34 | 0.36 |
| Normal | 16 | 1.02±0.40 | 1.98±0.40 | 2.38±0.35 | |||
| PLT | |||||||
| Increased | 28 | 1.36±0.55 | 0.057 | 2.24±0.50 | 0.217 | 2.43±0.32 | 0.63 |
| Normal | 13 | 1.05±0.34 | 2.04±0.44 | 2.48±0.42 | |||
| No | 19 | 1.25±0.73 | 2.09±0.46 | 2.38±0.35 | |||
| LDH | |||||||
| Upregulated | 24 | 1.54±0.82 | 0.004 | 2.41±0.43 | <0.001 | 2.55±0.29 | 0.016 |
| Normal | 17 | 0.86±0.49 | 1.85±0.37 | 2.29±0.37 | |||
| Uric acid | |||||||
| Upregulated | 10 | 1.30±0.74 | 0.759 | 2.24±0.51 | 0.664 | 2.45±0.46 | 0.961 |
| Normal | 31 | 1.24±0.43 | 2.16±0.49 | 2.44±0.31 | |||
| Lymphoma size of ≥6 cm | |||||||
| Yes | 22 | 1.53±0.84 | 0.011 | 2.51±0.34 | <0.001 | 2.53±0.29 | 0.078 |
| No | 19 | 0.95±0.55 | 1.79±0.31 | 2.34±0.38 | |||
| CD10 | |||||||
| + | 27 | 1.47±0.79 | 0.012 | 2.45±0.35 | <0.001 | 2.55±0.29 | 0.004 |
| − | 14 | 0.86±0.47 | 1.66±0.22 | 2.24±0.36 | |||
| Bcl-2 | |||||||
| + | 9 | 1.131±0.43 | 0.402 | 2.100±0.32 | 0.602 | 2.36±0.35 | 0.406 |
| − | 32 | 1.294±0.53 | 2.197±0.52 | 2.47±0.35 | |||
| Bcl-6 | |||||||
| + | 31 | 1.25±0.54 | 0.876 | 2.19±0.50 | 0.783 | 2.43±0.36 | 0.584 |
| − | 10 | 1.28±0.43 | 2.14±0.46 | 2.50±0.31 | |||
| Ki-67-positive rate | |||||||
| >75% | 35 | 1.26±0.51 | 0.868 | 2.19±0.46 | 0.599 | 2.46±0.36 | 0.449 |
| <75% | 6 | 1.23±0.57 | 2.08±0.64 | 2.34±0.26 | |||
| EBV | |||||||
| + | 8 | 1.23±0.54 | 0.883 | 2.001±0.59 | 0.262 | 2.43±0.35 | 0.91 |
| − | 33 | 1.26±0.51 | 2.22±0.46 | 2.45±0.35 | |||
| HBV | |||||||
| + | 7 | 1.02±0.38 | 0.173 | 2.17±0.33 | 0.952 | 2.56±0.53 | 0.525 |
| − | 34 | 1.31±0.52 | 2.18±0.52 | 2.42±0.30 | |||
LDH – lactate dehydrogenase; HIV – human immunodeficiency virus; EBV – Epstein-Barr virus; HBV – Hepatitis B virus; miRNA – microRNA; WBC – white blood cell; PLT – platelet; CD10 – cluster of differentiation 10.
Diagnostic thresholds of miRNA-21, miRNA-23a, and miRNA-125b expressions
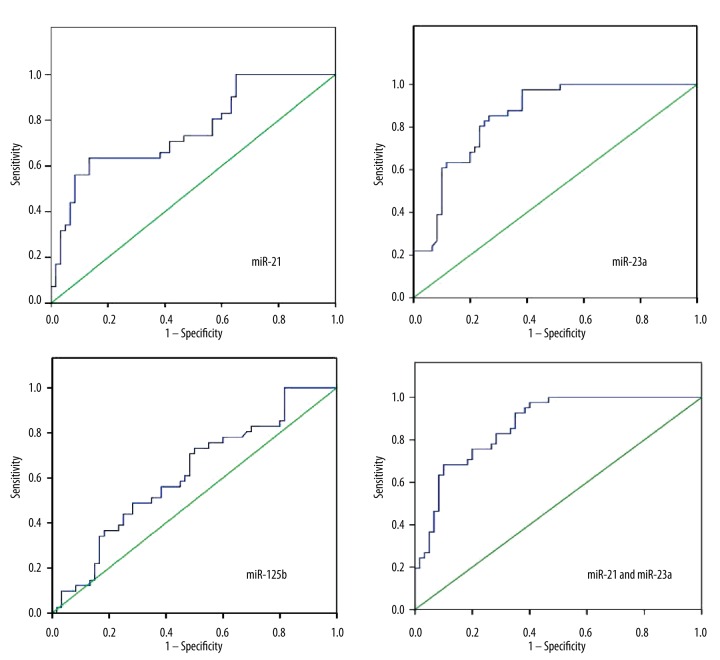
As shown in Figure 2, ROCs of miRNA-21, miRNA-23a, and miRA-125b expressions presented areas under the curve (AUC) of 0.759, 0.853, and 0.615; and miRNA-21 and miRNA-23a expression in combination had an AUC of 0.869, respectively. When compared with the reference value (AUC=0.5), AUCs of miRNA-21 and miRNA-23a were significantly higher (both P<0.05), while that of miRNA-125b was slightly (insignificantly) higher (P>0.05). Diagnostic threshold was 1.141 for miRNA-21 (sensitivity 63.4%, specificity 86.7%) and 1.376 for miRNA-23a (97.6% and 61.7%, respectively).
Figure 2.
ROC curves of miRNA-21, miRNA-23a, and miRNA-125b in isolation, and miRNA-21 and miRNA-23a in combination. ROC – receiver operating characteristic; miRNA – microRNA.
Expressions of miRNA-21, miRNA-23a, and miRA-125b before and after treatment
All the included patients received B-NHL-BFM-90 chemotherapy. After chemotherapy, miRNA-21, miRNA-23a, and miRNA-125b expressions were remeasured and compared with those before treatment (Figure 3). miRNA-21 expression and miRNA-23a expression were significantly decreased (miRNA-21: P<0.05; miRNA-23a: P<0.01), while miRNA-125b expression showed no significant change (P>0.05) after chemotherapy when compared with those before treatment.
Figure 3.
Pre- and post-chemotherapy expressions of miRNA-21, miRNA-23a, and miRNA-125b. miRNA – microRNA.
Short-term efficiency and expressions of miRNA-21, miRNA-23a, and miRA-125b and clinicopathological features
The associations between short-term efficacy after chemotherapy, and expressions of miRNA-21, miRNA-23a, and miRA-125b are presented in Table 4. The complete remission rate in patients with low miRNA-21 expression (56.7%) was significantly higher than that in patients with high miRNA-21 expression (18.2%) (P<0.05). Patients with high expression of miRNA-23a had significantly lower complete remission rate than patients with low expression of miRNA-23a (15.4% vs. 60.7%, P<0.05). No significant difference was observed between patients with high expression of miRNA-125b and patients with low expression of miRNA-125b in the complete remission rate (P>0.05). Among the clinicopathological features, advanced clinical stage, upregulated LDH, and lymphoma size of ≥6 cm were related to low complete remission rate (all P<0.05), while sex, B symptoms, uric acid level, anemia, WBC, PLT, CD10, Bcl-2 expression, Bcl-6 expression, Ki-67, EBV, and HBV had no significant association with complete remission rate (all P>0.05).
Table 4.
Associations of miRNA-2, miRNA-3a, and miRNA-125b expressions with short-term efficacy after chemotherapy.
| Cases | Cases of complete remission | Complete remission rate | P | ||
|---|---|---|---|---|---|
| MiR-21 | Low | 30 | 17 | 56.70% | 0.039 |
| High | 11 | 2 | 18.20% | ||
| MiR-23a | Low | 28 | 17 | 60.70% | 0.009 |
| High | 13 | 2 | 15.40% | ||
| MiR-125b | Low | 18 | 9 | 50.00% | 0.758 |
| High | 23 | 10 | 43.50% | ||
| Gender | Boy | 30 | 12 | 40.00% | 0.179 |
| Girl | 11 | 7 | 63.60% | ||
| Staging | I/II | 14 | 12 | 85.70% | < 0.001 |
| III/IV | 27 | 7 | 25.90% | ||
| B symptom | Yes | 22 | 8 | 36.40% | 0.168 |
| No | 19 | 11 | 57.90% | ||
| Anemia | Yes | 26 | 10 | 38.50% | 0.183 |
| No | 15 | 9 | 60.00% | ||
| WBC | Increased | 25 | 8 | 32.00% | 0.065 |
| Normal | 16 | 11 | 68.80% | ||
| PLT | Increased | 28 | 11 | 39.30% | 0.184 |
| Normal | 13 | 8 | 61.50% | ||
| LDH | Upregulated | 24 | 7 | 29.20% | 0.009 |
| Normal | 17 | 12 | 70.60% | ||
| Uric acid | Upregulated | 10 | 2 | 20.00% | 0.055 |
| Normal | 31 | 17 | 56.70% | ||
| Lymphoma size of ≥6 cm | Yes | 22 | 7 | 31.80% | 0.045 |
| No | 19 | 12 | 63.20% | ||
| CD10 | + | 27 | 10 | 37.00% | 0.097 |
| − | 14 | 9 | 64.30% | ||
| Bcl-2 | + | 9 | 3 | 33.30% | 0.376 |
| − | 32 | 16 | 50.00% | ||
| Bcl-6 | + | 31 | 12 | 45.20% | 0.085 |
| − | 10 | 7 | 50.00% | ||
| Ki-67 | >75% | 35 | 18 | 56.30% | 0.072 |
| <75% | 6 | 1 | 16.70% | ||
| EBV | + | 8 | 2 | 25.00% | 0.385 |
| − | 33 | 17 | 51.50% | ||
| HBV | + | 7 | 1 | 14.30% | 0.062 |
| − | 34 | 18 | 23.50% |
High expression level is defined as a level higher than the diagnostic threshold and low expression level is a level otherwise. miRNA – microRNA; LDH – lactate dehydrogenase; HIV – human immunodeficiency virus; EBV – Epstein-Barr virus; HBV – hepatitis B virus; miRNA – microRNA; WBC – white blood cell; PLT – platelet; CD10 – cluster of differentiation 10.
Expressions of miRNA-21, miRNA-23a, and miRA-125b and prognosis
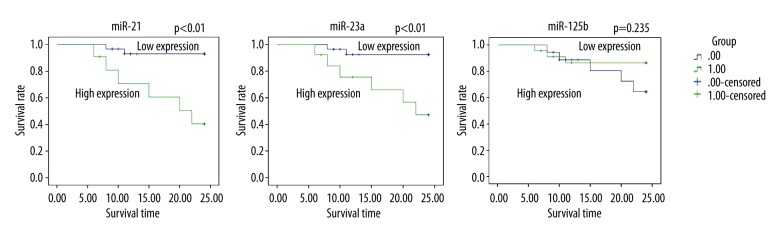
All the include patients were followed up for 6–24 moths, with a mean follow-up period of 18 months. Survival analyses by Kaplan-Meier method is illustrated in Figure 4. Patients with high expressions of miRNA-21 and miRNA-23a had significantly lower survival rate than patients with low expressions of miRNA-21 and miRNA-23a (both P<0.05). The survival rate did not vary with the expression of miRNA-125b (P>0.05).
Figure 4.
Expressions of miRNA-21, miRNA-23a, and miRNA-125b in relation to the prognosis of Burkitt lymphoma in children. miRNA – microRNA.
Discussion
Usually, miRNAs are regulated by RNA-binding proteins of high stability, making miRNAs applicable and reliable molecular biomarkers [29]. Circulating miRNAs have been shown to be stable blood-based markers for cancer detection [30]. In the present study, we recorded the serum level of miRNA-21, miRNA-23a, and miRNA-125b in children with BL, in order to explore the associations of expressions of miRNA-21, miRNA-23a, and miRNA-125b with the development of BL in children.
In our study, significant upregulation of serum miRNA-21 andmiRNA-23a level was found in the children with BL, but not in positive controls and negative controls. By statistical analyses, we also observed that expressions of miRNA-21, miRNA-23a, and miRNA-125b are closely associated with clinicopathologic features such as disease stage and LDH level. miRNA-21, miRNA-23a, and miRNA-125b appear to be important modulators of the cell cycle through different signaling pathways. Go et al. reported that miRNA-21 can activate the PI13/AKT pathway and thereby play an oncogenic role in diffuse large B cell lymphoma [31]. MiRNA-23a may regulate cell growth and cell apoptosis through targeting apoptotic protease activating factor-1 [32]. MiRNA-125b can target BAK1, leading to increased cell proliferation and blocked cell apoptosis [33]. On the basis of these results, we believe that miRNA-21, miRNA-23a, and miRNA-125b may be important participants in the development of BL. Concordantly, Sun found that circulating miRNA-21 is overexpressed in patients with B cell non-Hodgkin lymphoma and is closely associated with disease stage [34].
To evaluate the diagnostic and prognostic value of these 3 biomarkers for children, we calculated the AUC of diagnostic ROC and conducted a follow-up study. In our study, we found that the AUC of miRNA-21 was 0.759 and that of miRNA-23a was 0.853, both significantly higher than 0.5 by statistical analysis. For this reason, we believe that miRNA-21 and miRNA-23a may be useful diagnostic indicators. Consistent with previous studies, we found that miRNA-21 and miRNA-23a are biomarkers of diagnostic value [35,36]. In contrast with previous studies, miRNA-125b, although abnormally expressed in children with BL, had an AUC of only 0.615 and presented no significant diagnostic value in our study [37]. The exact reason for this contradiction is not unclear to us. Future studies may make efforts to verify the observations of our study. Another interesting result in our study is that miRNA-21 and miRNA-23a in combination had the highest diagnostic value, with high sensitivity and specificity. This finding, if validated in future studies, may contribute to raising the diagnostic accuracy.
After chemotherapy, we recorded the changes of expressions of miRNA-21, miRNA-23a, and miRNA-125b, finding that expressions of miRNA-21 and miRNA-23a were downregulated while miRNA-125b expression had no significant change. With this result, we again verified that miRNA-21 and miRNA-23a expressions are effective biomarkers in development of BL. We also found that after treatment, patients with low expressions of miRNA-21 and miRNA-23a had better short-term efficacy, but miRNA-125b showed no close association with short-term efficacy. This result suggests that miRNA-21 and miRNA-23a may be useful therapeutic targets, in agreement with previous studies [38,39]. During follow-ups, we observed that overexpression of miRNA-21 and miRNA-23a may be closely associated with poor prognosis. Cao et al. reported that miRNA-23a is a modulator in the transforming growth factor-β-induced epithelial-mesenchymal transition, which is an important mechanism for poor prognosis [40]. Several studies have demonstrated a positive association between upregulation of miRNA-23a and poor prognosis [41,42], and there is evidence that miRNA-21 overexpression is associated with poor prognosis in other diseases [43,44]. Li et al. suggested that miRNA-21 may be related to poor prognosis by its function as an apoptosis inhibitor [45]. There are limitations in our study. Our study is preliminary, and did not investigate the mechanism by which miRNA-21, miRNA-23a, and miRNA-125b are correlated with the development of BL in children. In addition, the sample size was relatively small, which may limit the reliability of our observations.
Conclusions
Our study strongly suggests that expressions of miRNA-21 and miRNA-23a are useful molecular biomarkers in the diagnosis and prognosis for BL in children. For the validation of the results in our study, more clinical observations with larger sample sizes are needed. In addition, future studies also should seek to define the mechanism by which these 3 miRNAs correlate with BL in children.
Footnotes
Competing interests
None.
Source of support: Departmental sources
References
- 1.Sandlund JT. Non-Hodgkin Lymphoma in Children. Curr Hematol Malig Rep. 2015;10:237–43. doi: 10.1007/s11899-015-0277-y. [DOI] [PubMed] [Google Scholar]
- 2.Molyneux EM, Rochford R, Griffin B, et al. Burkitt’s lymphoma. Lancet. 2012;379:1234–44. doi: 10.1016/S0140-6736(11)61177-X. [DOI] [PubMed] [Google Scholar]
- 3.Ogwang MD, Zhao W, Ayers LW, et al. Accuracy of Burkitt lymphoma diagnosis in constrained pathology settings: importance to epidemiology. Arch Pathol Lab Med. 2011;135:445–50. doi: 10.1043/2009-0443-EP.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woessmann W. XI. How to treat children and adolescents with relapsed non-Hodgkin lymphoma? Hematol Oncol. 2013;31(Suppl 1):64–68. doi: 10.1002/hon.2069. [DOI] [PubMed] [Google Scholar]
- 5.Farazi TA, Hoell JI, Morozov P, et al. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galasso M, Sana ME, Volinia S. Non-coding RNAs: A key to future personalized molecular therapy? Genome Med. 2010;2:12. doi: 10.1186/gm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan H, Lu H, Wang X, et al. MicroRNAs as potential biomarkers in cancer: Ppportunities and challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Iqbal J, Shen Y, Huang X, et al. Global microRNA expression profiling uncovers molecular markers for classification and prognosis in aggressive B-cell lymphoma. Blood. 2015;125:1137–45. doi: 10.1182/blood-2014-04-566778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrie AS, Cardigan RA, Williamson LM, et al. The dynamics of clot formation in fresh-frozen plasma. Vox Sang. 2008;94:306–14. doi: 10.1111/j.1423-0410.2008.01037.x. [DOI] [PubMed] [Google Scholar]
- 11.Lawrie CH, Soneji S, Marafioti T, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–61. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 12.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–75. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 13.Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117:3140–46. doi: 10.1182/blood-2010-09-308684. [DOI] [PubMed] [Google Scholar]
- 14.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 15.Kong KY, Owens KS, Rogers JH, et al. MIR-23A microRNA cluster inhibits B-cell development. Exp Hematol. 2010;38:629–40e1. doi: 10.1016/j.exphem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yong FL, Law CW, Wang CW. Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer. 2013;13:280. doi: 10.1186/1471-2407-13-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, Chen H, Si H, et al. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014;51:823–31. doi: 10.1007/s00592-014-0617-8. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Liu X, Xu W, et al. c-MYC-regulated miR-23a/24-2/27a cluster promotes mammary carcinoma cell invasion and hepatic metastasis by targeting Sprouty2. J Biol Chem. 2013;288:18121–33. doi: 10.1074/jbc.M113.478560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertus JL, Kluiver J, Weggemans C, et al. MiRNA profiling in B non-Hodgkin lymphoma: a MYC-related miRNA profile characterizes Burkitt lymphoma. Br J Haematol. 2010;149:896–99. doi: 10.1111/j.1365-2141.2010.08111.x. [DOI] [PubMed] [Google Scholar]
- 20.Bousquet M, Harris MH, Zhou B, et al. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–63. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Yan LX, Wu QN, et al. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011;71:3552–62. doi: 10.1158/0008-5472.CAN-10-2435. [DOI] [PubMed] [Google Scholar]
- 22.Yuxia M, Zhennan T, Wei Z. Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. J Cancer Res Clin Oncol. 2012;138:2045–50. doi: 10.1007/s00432-012-1285-0. [DOI] [PubMed] [Google Scholar]
- 23.Akbari Moqadam F, Lange-Turenhout EA, Aries IM, et al. MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia. Leuk Res. 2013;37:1315–21. doi: 10.1016/j.leukres.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 24.World Medical Association. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–94. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 25.Jaffe ES. The 2008 WHO classification of lymphomas: Implications for clinical practice and translational research. Hematology Am Soc Hematol Educ Program. 2009:523–31. doi: 10.1182/asheducation-2009.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czer LS, Chaux A, Matloff JM, et al. Ten-year experience with the St. Jude Medical valve for primary valve replacement. J Thorac Cardiovasc Surg. 1990;100:44–54. discussion 54–55. [PubMed] [Google Scholar]
- 27.Reiter A, Schrappe M, Tiemann M, et al. Improved treatment results in childhood B-cell neoplasms with tailored intensification of therapy: A report of the Berlin-Frankfurt-Munster Group Trial NHL-BFM 90. Blood. 1999;94:3294–306. [PubMed] [Google Scholar]
- 28.Cheson BD, Pfistner B, Juweid ME, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007;25:579–86. doi: 10.1200/JCO.2006.09.2403. [DOI] [PubMed] [Google Scholar]
- 29.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–56. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–18. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Go H, Jang JY, Kim PJ, et al. MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma. Oncotarget. 2015;6:15035–49. doi: 10.18632/oncotarget.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lian S, Shi R, Bai T, et al. Anti-miRNA-23a oligonucleotide suppresses glioma cells growth by targeting apoptotic protease activating factor-1. Curr Pharm Des. 2013;19:6382–89. doi: 10.2174/13816128113199990509. [DOI] [PubMed] [Google Scholar]
- 33.Wang YD, Cai N, Wu XL, et al. OCT4 promotes tumorigenesis and inhibits apoptosis of cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis. 2013;4:e760. doi: 10.1038/cddis.2013.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun C, MLuan CF. Overexpression of microRNA-21 in peripheral blood mononuclear cells of patients with B-cell non-Hodgkin’s lymphoma is associated with disease stage and treatment outcome. Eur Rev Med Pharmacol Sci. 2015;19:3397–402. [PubMed] [Google Scholar]
- 35.Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544–51. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 36.Chen G, Tang Y, Wu JH, et al. Role of microRNAs in diagnosis and treatment of the pathogenesis of gastric cancer. Int J Clin Exp Med. 2014;7:5947–57. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Wei X, Chen D, Lv T, et al. Serum MicroRNA-125b as a Potential Biomarker for Glioma Diagnosis. Mol Neurobiol. 2016;53(1):163–70. doi: 10.1007/s12035-014-8993-1. [DOI] [PubMed] [Google Scholar]
- 38.Zhong X, Chung AC, Chen HY, et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 2013;56:663–74. doi: 10.1007/s00125-012-2804-x. [DOI] [PubMed] [Google Scholar]
- 39.Wise DR, Thompson CB. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–33. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao M, Seike M, Soeno C, et al. MiR-23a regulates TGF-beta-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int J Oncol. 2012;41:869–75. doi: 10.3892/ijo.2012.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma G, Dai W, Sang A, et al. Upregulation of microRNA-23a/b promotes tumor progression and confers poor prognosis in patients with gastric cancer. Int J Clin Exp Pathol. 2014;7:8833–40. [PMC free article] [PubMed] [Google Scholar]
- 42.Cai S, Chen R, Li X, et al. Downregulation of microRNA-23a suppresses prostate cancer metastasis by targeting the PAK6-LIMK1 signaling pathway. Oncotarget. 2015;6:3904–17. doi: 10.18632/oncotarget.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu XG, Zhu WY, Huang YY, et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618–26. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 44.Gao W, Shen H, Liu L, et al. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol. 2011;137:557–66. doi: 10.1007/s00432-010-0918-4. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Huang H, Sun L, et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]