Abstract
Telomeres cap chromosome ends, protecting them from degradation, double-strand breaks, and end-to-end fusions. Telomeres are maintained by telomerase, a reverse transcriptase encoded by TERT, and an RNA template encoded by TERC. Loci in the TERT and adjoining CLPTM1L region are associated with risk of multiple cancers. We therefore investigated associations between variants in 22 telomere structure and maintenance gene regions and colorectal, breast, prostate, ovarian, and lung cancer risk. We performed subset-based meta-analyses of 204,993 directly-measured and imputed SNPs among 61,851 cancer cases and 74,457 controls of European descent. Independent associations for SNP minor alleles were identified using sequential conditional analysis (with gene-level P-value cutoffs ≤3.08×10−5). Of the thirteen independent SNPs observed to be associated with cancer risk, novel findings were observed for seven loci. Across the TERT-CLPTML1 region, rs12655062 was associated positively with prostate cancer, and inversely with colorectal and ovarian cancers, and rs115960372 was associated positively with prostate cancer. Across the TERC region, rs75316749 was positively associated with colorectal, breast, ovarian, and lung cancers. Across the DCLRE1B region, rs974404 and rs12144215 were inversely associated with prostate and lung cancers, and colorectal, breast, and ovarian cancers, respectively. Near POT1, rs116895242 was inversely associated with colorectal, ovarian, and lung cancers, and RTEL1 rs34978822 was inversely associated with prostate and lung cancers. The complex association patterns in telomere-related genes across cancer types may provide insight into mechanisms through which telomere dysfunction in different tissues influences cancer risk.
Keywords: telomere structure, telomere maintenance, cancer risk, GWAS, meta-analysis, lung cancer, breast cancer, ovarian cancer, prostate cancer, colorectal cancer
Introduction
Telomeres are complex nucleoprotein structures that cap chromosome ends (1,2), protecting them from degradation, double strand breaks, and end-to-end fusions (1,2). Thus, telomeres play an essential role in preserving genomic stability. Telomeres are maintained by the enzyme telomerase, which is made up of a reverse transcriptase encoded by TERT, and an RNA template encoded by TERC (1,2), with several other associated proteins encoded by DKC1, NOP10, NHP2, NAF1, and GAR1 (1). The telomere structure itself is composed of simple tandem TTAGGG repeats bound by six proteins (encoded by TERF1, TERF2, TINF2, TERF21P, ACD, and POT1), termed shelterin. Other proteins that interact with shelterin are encoded by OBFC1, RTEL1, DCLRE1B, TNKS, PINX1, and TEP1 (1). Germline SNPs in TERC, TERT, RTEL1, NAF1 (3), and OBFC1 (3,4) have been associated with telomere length in genome-wide association studies (GWAS). Additional genes associated with telomere length include: BICD1 (5), ACYP2, ZNF208, MPHOSPH6 (3), and DCAF4 (6).
Susceptibility loci for multiple cancer types have been identified in the TERT and adjoining CLPTM1L gene region in GWAS. Both increased and decreased risk associations have been reported for some loci for different cancers (7–9), suggesting complex patterns of associations across cancer types which could be due to tissue specificity or interactions with risk factors. Because properly functioning telomeres are vital for genomic stability and chromosomal integrity, genetic variants in other telomere structure and maintenance genes may affect cancer risk. Therefore, we sought to examine whether pleiotropic associations for variants in telomere structure and maintenance genes are observed across cancer types within the Genetic Associations and Mechanisms in Oncology Network (GAME-ON) (10) and the Genetic and Epidemiology of Colorectal Cancer Consortium (GECCO) (11).
GAME-ON was established by the National Cancer Institute (NCI) to foster collaborative post-GWAS research across consortia of colorectal, breast, prostate, ovarian, and lung cancers (10). The extensive genomic data available through GAME-ON and GECCO, including over 61,000 cases and 74,000 controls, were utilized to identify and systematically characterize patterns of associations between independent variants in 22 telomere structure and maintenance gene regions and risk of colorectal, breast, prostate, ovarian, and lung cancers.
Materials and Methods
Study Population
Our analysis included 61,851 cancer cases and 74,457 controls of European descent from 45 GWAS (12) (Table 1). Details of each study have been described previously (10–19) (Supplementary Table 1); at minimum, cases were frequency-matched to controls on age and sex. Each study obtained informed consent from participants; study procedures including certifications required for data sharing in accordance with National Institutes of Health policies were approved by all Institutional Review Boards.
Table 1.
Characteristics of genome-wide association studies included in consortium-based meta-analyses of colorectal, breast, prostate, ovarian, and lung cancers
| Cancer Type/Subtype-Consortium | Cases (N) | Controls (N) | GWAS (N) | Genotyping Platform | Covariates |
|---|---|---|---|---|---|
| Colorectal- GECCO | 10,314 | 12,857 | 13 | Illumina 300/240S, 300K, 550K, 610K, 730K; Affymetrix 100K, 500K | age, sex, PCA, center, batch effecta, smokingb |
| Colorectal- CORECT | 5,100 | 4,831 | 6 | Affeymetrix Axiom | age, sex, PCA |
| Breast- DRIVE | 15,748 | 18,084 | 11 | llumina 240K/317K/370K/550K/610K/610K+Cyto12/660K/670K/1.2M; Affymetrix 5.0/6.0 | age, PCA |
| ER- negative | 4,939 | 13,128 | 8 | age, PCA | |
| Prostate- ELLIPSE | 14,160 | 12,724 | 6 | Illumina 550K/610K/2.5M/iSELECT; Affymetrix GeneChip 5.0 | age, study |
| Aggressive | 4,450 | 12,724 | 6 | age, study, PCA | |
| Ovarian- FOCI | 4,369 | 9,123 | 3 | Illumina 317K/370K/550K/610K/670K/2.5M | site, PCA, age |
| Endometrioid | 715 | 9,123 | 3 | site, PCA, age | |
| Serous | 2,556 | 9,123 | 3 | site, PCA, age | |
| Lung- TRICL | 12,160 | 16,838 | 6 | Illumina 317K/370K/550K/610K | age, sex, PCA |
| Adenocarcinoma | 3,718 | 15,871 | 6 | age, sex, PCA | |
| Squamous | 3,422 | 16,015 | 6 | age, sex, PCA | |
|
| |||||
| Total | 61,851 | 74,457 | 45 | ||
Abbreviations: CORECT- ColoRectal Transdisciplinary Study; DRIVE- Discovery, Biology, and Risk of Inherited Variants in Breast Cancer; ELLIPSE- Elucidating Loci Involved in Prostate Cancer Susceptibility; ER-estrogen receptor; FOCI- Follow-up of Ovarian Cancer Genetic Association and Interaction Studies; GWAS- genome wide association studies; N- number; PCA- principal components analysis; TRICL- Transdisciplinary Research in Cancer of the Lung.
Adjusted for batch effect only in the Association STudy Evaluating RISK for Sporadic Colorectal Cancer (ASTERISK) study
Adjusted for smoking only in the Physician’s Health Study (PHS)
Consortium-based Imputation and Meta-analysis
Genotyping was performed using Illumina and Affymetrix GWAS platforms. Each consortium imputed unmeasured single nucleotide polymorphisms (SNPs) for their GWAS data from the 1000 Genomes (Phase 1) March 2012 Build 37 reference panel using MACH, IMPUTE, or Minimac (10–19) Supplementary Table 2. Within each consortium, per-allele odds ratios (ORs) and 95% confidence intervals (CIs) for each SNP and cancer risk were calculated using unconditional logistic regression. Study-specific results were combined using fixed-effects meta-analysis.
Gene Selection
We examined 204,993 SNPs within one mega-base upstream and downstream of the transcription start and end sites of the following genes, selected either because of their relevance to telomere structure and maintenance, or telomere length: ACD, ACYP2, BICD1, DCLRE1B, GAR1, MPHOSPH6, NAF1, NHP2, NOP10, OBFC1, PIK3C3, PINX1-TNKS, POT1, RTEL1, TEP1, TERC, TERF1, TERF2, TERF2IP, TERT-CLPTM1L, TINF2, and ZNF208. The chromosomal location and number of SNPs evaluated in each gene is in Supplementary Table 3 (20).
Cross-Cancer Association Analysis
ASSociation analysis based on SubSET (ASSET) meta-analysis allows for identification of associations that may be in the same, or opposite, direction for some cancer types versus others (21). We performed one-sided and two-sided ASSET analyses using summary data for each of the five cancer types, and repeated analyses additionally including the following cancer subtypes: estrogen receptor (ER) negative breast; aggressive prostate (defined as Gleason score ≥8, disease stage ‘distant’, prostate-specific antigen level >100 ng/ml, or death from prostate cancer (17)); endometrioid and serous ovarian; and adenocarcinoma and squamous lung. Other tumor subtypes were not independently evaluated due to low frequencies. ASSET takes into account matrices of overlapping cases and controls across datasets including overlap between cancer types and subtypes (Supplementary Table 4), and adjusts for correlations across studies. ASSET groups cancer types by the direction of their associations and identifies the strongest associations, so multiple testing penalties may be incurred, widening the CIs of summary results (21). A Manhattan plot of P-values from our two-sided unconditional ASSET analysis was produced in R Studio [http://www.rstudio.com]. Forest plots of two-sided unconditional ASSET meta-analysis results for individual SNPs were generated by cancer type and subtype. Because ASSET takes into account overlap between cancer types and subtypes, associations appearing statistically significant for a given cancer type (or subtype) may be included in the “null” category if the association is actually driven by that cancer’s subtype(s) included in the ‘positive” or “inverse” category. Statistically significant positive or inverse associations are only interpretable within ASSET if the overall one-sided (positive or negative) test is statistically significant.
Gene-level association tests to evaluate all SNPs within a gene and cancer risk after taking linkage disequilibrium (LD) into account were performed using VEGAS2 (22) on the overall two-sided unconditional ASSET meta-analysis P-values for all SNPs +/−50kb of each gene.
Identifying SNPs in Linkage Disequilibrium
Because GAME-ON and GECCO data included summary statistics for each SNP, not individual-level data, we could not calculate LD directly. Instead, we determined LD using individual-level data from European ancestry subjects in the Cancer Genetic Markers of Susceptibility (CGEMS) Project (23) and the Environmental and Genetics in Lung Cancer Etiology (EAGLE) study (24). To be comparable to the summary data used for our analyses, we imputed SNPs with IMPUTE2 (25) from 1000 Genomes (Phase 3) October 2014 Build 37 in CGEMS and EAGLE (26). Of 204,718 SNPs in the summary data, 7,015 SNPs could not be imputed in CGEMS and EAGLE because they were not present in 1000 Genomes (Phase 1) data used to impute the GAME-ON and GECCO data (12,13). Additionally, 8,977 SNPs failed quality-control measures (information score <0.3) and 96 were multi-allelic, leaving 188,630 markers in CGEMS and EAGLE for analysis. We identified sets of SNPs with r2>0.70 in CGEMS and EAGLE using Haploview (27). Given the complicated LD patterns in TERT-CLPTM1L and TERC, we generated LD plots of all significant SNPs from ASSET analyses, to the extent possible, with r2<0.70 (27).
Determining Gene-level P-value Thresholds
We used the Genetic type 1 Error Calculator (GEC) to calculate the effective number of independent tests (Me) and statistical significance P-value threshold for each gene (28). This method, developed to address the issue of multiple testing with SNPs in LD, utilizes eigenvalues derived from matrices of association test P-values between SNPs to calculate Me. For each gene, the P-value threshold required to keep type I error at 5% equals alpha divided by Me. Before applying the GEC, for simplicity we removed redundant SNPs (r2>0.98) from CGEMS and EAGLE using gPLINK version 1.07 [http://pngu.mgh.harvard.edu/purcell/plink/] ensuring that directly measured SNPs in our dataset were not eliminated, leaving 98,783 markers. Me and P-value thresholds for each gene are in Supplementary Table 3.
Conditional Analysis
To identify independent associations, we performed sequential conditional analysis using Yang et al.’s method for summary-level data (29). For each gene, SNPs were ranked by P-value, and in each step, a single SNP was added to the ASSET analysis, conditioning on SNPs that were most significantly associated in previous steps. This process was repeated until the two-sided P-value for the most significant SNP for a step remained below the Me P-value. To avoid collinearity, in each step, the program assesses r2 between the next SNP to add and the SNPs that are already included in the model, and skips SNPs that are correlated (in this case, r2>0.80). To evaluate if this resulted in over-fitting, we performed a sensitivity analysis by conducting sequential conditional analysis of TERT-CLPTM1L using pruned variants with r2≤0.70. No evidence of over-fitting was observed (data not shown).
For SNPs with two-sided P-values that reached multiple comparison-adjusted gene-level significance, we assessed whether both the positive and inverse results contributed to the association (versus the association being driven primarily by one-sided results) by evaluating whether the two-sided P-value was smaller than the most significant one-sided P-value. We used an arbitrary P-value cutoff of 0.01 for the contributing one-sided associations, and considered P-values between 0.01–0.05 as suggestive.
Functional annotations for SNPs with observed associations that have not been previously reported were obtained from HaploReg Version 4.1 on June 14th, 2016 (30). HaploReg is a data repository which integrates information on sequence conservation, regulatory protein binding, epigenomic evidence, expression quantitative trait loci, and regulatory motifs from several sources including the ENCODE project, the GRASP database, GTEx, SiPhy, and multiple other studies (30).
Results
We examined 204,993 SNPs in 22 telomere structure and maintenance gene regions and colorectal, breast, prostate, ovarian, and lung cancer risk in 61,851 cancer cases and 74,457 controls (Table 1). ASSET unconditional two-sided analysis combined P-values for each SNP are shown in the Manhattan plot (Supplementary Figure 1). VEGAS2 gene-based association tests evaluating all SNPs in each gene in aggregate and cancer risk were statistically significant for DCLRE1B (P=1.1×10−5), TERT-CLPTM1L (P=1.0×10−6), and RTEL1 (P=9.4×10−4). Using the per-gene P-value threshold for the effective number of independent tests, we observed significant associations with cancer risk for 89 DCLRE1B, 153 TERC, 1 GAR1, 95 TERT-CLPTM1L, 2 POT1, 1 TERF2, and 7 RTEL1 SNPs (Supplementary Table 5). After removing SNPs in LD at r2>0.70 with the lead SNP, 3 DCLRE1B, 19 TERC, 1 GAR1, 23 TERT-CLPTM1L, 2 POT1, 1 TERF2, and 2 RTEL1 SNPs remained (Table 2). Correlations between these SNPs (r2 and D′) are in Supplementary Table 6. Supplementary Table 7 includes r2 correlations between these SNPs and all other significantly associated SNPs by gene. Even after pruning, 3 SNP pairs in TERC remained correlated with r2>0.70 (rs75982374 and rs76925190; rs80304993 and rs969217; rs59758024 and rs9865021), as did 2 SNP pairs in TERT (rs35953391 and rs37004; rs3816659 and rs37005). LD between these highly correlated SNPs and the variants retained is in Supplementary Figure 2.
Table 2.
Unconditional ASSET two-sided meta-analysis results across five cancer typesa
| Gene (Chr.) | Ref: MA | MAF | Combined P-value | Positively Associated
|
Inversely Associated
|
Cancer types
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Position | OR (95% CI) | P-value | OR (95% CI) | P-value | Positively Associated | Negatively Associated | |||
| DCLRE1B (Chr. 1) | ||||||||||
| rs974404 | 114382025 | T:G | 0.449 | 9.19E-06e | 1.04 (1.01–1.07) | 2.47E-02f | 0.94 (0.91–0.97) | 2.43E-05e | Breast, Ovarian | Prostate, Lung |
| rs7523862b | 114443419 | G:A | 0.379 | 1.09E-05e | 0.94 (0.91–0.97) | 1.17E-05e | Prostate, Lung | |||
| rs12144215 | 114187155 | G:T | 0.131 | 1.50E-05e | 0.90 (0.87–0.94) | 2.11E-06e | Colorectal, Prostate | |||
| TERC (Chr. 3) | ||||||||||
| rs80304993 | 170097606 | G:A | 0.230 | 6.54E-15e | 0.82 (0.78–0.86) | 1.51E-15e | Prostate | |||
| rs71277158 | 169999216 | T:G | 0.162 | 8.88E-15e | 0.80 (0.76–0.84) | 3.64E-16e | Prostate | |||
| rs76925190 | 170066339 | A:C | 0.173 | 1.25E-14e | 0.80 (0.76–0.84) | 6.27E-16e | Prostate | |||
| rs75982374 | 170063227 | A:G | 0.140 | 4.65E-13e | 0.79 (0.74–0.84) | 2.62E-14e | Prostate | |||
| rs55953261 | 170121598 | G:A | 0.488 | 4.58E-09e | 0.89 (0.85–0.92) | 3.57E-09e | Prostate | |||
| rs77085460 | 170127536 | A:G | 0.063 | 7.30E-09e | 0.76 (0.70–0.83) | 3.81E-10e | Prostate | |||
| rs59758024c | 170119352 | A:T | 0.447 | 9.03E-09e | 1.12 (1.08–1.17) | 7.08E-09e | Prostate | |||
| rs75316749 | 168761423 | A:G | 0.041 | 1.38E-06e | 1.14 (1.08–1.20) | 1.38E-06e | Colorectal, Breast, Ovarian, Lung | |||
| rs75313056 | 170017609 | G:A | 0.082 | 1.51E-08e | 0.80 (0.74–0.86) | 9.54E-10e | Prostate | |||
| rs12487040 | 170103592 | T:C | 0.372 | 1.70E-08e | 1.05 (1.01–1.09) | 2.46E-02f | 0.89 (0.85–0.93) | 3.13E-08e | Breast, Ovarian | Prostate |
| rs10804842 | 170135700 | T:C | 0.234 | 1.73E-08e | 0.86 (0.82–0.90) | 1.94E-09e | Prostate | |||
| rs969217 | 170159134 | C:T | 0.391 | 2.73E-07e | 1.06 (1.02–1.11) | 2.62E-03 | 0.90 (0.87–0.94) | 5.47E-06e | Breast | Prostate |
| rs77964281 | 169916180 | T:C | 0.117 | 3.49E-07e | 0.85 (0.80–0.90) | 2.65E-08e | Prostate | |||
| rs62293480 | 170106672 | G:T | 0.388 | 1.35E-06e | 0.89 (0.84–0.93) | 5.97E-07e | Prostate | |||
| rs10936633 | 170158128 | G:A | 0.493 | 1.49E-06e | 0.90 (0.87–0.94) | 7.74E-07e | Prostate | |||
| rs9865021 | 170146881 | C:T | 0.487 | 2.48E-06e | 1.10 (1.06–1.14) | 2.11E-06e | Prostate | |||
| rs74677551 | 168861788 | T:G | 0.032 | 3.20E-06e | 1.15 (1.08–1.22) | 3.20E-06e | Colorectal, Breast, Prostate, Ovarian, Lung | |||
| rs9809168 | 168803900 | T:C | 0.033 | 1.20E-05e | 1.15 (1.08–1.22) | 1.20E-05e | Colorectal, Breast, Ovarian, Lung | |||
| rs2901621 | 170057704 | G:C | 0.098 | 1.77E-05e | 0.93 (0.91–0.96) | 3.52E-06e | Colorectal, Prostate | |||
| GAR1 (Chr. 4) | ||||||||||
| rs17042238d | 111745854 | A:G | 0.003 | 6.33E-06e | 0.04 (0.01–0.16) | 6.33E-06e | Prostate | |||
| TERT-CLPTM1L (Chr. 5) | ||||||||||
| rs37004b | 1356684 | C:T | 0.239 | 2.27E-11e | 0.84 (0.81–0.88) | 1.29E-12e | Lung | |||
| rs37005b | 1356450 | C:T | 0.460 | 1.98E-10e | 0.87 (0.84–0.91) | 9.85E-12e | Lung | |||
| rs3816659b | 1317820 | G:A | 0.441 | 2.44E-10e | 0.88 (0.85–0.91) | 9.97E-12e | Lung | |||
| rs2736100b | 1286516 | C:A | 0.500 | 3.38E-10e | 1.05 (1.01–1.09) | 1.72E-02f | 0.90 (0.86–0.93) | 7.54E-10e | Colorectal, Prostate | Lung |
| rs7725218b | 1282414 | G:A | 0.359 | 3.02E-09e | 1.12 (1.07–1.17) | 3.14E-07e | 0.90 (0.85–0.96) | 4.04E-04 | Lung | Prostate |
| rs35953391b | 1312329 | C:T | 0.201 | 6.44E-09e | 0.87 (0.83–0.91) | 1.43E-09e | Lung | |||
| rs2735940b | 1296486 | G:A | 0.499 | 7.44E-09e | 1.09 (1.05–1.14) | 1.91E-05 | 0.93 (0.90–0.96) | 1.70E-05 | Lung | Colorectal, Prostate |
| rs2736099b | 1287340 | G:A | 0.344 | 8.62E-09e | 1.12 (1.08–1.17) | 1.75E-07e | 0.95 (0.92–0.98) | 2.18E-03 | Lung | Colorectal, Breast, Prostate |
| rs35029535 | 1284976 | C:T | 0.352 | 5.54E-08e | 1.09 (1.03–1.15) | 2.09E-03 | 0.92 (0.89–0.95) | 1.28E-06e | Prostate | Breast, Ovarian, Lung |
| rs7713218 | 1283312 | G:A | 0.497 | 5.78E-08e | 1.10 (1.06–1.14) | 6.60E-07e | 0.95 (0.91–0.98) | 4.23E-03 | Ovarian, Lung | Colorectal, Prostate |
| rs10866498 | 1285162 | C:T | 0.472 | 6.36E-08e | 1.05 (1.01–1.09) | 6.76E-03 | 0.91 (0.88–0.94) | 4.57E-07e | Colorectal, Prostate | Ovarian, Lung |
| rs2735948b | 1299213 | G:A | 0.418 | 7.70E-08e | 0.88 (0.85–0.92) | 5.56E-09e | Lung | |||
| rs36019446 | 1339890 | A:G | 0.484 | 1.57E-07e | 0.88 (0.85–0.92) | 1.05E-08e | Lung | |||
| rs2736098b | 1294086 | C:T | 0.234 | 2.48E-07e | 1.08 (1.04–1.12) | 1.81E-04 | 0.93 (0.90–0.97) | 7.16E-05 | Prostate, Lung | Colorectal, Breast, Ovarian |
| rs7717443 | 1283486 | C:T | 0.483 | 5.37E-07e | 1.10 (1.06–1.14) | 2.24E-06e | 0.95 (0.91–0.989) | 1.31E-02f | Ovarian, Lung | Colorectal, Prostate |
| rs115960372 | 1518494 | C:T | 0.104 | 6.94E-07e | 1.19 (1.1–1.27) | 2.97E-06e | 0.90 (0.83–0.98) | 1.29E-02f | Prostate | Lung |
| rs2735944b | 1304432 | C:T | 0.132 | 1.27E-06e | 0.85 (0.80–0.90) | 1.38E-07e | Lung | |||
| rs2853677b | 1287194 | A:G | 0.400 | 1.33E-06e | 1.11 (1.06–1.16) | 1.54E-06e | 0.97 (0.93–1.000) | 4.99E-02f | Lung | Colorectal, Breast, Prostate |
| rs12655062 | 1890877 | G:A | 0.354 | 1.65E-06e | 1.12 (1.06–1.18) | 3.53E-05 | 0.95 (0.92–0.98) | 2.72E-03 | Prostate | Colorectal, Ovarian |
| rs2736109b | 1296759 | C:T | 0.392 | 2.99E-06e | 1.11 (1.06–1.16) | 5.88E-06e | 0.96 (0.93–0.996) | 3.08E-02f | Lung | Colorectal, Breast, Ovarian |
| rs33961405b | 1277577 | A:G | 0.491 | 1.20E-05e | 1.11 (1.06–1.16) | 4.55E-06e | Lung | |||
| rs55901723 | 1342154 | T:C | 0.232 | 2.14E-05 | 0.88 (0.84–0.93) | 2.94E-06e | Lung | |||
| rs6861230 | 304003 | T:C | 0.042 | 2.70E-05 | 1.25 (1.13–1.37) | 5.83E-06e | Breast, Ovarian | |||
| POT1 (Chr. 7) | ||||||||||
| rs116895242 | 123946403 | T:A | 0.041 | 5.21E-05 | 0.83 (0.77–0.90) | 6.99E-06e | Colorectal, Ovarian, Lung | |||
| rs74986217 | 123465182 | A:C | 0.041 | 2.54E-04e | 1.31 (1.16–1.48) | 2.17E-05e | Ovarian | |||
| TERF2 (Chr. 16) | ||||||||||
| rs117496043c | 69590365 | C:T | 0.003 | 4.28E-05 | 1.66 (1.33–2.06) | 6.14E-06e | Prostate | |||
| RTEL1 (Chr. 20) | ||||||||||
| rs34978822c | 62291599 | C:G | 0.015 | 2.14E-05 | 0.71 (0.62–0.82) | 3.17E-06e | Prostate, Lung | |||
| rs114220381c | 61477960 | T:A | 0.048 | 1.21E-04 | 1.31 (1.16–1.48) | 1.13E-05e | Prostate | |||
Abbreviations: Chr.- chromosome; CI- confidence interval; MA- Minor Allele; OR- odds ratio; Ref- reference; SNP- single nucleotide polymorphism.
Results are presented for SNPs after pruning at r2<0.70.
SNPs that are directly measured and not imputed.
ASSET meta-analytical results for these SNPs are based on 4 cancer types rather than all 5 studies.
ASSET meta-analytical results for these SNPs are based on 2 cancer types rather than all 5 studies.
Gene level P-value thresholds based on the number of effective tests are: DCLER1B P-value<2.65×10−5; TERC P-value<2.45×10−5; GAR1 P-value<2.44×10−5; TERT-CLPTM1 P-value<1.32×10−5; POT1 P-value<2.94×10−5; TERF2 P-value<3.08×10−5; RTEL1 P-value<1.86×10−5.
Positive or negative associations with P-values between 0.01 and 0.05 are considered to be suggestive.
For GAR1 and TERF2, only single SNPs reached gene-level significance, and the associations were entirely driven by prostate cancer. However, data were available only for prostate and ovarian cancers for the SNP in GAR1, and for colorectal, prostate, ovarian, and lung cancers for the SNP in TERF2. These two SNPs are “very rare” variants with minor allele frequencies (MAF) of 0.3%, making them difficult to impute. For POT1 and RTEL1, only one SNP in each gene was significantly associated in sequential conditional analyses. RTEL1 rs34978822 was associated with prostate and lung cancers (and was not investigated in breast cancer). rs34978822, and two SNPs in LD with it, are associated with chromatin structure changes in a large number of cell lines reported in HaploReg (30). POT1 rs116895242 was associated with colorectal, ovarian, and lung cancers (Table 2); this SNP creates six motif changes that may affect transcription factor binding (30).
Table 3 presents results from unconditional and sequential conditional analysis of DCLRE1B, TERC, and TERT-CLPTM1L gene regions, including all SNPs with ASSET two-sided results that reached gene-level P-value cutoffs. Sequential conditional analysis identified 11 independent SNPs associated with risk of multiple cancers. For all conditional results, two-sided P-values are smaller than one-sided P-values (data not shown).
Table 3.
Unconditional and conditional ASSET two-sided meta-analysis results across five cancer types
| Unconditional Results
|
Cancer types | Conditional Results
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Gene (Chr.) SNP |
Combined P-value |
Positively Associated | Inversely Associated | Combined P-value |
Positively Associated | Inversely Associated | ||||||
| OR (95% CI) |
P-value | OR (95% CI) |
P-value | Positively Associated |
Negatively Associated |
OR (95% CI) |
P-value | OR (95% CI) |
P-value | |||
| DCLRE1B (Chr. 1) | ||||||||||||
| rs974404a | 9.19e-06b | 1.04 (1.01–1.07) | 2.47E-02c | 0.94 (0.91–0.97) | 2.43E-05b | Breast, Ovarian | Prostate, Lung | |||||
| rs12144215 | 1.50E-05b | 0.90 (0.87–0.94) | 2.11E-06b | Lungd | Colorectal, Breastd, Prostate | 2.07E-05b | 1.05 (1.004–1.11) | 3.31E-02c | 0.93 (0.90–0.96) | 4.33E-05 | ||
| TERC (Chr. 3) | ||||||||||||
| rs80304993a | 6.54E-15b | 0.82 (0.78–0.86) | 1.51E-15b | Prostate | ||||||||
| rs62293480 | 1.35E-06b | 0.89 (0.84–0.93) | 5.97E-07b | Prostate | 2.16E-13b | 0.84 (0.81–0.88) | 1.44E-14b | |||||
| rs75316749 | 1.38E-06b | 1.14 (1.08–1.20) | 1.38E-06b | Colorectal, Breast, Ovary, Lung | 1.46E-06b | 1.14 (1.08–1.20) | 1.46E-06b | |||||
| TERT-CLPTM1L (Chr. 5) | ||||||||||||
| rs37004a,e | 2.27E-11b | 0.84 (0.81–0.88) | 1.29E-12b | Lung | ||||||||
| rs7717443 | 5.37E-07b | 1.10 (1.06–1.14) | 2.24E-06b | 0.95 (0.91–0.99) | 1.31E-02c | Ovary, Lung | Colorectal, Prostate | 1.26E-07b | 1.11 (1.06–1.15) | 5.31E-07b | 0.95 (0.91–0.990) | 1.19E-02c |
| rs10866498 | 6.36E-08b | 1.05 (1.01–1.09) | 6.76E-03 | 0.91 (0.88–0.94) | 4.57E-07b | Colorectal, Prostate | Ovary, Lung | 9.27E-18b | 1.07 (1.03–1.10) | 4.01E-05 | 0.88 (0.85–0.91) | 5.25E-15b |
| rs12655062 | 1.65E-06b | 1.12 (1.06–1.18) | 3.53E-05 | 0.95 (0.92–0.98) | 2.72E-03 | Prostate | Colorectal, Ovarian | 1.13E-06b | 1.12 (1.06–1.18) | 2.67E-05 | 0.95 (0.92–0.98) | 2.42E-03 |
| rs115960372 | 6.94E-07b | 1.19 (1.10–1.27) | 2.97E-06b | 0.90 (0.83–0.98) | 1.29E-02c | Prostate | Lung | 3.12E-06b | 1.17 (1.09–1.26) | 1.20E-05b | 0.91 (0.84–0.98) | 1.57E-02c |
| rs2736098e | 2.48E-07b | 1.08 (1.04–1.12) | 1.81E-04 | 0.93 (0.90–0.97) | 7.16E-05 | Prostate, Lung | Colorectal, Breast, Ovarian | 5.36E-06b | 1.08 (1.02–1.14) | 1.23E-02c | 0.93 (0.91–0.96) | 2.74E-05 |
| POT1 (Chr. 7) | ||||||||||||
| rs116895242 | 5.21E-05 | 0.83 (0.77–0.90) | 6.99E-06b | Colorectal, Ovarian, Lung | ||||||||
| RTEL1 (Chr. 20) | ||||||||||||
| rs34978822f | 2.14E-05 | 0.71 (0.62–0.82) | 3.17E-06b | Prostate, Lung | ||||||||
Abbreviations: Chr.- chromosome; CI- confidence interval; OR- odds ratio; SNP- single nucleotide polymorphism.
The most significant SNP is always conditioned on in the sequential conditional analysis (and therefore there are no conditional results for it)
Gene level P-value thresholds based on the number of effective tests are: DCLER1B P<2.65×10−5; TERC P<2.45×10−5; TERT-CLPTM1 P<1.32×10−5; POT1 P-value<2.94×10−5; RTEL1 P-value<1.86×10−5.
Positive or inverse associations with P-values between 0.01 and 0.05 are considered to be suggestive.
Associations with phenotypes were statistically significant in conditional analyses only
SNPs that are directly measured and not imputed.
ASSET meta-analytical results for these SNPs are based on 4 cancer types rather than all 5 studies.
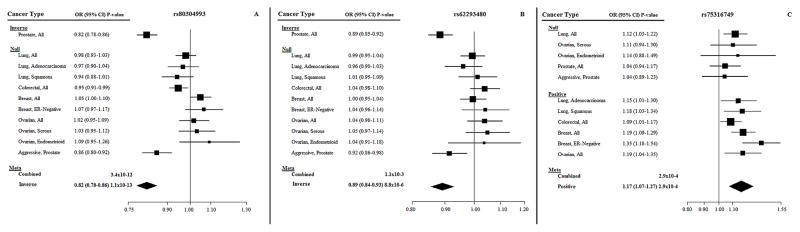
In TERC, three independent loci were identified (Table 3). We observed highly significant inverse associations with prostate cancer risk for the A allele of rs80304993 (P=1.51×10−15), and the T allele of rs62293480, particularly after conditioning on rs80304993 (Pconditional=1.44×10−14). Forest plots by cancer type and subtype (Figures 1A and 1B) show that these inverse associations were driven solely by overall prostate cancer (OR=0.82, 95% CI=0.78–0.86). In our sequential analysis, the next SNP (ranked by unconditional P-value) to add to the model conditioning on both rs80304993 and rs62293480 was rs4420873, but it was excluded due to collinearity (r2>0.80). Therefore, the next SNP, rs75316749, was evaluated in the model conditioning on rs80304993 and rs62293480, and had a combined conditional P-value of 1.46×10−6. Unlike the two other SNPs in TERC, rs75316749 was not associated with prostate cancer. The G allele was positively associated in conditional and unconditional analyses with colorectal, breast, ovarian, and lung cancers (Table 3), driven specifically by ER-negative breast cancer, and lung adenocarcinoma and squamous cancers, but not lung cancer overall (P=2.9×10−4; OR=1.17 95%CI=1.07–1.27; Figure 1C). While rs75316749 has been reported to result in a motif change and enhancer histone changes in breast and fat cell lines, SNPs in very high LD including rs115002293 and rs75963875 are associated with enhancer histone changes in a wide variety of cell lines, including breast and lung fibroblast cells (30).
Figure 1. Unconditional ASSET forest plots by cancer type and subtype for TERC SNPs rs80304993, rs62293480, and rs75316749.
(A) Forest plot associations for the A allele for rs80304993. (B) Forest plot associations for the T allele for rs62293480. (C) Forest plot associations for the G allele for rs75316749.
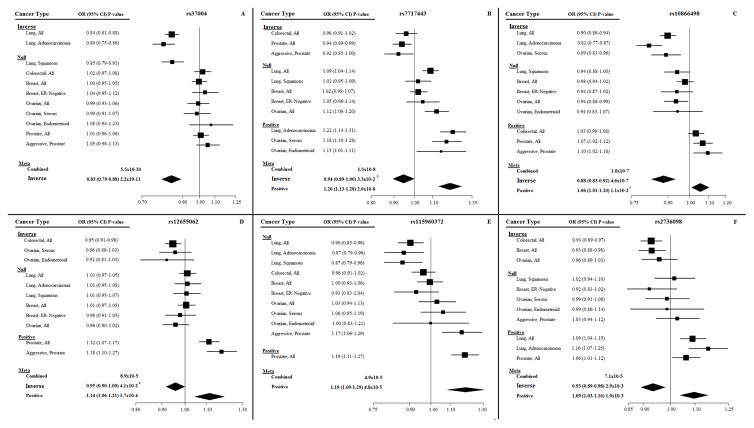
In the TERT-CLPTM1L region, six independent loci were identified (Table 3). The T allele of the SNP with the lowest P-value, rs37004, was inversely associated with lung cancer overall and specifically lung adenocarcinoma (P=2.2×10−11; OR=0.83 95%CI=0.79–0.88; Figure 2A). After conditioning on rs37004, rs7717443 had the lowest combined P-value (Pconditional=1.26×10−7). The T allele was associated with increased ovarian and lung cancer risks, and suggestive decreased colorectal and prostate cancer risks in conditional and unconditional analyses. The unconditional ASSET forest plot by cancer type and subtype for rs7717443 (Figure 2B) illustrates that the positive associations apply to serous and endometrioid ovarian cancer subtypes (but not overall ovarian cancer) and lung adenocarcinoma only (P=2.0×10−8; OR=1.20 95%CI=1.13–1.28), and inverse associations are for overall colorectal and prostate cancers, and aggressive prostate cancer (P=3.3×10−2; OR=0.94 95%CI=0.89–1.00). Next, after conditioning on both rs37004 and rs7717443, the combined P-value for rs10866498 was highly significant (9.27×10−18). In conditional and unconditional analyses, the T allele of rs10866498 was associated positively with colorectal and prostate cancers, and inversely with ovarian and lung cancers (Table 3). Associations with colorectal cancer, and with both overall and aggressive prostate cancers (P=0.01; OR=1.06 95%CI=1.01–1.10) were positive, and inverse associations were observed with overall lung cancer and lung adenocarcinoma, and serous ovarian cancer (P=4.6×10−7; OR=0.88 95%CI=0.83–0.92; Figure 2C). After conditioning on the top 3 TERT-CLPTM1L SNPs, rs12655062 was the next most significant (Pconditional=1.13×10−6). In both unconditional and conditional analyses, the rs12655062 A allele was associated positively with prostate and inversely with colorectal and ovarian cancers (Table 3). Positive associations were driven by both overall prostate cancer and aggressive prostate cancer (P=1.7×10−4; OR=1.14 95%CI=1.06–1.21), and inverse associations by overall colorectal cancer and endometrioid and serous ovarian cancers (P=4.1×10−2; OR=0.95 95%CI=0.90–1.00; Figure 2D). The rs12655062 A allele is associated with reduced expression of IRX4 and CTD02194D22.3 in prostate tissue (31), alters six motifs, and results in enhancer histone changes in breast and gastrointestinal cell lines (30). The next most significant SNP in sequential analysis after conditioning on the top four TERT-CLPTM1L SNPs was rs115960372 (Pconditional=3.12×10−6). The T allele was associated positively with prostate cancer and suggestively inversely associated with lung cancer in conditional and unconditional analyses (Table 3). This SNP results in changes to chromatin structure in several cell lines (including fetal, adult, and carcinoma lung cell lines) and two motif changes (30). The unconditional ASSET forest plot by cancer type and subtype revealed that positive associations were driven by overall prostate cancer (OR=1.19 95%CI=1.09–1.29); however, inverse associations were not significant in cancer subtype analyses (P=7.5×10−2; Figure 2E). The last significant SNP identified from sequential conditional analysis after conditioning on the above five TERT-CLPTM1L variants was rs2736098 (Pconditional=5.36×10−6). The T allele was suggestively positively associated with prostate and lung cancers, and inversely associated with colorectal, breast, and ovarian cancers, in both conditional and unconditional analyses (Table 3). Positive associations were driven not only by overall prostate and lung cancers, but also lung adenocarcinoma (P=1.9×10−3; OR=1.09 95%CI=1.03–1.16; Figure 2F).
Figure 2. Unconditional ASSET forest plots by cancer type and subtype for TERT-CLPTM1L SNPs rs37004, rs7717443, rs10866498, rs12655062, rs115960372, and rs2736098.
(A) Forest plot associations for the T allele for rs37004. (B) Forest plot associations for the T allele for rs7717443. (C) Forest plot associations for the T allele for rs10866498. (D) Forest plot associations for the A allele for rs12655062. (E) Forest plot associations for the T allele for rs115960372. (F) Forest plot associations for the T allele for rs2736098.
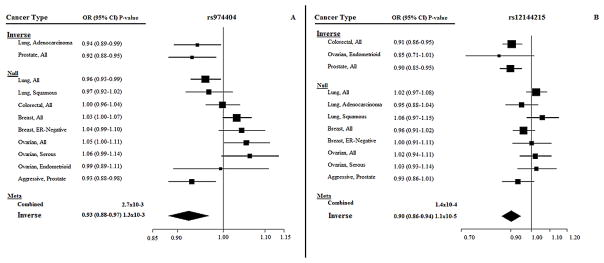
In DCLRE1B, rs974404 had the lowest combined P-value (P=9.19×10−6). The G allele was inversely associated with prostate and lung cancers, and suggestively positively associated with breast and ovarian cancers. The inverse associations were driven by overall prostate cancer and lung adenocarcinoma (P=1.3×10−3; OR=0.93 95%CI=0.88–0.97), but not by overall lung cancer, or squamous cell lung cancer; positive associations were no longer significant in analyses by cancer subtype (P=0.23) (Figure 3A). Considerable evidence supports that rs974404 and correlated SNPs alter gene function. rs974404 results in 27 altered motifs (30), and twelve SNPs in LD with rs974404 are associated with increased expression of DCLRE1B in whole blood (30).
Figure 3. Unconditional ASSET forest plots by cancer type and subtype for DCLRE1B SNPs rs974404 and rs12144215.
(A) Forest plot associations for the G allele for rs974404. (B) Forest plot associations for the T allele for rs12144215.
After conditioning on rs974404, the most significant SNP in DCLRE1B was rs12144215 (Punconditional=1.50×10−5; Pconditional=2.07×10−5). In unconditional analyses, the rs12144215 T allele was inversely associated with colorectal and prostate cancers, and after conditioning on rs974404, a suggestive positive association with lung cancer and a significant inverse association with breast cancer were additionally observed (Table 3). The unconditional inverse association was driven by overall colorectal and prostate cancers and the ovarian cancer endometrioid subtype (P=1.1×10−5; OR=0.90 95%CI=0.86–0.94; Figure 3B); the positive association with lung cancer observed in conditional analyses was no longer observed in unconditional analyses by cancer subtype (P=1.00). Several SNPs in LD with rs12144215 change chromatin structure in multiple cell lines, including mammary epithelial and lung (30).
Discussion
Our conditional subset-based meta-analysis of GWAS data from five different cancer types identified 13 independent SNPs in DCLRE1B, TERC, TERT-CLPTM1L, RTEL and POT1 gene regions that are associated with risk of multiple cancers. Across the DCLRE1B region, we identified two novel loci: rs974404, which is associated with increased DCLRE1B expression (30) and was associated with prostate and lung cancer risk, and rs12144215, which may be associated with chromatin structure alterations and was associated with colorectal, breast, and ovarian cancers risk. While the observed associations between two SNPs near the TERC gene, rs80304993 and rs62293480, and prostate cancer risk have been reported in GWAS previously (32), we show that the association between rs62293480 and prostate cancer is much more significant after conditioning on rs80304993 (Punconditional=1.35x−06, Pconditional=2.16x−13). We also report a novel finding in the TERC region; after conditioning on both rs80304993 and rs62293480, rs75316749 was associated with colorectal, breast, ovarian, and lung cancer risk. There is some evidence that this SNP and/or others in LD with it result in enhancer histone changes (30). Across the TERT-CLPTML1 regions, we detected six susceptibility loci where strong associations with lung and/or prostate cancer risk were generally observed. We report similar associations previously observed in GWAS for four TERT-CLPTM1L SNPs and lung and prostate cancer (7,9), but observe novel findings for two SNPs, rs12655062 and rs115960372. The rs12655062 variant is associated with reduced expression of the gene IRX4 in prostate tissue, and rs115960372 may alter chromatin structure in multiple tissue types (30). Our study demonstrated that for rs10866498, after controlling for the top two hits in TERT-CLPTM1L, the p-value for the inverse association with lung and ovarian cancer was even more significant (Punconditional=6.36×10−8, Pconditional=9.27×10−18). We also observed associations between rs116895242 in the POT1 region and colorectal, ovarian and lung cancer risk, and between rs34978822 in RTEL1 and prostate and lung cancer. There is limited evidence to support that these SNPs alter gene function (30).
DCLRE1B plays an important role in protecting telomeres by interacting with the shelterin complex to suppress DNA damage-sensing machinery during and after replication (20,33). The SNPs that we observed to be associated with risk of prostate and lung cancers (rs974404 in PTPN22), and colorectal, breast, and ovarian cancers (rs12144215 in MAGI3), have been previously associated in GWAS with rheumatoid arthritis and Grave’s disease, respectively (34,35). To date, only one SNP in the DCLRE1B gene, rs11552449, has been shown to be associated with breast cancer risk in a meta-analysis of nine GWAS and 41 studies in the Breast Cancer Association Consortium (P-value=1.8×10−8) (16). However, this SNP did not reach gene-level statistical significance in our analyses.
TERC is essential for telomerase expression because it encodes the RNA component of telomerase required for elongation of telomeric repeats (1,20). Variants in the 3q26 TERC region have been associated with risk of several different cancers in GWAS, including melanoma, glioma, bladder, colorectal, nasopharyngeal, chronic lymphatic leukemia, and multiple myeloma (36–42). In a GWAS of >25,000 prostate cancer cases and controls, Kote-Jarai et al. reported that rs10936632 was associated with a 10% decrease in prostate cancer risk (P-value 1.0×10−13) (32). In our unconditional ASSET analysis we also observed that rs10936632, which is in high LD (r2=0.97) with rs55953261, was significantly associated with reduced prostate cancer risk. It should be noted that 27% of prostate cancer cases and 26% of controls in Kote-Jarai et al. (32) were also included in our investigation.
Our additional TERC findings for rs80304993 and rs62293480 and prostate cancer risk have been observed previously in a multi-ethnic meta-analysis of GWAS (43). These SNPS are located in the SKIL gene, which regulates cell growth and differentiation (20). Our study findings for SNP rs75316749 and colorectal, breast, ovarian, and lung cancer risk are novel. SNP rs75316749 lies approximately 40kb 3′ of the MECOM gene which encodes a protein involved in hematopoiesis, apoptosis, development, and cell differentiation and proliferation (20).
The TERT gene, at 5p15.33, encodes the catalytic subunit of telomerase (1,20,33) and thus plays a vital role in maintaining telomerase expression and facilitating elongation of telomeric repeats. The 5′-end of TERT adjoins CLPTM1L, which is overexpressed in lung and pancreatic cancers (9,44,45). There is extensive LD between the two genes, and susceptibility loci in this combined gene region have been associated with multiple cancer types (7–9,46–48). The most commonly associated risk variants in the TERT-CLPTM1L regions are rs2736100 and rs401681, respectively. In GWAS, rs2736100 has been associated with lung, glioma, and testicular cancer risk (9,45) while rs401681 has been linked to lung, bladder, pancreas, prostate, and skin cancer risk (9,45). Our unconditional ASSET analyses corroborate the associations observed between these variants and lung cancer risk.
Mocellin et al. performed a systematic review of TERT-CLPTM1L polymorphisms and cancer risk in 85 studies including 27 GWAS of predominantly individuals of European ancestry (87%) (9). Of the 67 SNPs and 24 tumor types examined, statistically significant associations were reported for 22 SNPs, 19 of which were linked to lung cancer. In our investigation, unconditional ASSET analysis confirmed associations with lung cancer for 13 of these at our gene level cutoff (P-value <1.32×10−5) and for four more at P-value<0.05. Of particular interest from Mocellin et al.’s study was the highly significant association reported between rs2736098 and lung (4 studies, P-value=2.2×10−13) and bladder (3 studies, P-value=8.6×10−10) cancer risk and the association between rs451360 and lung cancer risk (2 studies, P-value=4×10−3) (9). Our findings are in agreement with these observations. In our analysis, rs37004, 2kb 5′ of CLPTM1L and in high LD (r2=0.89) with rs451360, was the SNP in the TERT-CLPTM1L region with the lowest P-value, due entirely to its association with lung cancer risk; rs2736098, located within TERT, was associated with lung cancer risk as well as prostate, colorectal, breast, and ovarian cancer. However, we did not observe the lung cancer association reported by Mocellin et al. for rs1801075 and we could not evaluate the association with rs4246742 because no data on lung cancer were available for this SNP.
Similar results were reported for these variants by Wang et al., who utilized the same ASSET meta-analytic approach that we used to examine common susceptibility alleles in TERT-CLPTM1L across six cancer types (lung, prostate, pancreatic, testicular, glioma, and bladder), in 34,248 cases and 45,036 controls (7). A large proportion of prostate (60.3%) and lung (46.9%) cancer cases from that study were also included in our investigation. Using sequential conditional ASSET analyses, Wang et al. identified five independent risk loci in individuals of European ancestry. These loci are included in the LD plot of our significant unconditional ASSET two-sided SNPs retained following LD pruning at r2>0.70 (Supplementary Figure 2). In one region, rs13170453 was associated positively with pancreatic and testicular cancer (P-value=4.38×10−13) and inversely with lung cancer risk (P-value=9.5×10−8). Our conditional ASSET findings for rs37004, which is in high LD (r2=0.88) with rs13170453, confirm the lung cancer association observed by Wang et al. for this SNP. In a second region, Wang et al. observed that rs2736098 was associated positively with lung, prostate, and bladder cancer (P-value=2.58×10−8) and inversely with pancreatic and testicular cancer (P-value=4.89×10−6). In our investigation, rs2736098, located within TERT, was similarly positively associated in conditional analyses with lung and prostate cancer, but inversely with colorectal, breast, and ovarian cancer. In a third region, Wang et al. reported rs4449583 as being associated positively with glioma, and inversely with testicular, prostate, and pancreatic cancers. In our unconditional ASSET analysis, this SNP was associated positively with ovarian and lung cancer, and inversely with prostate cancer. Cancer associations for the remaining two TERT-CLPTM1L regions including rs10069690 and rs13172201 in Wang’s study were not replicated in our investigation. Associations for these regions were also not confirmed in supplementary analyses conducted by Wang et al. across additional cancer types (esophageal, gastric, breast, endometrial, prostate, osteosarcoma, ovarian, renal, and additional prostate cancers) including 11,385 cases and 18,322 controls (7). We examined a larger region around TERT-CLPTM1L than did Wang et al. (chr5: 1,250,000–1,450,000), thus they did not assess rs12655062 and rs115960372 which lie outside of that region. Our significant conditional ASSET associations between TERT SNPs rs7717443 and rs10866498 and colorectal, prostate, ovarian, and lung cancer risk, which are not highly correlated with variants observed in Wang et al. have not been previously reported. In summary, our study confirms the Wang et al. findings for three of the five significant TERT-CLPTM1L SNPs that they reported among European subjects (conditionally for rs2736098 and rs37004 (r2=0.88 with rs13170453), and unconditionally for rs4449583); however, our study did not corroborate their findings for rs10069690 or rs13172201. Additionally, in our study, of the six conditionally significant TERT-CLPTM1L risk loci detected among European subjects, Wang et al. did not report findings for SNPs rs7717443 and rs10866498 nor did they examine associations for SNPs rs12655062 and rs115960372 which lied outside of the regions that they evaluated. Nonetheless, a Japanese fine-mapping study of 1,583 prostate cancer cases and 2,480 controls reported a highly significant association with rs115960372, (OR=1.31, P-value=7.76×10−10) which is in the LPCAT1 gene (49). The association between rs12655062, which is in the CTD-2194D22.4 gene, and colorectal, breast, and prostate cancer risk has not been previously reported.
Because associations with cancer risk may vary by histology, some studies have assessed SNPs across TERT-CLPTMIL in relation to cancer subtypes. Of particular interest were the lung cancer ASSET meta-analytic results reported by Wang et al. (50). Based on data from five GWAS, highly significant associations were reported between TERT-CLPTMIL rs7717443 (OR=1.24, P-value=4.90×10−10), rs10866498 (OR=0.81, P-value=3.28×10−11) and rs37004 (OR=0.78, P-value=2.52×10−12) and lung adenocarcinoma risk; an association between rs37004 (OR=0.82, P-value=7.94×10−8) and squamous lung cancer risk was also observed. In on our unconditional ASSET forest plot analysis, we observed similar associations between these variants and lung adenocarcinoma, but not with squamous lung cancer.
The associations reported here for variants in the GAR1, TERF2, POT1 and RTEL1 gene regions and colorectal, lung, breast, ovarian, and/or prostate cancers have not been reported in GWAS of these cancers. We advise caution in interpreting results for GAR1 and TERF2 variants with low MAF (0.3%). Although these SNPs passed imputation accuracy cutoffs in some consortium-specific meta-analyses, SNPs with such low MAFs are known to be difficult to impute accurately. The RTEL1 gene at 20p13.3 encodes a DNA helicase involved in stabilization, protection and elongation of telomeres (9,20). This gene interacts with shelterin complex proteins and variants in this gene have been associated in previous GWAS with high-grade glioma risk (37). POT1 at 7q31.33 and TERF2 at 16q22.1 are protein-coding genes that are components of the shelterin complex (20,33). Variants in POT1 have been previously associated with risk of chronic lymphocytic leukemia in GWAS (41).
To our knowledge, this is the largest meta-analysis of GWAS data on telomere structure and maintenance genes and cancer risk. With over 61,000 cancer cases and nearly 75,000 controls, our study is highly powered to detect significant associations for variants with common allele frequencies. Our study is unique in that we evaluated risk of multiple cancer types as well as risk of specific histologic or molecular subtypes of cancer and subtypes related to aggressiveness. Our subset-based meta-analysis also permitted us to examine the magnitude and direction of genetic associations allowing for heterogeneity of associations across cancer sites. Compared to traditional methods, ASSET helps minimize false-positives through multiple testing penalties and improves detection power (21). We were able to determine independent associations between SNPs and cancer types by conditioning on the effects of SNPs with lower P-values. Because there is considerable evidence linking TERT-CLPTM1L variants to risk of many different cancer types, and several other telomere structure and maintenance genes have been implicated in GWAS of various cancer types, we used gene-level P-value thresholds to define statistical significance. Although we were able to interrogate a very large number of SNPs in telomere structure and maintenance genes, we did not assess SNPs across all known telomere structure and maintenance genes, and most of the SNPs (97.5%) we examined were imputed. Our study was not well-designed to examine imputed rare variants since these SNPs may be poorly represented or poorly tagged on genotyping arrays. While we were able to use the available aggregate data to evaluate whether variation in all SNPs combined in each gene was associated with risk, we could not evaluate haplotypes.
In summary, our results indicate that patterns of association in telomere structure and maintenance genes observed across cancer types and subtypes are complex. These findings may provide insight into the mechanisms through which telomere dysfunction in different tissues influences cancer risk. In our investigation, seven of the thirteen conditional associations identified were novel. While we observed suggestive pleiotropic associations within the DCLRE1B, TERC, TERT-CLPTM1L, POT1 and RTEL1 gene regions, fine-mapping studies with the ability to assess haplotypes are needed to evaluate the relationship between alleles at different loci in order to help identify potential variants that may have gone undetected. Replication and mechanistic studies are also needed to help provide insight regarding the function and variability of risk across cancers for these telomere structure and maintenance SNPs.
Supplementary Material
Supplementary Figure 1. Manhattan plot of associations between 204,993 SNPs in telomere structure and maintenance genes and five cancer types.
The dotted line indicates the genome-wide significance level (P-value=5×10−8), and the individual solid lines indicate the gene-level P-value cutoffs taking into account the number of effective tests. Genes are evaluated in the order listed in the key.
Supplementary Figure 2. TERT-CLPTM1L and TERC LD plots for statistically significant ASSET variants that were retained following LD pruning at r2>0.70.
Associations between alleles across variants are based on CGEMS and EAGLE datasets. SNPs circled in black are variants identified as statistically significant in ASSET conditional analyses. aSNPs identified as statistically significant in ASSET conditional analyses in the Wang et al. study among individuals of European ancestry [7]. A) LD plot for TERT-CLPTM1L variants. B) LD plot for TERC variant.
Novelty & Impact Statements.
Utilizing the novel ASSociation analysis based on SubSET (ASSET) meta-analytic approach, we examined associations between >200,000 variants in 22 telomere structure and maintenance gene regions and colorectal, breast, prostate, ovarian, and lung cancer risk. We observed pleiotropic associations across cancer types in the DCLRE1B, TERC, TERT-CLPTM1L, POT1, and RTEL1 gene regions. Additional studies clarifying the mechanisms through which these complex association patterns in telomere-related genes influence cancer risk are needed.
Acknowledgments
ASTERISK: We are very grateful to Dr. Bruno Buecher without whom this project would not have existed. We also thank all those who agreed to participate in this study, including the patients and the healthy control persons, as well as all the physicians, technicians and students.
DACHS: We thank all participants and cooperating clinicians, and Ute Handte-Daub, Utz Benscheid, Muhabbet Celik and Ursula Eilber for excellent technical assistance.
GECCO: The authors would like to thank all those at the GECCO Coordinating Center for helping bring together the data and people that made this project possible. The authors acknowledge Dave Duggan and team members at TGEN (Translational Genomics Research Institute), the Broad Institute, and the Génome Québec Innovation Center for genotyping DNA samples of cases and controls, and for scientific input for GECCO.
HPFS, NHS and PHS: We would like to acknowledge Patrice Soule and Hardeep Ranu of the Dana Farber Harvard Cancer Center High-Throughput Polymorphism Core who assisted in the genotyping for NHS, HPFS, and PHS under the supervision of Dr. Immaculata Devivo and Dr. David Hunter, Qin (Carolyn) Guo and Lixue Zhu who assisted in programming for NHS and HPFS, and Haiyan Zhang who assisted in programming for the PHS. We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-Up Study, for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
PLCO: The authors thank Drs. Christine Berg and Philip Prorok, Division of Cancer Prevention, National Cancer Institute, the Screening Center investigators and staff or the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, Mr. Tom Riley and staff, Information Management Services, Inc., Ms. Barbara O’Brien and staff, Westat, Inc., and Drs. Bill Kopp and staff, SAIC-Frederick. Most importantly, we acknowledge the study participants for their contributions to making this study possible. The statements contained herein are solely those of the authors and do not represent or imply concurrence or endorsement by NCI.
PMH: The authors would like to thank the study participants and staff of the Hormones and Colon Cancer study.
WHI: The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Short%20List.pdf
COGS: This study would not have been possible without the contributions of the following: Per Hall (COGS); Douglas F. Easton, Paul Pharoah, Kyriaki Michailidou, Manjeet K. Bolla, Qin Wang (BCAC), Andrew Berchuck (OCAC), Rosalind A. Eeles, Douglas F. Easton, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Georgia Chenevix-Trench, Antonis Antoniou, Lesley McGuffog, Fergus Couch and Ken Offit (CIMBA), Joe Dennis, Alison M. Dunning, Andrew Lee, and Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, Javier Benitez, Anna Gonzalez-Neira and the staff of the CNIO genotyping unit, Jacques Simard and Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, Stig E. Bojesen, Sune F. Nielsen, Borge G. Nordestgaard, and the staff of the Copenhagen DNA laboratory, and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility.
Funding for the iCOGS infrastructure came from: the European Community’s Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 – the GAME-ON initiative), the Department of Defense (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund.
Funding Sources
This work was supported by the National Institutes of Health (NIH), National Cancer Institute (NCI) (R01 CA151989 supplement, to JA Doherty).
The scientific development and funding for this project were in part supported by the US NCI GAME-ON Post-GWAS Initiative (U19-CA148112); TRICL (Transdisciplinary Research for Cancer of Lung): NIH U19 CA148127-01 (PI: Amos), Canadian Cancer Society Research Institute (no. 020214, PI: Hung); DRIVE (Discovery, Biology, and Risk of Inherited Variants in Breast Cancer): NIH U19 CA148065; CORECT (ColoRectal Transdisciplinary Study): NIH U19 CA148107; R01 CA81488, P30 CA014089; ELLIPSE (ELLIPSE, Elucidating Loci in Prostate Cancer Susceptibility): This work was support by the GAMEON U19 initiative for prostate cancer (ELLIPSE), U19 CA148537; FOCI (Transdisciplinary Cancer Genetic Association and Interacting Studies): NIH U19 CA148112-01 (PI: Sellers), R01-CA122443, P50-CA136393, P30-CA15083 (PI: Goode), Cancer Research UK (C490/A8339, C490/A16561, C490/A10119, C490/A10124 (PI: Pharoah); GECCO: NCI, NIH, U.S. Department of Health and Human Services (DHHS) (U01 CA137088; R01 CA059045); ASTERISK: a Hospital Clinical Research Program (PHRC) and supported by the Regional Council of Pays de la Loire, the Groupement des Entreprises Françaises dans la Lutte contre le Cancer (GEFLUC), the Association Anne de Bretagne Génétique and the Ligue Régionale Contre le Cancer (LRCC); COLO2&3: NIH (R01 CA60987). DACHS: German Research Council (Deutsche Forschungsgemeinschaft, BR 1704/6-1, BR 1704/6-3, BR 1704/6-4 and CH 117/1-1), and the German Federal Ministry of Education and Research (01KH0404 and 01ER0814). DALS: NIH (R01 CA48998 to M. L. Slattery); HPFS is supported by the NIH (P01 CA 055075, UM1 CA167552, R01 137178, R01 CA151993 and P50 CA127003), NHS by the NIH (UM1 CA186107, R01 CA137178, P01 CA87969, R01 CA151993 and P50 CA127003,) and PHS by the NIH (R01 CA042182); MEC: NIH (R37 CA54281, P01 CA033619, and R01 CA63464); OFCCR: NIH, through funding allocated to the Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783); additional funding toward genetic analyses of OFCCR includes the Ontario Research Fund, the Canadian Institutes of Health Research, and the Ontario Institute for Cancer Research, through generous support from the Ontario Ministry of Research and Innovation. PLCO: Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, NCI, NIH, DHHS; additionally, a subset of control samples were genotyped as part of the Cancer Genetic Markers of Susceptibility (CGEMS) Prostate Cancer GWAS (Yeager, M et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet 2007 May;39(5):645–9), Colon CGEMS pancreatic cancer scan (PanScan) (Amundadottir, L et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009 Sep;41(9):986–90, and Petersen, GM et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010 Mar;42(3):224–8), and the Lung Cancer and Smoking study (Landi MT, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009 Nov;85(5):679–91); the prostate and PanScan study datasets were accessed with appropriate approval through the dbGaP online resource (http://cgems.cancer.gov/data/) accession numbers phs000207.v1.p1 and phs000206.v3.p2, respectively, and the lung datasets were accessed from the dbGaP website (http://www.ncbi.nlm.nih.gov/gap) through accession number phs000093.v2.p2; funding for the Lung Cancer and Smoking study was provided by NIH, Genes, Environment and Health Initiative (GEI) Z01 CP 010200, NIH U01 HG004446, and NIH GEI U01 HG 004438. For the lung study, the GENEVA Coordinating Center provided assistance with genotype cleaning and general study coordination, and the Johns Hopkins University Center for Inherited Disease Research conducted genotyping. PMH: NIH (R01 CA076366 to P.A. Newcomb); VITAL: NIH (K05 CA154337); WHI: The WHI program is funded by the National Heart, Lung, and Blood Institute, NIH, DHHS through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C; and MD Anderson: NCI, NIH K07CA160753 (PI: Pande).
Abbreviations used
- ASSET
ASSociation analysis based on SubSET
- CGEMS
Cancer Genetic Markers of Susceptibility Project
- CI
confidence interval
- EAGLE
Environmental and Genetics in Lung Cancer Etiology study
- ER
estrogen receptor
- GAME-ON
Genetic Associations and Mechanisms in Oncology Network
- GEC
Genetic type 1 Error Calculator
- GECCO
Genetic and Epidemiology of Colorectal Cancer Consortium
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- MAF
minor allele frequency
- Me
effective number of independent tests
- OR
odds ratio
- SNPs
single nucleotide polymorphisms
Footnotes
Author’s disclosures of potential conflicts of interest: non for all authors.
References
- 1.Robles-Espinoza CD, del Castillo Velasco-Herrera M, Hayward NK, Adams DJ. Telomere-Regulating Genes and the Telomere Interactome in Familial Cancers. Mol Cancer Res. 2015;13:211–222. doi: 10.1158/1541-7786.MCR-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabello L, Yu K, Kraft P, De Vivo I, Hunter DJ, Prescott J, Wong JY, Chatterjee N, Hayes RB, Savage SA. The association of telomere length and genetic variation in telomere biology genes. Hum Mutat. 2010;31:1050–1058. doi: 10.1002/humu.21314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Codd V, Nelson CP, Albrecht E, Mangino M, Deelen J, Buxton JL, Hottenga JJ, Fischer K, Esko T, Surakka I, Broer L, Nyholt DR, Mateo Leach I, Salo P, Hägg S, Matthews MK, Palmen J, Norata GD, O’Reilly PF, Saleheen D, Amin N, Balmforth AJ, Beekman M, de Boer RA, Böhringer S, Braund PS, Burton PR, de Craen AJ, Denniff M, Dong Y, Douroudis K, Dubinina E, Eriksson JG, Garlaschelli K, Guo D, Hartikainen AL, Henders AK, Houwing-Duistermaat JJ, Kananen L, Karssen LC, Kettunen J, Klopp N, Lagou V, van Leeuwen EM, Madden PA, Mägi R, Magnusson PK, Männistö S, McCarthy MI, Medland SE, Mihailov E, Montgomery GW, Oostra BA, Palotie A, Peters A, Pollard H, Pouta A, Prokopenko I, Ripatti S, Salomaa V, Suchiman HE, Valdes AM, Verweij N, Viñuela A, Wang X, Wichmann HE, Widen E, Willemsen G, Wright MJ, Xia K, Xiao X, van Veldhuisen DJ, Catapano AL, Tobin MD, Hall AS, Blakemore AI, van Gilst WH, Zhu H, Erdmann J, Reilly MP, Kathiresan S, Schunkert H, Talmud PJ, Pedersen NL, Perola M, Ouwehand W, Kaprio J, Martin NG, van Duijn CM, Hovatta I, Gieger C, Metspalu A, Boomsma DI, Jarvelin MR, Slagboom PE, Thompson JR, Spector TD, van der Harst P, Samani NJ Consortium C. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45:422–427. doi: 10.1038/ng.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy D, Neuhausen SL, Hunt SC, Kimura M, Hwang SJ, Chen W, Bis JC, Fitzpatrick AL, Smith E, Johnson AD, Gardner JP, Srinivasan SR, Schork N, Rotter JI, Herbig U, Psaty BM, Sastrasinh M, Murray SS, Vasan RS, Province MA, Glazer NL, Lu X, Cao X, Kronmal R, Mangino M, Soranzo N, Spector TD, Berenson GS, Aviv A. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc Natl Acad Sci U S A. 2010;107:9293–9298. doi: 10.1073/pnas.0911494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangino M, Brouilette S, Braund P, Tirmizi N, Vasa-Nicotera M, Thompson JR, Samani NJ. A regulatory SNP of the BICD1 gene contributes to telomere length variation in humans. Hum Mol Genet. 2008;17:2518–2523. doi: 10.1093/hmg/ddn152. [DOI] [PubMed] [Google Scholar]
- 6.Mangino M, Christiansen L, Stone R, Hunt SC, Horvath K, Eisenberg DT, Kimura M, Petersen I, Kark JD, Herbig U, Reiner AP, Benetos A, Codd V, Nyholt DR, Sinnreich R, Christensen K, Nassar H, Hwang SJ, Levy D, Bataille V, Fitzpatrick AL, Chen W, Berenson GS, Samani NJ, Martin NG, Tishkoff S, Schork NJ, Kyvik KO, Dalgård C, Spector TD, Aviv A. DCAF4, a novel gene associated with leucocyte telomere length. J Med Genet. 2015;52:157–162. doi: 10.1136/jmedgenet-2014-102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Zhu B, Zhang M, Parikh H, Jia J, Chung CC, Sampson JN, Hoskins JW, Hutchinson A, Burdette L, Ibrahim A, Hautman C, Raj PS, Abnet CC, Adjei AA, Ahlbom A, Albanes D, Allen NE, Ambrosone CB, Aldrich M, Amiano P, Amos C, Andersson U, Andriole G, Jr, Andrulis IL, Arici C, Arslan AA, Austin MA, Baris D, Barkauskas DA, Bassig BA, Beane Freeman LE, Berg CD, Berndt SI, Bertazzi PA, Biritwum RB, Black A, Blot W, Boeing H, Boffetta P, Bolton K, Boutron-Ruault MC, Bracci PM, Brennan P, Brinton LA, Brotzman M, Bueno-de-Mesquita HB, Buring JE, Butler MA, Cai Q, Cancel-Tassin G, Canzian F, Cao G, Caporaso NE, Carrato A, Carreon T, Carta A, Chang GC, Chang IS, Chang-Claude J, Che X, Chen CJ, Chen CY, Chen CH, Chen C, Chen KY, Chen YM, Chokkalingam AP, Chu LW, Clavel-Chapelon F, Colditz GA, Colt JS, Conti D, Cook MB, Cortessis VK, Crawford ED, Cussenot O, Davis FG, De Vivo I, Deng X, Ding T, Dinney CP, Di Stefano AL, Diver WR, Duell EJ, Elena JW, Fan JH, Feigelson HS, Feychting M, Figueroa JD, Flanagan AM, Fraumeni JF, Jr, Freedman ND, Fridley BL, Fuchs CS, Gago-Dominguez M, Gallinger S, Gao YT, Gapstur SM, Garcia-Closas M, Garcia-Closas R, Gastier-Foster JM, Gaziano JM, Gerhard DS, Giffen CA, Giles GG, Gillanders EM, Giovannucci EL, Goggins M, Gokgoz N, Goldstein AM, Gonzalez C, Gorlick R, Greene MH, Gross M, Grossman HB, Grubb R, 3rd, Gu J, Guan P, Haiman CA, Hallmans G, Hankinson SE, Harris CC, Hartge P, Hattinger C, Hayes RB, He Q, Helman L, Henderson BE, Henriksson R, Hoffman-Bolton J, Hohensee C, Holly EA, Hong YC, Hoover RN, Hosgood HD, 3rd, Hsiao CF, Hsing AW, Hsiung CA, Hu N, Hu W, Hu Z, Huang MS, Hunter DJ, Inskip PD, Ito H, Jacobs EJ, Jacobs KB, Jenab M, Ji BT, Johansen C, Johansson M, Johnson A, Kaaks R, Kamat AM, Kamineni A, Karagas M, Khanna C, Khaw KT, Kim C, Kim IS, Kim JH, Kim YH, Kim YC, Kim YT, Kang CH, Jung YJ, Kitahara CM, Klein AP, Klein R, Kogevinas M, Koh WP, Kohno T, Kolonel LN, Kooperberg C, Kratz CP, Krogh V, Kunitoh H, Kurtz RC, Kurucu N, Lan Q, Lathrop M, Lau CC, Lecanda F, Lee KM, Lee MP, Le Marchand L, Lerner SP, Li D, Liao LM, Lim WY, Lin D, Lin J, Lindstrom S, Linet MS, Lissowska J, Liu J, Ljungberg B, Lloreta J, Lu D, Ma J, Malats N, Mannisto S, Marina N, Mastrangelo G, Matsuo K, McGlynn KA, McKean-Cowdin R, McNeill LH, McWilliams RR, Melin BS, Meltzer PS, Mensah JE, Miao X, Michaud DS, Mondul AM, Moore LE, Muir K, Niwa S, Olson SH, Orr N, Panico S, Park JY, Patel AV, Patino-Garcia A, Pavanello S, Peeters PH, Peplonska B, Peters U, Petersen GM, Picci P, Pike MC, Porru S, Prescott J, Pu X, Purdue MP, Qiao YL, Rajaraman P, Riboli E, Risch HA, Rodabough RJ, Rothman N, Ruder AM, Ryu JS, Sanson M, Schned A, Schumacher FR, Schwartz AG, Schwartz KL, Schwenn M, Scotlandi K, Seow A, Serra C, Serra M, Sesso HD, Severi G, Shen H, Shen M, Shete S, Shiraishi K, Shu XO, Siddiq A, Sierrasesumaga L, Sierri S, Loon Sihoe AD, Silverman DT, Simon M, Southey MC, Spector L, Spitz M, Stampfer M, Stattin P, Stern MC, Stevens VL, Stolzenberg-Solomon RZ, Stram DO, Strom SS, Su WC, Sund M, Sung SW, Swerdlow A, Tan W, Tanaka H, Tang W, Tang ZZ, Tardon A, Tay E, Taylor PR, Tettey Y, Thomas DM, Tirabosco R, Tjonneland A, Tobias GS, Toro JR, Travis RC, Trichopoulos D, Troisi R, Truelove A, Tsai YH, Tucker MA, Tumino R, Van Den Berg D, Van Den Eeden SK, Vermeulen R, Vineis P, Visvanathan K, Vogel U, Wang C, Wang C, Wang J, Wang SS, Weiderpass E, Weinstein SJ, Wentzensen N, Wheeler W, White E, Wiencke JK, Wolk A, Wolpin BM, Wong MP, Wrensch M, Wu C, Wu T, Wu X, Wu YL, Wunder JS, Xiang YB, Xu J, Yang HP, Yang PC, Yatabe Y, Ye Y, Yeboah ED, Yin Z, Ying C, Yu CJ, Yu K, Yuan JM, Zanetti KA, Zeleniuch-Jacquotte A, Zheng W, Zhou B, Mirabello L, Savage SA, Kraft P, Chanock SJ, Yeager M, Landi MT, Shi J, Chatterjee N, Amundadottir LT. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the TERT-CLPTM1L region on chromosome 5p15. 33. Hum Mol Genet. 2014;23:6616–6633. doi: 10.1093/hmg/ddu363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, Hillman KM, Mai PL, Lawrenson K, Stutz MD, Lu Y, Karevan R, Woods N, Johnston RL, French JD, Chen X, Weischer M, Nielsen SF, Maranian MJ, Ghoussaini M, Ahmed S, Baynes C, Bolla MK, Wang Q, Dennis J, McGuffog L, Barrowdale D, Lee A, Healey S, Lush M, Tessier DC, Vincent D, Bacot F, Vergote I, Lambrechts S, Despierre E, Risch HA, González-Neira A, Rossing MA, Pita G, Doherty JA, Alvarez N, Larson MC, Fridley BL, Schoof N, Chang-Claude J, Cicek MS, Peto J, Kalli KR, Broeks A, Armasu SM, Schmidt MK, Braaf LM, Winterhoff B, Nevanlinna H, Konecny GE, Lambrechts D, Rogmann L, Guénel P, Teoman A, Milne RL, Garcia JJ, Cox A, Shridhar V, Burwinkel B, Marme F, Hein R, Sawyer EJ, Haiman CA, Wang-Gohrke S, Andrulis IL, Moysich KB, Hopper JL, Odunsi K, Lindblom A, Giles GG, Brenner H, Simard J, Lurie G, Fasching PA, Carney ME, Radice P, Wilkens LR, Swerdlow A, Goodman MT, Brauch H, Garcia-Closas M, Hillemanns P, Winqvist R, Dürst M, Devilee P, Runnebaum I, Jakubowska A, Lubinski J, Mannermaa A, Butzow R, Bogdanova NV, Dörk T, Pelttari LM, Zheng W, Leminen A, Anton-Culver H, Bunker CH, Kristensen V, Ness RB, Muir K, Edwards R, Meindl A, Heitz F, Matsuo K, du Bois A, Wu AH, Harter P, Teo SH, Schwaab I, Shu XO, Blot W, Hosono S, Kang D, Nakanishi T, Hartman M, Yatabe Y, Hamann U, Karlan BY, Sangrajrang S, Kjaer SK, Gaborieau V, Jensen A, Eccles D, Høgdall E, Shen CY, Brown J, Woo YL, Shah M, Azmi MA, Luben R, Omar SZ, Czene K, Vierkant RA, Nordestgaard BG, Flyger H, Vachon C, Olson JE, Wang X, Levine DA, Rudolph A, Weber RP, Flesch-Janys D, Iversen E, Nickels S, Schildkraut JM, Silva Idos S, Cramer DW, Gibson L, Terry KL, Fletcher O, Vitonis AF, van der Schoot CE, Poole EM, Hogervorst FB, Tworoger SS, Liu J, Bandera EV, Li J, Olson SH, Humphreys K, Orlow I, Blomqvist C, Rodriguez-Rodriguez L, Aittomäki K, Salvesen HB, Muranen TA, Wik E, Brouwers B, Krakstad C, Wauters E, Halle MK, Wildiers H, Kiemeney LA, Mulot C, Aben KK, Laurent-Puig P, Altena AM, Truong T, Massuger LF, Benitez J, Pejovic T, Perez JI, Hoatlin M, Zamora MP, Cook LS, Balasubramanian SP, Kelemen LE, Schneeweiss A, Le ND, Sohn C, Brooks-Wilson A, Tomlinson I, Kerin MJ, Miller N, Cybulski C, Henderson BE, Menkiszak J, Schumacher F, Wentzensen N, Le Marchand L, Yang HP, Mulligan AM, Glendon G, Engelholm SA, Knight JA, Høgdall CK, Apicella C, Gore M, Tsimiklis H, Song H, Southey MC, Jager A, den Ouweland AM, Brown R, Martens JW, Flanagan JM, Kriege M, Paul J, Margolin S, Siddiqui N, Severi G, Whittemore AS, Baglietto L, McGuire V, Stegmaier C, Sieh W, Müller H, Arndt V, Labrèche F, Gao YT, Goldberg MS, Yang G, Dumont M, McLaughlin JR, Hartmann A, Ekici AB, Beckmann MW, Phelan CM, Lux MP, Permuth-Wey J, Peissel B, Sellers TA, Ficarazzi F, Barile M, Ziogas A, Ashworth A, Gentry-Maharaj A, Jones M, Ramus SJ, Orr N, Menon U, Pearce CL, Brüning T, Pike MC, Ko YD, Lissowska J, Figueroa J, Kupryjanczyk J, Chanock SJ, Dansonka-Mieszkowska A, Jukkola-Vuorinen A, Rzepecka IK, Pylkäs K, Bidzinski M, Kauppila S, Hollestelle A, Seynaeve C, Tollenaar RA, Durda K, Jaworska K, Hartikainen JM, Kosma VM, Kataja V, Antonenkova NN, Long J, Shrubsole M, Deming-Halverson S, Lophatananon A, Siriwanarangsan P, Stewart-Brown S, Ditsch N, Lichtner P, Schmutzler RK, Ito H, Iwata H, Tajima K, Tseng CC, Stram DO, van den Berg D, Yip CH, Ikram MK, Teh YC, Cai H, Lu W, Signorello LB, Cai Q, Noh DY, Yoo KY, Miao H, Iau PT, Teo YY, McKay J, Shapiro C, Ademuyiwa F, Fountzilas G, Hsiung CN, Yu JC, Hou MF, Healey CS, Luccarini C, Peock S, Stoppa-Lyonnet D, Peterlongo P, Rebbeck TR, Piedmonte M, Singer CF, Friedman E, Thomassen M, Offit K, Hansen TV, Neuhausen SL, Szabo CI, Blanco I, Garber J, Narod SA, Weitzel JN, Montagna M, Olah E, Godwin AK, Yannoukakos D, Goldgar DE, Caldes T, Imyanitov EN, Tihomirova L, Arun BK, Campbell I, Mensenkamp AR, van Asperen CJ, van Roozendaal KE, Meijers-Heijboer H, Collée JM, Oosterwijk JC, Hooning MJ, Rookus MA, van der Luijt RB, Os TA, Evans DG, Frost D, Fineberg E, Barwell J, Walker L, Kennedy MJ, Platte R, Davidson R, Ellis SD, Cole T, Bressac-de Paillerets B, Buecher B, Damiola F, Faivre L, Frenay M, Sinilnikova OM, Caron O, Giraud S, Mazoyer S, Bonadona V, Caux-Moncoutier V, Toloczko-Grabarek A, Gronwald J, Byrski T, Spurdle AB, Bonanni B, Zaffaroni D, Giannini G, Bernard L, Dolcetti R, Manoukian S, Arnold N, Engel C, Deissler H, Rhiem K, Niederacher D, Plendl H, Sutter C, Wappenschmidt B, Borg A, Melin B, Rantala J, Soller M, Nathanson KL, Domchek SM, Rodriguez GC, Salani R, Kaulich DG, Tea MK, Paluch SS, Laitman Y, Skytte AB, Kruse TA, Jensen UB, Robson M, Gerdes AM, Ejlertsen B, Foretova L, Savage SA, Lester J, Soucy P, Kuchenbaecker KB, Olswold C, Cunningham JM, Slager S, Pankratz VS, Dicks E, Lakhani SR, Couch FJ, Hall P, Monteiro AN, Gayther SA, Pharoah PD, Reddel RR, Goode EL, Greene MH, Easton DF, Berchuck A, Antoniou AC, Chenevix-Trench G, Dunning AM Australian Cancer Study; Australian Ovarian Cancer Study; Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab); Gene Environment Interaction and Breast Cancer (GENICA); Swedish Breast Cancer Study (SWE-BRCA); Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON); Epidemiological study of BRCA1 & BRCA2 Mutation Carriers (EMBRACE); Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers (GEMO) Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–384. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mocellin S, Verdi D, Pooley KA, Landi MT, Egan KM, Baird DM, Prescptt J, De Vivo I, Nitti D. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst. 2012;104:840–854. doi: 10.1093/jnci/djs222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Research Genetic Associations and Mechanisms in Oncology (GAME-ON) A Network of Consortia for Post-Genome Wide Association (Post-GWA) National Cancer Institute Epidemiology and Genomics Research; 2015. [Accessed 11 Jan 2016]. ( http://epi.grants.cancer.gov/gameon/) [Google Scholar]
- 11.Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, Berndt SI, Bézieau S, Brenner H, Butterbach K, Caan BJ, Campbell PT, Carlson CS, Casey G, Chan AT, Chang-Claude J, Chanock SJ, Chen LS, Coetzee GA, Coetzee SG, Conti DV, Curtis KR, Duggan D, Edwards T, Fuchs CS, Gallinger S, Giovannucci EL, Gogarten SM, Gruber SB, Haile RW, Harrison TA, Hayes RB, Henderson BE, Hoffmeister M, Hopper JL, Hudson TJ, Hunter DJ, Jackson RD, Jee SH, Jenkins MA, Jia WH, Kolonel LN, Kooperberg C, Küry S, Lacroix AZ, Laurie CC, Laurie CA, Le Marchand L, Lemire M, Levine D, Lindor NM, Liu Y, Ma J, Makar KW, Matsuo K, Newcomb PA, Potter JD, Prentice RL, Qu C, Rohan T, Rosse SA, Schoen RE, Seminara D, Shrubsole M, Shu XO, Slattery ML, Taverna D, Thibodeau SN, Ulrich CM, White E, Xiang Y, Zanke BW, Zeng YX, Zhang B, Zheng W, Hsu L Colon Cancer Family Registry and the Genetics and Epidemiology of Colorectal Cancer Consortium. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144:799–807. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kachuri L, Amos CI, McKay JD, Johansson M, Vineis P, Bueno-de-Mesquita HB, Boutron-Ruault MC, Johansson M, Quirós JR, Sieri S, Travis RC, Weiderpass E, Le Marchand L, Henderson BE, Wilkens L, Goodman GE, Chen C, Doherty JA, Christiani DC, Wei Y, Su L, Tworoger S, Zhang X, Kraft P, Zaridze D, Field JK, Marcus MW, Davies MP, Hyde R, Caporaso NE, Landi MT, Severi G, Giles GG, Liu G, McLaughlin JR, Li Y, Xiao X, Fehringer G, Zong X, Denroche RE, Zuzarte PC, McPherson JD, Brennan P, Hung RJ. Cross Cancer Genomic Investigation of Inflammation Pathway Based on 64,411 cases and 89,922 Controls for Five Common Cancers. Hum Mol Genet. 2016;37:96–105. [Google Scholar]
- 13.Wang H, Burnett T, Kono S, Haiman CA, Iwasaki M, Wilkens LR, Loo LW, Van Den Berg D, Kolonel LN, Henderson BE, Keku TO, Sandler RS, Signorello LB, Blot WJ, Newcomb PA, Pande M, Amos CI, West DW, Bézieau S, Berndt SI, Zanke BW, Hsu L, Lindor NM, Haile RW, Hopper JL, Jenkins MA, Gallinger S, Casey G, Stenzel SL, Schumacher FR, Peters U, Gruber SB, Tsugane S, Stram DO, Le Marchand L Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO), Colon Cancer Family Registry (CCFR), Colorectal Transdisciplinary Study (CORECT) Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat Commun. 2014;5:4613. doi: 10.1038/ncomms5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siddiq A, Couch FJ, Chen GK, Lindström S, Eccles D, Millikan RC, Michailidou K, Stram DO, Beckmann L, Rhie SK, Ambrosone CB, Aittomäki K, Amiano P, Apicella C, Baglietto L, Bandera EV, Beckmann MW, Berg CD, Bernstein L, Blomqvist C, Brauch H, Brinton L, Bui QM, Buring JE, Buys SS, Campa D, Carpenter JE, Chasman DI, Chang-Claude J, Chen C, Clavel-Chapelon F, Cox A, Cross SS, Czene K, Deming SL, Diasio RB, Diver WR, Dunning AM, Durcan L, Ekici AB, Fasching PA, Feigelson HS, Fejerman L, Figueroa JD, Fletcher O, Flesch-Janys D, Gaudet MM, Gerty SM, Rodriguez-Gil JL, Giles GG, van Gils CH, Godwin AK, Graham N, Greco D, Hall P, Hankinson SE, Hartmann A, Hein R, Heinz J, Hoover RN, Hopper JL, Hu JJ, Huntsman S, Ingles SA, Irwanto A, Isaacs C, Jacobs KB, John EM, Justenhoven C, Kaaks R, Kolonel LN, Coetzee GA, Lathrop M, Le Marchand L, Lee AM, Lee IM, Lesnick T, Lichtner P, Liu J, Lund E, Makalic E, Martin NG, McLean CA, Meijers-Heijboer H, Meindl A, Miron P, Monroe KR, Montgomery GW, Müller-Myhsok B, Nickels S, Nyante SJ, Olswold C, Overvad K, Palli D, Park DJ, Palmer JR, Pathak H, Peto J, Pharoah P, Rahman N, Rivadeneira F, Schmidt DF, Schmutzler RK, Slager S, Southey MC, Stevens KN, Sinn HP, Press MF, Ross E, Riboli E, Ridker PM, Schumacher FR, Severi G, Dos Santos Silva I, Stone J, Sund M, Tapper WJ, Thun MJ, Travis RC, Turnbull C, Uitterlinden AG, Waisfisz Q, Wang X, Wang Z, Weaver J, Schulz-Wendtland R, Wilkens LR, Van Den Berg D, Zheng W, Ziegler RG, Ziv E, Nevanlinna H, Easton DF, Hunter DJ, Henderson BE, Chanock SJ, Garcia-Closas M, Kraft P, Haiman CA, Vachon CM Australian Breast Cancer Tissue Bank Investigators, Familial Breast Cancer Study, GENICA Consortium. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5573–5384. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, Le Marchand L, Buring JE, Eccles D, Miron P, Fasching PA, Brauch H, Chang-Claude J, Carpenter J, Godwin AK, Nevanlinna H, Giles GG, Cox A, Hopper JL, Bolla MK, Wang Q, Dennis J, Dicks E, Howat WJ, Schoof N, Bojesen SE, Lambrechts D, Broeks A, Andrulis IL, Guénel P, Burwinkel B, Sawyer EJ, Hollestelle A, Fletcher O, Winqvist R, Brenner H, Mannermaa A, Hamann U, Meindl A, Lindblom A, Zheng W, Devillee P, Goldberg MS, Lubinski J, Kristensen V, Swerdlow A, Anton-Culver H, Dörk T, Muir K, Matsuo K, Wu AH, Radice P, Teo SH, Shu XO, Blot W, Kang D, Hartman M, Sangrajrang S, Shen CY, Southey MC, Park DJ, Hammet F, Stone J, Veer LJ, Rutgers EJ, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Peto J, Schrauder MG, Ekici AB, Beckmann MW, Dos Santos Silva I, Johnson N, Warren H, Tomlinson I, Kerin MJ, Miller N, Marme F, Schneeweiss A, Sohn C, Truong T, Laurent-Puig P, Kerbrat P, Nordestgaard BG, Nielsen SF, Flyger H, Milne RL, Perez JI, Menéndez P, Müller H, Arndt V, Stegmaier C, Lichtner P, Lochmann M, Justenhoven C, Ko YD, Muranen TA, Aittomäki K, Blomqvist C, Greco D, Heikkinen T, Ito H, Iwata H, Yatabe Y, Antonenkova NN, Margolin S, Kataja V, Kosma VM, Hartikainen JM, Balleine R, Tseng CC, Berg DV, Stram DO, Neven P, Dieudonné AS, Leunen K, Rudolph A, Nickels S, Flesch-Janys D, Peterlongo P, Peissel B, Bernard L, Olson JE, Wang X, Stevens K, Severi G, Baglietto L, McLean C, Coetzee GA, Feng Y, Henderson BE, Schumacher F, Bogdanova NV, Labrèche F, Dumont M, Yip CH, Taib NA, Cheng CY, Shrubsole M, Long J, Pylkäs K, Jukkola-Vuorinen A, Kauppila S, Knight JA, Glendon G, Mulligan AM, Tollenaar RA, Seynaeve CM, Kriege M, Hooning MJ, van den Ouweland AM, van Deurzen CH, Lu W, Gao YT, Cai H, Balasubramanian SP, Cross SS, Reed MW, Signorello L, Cai Q, Shah M, Miao H, Chan CW, Chia KS, Jakubowska A, Jaworska K, Durda K, Hsiung CN, Wu PE, Yu JC, Ashworth A, Jones M, Tessier DC, González-Neira A, Pita G, Alonso MR, Vincent D, Bacot F, Ambrosone CB, Bandera EV, John EM, Chen GK, Hu JJ, Rodriguez-Gil JL, Bernstein L, Press MF, Ziegler RG, Millikan RM, Deming-Halverson SL, Nyante S, Ingles SA, Waisfisz Q, Tsimiklis H, Makalic E, Schmidt D, Bui M, Gibson L, Müller-Myhsok B, Schmutzler RK, Hein R, Dahmen N, Beckmann L, Aaltonen K, Czene K, Irwanto A, Liu J, Turnbull C, Rahman N, Meijers-Heijboer H, Uitterlinden AG, Rivadeneira F, Olswold C, Slager S, Pilarski R, Ademuyiwa F, Konstantopoulou I, Martin NG, Montgomery GW, Slamon DJ, Rauh C, Lux MP, Jud SM, Bruning T, Weaver J, Sharma P, Pathak H, Tapper W, Gerty S, Durcan L, Trichopoulos D, Tumino R, Peeters PH, Kaaks R, Campa D, Canzian F, Weiderpass E, Johansson M, Khaw KT, Travis R, Clavel-Chapelon F, Kolonel LN, Chen C, Beck A, Hankinson SE, Berg CD, Hoover RN, Lissowska J, Figueroa JD, Chasman DI, Gaudet MM, Diver WR, Willett WC, Hunter DJ, Simard J, Benitez J, Dunning AM, Sherman ME, Chenevix-Trench G, Chanock SJ, Hall P, Pharoah PD, Vachon C, Easton DF, Haiman CA, Kraft P Gene ENvironmental Interaction and breast CAncer (GENICA) Network, kConFab Investigators, Familial Breast Cancer Study (FBCS), Australian Breast Cancer Tissue Bank (ABCTB) Investigators. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–398. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michailidou K, En SE, Bolla MK, Wang Q, Dicks E, Lee A, Turnbull C, Rahman N, Fletcher O, Peto J, Gibson L, Dos Santos Silva I, Nevanlinna H, Muranen TA, Aittomäki K, Blomqvist C, Czene K, Irwanto A, Liu J, Waisfisz Q, Meijers-Heijboer H, Adank M, van der Luijt RB, Hein R, Dahmen N, Beckman L, Meindl A, Schmutzler RK, Müller-Myhsok B, Lichtner P, Hopper JL, Southey MC, Makalic E, Schmidt DF, Uitterlinden AG, Hofman A, Hunter DJ, Chanock SJ, Vincent D, Bacot F, Tessier DC, Canisius S, Wessels LF, Haiman CA, Shah M, Luben R, Brown J, Luccarini C, Schoof N, Humphreys K, Li J, Nordestgaard BG, Nielsen SF, Flyger H, Couch FJ, Wang X, Vachon C, Stevens KN, Lambrechts D, Moisse M, Paridaens R, Christiaens MR, Rudolph A, Nickels S, Flesch-Janys D, Johnson N, Aitken Z, Aaltonen K, Heikkinen T, Broeks A, Veer LJ, van der Schoot CE, Guénel P, Truong T, Laurent-Puig P, Menegaux F, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Zamora MP, Perez JI, Pita G, Alonso MR, Cox A, Brock IW, Cross SS, Reed MW, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Henderson BE, Schumacher F, Le Marchand L, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Lindblom A, Margolin S, Hooning MJ, Hollestelle A, van den Ouweland AM, Jager A, Bui QM, Stone J, Dite GS, Apicella C, Tsimiklis H, Giles GG, Severi G, Baglietto L, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Brenner H, Müller H, Arndt V, Stegmaier C, Swerdlow A, Ashworth A, Orr N, Jones M, Figueroa J, Lissowska J, Brinton L, Goldberg MS, Labrèche F, Dumont M, Winqvist R, Pylkäs K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Brüning T, Radice P, Peterlongo P, Manoukian S, Bonanni B, Devilee P, Tollenaar RA, Seynaeve C, van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Durda K, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Bogdanova NV, Antonenkova NN, Dörk T, Kristensen VN, Anton-Culver H, Slager S, Toland AE, Edge S, Fostira F, Kang D, Yoo KY, Noh DY, Matsuo K, Ito H, Iwata H, Sueta A, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Shu XO, Lu W, Gao YT, Cai H, Teo SH, Yip CH, Phuah SY, Cornes BK, Hartman M, Miao H, Lim WY, Sng JH, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Shen CY, Hsiung CN, Wu PE, Ding SL, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Blot WJ, Signorello LB, Cai Q, Zheng W, Deming-Halverson S, Shrubsole M, Long J, Simard J, Garcia-Closas M, Pharoah PD, Chenevix-Trench G, Dunning AM, Benitez J, Easton DF Breast and Ovarian Cancer Susceptibility Collaboration, Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON), kConFab Investigators; Australian Ovarian Cancer Study Group, GENICA (Gene Environment Interaction and Breast Cancer in Germany) Network. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amin Al Olama A, Kote-Jarai Z, Schumacher FR, Wiklund F, Berndt SI, Benlloch S, Giles GG, Severi G, Neal DE, Hamdy FC, Donovan JL, Hunter DJ, Henderson BE, Thun MJ, Gaziano M, Giovannucci EL, Siddiq A, Travis RC, Cox DG, Canzian F, Riboli E, Key TJ, Andriole G, Albanes D, Hayes RB, Schleutker J, Auvinen A, Tammela TL, Weischer M, Stanford JL, Ostrander EA, Cybulski C, Lubinski J, Thibodeau SN, Schaid DJ, Sorensen KD, Batra J, Clements JA, Chambers S, Aitken J, Gardiner RA, Maier C, Vogel W, Dörk T, Brenner H, Habuchi T, Ingles S, John EM, Dickinson JL, Cannon-Albright L, Teixeira MR, Kaneva R, Zhang HW, Lu YJ, Park JY, Cooney KA, Muir KR, Leongamornlert DA, Saunders E, Tymrakiewicz M, Mahmud N, Guy M, Govindasami K, O’Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English D, Virtamo J, Le Marchand L, Campa D, Kaaks R, Lindstrom S, Diver WR, Gapstur S, Yeager M, Cox A, Stern MC, Corral R, Aly M, Isaacs W, Adolfsson J, Xu J, Zheng SL, Wahlfors T, Taari K, Kujala P, Klarskov P, Nordestgaard BG, Røder MA, Frikke-Schmidt R, Bojesen SE, FitzGerald LM, Kolb S, Kwon EM, Karyadi DM, Orntoft TF, Borre M, Rinckleb A, Luedeke M, Herkommer K, Meyer A, Serth J, Marthick JR, Patterson B, Wokolorczyk D, Spurdle A, Lose F, McDonnell SK, Joshi AD, Shahabi A, Pinto P, Santos J, Ray A, Sellers TA, Lin HY, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Tsuchiya N, Narita S, Cao GW, Slavov C, Mitev V, Chanock S, Gronberg H, Haiman CA, Kraft P, Easton DF, Eeles RA UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; UK ProtecT Study Collaborators; Australian Prostate Cancer Bioresource; PRACTICAL Consortium. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet Mol. 2013;22:408–415. doi: 10.1093/hmg/dds425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pharoah PDP, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H, Weber R, Karevan R, Larson MC, Song H, Tessier DC, Bacot F, Vincent D, Cunningham JM, Dennis J, Dicks E, Aben KK, Anton-Culver H, Antonenkova N, Armasu SM, Baglietto L, Bandera EV, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brenton JD, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Campbell I, Carney ME, Carvalho RS, Chang-Claude J, Chen YA, Chen Z, Chow WH, Cicek MS, Coetzee G, Cook LS, Cramer DW, Cybulski C, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dörk T, du Bois A, Dürst M, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher D, Flanagan J, Gao YT, Garcia-Closas M, Gentry-Maharaj A, Giles G, Gjyshi A, Gore M, Gronwald J, Guo Q, Halle MK, Harter P, Hein A, Heitz F, Hillemanns P, Hoatlin M, Høgdall E, Høgdall CK, Hosono S, Jakubowska A, Jensen A, Kalli KR, Karlan BY, Kelemen LE, Kiemeney LA, Kjaer SK, Konecny GE, Krakstad C, Kupryjanczyk J, Lambrechts D, Lambrechts S, Le ND, Lee N, Lee J, Leminen A, Lim BK, Lissowska J, Lubiński J, Lundvall L, Lurie G, Massuger LF, Matsuo K, McGuire V, McLaughlin JR, Menon U, Modugno F, Moysich KB, Nakanishi T, Narod SA, Ness RB, Nevanlinna H, Nickels S, Noushmehr H, Odunsi K, Olson S, Orlow I, Paul J, Pejovic T, Pelttari LM, Permuth-Wey J, Pike MC, Poole EM, Qu X, Risch HA, Rodriguez-Rodriguez L, Rossing MA, Rudolph A, Runnebaum I, Rzepecka IK, Salvesen HB, Schwaab I, Severi G, Shen H, Shridhar V, Shu XO, Sieh W, Southey MC, Spellman P, Tajima K, Teo SH, Terry KL, Thompson PJ, Timorek A, Tworoger SS, van Altena AM, van den Berg D, Vergote I, Vierkant RA, Vitonis AF, Wang-Gohrke S, Wentzensen N, Whittemore AS, Wik E, Winterhoff B, Woo YL, Wu AH, Yang HP, Zheng W, Ziogas A, Zulkifli F, Goodman MT, Hall P, Easton DF, Pearce CL, Berchuck A, Chenevix-Trench G, Iversen E, Monteiro AN, Gayther SA, Schildkraut JM, Sellers TA Australian Cancer Study; Australian Ovarian Cancer Study Group. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45:362–370. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeböller H, Risch A, McKay JD, Wang Y, Dai J, Gaborieau V, McLaughlin J, Brenner D, Narod SA, Caporaso NE, Albanes D, Thun M, Eisen T, Wichmann HE, Rosenberger A, Han Y, Chen W, Zhu D, Spitz M, Wu X, Pande M, Zhao Y, Zaridze D, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Krokan HE, Gabrielsen ME, Skorpen F, Vatten L, Njølstad I, Chen C, Goodman G, Lathrop M, Benhamou S, Vooder T, Välk K, Nelis M, Metspalu A, Raji O, Chen Y, Gosney J, Liloglou T, Muley T, Dienemann H, Thorleifsson G, Shen H, Stefansson K, Brennan P, Amos CI, Houlston R, Landi MT Transdisciplinary Research in Cancer of the Lung (TRICL) Research Team. Influence of common genetic variation on lung cancer risk : meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980–4995. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.PubMed Gene. National Center for Biotechnology Information, U.S. National Library of Medicine; n.d. [Accessed 20 Jan 2015]. ( www.ncbi.nlm.nih.gov/gene) [Google Scholar]
- 21.Bhattacharjee S, Rajaraman P, Jacobs KB, Wheeler WA, Melin BS, Hartge P, Yeager M, Chung CC, Chanock SJ, Chatterjee N GliomaScan Consortium. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet. 2012;90:821–835. doi: 10.1016/j.ajhg.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra A, Macgregor S. VEGAS2: Software for More Flexible Gene-Based Testing. Twin Res Hum Genet. 2015;18:86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genetic Markers of Susceptibility (CGEMS) Project. National Cancer Institute Division of Cancer Epidemiology & Genetics; n.d. [Accessed 10 March 2015]. ( http://dceg.cancer.gov/research/how-we-study/genomic-studies/cgems-summary) [Google Scholar]
- 24.Environment and Genes in Lung cancer Etiology (EAGLE) National Cancer Institute Division of Cancer Epidemiology & Genetics; n.d. [Accessed 10 March 2015]. ( http://dceg.cancer.gov/research/cancer-types/lung/environment-genes-lung-cancer-etiology-eagle) [Google Scholar]
- 25.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 26.1000 Genomes. [Accessed 10 March 2015];A Deep Catalog of Human Genetic Variation. 2008–2012 ( http://www.1000genomes.org/)
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Li MX, Yeung JMY, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, Frayling TM, McCarthy MI, Hirschhorn JN, Goddard ME, Visscher PM Genetic Investigation of ANthropometric Traits (GIANT) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward L, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database Issue):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, Campa D, Riboli E, Key T, Gronberg H, Hunter DJ, Kraft P, Thun MJ, Ingles S, Chanock S, Albanes D, Hayes RB, Neal DE, Hamdy FC, Donovan JL, Pharoah P, Schumacher F, Henderson BE, Stanford JL, Ostrander EA, Sorensen KD, Dörk T, Andriole G, Dickinson JL, Cybulski C, Lubinski J, Spurdle A, Clements JA, Chambers S, Aitken J, Gardiner RA, Thibodeau SN, Schaid D, John EM, Maier C, Vogel W, Cooney KA, Park JY, Cannon-Albright L, Brenner H, Habuchi T, Zhang HW, Lu YJ, Kaneva R, Muir K, Benlloch S, Leongamornlert DA, Saunders EJ, Tymrakiewicz M, Mahmud N, Guy M, O’Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English DR, Wahlfors T, Tammela TL, Klarskov P, Nordestgaard BG, Røder MA, Tybjærg-Hansen A, Bojesen SE, Travis R, Canzian F, Kaaks R, Wiklund F, Aly M, Lindstrom S, Diver WR, Gapstur S, Stern MC, Corral R, Virtamo J, Cox A, Haiman CA, Le Marchand L, Fitzgerald L, Kolb S, Kwon EM, Karyadi DM, Orntoft TF, Borre M, Meyer A, Serth J, Yeager M, Berndt SI, Marthick JR, Patterson B, Wokolorczyk D, Batra J, Lose F, McDonnell SK, Joshi AD, Shahabi A, Rinckleb AE, Ray A, Sellers TA, Lin HY, Stephenson RA, Farnham J, Muller H, Rothenbacher D, Tsuchiya N, Narita S, Cao GW, Slavov C, Mitev V, Easton DF, Eeles RA UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; UK ProtecT Study Collaborators, The Australian Prostate Cancer BioResource; PRACTICAL Consortium. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43:785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diotti R, Loayza D. Shelterin complex and associated factors at human telomeres. Nucleus. 2011;2:119–135. doi: 10.4161/nucl.2.2.15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlton VEH, Hu X, Chokkalingam AP, Schrodi SJ, Brandon R, Alexander HC, Chang M, Catanese JJ, Leong DU, Ardlie KG, Kastner DL, Seldin MF, Criswell LA, Gregersen PK, Beasley E, Thomson G, Amos CI, Begovich AB. PTPN22 genetic variation: evidence for multiple variants associated with rheumatoid arthritis. Am J Hum Genet. 2005;77:567–581. doi: 10.1086/468189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue L, Pan C, Gu Z, Zhao S, Han B, Liu W, Yang S, Yu S, Sun Y, Liang J, Gao G, Zhang X, Yuan G, Li C, Du W, Chen G, Chen J, Song H. Genetic heterogeneity of susceptibility gene in different ethnic populations: Refining association study of PTPN22 for Graves’ disease in a Chinese Han population. PLoS One. 2013;8:e84514. doi: 10.1371/journal.pone.0084514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song F, Amos C, Lee J, Lian C, Fang S, Liu H, MacGregor S, Iles MM, Law MH, Lindeman NI, Montgomery GW, Duffy DL, Cust AE, Jenkins MA, Whiteman DC, Kefford RF, Giles GG, Armstrong BK, Aitken JF, Hopper JL, Brown KM, Martin NG, Mann GJ, Bishop DT, Bishop JA, Kraft P, Qureshi AA, Kanetsky PA, Hayward NK, Hunter DJ, Wei Q, Han J GenoMEL consortium. Identification of a melanoma susceptibility locus and somatic mutation in TET2. Carcinogenesis. 2014;35:2097–2101. doi: 10.1093/carcin/bgu140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, Kollmeyer TM, Kosel ML, LaChance DH, McCoy L, O’Neill BP, Patoka J, Pico AR, Prados M, Quesenberry C, Rice T, Rynearson AL, Smirnov I, Tihan T, Wiemels J, Yang P, Wiencke JK. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Figueroa JD, Ye Y, Siddiq A, Garcia-Closas M, Chatterjee N, Prokunina-Olsson L, Cortessis VK, Kooperberg C, Cussenot O, Benhamou S, Prescott J, Porru S, Dinney CP, Malats N, Baris D, Purdue M, Jacobs EJ, Albanes D, Wang Z, Deng X, Chung CC, Tang W, Bas Bueno-de-Mesquita H, Trichopoulos D, Ljungberg B, Clavel-Chapelon F, Weiderpass E, Krogh V, Dorronsoro M, Travis R, Tjønneland A, Brenan P, Chang-Claude J, Riboli E, Conti D, Gago-Dominguez M, Stern MC, Pike MC, Van Den Berg D, Yuan JM, Hohensee C, Rodabough R, Cancel-Tassin G, Roupret M, Comperat E, Chen C, De Vivo I, Giovannucci E, Hunter DJ, Kraft P, Lindstrom S, Carta A, Pavanello S, Arici C, Mastrangelo G, Kamat AM, Lerner SP, Barton Grossman H, Lin J, Gu J, Pu X, Hutchinson A, Burdette L, Wheeler W, Kogevinas M, Tardón A, Serra C, Carrato A, García-Closas R, Lloreta J, Schwenn M, Karagas MR, Johnson A, Schned A, Armenti KR, Hosain GM, Andriole G, Jr, Grubb R, 3rd, Black A, Ryan Diver W, Gapstur SM, Weinstein SJ, Virtamo J, Haiman CA, Landi MT, Caporaso N, Fraumeni JF, Jr, Vineis P, Wu X, Silverman DT, Chanock S, Rothman N. Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum Mol Genet. 2014;23:1387–1398. doi: 10.1093/hmg/ddt519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Chandler I, Vijayakrishnan J, Sullivan K, Penegar S, Carvajal-Carmona L, Howarth K, Jaeger E, Spain SL, Walther A, Barclay E, Martin L, Gorman M, Domingo E, Teixeira AS, Kerr D, Cazier JB, Niittymäki I, Tuupanen S, Karhu A, Aaltonen LA, Tomlinson IP, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Cetnarskyj R, Porteous ME, Pharoah PD, Koessler T, Hampe J, Buch S, Schafmayer C, Tepel J, Schreiber S, Völzke H, Chang-Claude J, Hoffmeister M, Brenner H, Zanke BW, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG Colorectal Cancer Association Study Consortium, CoRGI Consortium. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bei J-X, Li Y, Jia WH, Feng BJ, Zhou G, Chen LZ, Feng QS, Low HQ, Zhang H, He F, Tai ES, Kang T, Liu ET, Liu J, Zeng YX. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42:599–603. doi: 10.1038/ng.601. [DOI] [PubMed] [Google Scholar]
- 41.Speedy HE, Di Bernardo MC, Sava GP, Dyer MJ, Holroyd A, Wang Y, Sunter NJ, Mansouri L, Juliusson G, Smedby KE, Roos G, Jayne S, Majid A, Dearden C, Hall AG, Mainou-Fowler T, Jackson GH, Summerfield G, Harris RJ, Pettitt AR, Allsup DJ, Bailey JR, Pratt G, Pepper C, Fegan C, Rosenquist R, Catovsky D, Allan JM, Houlston RS. A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2014;46:56–60. doi: 10.1038/ng.2843. [DOI] [PubMed] [Google Scholar]
- 42.Chubb D, Weinhold N, Broderick P, Chen B, Johnson DC, Försti A, Vijayakrishnan J, Migliorini G, Dobbins SE, Holroyd A, Hose D, Walker BA, Davies FE, Gregory WA, Jackson GH, Irving JA, Pratt G, Fegan C, Fenton JA, Neben K, Hoffmann P, Nöthen MM, Mühleisen TW, Eisele L, Ross FM, Straka C, Einsele H, Langer C, Dörner E, Allan JM, Jauch A, Morgan GJ, Hemminki K, Houlston RS, Goldschmidt H. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13. 1 influences multiple myeloma risk. Nat Genet. 2013;45:1221–1225. doi: 10.1038/ng.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han Y, Hazelett DJ, Wiklund F, Schumacher FR, Stram DO, Berndt SI, Wang Z, Rand KA, Hoover RN, Machiela MJ, Yeager M, Burdette L, Chung CC, Hutchinson A, Yu K, Xu J, Travis RC, Key TJ, Siddiq A, Canzian F, Takahashi A, Kubo M, Stanford JL, Kolb S, Gapstur SM, Diver WR, Stevens VL, Strom SS, Pettaway CA, Al Olama AA, Kote-Jarai Z, Eeles RA, Yeboah ED, Tettey Y, Biritwum RB, Adjei AA, Tay E, Truelove A, Niwa S, Chokkalingam AP, Isaacs WB, Chen C, Lindstrom S, Le Marchand L, Giovannucci EL, Pomerantz M, Long H, Li F, Ma J, Stampfer M, John EM, Ingles SA, Kittles RA, Murphy AB, Blot WJ, Signorello LB, Zheng W, Albanes D, Virtamo J, Weinstein S, Nemesure B, Carpten J, Leske MC, Wu SY, Hennis AJ, Rybicki BA, Neslund-Dudas C, Hsing AW, Chu L, Goodman PJ, Klein EA, Zheng SL, Witte JS, Casey G, Riboli E, Li Q, Freedman ML, Hunter DJ, Gronberg H, Cook MB, Nakagawa H, Kraft P, Chanock SJ, Easton DF, Henderson BE, Coetzee GA, Conti DV, Haiman CA. Integration of multiethnic fine-mapping and genomic annotation to prioritize candidate functional SNPs at prostate cancer susceptibility regions. Hum Mol Genet. 2015;24:5603–5618. doi: 10.1093/hmg/ddv269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia J, Bosley AD, Thompson A, Hoskins JW, Cheuk A, Collins I, Parikh H, Xiao Z, Ylaya K, Dzyadyk M, Cozen W, Hernandez BY, Lynch CF, Loncarek J, Altekruse SF, Zhang L, Westlake CJ, Factor VM, Thorgeirsson S, Bamlet WR, Hewitt SM, Petersen GM, Andresson T, Amundadottir LT. Clptm1l promotes growth and enhances aneuploidy in pancreatic cancer cells. Cancer Res. 2014;74:2785–2795. doi: 10.1158/0008-5472.CAN-13-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung RJ, Ulrich CM, Goode EL, Brhane Y, Muir K, Chan AT, Marchand LL, Schildkraut J, Witte JS, Eeles R, Boffetta P, Spitz MR, Poirier JG, Rider DN, Fridley BL, Chen Z, Haiman C, Schumacher F, Easton DF, Landi MT, Brennan P, Houlston R, Christiani DC, Field JK, Bickeböller H, Risch A, Kote-Jarai Z, Wiklund F, Grönberg H, Chanock S, Berndt SI, Kraft P, Lindström S, Al Olama AA, Song H, Phelan C, Wentzensen N, Peters U, Slattery ML, Sellers TA, Casey G, Gruber SB, Hunter DJ, Amos CI, Henderson B GECCO, FOCI, CORECT, DRIVE, GAME-ON Network. Cross Cancer Genomic Investigation of Inflammation Pathway for Five Common Cancers: Lung, Ovary, Prostate, Breast, and Colorectal Cancer. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/djv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, Li Z, Deng Q, Wang J, Wu W, Jin G, Jiang Y, Yu D, Zhou G, Chen H, Guan P, Chen Y, Shu Y, Xu L, Liu X, Liu L, Xu P, Han B, Bai C, Zhao Y, Zhang H, Yan Y, Ma H, Chen J, Chu M, Lu F, Zhang Z, Chen F, Wang X, Jin L, Lu J, Zhou B, Lu D, Wu T, Lin D, Shen H. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12. 2 in Han Chinese. Nat Genet. 2011;43:792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 47.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, Idbaih A, Zelenika D, Andersson U, Henriksson R, Bergenheim AT, Feychting M, Lönn S, Ahlbom A, Schramm J, Linnebank M, Hemminki K, Kumar R, Hepworth SJ, Price A, Armstrong G, Liu Y, Gu X, Yu R, Lau C, Schoemaker M, Muir K, Swerdlow A, Lathrop M, Bondy M, Houlston RS. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, Ricketts M, Linger R, Nsengimana J, Deloukas P, Huddart RA, Bishop DT, Easton DF, Stratton MR, Rahman N UK Testicular Cancer Collaboration. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–607. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen HH, Takata R, Akamatsu S, Shigemizu D, Tsunoda T, Furihata M, Takahashi A, Kubo M, Kamatani N, Ogawa O, Fujioka T, Nakamura Y, Nakagawa H. IRX4 at 5p15 suppresses prostate cancer growth through the interaction with vitamin D receptor, conferring prostate cancer susceptibility. Hum Mol Genet. 2010;21:2076–2085. doi: 10.1093/hmg/dds025. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Wei Y, Gaborieau V, Shi J, Han Y, Timofeeva MN, Su L, Li Y, Eisen T, Amos CI, Landi MT, Christiani DC, McKay JD, Houlston RS. Deciphering associations for lung cancer risk through imputation and analysis of 12316 cases and 16831 controls. Eur J Hum Genet. 2015;23:1723–1728. doi: 10.1038/ejhg.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Manhattan plot of associations between 204,993 SNPs in telomere structure and maintenance genes and five cancer types.
The dotted line indicates the genome-wide significance level (P-value=5×10−8), and the individual solid lines indicate the gene-level P-value cutoffs taking into account the number of effective tests. Genes are evaluated in the order listed in the key.
Supplementary Figure 2. TERT-CLPTM1L and TERC LD plots for statistically significant ASSET variants that were retained following LD pruning at r2>0.70.
Associations between alleles across variants are based on CGEMS and EAGLE datasets. SNPs circled in black are variants identified as statistically significant in ASSET conditional analyses. aSNPs identified as statistically significant in ASSET conditional analyses in the Wang et al. study among individuals of European ancestry [7]. A) LD plot for TERT-CLPTM1L variants. B) LD plot for TERC variant.