Abstract
Activation of complex molecular programs in specific cell lineages governs mammalian heart development, from a primordial linear tube to a four-chamber organ. To characterize lineage-specific, temporal-spatial developmental programs, we performed single-cell RNA sequencing of >1200 murine cells isolated at seven time points spanning E9.5 (primordial heart tube) to P21 (mature heart). Using unbiased transcriptional data we classified cardiomyocytes (CM), endothelial cells (EC), and fibroblast-enriched cells, thus identifying markers for temporal and chamber-specific developmental programs. Harnessing these datasets, we defined developmental ages of human and mouse pluripotent stem cell-derived CMs and characterized lineage-specific maturation defects in hearts of mice with heterozygous mutations in Nkx2.5 that cause human heart malformations. This spatial-temporal transcriptome analysis of heart development reveals lineage-specific gene programs underlying normal cardiac development and congenital heart disease.
Introduction
Formation and maturation of the mammalian heart is governed by signaling interactions among distinct cell lineages during development that transform a linear heart tube into four-chamber heart. Transcriptional profiles of human, mouse, and chick hearts have identified dynamic changes in RNA expression throughout cardiogenesis Jensen et al. (2013); (Wagner and Siddiqui, 2007). Understanding how these relate to cellular differentiation and maturation remains incompletely understood, in part because of the inability of conventional transcriptome analysis experiments to de-convolute the multiple cell populations within the heart. Recent advances in single-cell transcription profiling approaches enable rapid capture and profiling of hundreds of cells (Buettner et al., 2015; Streets and Huang, 2014; Trapnell et al., 2014; Treutlein et al., 2014) that allow separation and characterization of lineages and cell lineage subtypes, independent of preexisting cell markers. Longitudinal assessments of transcriptional maps provide information that can be assembled into a transcriptional atlas across development or in response to disease.
To investigate cellular heterogeneity during cardiogenesis, we characterized the developmental temporal and spatial transcriptomes of heart development at single cell resolution. Wildtype (WT) murine cells isolated from the left atria (LA), primordial ventricle, and subsequently left (LV) and right (RV) ventricles at time points spanning embryonic to post-natal cardiac development were studied using single-cell RNA sequencing (RNA-seq). Cells were classified using unbiased transcriptome-wide analysis without a priori selection of cell-specific markers. We identified transcripts that delineated atrial and ventricular CMs, and ECs and fibroblast-enriched cell lineages. We also characterized CMs derived from stem cells and CMs and ECs derived from a congenital heart disease mouse model with haploinsufficiency of Nkx2.5, a critical transcription factor that is expressed early in cardiogenesis (Ashraf et al., 2014; Biben et al., 2000; Bruneau et al., 2001).
Single cell transcriptional data of distinct cardiac lineages provided insights into lineage specific markers and subpopulations that contribute to normal and aberrant cardiac development and provided a robust benchmark to assess the maturity of stem cell-derived CMs.
Results
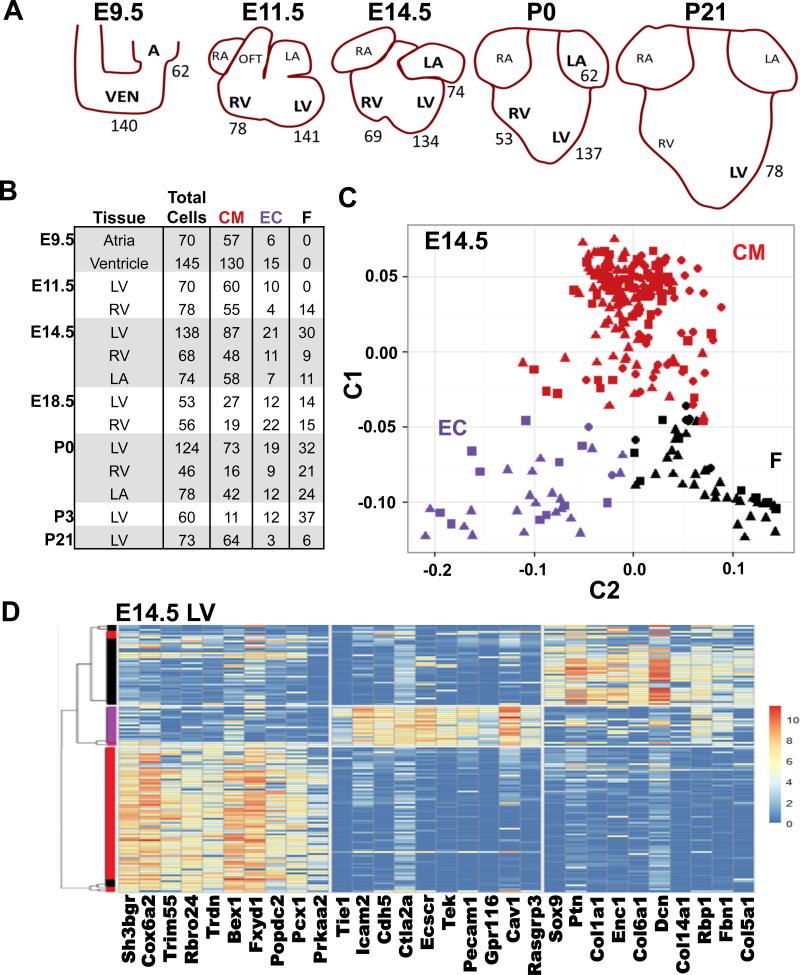
We captured mouse (SV129) cardiac cells during important embryonic (E) developmental milestones (Fig 1A): E9.5 during heart looping, when the atria and ventricle become morphologically distinct; E11.5 when left and right cardiac chambers form, E14.5 when septation occurs with development of compact and trabecular layers of myocardium; E18.5 when the heart undergoes metabolic remodeling. Cells were also captured at birth (P0) when the left atria and ventricle experience hemodynamic changes due to higher systemic pressures, and at P3, P7, and P21, when the heart transitions from proliferative growth (P3) to hypertrophic growth (P21).
Figure 1.
Single-cell RNA-seq of CMs, ECs, and fibroblast-enriched cells in the developing heart. A) Schematic representation of developing heart from which single cells were harvested (denoted by bold lettering). Numbers indicate numbers of captured wildtype cells. B) The number single cells captured per tissue and time point and their lineage as determined by PCA. CM, cardiomyocytes, EC, endothelial cells, F, fibroblast-enriched cells. C) PCA components C1 and C2 separates E14.5 cells into three subgroups, each that robustly expressed genes associated with CMs (red), ECs (E, purple), or Fs (black). The percent of variance explained each principle component is provided in Table S2. D) Unsupervised clustering (SC3) of E14.5 cells separates CMs, ECs, and fibroblast-enriched cells. The dendrogram was generated using consensus clustering with k=3. The heatmap depicts the top ten most significant genes with enriched expression in each cluster. See also Fig S1, Fig S4, Table S1, Table S2, and Table S3.
The common atria and ventricle (E.9.5) were carefully dissected away from the adjacent atrioventricular canal or outflow tract (OFT), and at E14.5-P21, the free wall was dissected from the LV and RV to exclude any valve or septal tissue. Tissues were digested into single cell suspensions and isolated in a microfluidic device, after which single-cell RNA-seq analyses were performed. To assess technical variation and the dynamic range of transcript data we compared and observed good correlation of data derived from pooled single cell RNA-seq and whole LV RNA-seq. (Fig S1). Consistent with studies of single cells from other tissues, we found high single-transcript sensitivity and detected transcripts in a broad dynamic range (~105) between the most and least abundant transcripts.
Unbiased transcriptome-wide identification of cell populations
We studied over 1200 single cells isolated at different stages and chambers of murine heart development (Fig 1A-B, Table S1). To demonstrate that single cell RNA-seq could distinguish distinct cell lineages in the heart, we employed Principal Component Analysis (PCA) of all single cells from LV, RV, and LA tissues captured at E14.5 (261 cells) when the four-chambered heart is formed. Informative PCA components (Table S2) clustered cells into three groups (Fig 1C). Examining transcripts that were enriched in each of the three groups revealed that this unbiased clustering method faithfully identified previous markers known to be expressed in CMs (red), ECs (purple), or fibroblast-enriched cells (black, e.g., non-CM, non-ECs). For example, among the top 30 genes in PCA component 1 that explain the largest proportion of the variation (Table S2; see Methods), CMs contained 11 sarcomere proteins genes (e.g. Myh6, Myh7, Ttn), ECs contained 14 genes encoding cell surface markers (e.g. Pecam1, Cd93), and fibroblast-enriched cells contained nine genes encoding extracellular matrix components (e.g. Col1a2, Col1a1).
To independently confirm cell classifications, unsupervised clustering was undertaken using the Single-Cell Consensus Clustering (SC3) tool (Kiselev VY, 2016). SC3 uses a consensus clustering approach that integrates several methods for clustering of single-cell RNA-seq data to generate user-defined numbers of clusters (k). Assessments of variable numbers of clusters demonstrated that k=3 and k=6 maximized the silhouette width metric, which quantifies the ratio of each cell's transcriptional similarities with other co-clustered cells to the transcriptional differences with cells outside of its cluster (Rousseeuw, 1987). This independent approach yielded cell clusters that closely mirrored PCA method (Fig 1B) with enriched expression of genes associated with CMs, endothelial cells, or fibroblast-enriched cells (Fig 1D).
We undertook “perfect marker” gene analysis of PCA-separated cells (Treutlein et al., 2014) to identify markers for CMs, ECs, and fibroblast-enriched cells (Table S3). This approach identifies genes with high expression in specified cells and low expression in all other cells. “Perfect marker” gene analyses of PCA-separated CMs identified eight sarcomere protein genes (Table S3, bolded) and Sh3bgr, which encodes a SH3 domain binding glutamate-rich protein encoded within the trisomy chromosome 21 region that contributes to congenital heart defects. Morpholino-mediated knockdown of Sh3bgr in Xenopus, results in disrupted segmentation of somites and heart malformations (Jang et al., 2015). Eight “perfect marker” genes for ECs encoded prototypic molecules (Pecam1, Cdh5) and two additional genes: Klhl4 and Gpr116 (Table S3, italicized). Klhl4 encodes an actin-binding kelch domain protein with unknown functions that has been implicated in developmental anomalies (Braybrook et al., 2001; Cheroki et al., 2008). Notably, an automated in situ hybridization screen (Gene Expression Database (Diez-Roux et al., 2011; Smith et al., 2014) of the E14.5 heart also identified Klhl4 in the endocardium. Gpr116 encodes a highly conserved G-protein coupled adhesion receptor that is widely expressed in vascular beds, that when depleted from ECs produced vascular leakage (Niaudet et al., 2015; Yang et al., 2013). Six of ten “perfect” marker genes identified in fibroblast-enriched cells encoded ECM proteins (Table S3, underlined).
We then evaluated the distribution and maturation of CMs, ECs, and fibroblast-enriched cells throughout development. At E9.5, no fibroblast-enriched cells were identified in atria, ventricle or outflow tract, but subsequently, the proportion of CMs in these region decreased, while fibroblast-enriched cells increased (Fig 1B). At P0 approximately, 30% of all LV cells and 45% of RV cells were fibroblast-enriched cells, a finding that is consistent with prior reports that cardiac fibroblasts appear by E12.5 and increase in number through P1 (Ieda et al., 2009). The proportion of ECs remained relatively constant from E9.5 (10%) to P0 (15%). At P21, the larger chip size used to capture CMs excluded many smaller non-CMs (Table S1), including hematopoietic-derived cells that comprise 5-10% of non-CM cells in the adult mouse heart (Pinto et al., 2016). These cells were also not captured in embryonic hearts, likely due to their low abundance.
Among developing ventricular CMs, RNAseq profiles were indicative of chamber myocardium and included two distinct sub-populations. One sub-population, (identified through consensus clustering using SC3 k=6), expressed transcripts associated with cell division (Fig S2A). Using hierarchical clustering of the expression of cell cycle genes (e.g., Prc1, Ccna2, Cdca8, Cdca3, Top2a, Ccnb2, Mki67, Ccnb1) we identified the proportion of CMs with proliferative capacity during development (Fig S2B). Approximately 60% of E9.5 and E14.5 CMs, but only 20% of P0 and P7 CMs, expressed these cell cycle transcripts (Fig S2B). At P21, this subpopulation was absent.
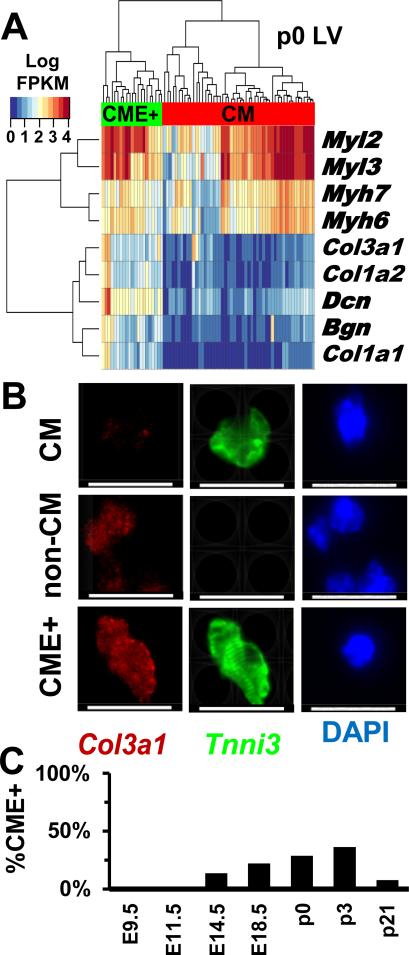
A second CM subpopulation was identified by hierarchical clustering (Fig 2A) of marker genes for CMs and fibroblast-enriched cells (identified in Fig 1C and listed in Table S2). This CM subpopulation expressed transcripts encoding extracellular matrix (ECM) proteins (denoted CME+) that are typically associated with cardiac fibroblasts (e.g. Col1a1, Col3a1, Dcn). RaceID, which identifies rare cell populations in complex data, (Grun et al., 2015) also defined cluster of cells that expressed both sarcomere components and ECM (Fig S2C: cluster 7). To determine if these cells were more similar to either CMs or fibroblast-enriched cells, gene expressions for all CMs, fibroblast-enriched cells, or CME+ were averaged at E14.5, E18.5, or P0 (Fig S1C). Comparisons of the mean gene expression values and Pearson correlations indicated that the average transcriptome of CME+ cells were more similar to CMs (E14.5, r=0.77; E18.5, r=0.96; P0, r=0.88) than fibroblast-enriched cells (E14.5, r=0.61; E18.5, r=0.66; P0, r=0.43). To independently confirm cell identities, preparations of single cell suspensions from P0 and P3 LV tissues were studied by immunohistochemistry (Fig 2B, Fig S2D) to detect the sarcomere protein troponin I (Tnni3), and collagen (Col3a1). Cell diameters (long axis) were compared (n=145) from CM and non-CMs expressing Tnni3 and/or Col3a1. In the P3 LV, 47% of CMs expressed Col3a1, similar to the proportion observed in RNA-seq data (36%). CMs and CME+ cells had a similar mean length (17.8±3.7 μM vs 17.5±3.3 μM) compared to all non-CMs (10.0±3.2 μM) and non- CM that expressed Col3a1 (8.2±2.3 μM) (Fig S2E). Based on the comparable cell sizes, organized sarcomeres, and expression of Col3a1 and other ECM proteins, we deduced that CME+ cells are a subpopulation of CMs, and not fibroblasts that express sarcomere components, such as myofibroblasts (Mayer and Leinwand, 1997). Throughout LV development, CEM+ cells comprised variable proportions of all CMs (Fig 2C): 9% at E14.5, 22% at E18.5, 29% at P0, 36% at P3, and 1% at P21.
Figure 2.
Subpopulations of CMs with distinct gene expression profiles. A) Hierarchical clustering distinguishes CMs (red) from CMs expressing extracellular matrix genes (CME+, green) isolated from the LV at p0. B) Micrographs of p3 LV cells stained with Tnni3 (green), Col3 (red), or DAPI (blue). Scale bar- 20μM. C) The proportion of LV CMs and CME+ cells varies during development. See also Fig S1 and Fig S2.
Developmental transcription profiles of non-CM cells
Despite increased numbers of fibroblast-enriched cells throughout cardiac development, we observed minimal changes in the transcriptional profiles of non-CMs from E9.5 to P21 (Fig S3A) possibly due to limited numbers of captured cells (Fig 1B). By contrast, the transcriptional profiles of ventricular ECs (Fig S3B) showed a distinct temporal progression with three stages of evolution. During heart looping and chamber formation (E9.5, E11.5), unsupervised clustering using SC3 followed by gene-enrichment analysis (Fig S3C) identified high expression levels of Hapln1 (hyaluronan and proteoglycan link protein 1), and Hmga1 (high mobility group AT-hook protein). Hapln1 is an important regulator of developmental extracellular matrix interactions (DeLaughter et al., 2013) while Hmga1 regulates chromatin organization and promote endothelial-mesenchymal transformation (Benecke and Eilebrecht, 2015). Expression of these genes during chamber formation is consistent with matrix remodeling that participates in early ventricular trabeculation. Later in development (E14.5, E18.5), ECs showed increased expression of Fabp4 (fatty acid binding protein-4), a molecule that contributes to cardiac metabolic shifts from glycolysis to oxidative phosphorylation (Vander Heiden et al., 2010). From P0 through to P21 the EC transcriptional program remained unchanged.
Developmental transcriptional profiles of CMs
E9.5 CMs isolated from the common atria or the primordial ventricle had differential expression of 563 genes (Table S4) and PCA analyses identified gene subsets that reliably discriminated atria from ventricular CMs (Fig S4A, Table S2). For example, E9.5 ventricular CMs expressed high levels of Hey2 and Pln while E9.5 atrial CMs expressed high levels of COUP-TFII (Fig S4B). As these profiles remained distinct over time, we characterized the individual temporal transcriptional programs governing the maturation of atrial or ventricular CMs separately.
Atrial CMs
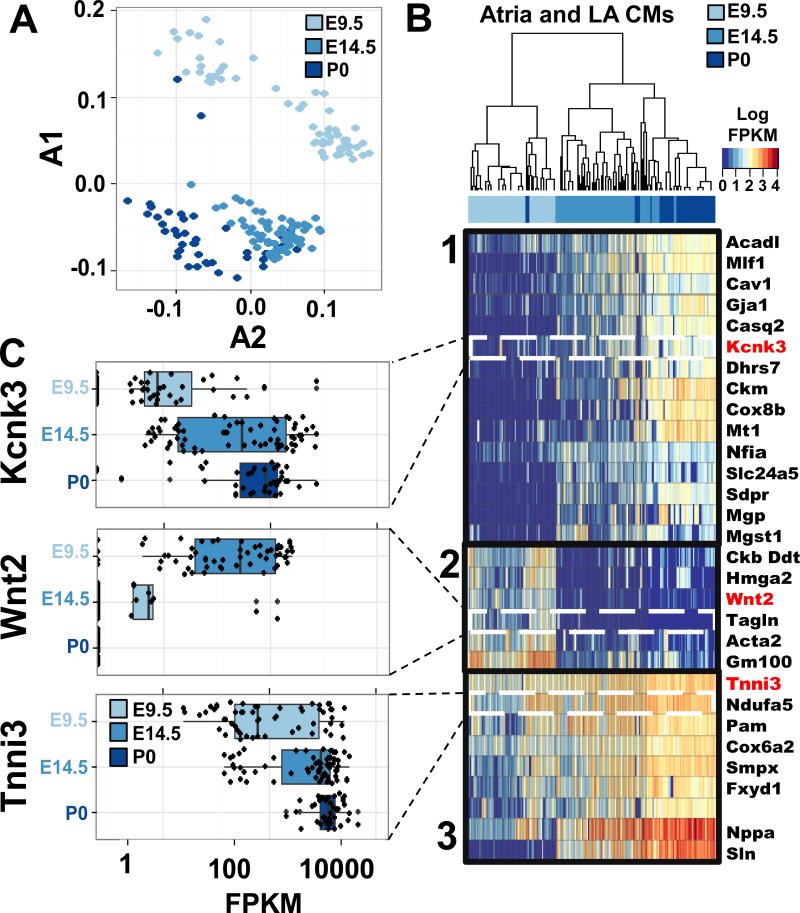
Maturation of atrial CMs resulted in both decreased and increased expression of genes from E9.5 to E14.5 and from E14.5 to P0 (Table S4). PCA suggested progressive transcriptional states (Fig 3A, Table S2) that we characterized by hierarchical clustering of the genes associated with each state. Three distinct gene expression patterns were identified (Fig 3B, C). Pattern 1 consisted of genes with binary expression (i.e., on or off) in an increasingly larger proportion of cells. This pattern included genes that regulate many aspects of adult atrial CM physiology, including cell-cell adhesion (Gja1) (Desplantez et al., 2012; Luo et al., 2007), calcium buffering (Csq2, encoding calsequestrin), and electrophysiologic conductance (Kcnk3; (Liang et al., 2014). Pattern 2 identified genes that were expressed early and globally in atrial development but were silent at E14.5 and thereafter. These genes included Wnt2, a regulator of early atrial development (Tian et al., 2010), genetic markers of immature CMs (Acta2), and a regulator of chromatin remodeling that may promote early CM differentiation (Hmga2; (Monzen et al., 2008). Pattern 3 genes included globally expressed genes with increased expression as development progressed. This group includes genes with fundamental CM function including sarcomere proteins (Tnni3).
Figure 3.
Distinct temporal and chamber gene expression patterns in CMs. A) PCA of gene expression in E9.5 common atria (light blue), E14.5 LA (blue), and P0 LA (dark blue) demonstrate that atrial CMs exhibited stepwise, temporal expression patterns. The percent of variance explained each principle component is provided in Table S2. B) Hierarchical clustering of E9.5 common atria (light blue), E14.5 LA (blue), and P0 LA (dark blue) CMs show three temporal gene expression programs. C) Representative genes depicting single cell gene expression (black dots) from each of the three temporal atrial CM gene expression programs. See also Fig S4, Table S2, and Table S5.
Right versus Left Ventricular CMs
Before considering the developmental programs in maturing ventricular CMs, we used PCA to evaluate the transcriptomes of E11.5 CMs within the developing right and left ventricular chambers (Fig S4A, Table S2). We identified 279 genes significantly differentially expressed (p<1×10−−7) between LV and RV CMs using Mann-Whitney-Wilcoxon rank sum test (Table S5). Included in this subset are genes previously described as regulators of ventricular chamber formation (Fig S4C), including Hand1 and Tbx5, which had restricted expression in LV CMs (Liberatore et al., 2000; Thomas et al., 1998). By contrast, Tbx5-repressed genes had higher expression in RV CMs, findings that support prior studies indicating that Tbx5 is critical for the early establishment of chamber identity (Bruneau et al., 2001). Genes enriched in RV CMs also included proteins associated with the ECM, microfibrillar-associated protein 2 (Mfap2) which binds active Tgfβ-1 Tgfβ-2 and Bmp2 (Combs et al., 2013), and a cell adhesion, melanoma cell adhesion molecule (Mcam) that marks neural crest-derived lineages (Medic and Ziman, 2010; Pujades et al., 2002).
Gene ontology and KEGG pathway analyses (Fig S4D) of 138 genes that were significantly enriched in RV CMs identified extracellular matrix organization (n=11, p=2×10−7), cell adhesion (n=19, p=1.2×10−6), and glycolysis (n=7, p=1.9×10−6) or enrichment in pathways involved in dilated cardiomyopathy (n=7, p=7×10−5) and arrhythmogenic right ventricular cardiomyopathy (n=7, p=1.8×10−4). Although parallel analyses of 141 genes enriched in the LV CMs (Table S4) were less informative of specific pathways, we observed several genes that participate in processes implicated in congenital heart disease (e.g., retinoic acid signaling (Nash et al., 2015); histone modification in response to Nodal signaling (Dahle et al., 2010; Homsy et al., 2015). Notably, these right/left differences were not observed when comparing ECs or fibroblast-enriched cells. Thus, we suggest that developmental pathways that regulate left and right chamber ventricular formation are expressed primarily in CMs rather than in non-myocytes cells.
Between E14.5 and P0, LV and RV CMs became transcriptionally more similar to one another (Fig S4 E-G, Table S2) and expressed genes that characterize CM maturation (e.g. Myh6, Myl3). Taken together we propose that ventricular CMs exhibit distinct transcriptional programs underlying LV and RV chamber morphogenesis after which transcriptional convergence promotes CM maturation.
Developmental Transcriptional Programs in Ventricular CMs
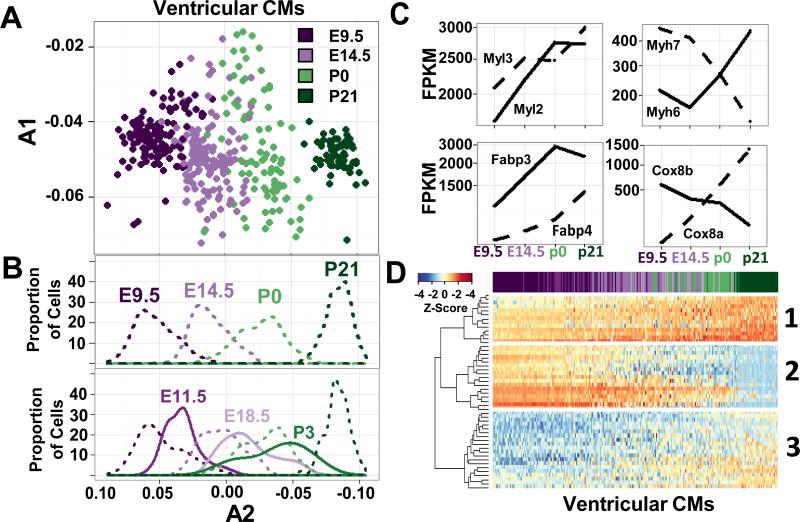
Similar to atrial myocytes, we observed a temporal evolution of ventricular CM gene expression (Fig 4A). PCA analyses of CMs isolated from four time points (E9.5, E14.5, P0, P21) were undertaken using 997 genes expressed in at least 75% of CMs (Table S6). PCA component A2 (Fig 4A, Table S2) separated cells in overlapping, stepwise clusters ordered by age that were visualized using a density plot (Fig 4B). Among the genes that demarcated the transition from least to most mature ventricular CMs, ≥ 11 genes are involved in mitochondrial fatty acid beta-oxidation (e.g. Fabp3, Fapb4, Uqcr11, Cox6c) and accounts for the developmental metabolic switch from embryonic glycolysis to mitochondrial fatty acid beta-oxidation of adult CMs (Vander Heiden et al., 2010). Indeed, ventricular CMs expressed reciprocally increased and decreased transcript levels for Fabp3 and Fabp4, Myh7 and Myh6, and prenatal Cox8a but post-natal Cox8b provided evidence of progressive maturation (Fig 4C). To examine whether CMs remained this distinct between all stages of cardiogenesis, the PCA was expanded to include 3 intermediate time points (E11.5, E18.5, P3). From E9.5 to P21 (Fig 4B) ventricular CMs transcriptomes exhibited a continuum of maturation, but also demarcation from the prior and subsequent stage. This resulted in a progressive wave of ventricular maturation, a developmental pattern that correlated with progressive cell cycle exit (Fig S2A).
Figure 4.
Temporal gene expression profiles of ventricular CMs. A) PCA of ventricular CMs isolated at E9.5 (dark purple), E14.5 (light purple), P0 (light green), and P21 (dark green). The percent of variance explained each principle component is provided in Table S2. B) (Top panel) A density plot depicting the proportion of ventricular CMs isolated at four time points (E9.5, E14.5, P0, P21) on component A2. These time points were evenly distributed along A2. (Bottom panel) Density plot depicting the proportion of cells with a given value of component A2 from PCA of CMs isolated at seven time points (E9.5, E11.5, E14.5, E18.5, P0, P3, P21), show considerable overlap in temporal development from E14.5-P3. C) Box plots depict mean fragments per kilobase of transcript per million mapped reads (FPKM) of genes with notable isoform switches in CMs isolated at E9.5, E14.5, P0, or P21. D) Hierarchical clustering of the top 50 genes in component A2 from B. The genes clustered into three patterns (detailed in text). Columns represent individual single cells ordered by value of component A2 from B and colored by age at time of isolation. Z-score reflected normalization by column. See also Fig S4, Fig S6, Table S2, Table S6, and Table S7.
Within the developmental waves, density plots revealed three stages of CMs maturation between E9.5 and P21 (Fig 4B). CMs on each extreme of this developmental axis are derived primarily from E9.5/E11.5 cells and from P21 cells. Between these extremes we observed a heterogeneous population of CMs comprised of cells isolated from E14.5-P3. This same clustering was also observed when unsupervised clustering using the SC3 tool was performed on ventricular myocytes (Fig S5A). To examine how gene expression changes between these stages of CM maturation, hierarchical clustering was performed on the 50 genes that explained most of the component A2 variance in Fig 4A. Cells ordered by their expression of component A2 (Fig 4D) also yielded three clusters with distinct patterns of gene expression. Parallel analyses of 400 genes confirmed these patterns (Fig S5B and Table S7). Cluster 1 consisted of genes whose expression began on or before E9.5 and increased steadily until P21, such as Myl2, Myh6, Fabp3, and Cox6c (Table S7). The second cluster was defined by genes whose expression decreased as development proceeded, either at E14.5 (Cdk1b, Pgam1 encoding phosphoglycerate mutase 1) or at P21 (Myl7 and Hmgn2 (encoding high mobility group nucleosomal binding domain 2)). Cluster 3 genes were lowly expressed early (E9.5-E11.5) but had increasing expression at E14.5 (Oxct1 encoding succinyl-CoA:3-ketoacid-coenzyme A transferase 1, Ryr2 encoding ryanodine receptor 2) or at p21 (S100a2 encoding S100 calcium binding protein A2). Thus, the genes observed in these three clusters implicate different biological processes critical in heart maturation; sarcomere structure and fatty acid oxidation in cluster 1; glycolysis and proliferation in cluster 2; mitochondrial and calcium handling in cluster 3.
The heterogeneity in maturity of individual CMs, isolated at the same time point, raised the question of whether whole tissue samples at harvested at specified distinct embryonic times could be distinguished based on gene expression. To assess this, we processed whole LV free wall at E14.5 and E18.5 for RNA-seq similar to single cell methods. Analyses were restricted to 997 genes expressed at ≥ 1 FPKM (fragments per kilobase of transcript per million mapped reads) in 75% of single CMs harvested at E9.5, E11.5, E14.5, E18.5, P0, P3 and P21 (Table S6). A density plot was generated from the most informative temporal component (A2, Fig 5A). LV free wall tissue from E14.5 and E18.5 mice mapped to the middle of the distribution of single cells, confirming that the maturation profiles from many single cells accurately captured in vivo developmental transcriptional profiles and also confirmed that whole tissue RNA-seq data averages the expression of CMs with different maturation stages.
Figure 5.
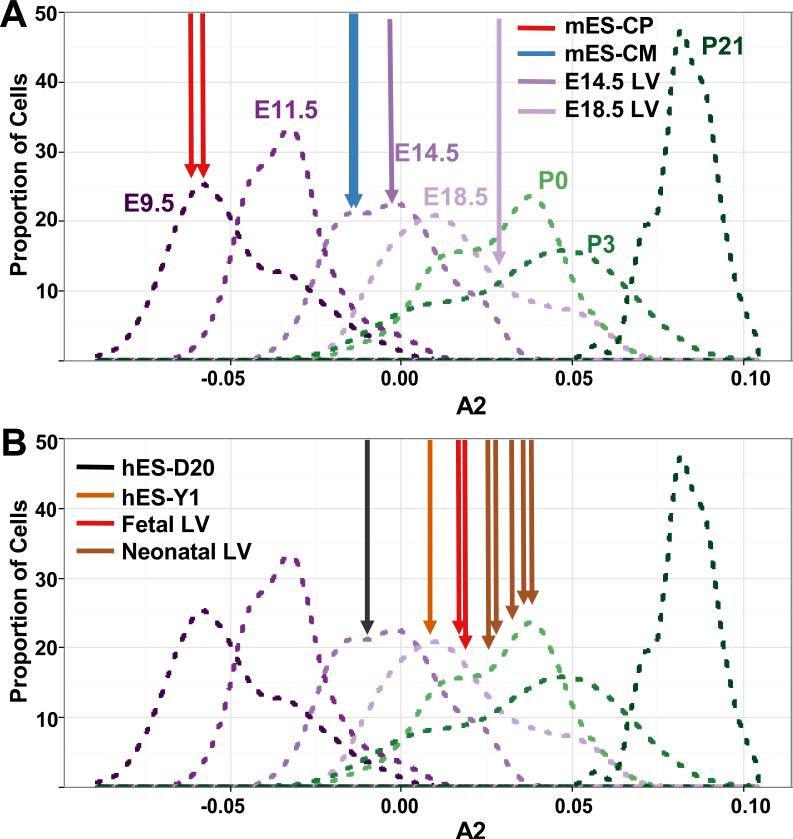
Developmental ages of CMs derived from human and mouse stem cell and LV tissues (depicted using density plots as described in Fig. 4B). A) Murine whole tissue samples from E14.5 (purple arrow) and E18.5 LV (light purple arrow) have A2 values near the mean of the distribution of E14.5 and E18.5 LV single cells respectively. Murine embryonic stem cell-derived cardiac progenitors (mES-CP, red arrow) correspond to E9.5 CMs and murine embryonic stem cell-derived CM lines (mES-CM, blue arrow) correspond to E14.5 CMs. mES transcriptome data are from two independent clonal lines (Wamstad Cell 2012). B) Similar analyses of transcriptional analyses from pooled human embryonic stem cell-derived CMs (Kuppusamy, PNAS 2015) cultured for 20 days (hES-D20, black arrow) or one year (hES-Y1, orange arrow) indicate that these respectively correspond to mouse E14.5 and E18.5 CMs. The transcriptome of the free wall of human fetal LV (ages 8 and 12 weeks, red arrow, n=2) and human neonatal LV or RV (brown arrow; n=5) are shown for comparison. See also Table S6.
Developmental Ages of Stem Cell-derived CMs
Applying the method described above (Fig 4B), we used published RNA-seq datasets that were processed through our pipeline to determine the developmental ages of mouse and human ES cells. Two replicate RNA-seq datasets from cardiac progenitors (mES-CP) and CMs (mES-CM) derived from murine ES-cells (Wamstad et al., 2012) showed high concordance. Expression data from mES-CP corresponded to E9.5 single-cell CMs, while data from mES-CM cells corresponded to E14.5 cells (Fig 5A).
Similar analyses of human ES cell-derived (hES) CMs were performed using 847 genes with human orthologs (Bick et al., 2012). RNA-seq datasets from hES-derived CMs cultured for 20 days (hES-D20) or approximately one (13.5 months) year (hES-Y1) (Kuppusamy et al., 2015) showed high concordance. The hES-D20 cells corresponded to E14.5 ventricular CMs while hES-Y1 cells corresponded to E18.5 ventricular CMs. To confirm that these conclusions did not reflect differences in mouse and human development, we evaluated RNA-seq data from fetal and neonatal human tissues, including 2 LV tissues from 8 and 12 weeks of gestation and 5 tissue samples from post-natal infant hearts. Human fetal heart samples corresponded to late E18.5, while neonatal samples corresponded to P0 to P3 (Fig 5B). Together these findings support the conclusion by Kuppusamy and colleagues that prolonged culturing increases CMs maturation, but also indicated that hES-Y1 CMs remain more immature than P21 cells (Fig 5B).
Nkx2.5 Haploinsufficiency Impairs Maturation of Distinct Cardiac Cell Lineages
Dominant mutations in NKX2.5 cause human cardiac septation defects (Elliott et al., 2003; Schott et al., 1998; Yuan et al., 2015). Nkx2.5+/− mice recapitulate many of these cardiac phenotypes albeit with incomplete penetrance (Ashraf et al., 2014; Biben et al., 2000; Bruneau et al., 2001). We hypothesized that haploinsufficiency of transcription factors Nkx2.5 cause heart malformations by altering developmental transcription in specific cardiac lineages. To test this model we isolated cells from the LV at E14.5, P0, P7, and P21 in Nkx2.5+/− mice and compared PCA of single-cell RNA-seq data to that from WT mice (Table S4).
Nkx2.5+/− CMs expressed lower levels of Myh7 (−5.0 fold, p=2.3×10−11 compared to WT) implying a potential defect in CM maturation. We therefore compared the maturity of Nkx2.5+/− cardiac cell lineages using single cell transcriptomes from mutant and WT CMs isolated at E14.5, P0, P7 and P21 (Fig 6A). To better visualize maturation differences we compared density plots of cell distributions in mutant (solid waves) and WT (dashed waves) mice derived from PCA components A2 (Fig 6B). Nkx2.5+/− and WT CMs isolated at p0 or p21 had similar developmental distributions but E14.5 Nkx2.5+/− CMs were skewed towards more immature CM populations. Comparing the mean FPKM (Student's T-test), 420 genes were differentially expressed between Nkx2.5+/− and WT CMs of which several were associated with myocyte maturation including Ttn and Myh6 (Table S4).
Figure 6.
Maturation defects in Nkx2.5+/− CMs. A) Developmental profile of E9.5 VEN and E14.5, P0, and P21 LV CMs from WT (dashed line) and Nkx2.5+/− mice (solid line) depicted using density plots as described in Fig. 4B. At E14.5 (2nd panel), Nkx2.5+/− CMs are less mature than E14.5 WT cells. This immaturity decreases gradually from P0 (3rd panel) to P21 (4th panel). B) Using hierarchical clustering, the proportion Nkx2.5+/− CMs (CM) that cluster with similarly aged WT cells is significantly different at all time points (Fisher's exact p-values: E14.5= 2.1×10−25, P0= 0.0012, P7= 0.010, P21= 0.0071). See also Fig S5 and Table S4.
To more precisely quantitate the maturity defect in Nkx2.5+/− CMs, we defined the top 20 genes contained in PCA component A2 that distinguished E14.5 mutant from WT CMs (Fig 6A; Table S2) and used these genes for hierarchical clustering of CMs. Two populations of WT CMs were identified: cells that were more mature (52%, 10/19) or less mature (48%, 9/19) based on gene expression (Myl2, Myl3, Fabp3; Fig 6B). At P7 (60%; 12/20) and P21 (97%; 62/64) increasing numbers of WT CMs were more mature.
In contrast, P0 Nkx2.5+/− CMs clustered predominantly as less mature (87%; 13/15) rather than more (2/15) mature cells (compared to WT, p=0.012; Fisher exact test). P7 gene expression profiles of Nkx2.5+/− CMs were more comparable to P0 WT CMs and only 20% of CMs exhibited mature gene expression patterns, in comparison to 60% of WT CMs (p=0.01) (Fig 6B, Fig S5C). At P21 most (78%, 21/27) of Nkx2.5+/− CMs were mature, albeit a significantly less proportion than in WT LV (97% 62/64, p=0.0071, Fisher exact) (Fig 6B).
Prior studies demonstrate Nkx2.5 expression principally in CMs (Kasahara et al., 1998), which we confirmed by single cells analyses (Fig S5D). As recent evidence indicates developmental cross-talk between CMs and ECs lineages (Palencia-Desai et al., 2015), we also assessed the maturation of ECs from Nkx2.5+/− mice (Fig S3D). Transcriptional profiles of Nkx2.5+/− ECs were comparable to WT ECs at E14.5, but significantly delayed at P0 (Nkx2.5+/− 12/14 immature ECs versus WT 4/19; p=0.038, Fisher's exact test).
Together these data indicate that that Nkx2.5+/− haploinsufficiency delays maturation in both CMs and ECs and provides further support that the myocardium is crucial for endocardial differentiation.
Discussion
Using single cell RNA-seq we performed a high-resolution time course analysis of cardiac development. Our efforts leveraged unbiased, single-cell analyses to separate and characterize over 1200 cardiac cells from three distinct lineages. In addition to characterizing longitudinal developmental programs we identified distinct transcriptional profiles for CM subpopulations and chamber-specific profiles. Leveraging these analyses, we established developmental ages of CMs derived from stem cells, and characterized lineage-specific consequences of haploinsufficiency of the transcription factor Nkx2-5.
We identified considerable transcriptional heterogeneity among CMs. Among E14.5 ventricular CMs, a subpopulation (CME+) expressed extracellular matrix proteins that are emblematic of fibroblasts, even though their overall transcriptome typified CMs. The proportion of this CM subpopulation increased until P3, after which CME+ cells were rarely identified. We speculate that CME+ cells provide a developmental scaffold for the formation and maintenance of heart architecture and may be disease-responsive, therein accounting for increased cardiomyocyte expression of extracellular matrix proteins (Ambrosino et al., 2006) due to increased proportions of CME+ cells rather than global transcriptional changes in all CMs.
Another CM subpopulation was distinguished by the expression of cell cycle genes, which we presumed implies active proliferation. The transcriptional profiles of these cells were not suggestive of stem cells nor recently described cardiomyoblasts (Jain et al., 2015); earlier developmental time points are likely needed to capture these cells. Instead, cell cycle gene expression was found in 60% of E9.5 CMs. With developmental progression, these transcripts rapidly declined. At P21 no CMs expressed cell cycle genes.
Notably, the transcriptomes of proliferating and non-proliferating CMs were remarkably similar, indicating that these cells had comparable developmental maturity. Consistent with this, genes that demarcated distinct waves of development by CMs did not include cell cycle genes. We suggest that throughout embryogenesis newly born CMs ‘kept pace’ with differentiating cells, perhaps due to a rich milieu of intrinsic and secreted factors that promoted maturation. Identification of these molecules could be important to accelerate in vitro maturation of stem-cell derived CMs.
Ventricular CMs exhibited overlapping, stepwise developmental patterns that we grouped into three transcriptional stages of maturation. At E9.5-E11.5 CM expressed genes involved in proliferation (e.g., cell cycle genes, Fig. S2A) and morphogenesis (e.g., Sall4, a Tbx5-regulated transcription factor that promotes ventricular morphogenesis and chamber maturation (Koshiba-Takeuchi et al., 2006). CMs subsequently matured through sequential overlapping waves of gene expression (Fig 4B) between E14.5-P3. These transitional CM expressed genes involved in cellular growth (e.g., Tcea1, encoding transcription elongation factor), CM differentiation (Cbx3 an epigenetic silencer that promotes pluripotent cells differentiation; Casq2, calsequestrin 2; Ckm, creatinine kinase) and the metabolic switch from glycolysis (reduced Pgam1 expression) to fatty acid oxidation (increased Fabp3, Fabp4, encoding fatty acid binding protein; Fig 4C) that characterizes mature CMs (Vander Heiden et al., 2010). Further maturation occurred by P21 when there was decreased expression of calmodulin-interacting proteins, Bex1 and Bex4, that promote muscle regeneration (Koo et al., 2007).
Using these maturation stages we characterized stem-cell derived CMs, which are advancing understanding of human heart disease and propelling the development of new therapeutics, despite the limitation that these cells fail to recapitulate adult CM gene expression even after a year in culture (Kuppusamy et al., 2015). While a few markers have been previously used to gauge the maturation of stem-cell derived CMs (Li et al., 2015), our spatio-temporal CM transcriptional profile of single cells and in vivo mouse and human tissues precisely defined the developmental “age” of stem cells and derived CMs (Fig 5). We observed remarkably homogeneity and consistent immaturity (≤ E18.5) in both mouse and human CMs derived from stem cells. Knowledge of the repertoire of genes expressed through development should advance chemical or genomic strategies to improve both the maturation and subpopulation diversity of stem-cell derived CMs and cardiac lineages.
Single cell-derived transcriptome maps also uncovered lineage-specific perturbations due to haploinsufficiency of the cardiac transcription factor Nkx2.5+/−, a cause of human congenital heart disease. Our data extends earlier transcriptional analyses of Nkx2.5+/− mouse heart tissues (Bouveret et al., 2015) by delineating lineage-specific and temporal changes that persisted late into development. In comparison to WT cells, we demonstrate that developmental immaturity of E14.5 Nkx2.5+/− LV CMs and P0 ECs (Fig 6, Fig S3D). As Nkx2.5 is not expressed outside of CMs, we suggest that the dysregulated EC gene expression in Nkx2.5+/− hearts indicates non-autonomous signals that are evoked by haploinsufficiency of this transcription factor.
In conclusion, we demonstrate that single cell RNA expression analyses improved delineation of lineage-specific and spatio-temporal developmental transcriptional programs. Expansion of these analyses to include earlier developmental time points and additional lineages is expected to expand insights into molecular pathways that are critical for heart development.
Methods
Mice and Dissection
Pregnant female 129SV mice (Taconic) were used for tissue dissection. The construction of Nkx2.5-del (Tanaka et al., 1999) mice was previously described. Nkx2.5 heterozygous males from a mixed background were crossed with 129SV females. Embryonic age was confirmed by ascertaining the Thieler stage of each pup. Tissue derived from one pup was used for each individual chip. Hearts were excised into PBS and incubated with agitation at 37° for 20-60 minutes, depending on stage, in digestion buffer consisting of PBS, 0.70% weight/volume % collagenase B, and 0.75% weight/volume % collagenase D (Roche).
Cell Capture
Fluidigm integrated fluidic circuits (IFC) were used to capture single cells. After capture, the IFC lysed individual cells, reverse transcribed RNAs and amplified complementary DNAs. The resulting cDNAs from individual cells were barcoded, indexed and sequenced to an average depth of 1.5 million reads per library. IFCs were selected to capture all major cell populations from all cell size ranges observed using IFCs which capture cells of different sizes: 5-10uM (embryonic), 10-17uM (embryonic, neonatal), 17-25uM (3 weeks of age). No batch effects were observed between chips of the same size. Captured cells were imaged on chip to confirm the number of cells per site and the viability of the cell was confirmed using a LIVE/DEAD cell assay (Life Technologies). Only single, viable cells were used for further analysis. Onboard cell lyses, in vitro transcription and cDNA synthesis were performed using IFC RNA-seq reagent kit (Fluidigm) and SMARTer Ultra Low RNA kit (Clonetech) reagents following the manufacturer's protocol.
Library Construction and DNA Sequencing
Libraries were constructed from cDNA yielded from IFCs in 96 well plates using Nextera XT DNA sample preparation kit (Illumina). cDNA concentrations before and after library prep was confirmed using High-Sensitivity DNA Tapes (Agilent) run on an Agilent Tape Station. For whole tissue experiments, RNA was extracted from whole tissue using phenol-chloroform. Reverse transcription, initial cDNA synthesis and cDNA library preparation were performed using the same reagents as for single cells. cDNA size distribution of single-cell, whole tissue and pooled libraries was performed using High-Sensitivity DNA Tapes (Agilent) on an Agilent Tape Station. Up to 96 single-cell libraries were pooled together for 50 bp, paired-end DNA sequencing on a single lane of an Illumina HiSeq2500 following the manufacturer's standard protocol. Single-cell libraries with >400,000 reads were used for further analysis. Reads were aligned to the mouse genome (mm10) using Tophat (Trapnell et al., 2009).
Statistical Analysis of Single Cell Transcriptomes
PCA, Mann-Whitney-Wilcoxon rank sum test, hierarchical clustering, and Student's t-tests were performed using custom scripts in R. PCA was performed on Log10 FPKM. Principal components with eigenvalues greater than 1 were evaluated to cluster cells. Genes identified by these components were used to perform hierarchical clustering. Euclidean distances were calculated from Log10 FPKM and clustered by the complete linkage method. Consensus clustering was performed using Single-Cell Consensus Clustering (SC3) package (Kiselev VY, 2016). Only cells expressing >3000 genes were used for clustering. The k and distance metrics were chosen to maximize the silhouette width. Dendrograms were derived from consensus clustering and overlaid over heatmaps depicting marker genes expression also calculated with the SC3 tool. RaceID was performed as described (Grun et al., 2015). Student's t-test were 2 tailed and assuming unequal variance.
Gene ontology and KEGG pathway enrichment analysis were performed using DAVID software (Huang et al., 2007). “Perfect marker” gene analysis was performed as described (Treutlein et al., 2014). Briefly, a “perfect marker” gene was defined for each cell lineage such that its expression was set at Log10(FPKM)=10 in all cells of the desired lineage (i.e. myocytes) and 0 in all other cells. A pairwise Pearson's correlation was calculated comparing the expression profile across all cells for a given gene to that of the perfect marker gene.
Stem-cell derived CM analysis
We derived a gene expression axis correlating with developmental time from the set of 997 genes expressed in 75% of ventricular myocytes at E9.5, E11.5, E14.5, E18.5, P0, P3 and P21. For comparisons to human cells, we limited this list to the 847 genes with human orthologs (Bick et al., 2012). Sequence from short-read files were downloaded from NCBI GEO databank (GEO ID GSE47948 and GSE62913 respectively) and rerun through our RNA-Seq pipeline (Li et al., 2015; Wamstad et al., 2012). Human whole tissue isolation, cDNA library construction and mRNA-seq analysis were performed as previously described in (McKean et al., 2016).
Immunohistochemistry
Cells isolated from p0 or p3 LVs were plated onto laminin coated chamber slides and incubated at 37°C for 1 hour. Cells were fixed with 4% paraformaldehyde for 20 minutes and washed with phosphate buffered saline (PBS) subsequent to permeabilization with 0.1% (vol/vol) Triton-X100 in PBS. 10% (vol/vol) donkey serum was used to block nonspecific binding. Cells were incubated with two primary antibodies, rabbit anti-Tnni3 (Abcam-ab56357), or goat anti-Col3a1 (Abcam-ab7778) diluted 1:250 in blocking solution for 1 hour at room temperature. Cells were washed in PBS and fluorophore-conjugated secondary antibody (Molecular Probes) diluted 1:1000 in PBS for one hour. Slides were washed and then mounted and counterstained using VECTASHIELD antifade mounting medium with DAPI (Vector Laboratories).
Supplementary Material
Highlights.
Single-cell RNA-seq data characterizing >1200 murine cells from E9.5-P21 hearts
Dynamic temporospatial gene expression defines distinct cardiomyocyte populations
Human/mouse stem cell-derived cardiomyocytes are developmentally immature
Nkx2.5+/− mice have lineage-specific maturation defects in cardiac cells
Acknowledgements
This work was supported by grants from the NIGMS T32GM007753 (A.G.B.), John S. LaDue Memorial Fellowship at Harvard Medical School (J.H.) NHLBI Bench to Bassinet Program (U01HL098179/UM1HL098179, B.G.B., 2UM1HL098147, C.E.S.; 2UM1HL098166, W.P., J.G.S., and C.E.S.) the Howard Hughes Medical Institute (CES), NIMH (R01 MH101528-01, J.G.), NIH (HL125807, J.T.H.), I.S.K was supported by the Foundation for Anaesthesia Education and Research. The authors thank David Conner, and Steve DePalma for helpful discussions, advice, and technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, Methodology and Writing – Original Draft, D.M.D., A.G.B., J.G.S., and C.E.S.; Investigation, D.M.D., A.G.B., H.W., and J.M.G.; Software, Formal Analysis and Visualization, D.M.D. and A.G.B; Resources, D.M., I.K., J.T.H., J.G.; Review & Editing, H.W., D.M., I.K., J.T.H., J.H., J.G., W.P., B.G.B. C.E.S.; Supervision and Funding Acquisition, J.G., W.P., B.G.B, J.G.S., C.E.S.
Accession Numbers
All single cell sequencing data reported in this article have been deposited in the GNomEx database under accession numbers 272R, 274R, 275-292R, 439R, and 440R can be found at https://b2b.hci.utah.edu/gnomex/ by using the guest login.
References
- Ambrosino C, Iwata T, Scafoglio C, Mallardo M, Klein R, Nebreda AR. TEF-1 and C/EBPbeta are major p38alpha MAPK-regulated transcription factors in proliferating cardiomyocytes. Biochem J. 2006;396:163–172. doi: 10.1042/BJ20051502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf H, Pradhan L, Chang EI, Terada R, Ryan NJ, Briggs LE, Chowdhury R, Zarate MA, Sugi Y, Nam HJ, et al. A mouse model of human congenital heart disease: high incidence of diverse cardiac anomalies and ventricular noncompaction produced by heterozygous Nkx2-5 homeodomain missense mutation. Circ Cardiovasc Genet. 2014;7:423–433. doi: 10.1161/CIRCGENETICS.113.000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benecke AG, Eilebrecht S. RNA-Mediated Regulation of HMGA1 Function. Biomolecules. 2015;5:943–957. doi: 10.3390/biom5020943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Koentgen F, Robb L, Feneley M, et al. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circulation research. 2000;87:888–895. doi: 10.1161/01.res.87.10.888. [DOI] [PubMed] [Google Scholar]
- Bick AG, Calvo SE, Mootha VK. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012;336:886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret R, Waardenberg AJ, Schonrock N, Ramialison M, Doan T, de Jong D, Bondue A, Kaur G, Mohamed S, Fonoudi H, et al. NKX2-5 mutations causative for congenital heart disease retain functionality and are directed to hundreds of targets. Elife. 2015;4 doi: 10.7554/eLife.06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook C, Warry G, Howell G, Arnason A, Bjornsson A, Moore GE, Ross MT, Stanier P. Identification and characterization of KLHL4, a novel human homologue of the Drosophila Kelch gene that maps within the X-linked cleft palate and Ankyloglossia (CPX) critical region. Genomics. 2001;72:128–136. doi: 10.1006/geno.2000.6478. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nature biotechnology. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- Cheroki C, Krepischi-Santos AC, Szuhai K, Brenner V, Kim CA, Otto PA, Rosenberg C. Genomic imbalances associated with mullerian aplasia. J Med Genet. 2008;45:228–232. doi: 10.1136/jmg.2007.051839. [DOI] [PubMed] [Google Scholar]
- Combs MD, Knutsen RH, Broekelmann TJ, Toennies HM, Brett TJ, Miller CA, Kober DL, Craft CS, Atkinson JJ, Shipley JM, et al. Microfibril-associated glycoprotein 2 (MAGP2) loss of function has pleiotropic effects in vivo. J Biol Chem. 2013;288:28869–28880. doi: 10.1074/jbc.M113.497727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahle O, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal. 2010;3:ra48. doi: 10.1126/scisignal.2000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaughter DM, Christodoulou DC, Robinson JY, Seidman CE, Baldwin HS, Seidman JG, Barnett JV. Spatial transcriptional profile of the chick and mouse endocardial cushions identify novel regulators of endocardial EMT in vitro. J Mol Cell Cardiol. 2013;59:196–204. doi: 10.1016/j.yjmcc.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplantez T, McCain ML, Beauchamp P, Rigoli G, Rothen-Rutishauser B, Parker KK, Kleber AG. Connexin43 ablation in foetal atrial myocytes decreases electrical coupling, partner connexins, and sodium current. Cardiovasc Res. 2012;94:58–65. doi: 10.1093/cvr/cvs025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, Grossfeld P, Fatkin D, Jones O, Hayes P, et al. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. Journal of the American College of Cardiology. 2003;41:2072–2076. doi: 10.1016/s0735-1097(03)00420-0. [DOI] [PubMed] [Google Scholar]
- Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Li D, Gupta M, Manderfield LJ, Ifkovits JL, Wang Q, Liu F, Liu Y, Poleshko A, Padmanabhan A, et al. HEART DEVELOPMENT. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science. 2015;348:aaa6071. doi: 10.1126/science.aaa6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang DG, Sim HJ, Song EK, Medina-Ruiz S, Seo JK, Park TJ. A thioredoxin fold protein Sh3bgr regulates Enah and is necessary for proper sarcomere formation. Dev Biol. 2015;405:1–9. doi: 10.1016/j.ydbio.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen B, Wang T, Christoffels VM, Moorman AF. Evolution and development of the building plan of the vertebrate heart. Biochimica et biophysica acta. 2013;1833:783–794. doi: 10.1016/j.bbamcr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circulation research. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- Kiselev VY KK, Schaub MT, Andrews T, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, Hemberg M. SC3 - consensus clustering of single-cell RNA-Seq data. bioRxiv. 2016 doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JH, Smiley MA, Lovering RM, Margolis FL. Bex1 knock out mice show altered skeletal muscle regeneration. Biochem Biophys Res Commun. 2007;363:405–410. doi: 10.1016/j.bbrc.2007.08.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K, Takeuchi JK, Arruda EP, Kathiriya IS, Mo R, Hui CC, Srivastava D, Bruneau BG. Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nat Genet. 2006;38:175–183. doi: 10.1038/ng1707. [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, Pabon L, Zhu WZ, Tulloch NL, Yang X, et al. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci U S A. 2015;112:E2785–2794. doi: 10.1073/pnas.1424042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Plonowska K, Kuppusamy R, Sturzu A, Wu SM. Identification of cardiovascular lineage descendants at single-cell resolution. Development. 2015;142:846–857. doi: 10.1242/dev.116897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Soka M, Christensen AH, Olesen MS, Larsen AP, Knop FK, Wang F, Nielsen JB, Andersen MN, Humphreys D, et al. Genetic variation in the two-pore domain potassium channel, TASK-1, may contribute to an atrial substrate for arrhythmogenesis. J Mol Cell Cardiol. 2014;67:69–76. doi: 10.1016/j.yjmcc.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Liberatore CM, Searcy-Schrick RD, Yutzey KE. Ventricular expression of tbx5 inhibits normal heart chamber development. Dev Biol. 2000;223:169–180. doi: 10.1006/dbio.2000.9748. [DOI] [PubMed] [Google Scholar]
- Luo MH, Li YS, Yang KP. Fibrosis of collagen I and remodeling of connexin 43 in atrial myocardium of patients with atrial fibrillation. Cardiology. 2007;107:248–253. doi: 10.1159/000095501. [DOI] [PubMed] [Google Scholar]
- Mayer DC, Leinwand LA. Sarcomeric gene expression and contractility in myofibroblasts. J Cell Biol. 1997;139:1477–1484. doi: 10.1083/jcb.139.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean DM, Homsy J, Wakimoto H, Patel N, Gorham J, DePalma SR, Ware JS, Zaidi S, Ma L, Patel V, et al. Loss of RNA expression and allele-specific expression associated with congenital heart disease. Nat Commun. 2016;7:12824. doi: 10.1038/ncomms12824. doi: 10.1038/ncomms12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medic S, Ziman M. PAX3 expression in normal skin melanocytes and melanocytic lesions (naevi and melanomas). PLoS One. 2010;5:e9977. doi: 10.1371/journal.pone.0009977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzen K, Ito Y, Naito AT, Kasai H, Hiroi Y, Hayashi D, Shiojima I, Yamazaki T, Miyazono K, Asashima M, et al. A crucial role of a high mobility group protein HMGA2 in cardiogenesis. Nat Cell Biol. 2008;10:567–574. doi: 10.1038/ncb1719. [DOI] [PubMed] [Google Scholar]
- Nash D, Arrington CB, Kennedy BJ, Yandell M, Wu W, Zhang W, Ware S, Jorde LB, Gruber PJ, Yost HJ, et al. Shared Segment Analysis and Next-Generation Sequencing Implicates the Retinoic Acid Signaling Pathway in Total Anomalous Pulmonary Venous Return (TAPVR). PLoS One. 2015;10:e0131514. doi: 10.1371/journal.pone.0131514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet C, Hofmann JJ, Mae MA, Jung B, Gaengel K, Vanlandewijck M, Ekvarn E, Salvado MD, Mehlem A, Al Sayegh S, et al. Gpr116 Receptor Regulates Distinctive Functions in Pneumocytes and Vascular Endothelium. PLoS One. 2015;10:e0137949. doi: 10.1371/journal.pone.0137949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palencia-Desai S, Rost MS, Schumacher JA, Ton QV, Craig MP, Baltrunaite K, Koenig AL, Wang J, Poss KD, Chi NC, et al. Myocardium and BMP signaling are required for endocardial differentiation. Development. 2015;142:2304–2315. doi: 10.1242/dev.118687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D'Antoni ML, Debuque R, Chandran A, Wang L, Arora K, Rosenthal NA, et al. Revisiting Cardiac Cellular Composition. Circulation research. 2016;118:400–409. doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujades C, Guez-Guez B, Dunon D. Melanoma Cell Adhesion Molecule (MCAM) expression in the myogenic lineage during early chick embryonic development. Int J Dev Biol. 2002;46:263–266. doi: 10.1387/ijdb.011493. [DOI] [PubMed] [Google Scholar]
- Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics. 1987;20:53–65. [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Smith CM, Finger JH, Hayamizu TF, McCright IJ, Xu J, Berghout J, Campbell J, Corbani LE, Forthofer KL, Frost PJ, et al. The mouse Gene Expression Database (GXD): 2014 update. Nucleic Acids Res. 2014;42:D818–824. doi: 10.1093/nar/gkt954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streets AM, Huang Y. How deep is enough in single-cell RNA-seq? Nature biotechnology. 2014;32:1005–1006. doi: 10.1038/nbt.3039. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Thomas T, Yamagishi H, Overbeek PA, Olson EN, Srivastava D. The bHLH factors, dHAND and eHAND, specify pulmonary and systemic cardiac ventricles independent of left-right sidedness. Dev Biol. 1998;196:228–236. doi: 10.1006/dbio.1998.8849. [DOI] [PubMed] [Google Scholar]
- Tian Y, Yuan L, Goss AM, Wang T, Yang J, Lepore JJ, Zhou D, Schwartz RJ, Patel V, Cohen ED, et al. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Dev Cell. 2010;18:275–287. doi: 10.1016/j.devcel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Siddiqui MA. Signal transduction in early heart development (I): cardiogenic induction and heart tube formation. Experimental biology and medicine. 2007;232:852–865. [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Hilton MB, Seaman S, Haines DC, Nagashima K, Burks CM, Tessarollo L, Ivanova PT, Brown HA, Umstead TM, et al. Essential regulation of lung surfactant homeostasis by the orphan G protein-coupled receptor GPR116. Cell Rep. 2013;3:1457–1464. doi: 10.1016/j.celrep.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Qiu XB, Li RG, Qu XK, Wang J, Xu YJ, Liu X, Fang WY, Yang YQ, Liao DN. A novel NKX2-5 loss-of-function mutation predisposes to familial dilated cardiomyopathy and arrhythmias. Int J Mol Med. 2015;35:478–486. doi: 10.3892/ijmm.2014.2029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.