SUMMARY
Macrophages activated by the TLR4 agonist LPS undergo dramatic changes in their metabolic activity. We here show that LPS induces expression of the key metabolic regulator Pyruvate Kinase M2 (PKM2). Activation of PKM2 using two well-characterized small molecules, DASA-58 and TEPP-46, inhibited LPS-induced Hif-1α and IL-1β, as well as the expression of a range of other Hif-1α-dependent genes. Activation of PKM2 attenuated an LPS-induced pro-inflammatory M1 macrophage phenotype while promoting traits typical of an M2 macrophage. We show that LPS-induced PKM2 enters into a complex with Hif-1α, which can directly bind to the IL-1β promoter, an event that is inhibited by activation of PKM2. Both compounds inhibited LPS- induced glycolytic reprogramming and succinate production. Finally, activation of PKM2 by TEPP-46 in vivo inhibited LPS and Salmonella typhimurium-induced IL-1β production, whilst boosting production of IL-10. PKM2 is therefore a critical determinant of macrophage activation by LPS, promoting the inflammatory response.
INTRODUCTION
Insights into the critical role of metabolic reprogramming in host defense and inflammation have recently emerged (Krawczyk et al., 2010; O’Neill and Hardie, 2013). In macrophages activated with the TLR4 agonist LPS there is a high demand for biosynthetic precursors including proteins, lipids, and nucleic acids as well as increased energy demand. These events are required for the cells to respond to infection or tissue injury during inflammation. The metabolic changes that occur resemble the well-described Warburg effect (also known as aerobic glycolysis) first reported in tumor cells (Altenberg and Greulich, 2004; Warburg, 1923). These alterations include increased glucose uptake, an elevated rate of glycolysis, and an up-regulated pentose phosphate pathway in conjunction with a reduced level of oxidative phosphorylation via the tricarboxylic acid (TCA) cycle (Tannahill et al., 2013).
Increased levels of the TCA cycle intermediate succinate as a consequence of this metabolic switch in highly glycolytic activated ‘M1’ macrophages has revealed that succinate facilitates the signal leading to stabilization of Hypoxia Inducible Factor-1α (Hif-1α) (Koivunen et al., 2007; Tannahill et al., 2013). This phenomenon, which has been described as ‘pseudo hypoxia’ takes place when excess succinate is transported out of the mitochondria into the cytosol where it can impair the activity of prolylhydroxylases (PHD) leading to stabilization and activation of Hif-1α (Selak et al., 2005). Hif-1α then positively regulates the proinflammatory cytokine IL-1β and other Hif-dependent genes including those encoding enzymes in glycolysis (Tannahill et al., 2013).
There is only limited information on the precise molecular determinants of this profound change in metabolism.
Numerous studies have reported a key role for PKM2 in aerobic glycolysis of tumors (Christofk et al., 2008; Mazurek et al., 2005; Tamada et al., 2012). Mammalian cells contain two PK genes. One encodes PKL and PKR, which are exclusively expressed in liver and red blood cells, respectively. The second gene encodes PKM1 and PKM2. These are generated by exclusive alternative splicing of the Pkm pre- mRNA (Chen et al., 2012; Takenaka et al., 1991). In glycolysis pyruvate kinase is the rate-limiting enzyme that converts phosphoenolpyruvic acid (PEP) to pyruvate, a reaction that in differentiated tissue is mediated by the enzymatically active isoform PKM1. PKM2 on the other hand is up- regulated in tumors (Altenberg and Greulich, 2004) and exists primarily as an enzymatically inactive monomer or dimer. The PKM2 dimer can translocate to the nucleus where it will interact with Hif-1α and regulate expression of numerous pro-glycolytic enzymes (Luo et al., 2011), an event that is dependent on ERK1/2 activity and regulated by Jumoji C domain-containing dioxygenase (JMJD5) (Wang et al., 2014; Yang et al., 2012). Other nuclear functions of dimeric PKM2 includes the ability to act as a protein kinase, activating transcription of mek5 through phosphorylation of STAT3(Gao et al., 2012) as well as promoting β-catenin translocation, leading to expression of cyclinD1 and c-myc(Yang et al., 2011). Current evidence indicates that nuclear PKM2 thereby drives the Warburg effect in tumors (Yang and Lu, 2013).
The enzymatic pyruvate kinase activity of PKM2 can be allosterically triggered endogenously by Fructose-1,6-bisphosphate (FBP), serine, and succinylaminoimidazolecarboxamide ribose-5′-phosphate (SAICAR) (Chaneton et al., 2012; Jurica et al., 1998; Keller et al., 2012). PKM2 can also be activated using the highly specific small molecule activators DASA-58 and TEPP-46 (Anastasiou et al., 2012). These will bind PKM2 promoting tetramers to form, leading to an enzyme with a high pyruvate kinase activity, allowing PKM2 to mediate the last step of glycolysis by promoting the flux through glycolysis.
We provide evidence here for PKM2 induction in response to LPS in macrophages. This event is required for macrophage activation by LPS and we identify PKM2 as a critical modulator of IL-1β production, macrophage polarization, glycolytic reprogramming and Warburg metabolism in LPS-activated macrophages. PKM2 therefore presents itself as a therapeutic target in both cancer and inflammation.
RESULTS
Tetramerization of PKM2 using small molecule activators inhibits LPS-induced activation of primary bone marrow derived macrophages (BMDMs)
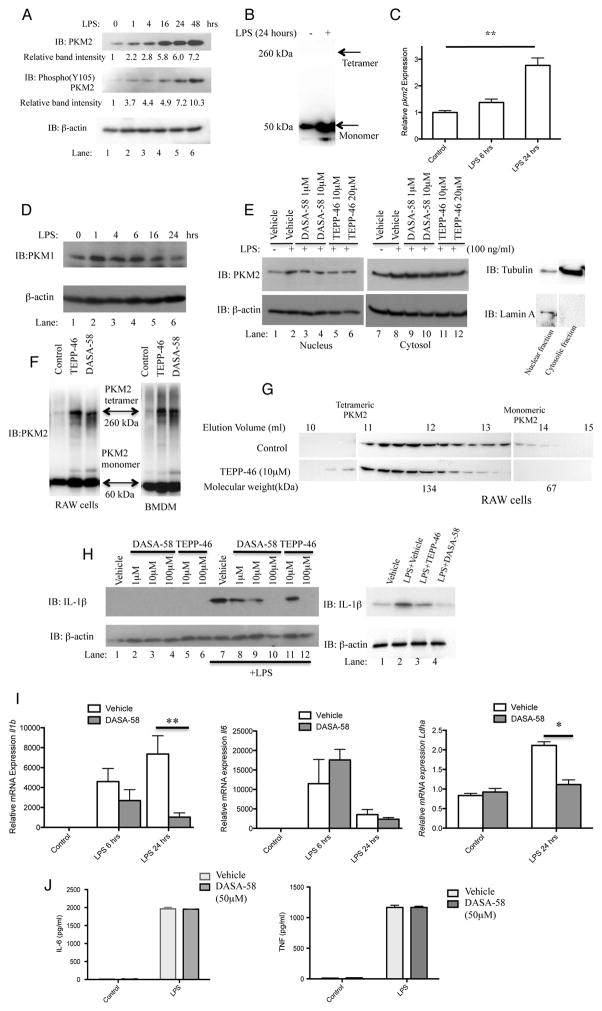
We hypothesized that similar to tumor cells, PKM2 would be required for Warburg metabolism in LPS-activated macrophages. LPS induced high levels of PKM2 in BMDMs up to 48 hours (Figure 1A). Disuccinimidyl suberate (DSS) crosslinking of lysates from LPS treated BMDMs suggest that LPS induced PKM2 is primarily of a monomeric configuration (Figure 1B). Furthermore, LPS strongly induced Pkm2 mRNA expression (Figure 1C), with expression levels increasing approximately 2.8 fold after 24 hours of LPS.
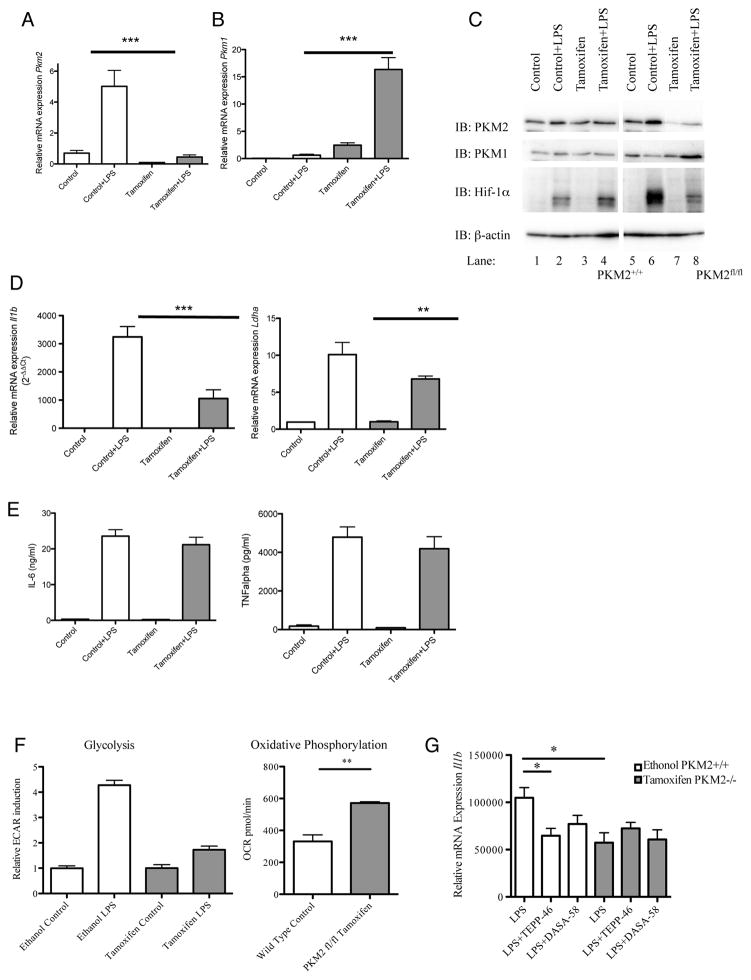
Figure 1. Tetramerization of LPS-induced PKM2 in primary BMDMs inhibits the Hif-1α targets IL-1β and Ldha.
LPS-stimulated BMDMs were assayed for expression of PKM2, Y105 phosphorylated PKM2 and β-actin by Western Blotting (A) and Pkm2 mRNA by qRT-PCR (C). (B) Crosslinking (500μM DSS) and western blot of endogenous PKM2 in BMDMs ±LPS (24 hrs). (D) LPS did not significantly affect expression of PKM1 in BMDMs. (E) BMDMs pretreated with ± DASA-58 or ±TEPP-46 as indicated, followed by LPS for 24 hours. Cytosolic and nuclear fractions were isolated and PKM2, β-actin, Lamin A, and Tubulin were detected by western blotting. (F) DSS crosslinking and western blotting of PKM2 in BMDMs and RAW macrophages after treatment ±100μM TEPP-58 or ±50μM DASA-46. (G) RAW macrophages treated with ±10μM TEPP-58 or DMSO (1h), followed by LPS. Protein separated by size exclusion chromatography and blotted for PKM2. (H) BMDMs (left) or PECs (right) were pretreated ±DASA-58 or TEPP-46 (30 min), followed by stimulation with LPS for 24 hours. Cell lysates were analyzed for pro-IL-1β or β-actin expression by western blotting. (I) Il1b (left panel), Il6 (middle panel) and Ldha (right panel) mRNA and (J) IL-6 (left) and TNF (right) protein expression were measured in BMDMs treated with ±DASA-58 and LPS for 6–24 hours. Data represents Mean ± SEM, n=3, **p<0.01.
Phosphorylation of PKM2 on Tyrosine 105 is indicative of monomer/dimer formation, as it prevents PKM2 tetramer configuration, further promoting the Warburg effect (Hitosugi et al., 2009). LPS- induced expression of PKM2 causes concurrent phosphorylation of Tyrosine 105 to an extent comparable or greater to the increased expression levels. This phosphorylation is evident after just one hour, with the strongest induction after 48 hours (Figure 1A middle panel, 10.3 fold increase in relative band intensity). This phosphorylation event suggests that PKM2 in LPS activated macrophages primarily forms an enzymatically inactive dimer or monomer. This is further supported by results from the cross-linking experiments shown in Figure 1B, where DSS cross-linked lysates from LPS treated BMDMs failed to reveal any high molecular weight PKM2 complexes.
Since PKM1 is the second alternatively spliced gene product of the Pkm gene we also examined protein expression of PKM1 in response to LPS. As shown in Figure 1D, PKM1 expression levels did not increase substantially in response to LPS but instead a slight decrease in PKM1 protein was observed after 24 hours of LPS, confirming that in relation to the Pkm gene, the effect of LPS is specific to PKM2.
In order to examine the functional relevance of PKM2 for LPS action we next examined the effect of two highly specific small-molecule PKM2 activators, TEPP-46 and DASA-58. These tool compounds were used to probe PKM2 function. They promote the formation of active PKM2 tetramers thereby boosting PKM2 enzymatic activity and limiting tumor growth in vivo (Anastasiou et al., 2012).
We confirmed the targeting of PKM2 by DASA-58 and TEPP-46 by assessing the effect of PKM2 activation on the subcellular localization of PKM2. As depicted in Figure 1E, LPS caused an increased expression level of PKM2 in the cytosol (compare lane 7 to lane 8) as well as in the nucleus (compare lane 1 to lane 2). However, driving PKM2 into tetramers using DASA-58 (compare lane 3–4 to lane 2) and TEPP-46 (compare lane 5–6 to lane 2) inhibited LPS-induced nuclear translocation. However such reduction of PKM2 in the cytosolic fraction was not evident in response to either compound. Fractions from vehicle treated samples were immunoblotted for Tubulin and Lamin A to control for preparation purity.
Since previous reports have established that PKM2 enters the nucleus as a dimer, we then carried out further DSS cross-linking studies in order to support a model for nuclear exclusion of PKM2 following treatment with our small molecule activators. RAW cells as well as primary BMDMs (Figure 1F) were treated with TEPP-46 and DASA-58 as indicated. Lysates were cross-linked using DSS and endogenous PKM2 was subsequently detected. The blot clearly reveals a high molecular weight PKM2-containing complex of approximately 250 kDa following treatment with TEPP-46 and DASA-58, pointing to a tetramer formation of PKM2. In addition, lysates from RAW cells treated with TEPP-46 were separated through size exclusion using fast protein liquid chromatography (FPLC) (Figure 1G). Fractions collected were analyzed by western blotting detecting PKM2 and the result reveals a shift in the molecular weight of PKM2 with the appearance of a band corresponding to approximately 250kDa (Figure 1G, elution volume 10–11 ml), and a reduction of the smaller molecular weight complexes (elution volume 13–14) again suggesting a tetrameric formation of endogenous PKM2 following TEPP-46 treatment.
We know from a previous study that LPS-induced transcription of the proinflammatory cytokine IL-1β is an event closely linked to the Warburg metabolism of activated macrophages (Tannahill et al., 2013). Hence we next examined the effect of activation of LPS-induced PKM2 using TEPP-46 and DASA-58 on the production of pro-IL-1β in activated BMDMs and peritoneal (PEC) cells. As shown in Figure 1H, DASA-58 and TEPP-46 inhibited the induction of pro-IL-1β in LPS-activated BMDMs. Both compounds also inhibited LPS-induced pro-IL-1β in PECs (Figure 1H). As shown in Figure 1I, activation of PKM2 using DASA-58 also inhibited LPS-induced expression of Il1b mRNA but importantly had no effect on LPS-induced Il6 mRNA levels. PKM2 activation also significantly inhibited LPS-induced expression of the Hif-1α target gene Ldha. Furthermore, LPS-induced IL-6 and TNF protein expression (Figure 1J) were unaffected by activation of PKM2 consistent with these cytokine responses not directly requiring Warburg metabolism (Tannahill et al., 2013).
To further examine the effects of the specific small molecule PKM2 activator in an unbiased fashion, we undertook a whole genome transcriptomics analysis of LPS and LPS+DASA-58 treated BMDMs. We identified a signature set of 100 genes (Figure S1, Table S1) for which the activation of PKM2 induced a markedly distinct response from that observed during LPS treatment alone. A general enrichment analysis of this gene-set for biological pathways and processes (using MetaCore) identified significant regulation in predominantly immune response pathways (Tables S2, S3). These included Interferon signaling, FcγR- mediated phagocytosis and IL4 signaling. To examine specifically for signatures of metabolic pathways, we analyzed enrichment of metabolic networks and found 48 pathways to be significantly enriched (FDR < 0.05) with Glycolysis being the top ranking metabolic pathway (Table S4). The key glycolytic enzymes underlying this signature are PFKP, ALDOC and TPI1, all three of which directly regulate the levels of FBP in this pathway. Phosphofructokinase (PFKP), which catalyzes the irreversible conversion of fructose-6-phosphate to FBP, is relatively down-regulated by activation of PKM2 whereas the enzymes downstream of this metabolite, namely ALDOC and TPI1, are significantly up- regulated by DASA-58. This suggested that we would expect to find significantly lower levels of FBP metabolite during PKM2 activation compared to LPS stimulation alone. To assess candidate transcription factors that are likely perturbed by activation of PKM2, we analyzed the signature set for enrichment of target genes of known transcription factors (TFs). We found enrichment for several interferon associated factors (IRF1, IRF8, ISGF3) as well as NRF2 and Hif-1α amongst the top 10 candidate TFs (Table S5). Several of the well-known targets of HIF1α, such as Vimentin, Plaur (uPAR), Stap1 (Art-27), Sort1, and PFKP were either down regulated or not expressed when BMDMs were stimulated with LPS in the presence of DASA-58.
These results suggest that activation of PKM2 in macrophages inhibited LPS-induced expression of pro-glycolytic and Hif-1α dependent genes.
Activation of PKM2 attenuates LPS-induced M1 macrophage polarization in BMDMs
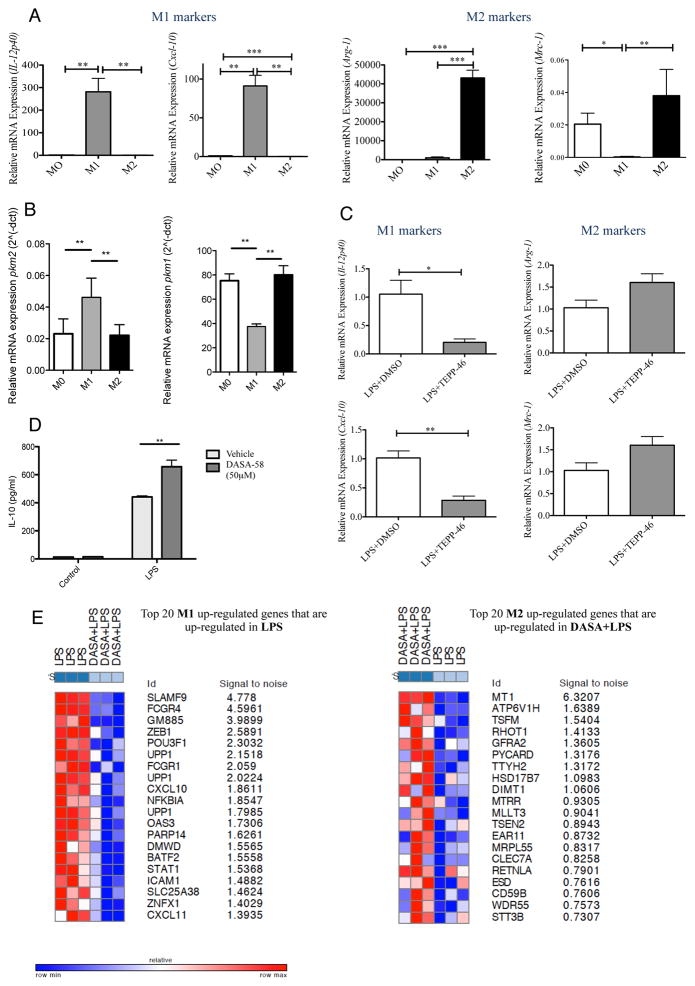
To further investigate the effect of activation of PKM2 on the pro- or anti-inflammatory phenotype of activated macrophages we set up polarizing conditions to generate M0 (undifferentiated), M1 (classically activated) or M2 (alternatively activated) macrophages as described. Figure 2A validates the polarizing conditions showing an upregulation of the markers Interleukin-12 p40 (Il12p40) and C-X-C chemokine 10 (Cxcl-10) in M1 macrophages and an increased expression of Arginase-1 (Arg-1) and Manose Receptor C, type 1 (Mrc-1) during M2 polarizing conditions. We next measured expression of PKM2 in these distinct macrophage populations and as Figure 2B shows, PKM2 expression is upregulated in M1 macrophages. In addition we see a clear down regulation of PKM1 mRNA in these M1 macrophages.
Figure 2. Activation of PKM2 using TEPP-46 attenuates the M1 attributes of LPS-activated BMDMs.
BMDMs were stimulated with 100ng/ml LPS or 20ng/ml IL-4 for M1 and M2 polarization respectively. 24 hours after stimulation RNA was extracted. (A) Left to right, expression of il12-p40, cxcl-10, arginase-1 and mrc-1 were analyzed to assess differentiation into M1 and M2 macrophages. (B) Expression of pkm2 (left) and pkm1 (right) was measured for each polarizing condition. (C) BMDMs were pretreated with 100μM TEPP-46 or DMSO (1hr), followed by LPS (24h). RNA was extracted and expression of il12-p40, cxcl-10, arginase-1 and mrc-1 was analyzed. (D) IL-10 expression measured in supernatants from BMDMs ± DASA-58 (50μM, 30min) followed by LPS (24h). Data represents Mean ± SEM, n=3, **p<0.01. (E) Transcriptomics dataset profile GSE53053 used to identify genes that were significantly up- or down- regulated in M1 or M2 polarizing conditions (Two tail Two sample T-test; nominal p-value <=0.05, Absolute log2-fold-change >= 2-fold). A chi-square test was performed to identify sets of up- or down- regulated genes with significant patterns from microarray data (Figure S1) comparing LPS+DASA-58 to LPS treated macrophages.
In order to examine the effect of our tool compounds on the expression of M1 and M2 signature gene subsets we next treated LPS activated M1 macrophages with TEPP-46. As seen in Figure 2C, the expression of the M1 markers Il12p40 and Cxcl-10 are significantly down regulated following TEPP-46 treatment. Similarly, although not significant we consistently observed a boost in the expression of the M2 markers Arg-1 and Mrc-1 by TEPP-46 in M1 polarized macrophages.
Interestingly, activation of PKM2 boosted LPS-induced expression of the anti-inflammatory M2 cytokine Interleukin-10 (IL-10) in BMDMs (Figure 2D) further supporting an attenuation of the M1 phenotype after activation of PKM2 in favor of a more M2-like macrophage.
To conclusively establish whether addition of DASA-58 prior to LPS treatment of macrophages led to an attenuation of LPS induced M1 traits and these cells becoming more M2-like, we first downloaded and analyzed a transcriptomics profile of bone-marrow derived macrophages treated with either IL4 or a combination of LPS+Interferon-γ (IFNγ)(GSE53053). Using this dataset, we identified genes that were significantly up- or down- regulated in either M1 or M2 polarizing conditions (Two tail Two sample T-test; nominal p-value <=0.05, Absolute log2-fold-change >= 2-fold). Next we used a chi-square test to assess which set of up- or down- regulated genes showed significant patterns in our own microarray data (from Supplemental Figure 1) comparing LPS+DASA-58 treated macrophages to LPS treated cells. We found that genes upregulated in M1 cells were significantly upregulated by LPS treatment compared to LPS+DASA-58 (Figure 2E). In addition to this, genes upregulated in the M2 signature set were also enriched for genes upregulated by DASA+LPS compared to LPS alone but to a lesser extent with fewer number and lesser degree of fold-change.
This effect on expression of M1 and M2 markers, together with our observation that DASA-58 causes an increased expression of LPS- induced IL-10 suggests that activation of PKM2 in LPS activated macrophages may help dampening the pro-inflammatory phenotype of the cell.
LPS promotes binding of PKM2 and Hif-1α to the IL-1β promoter
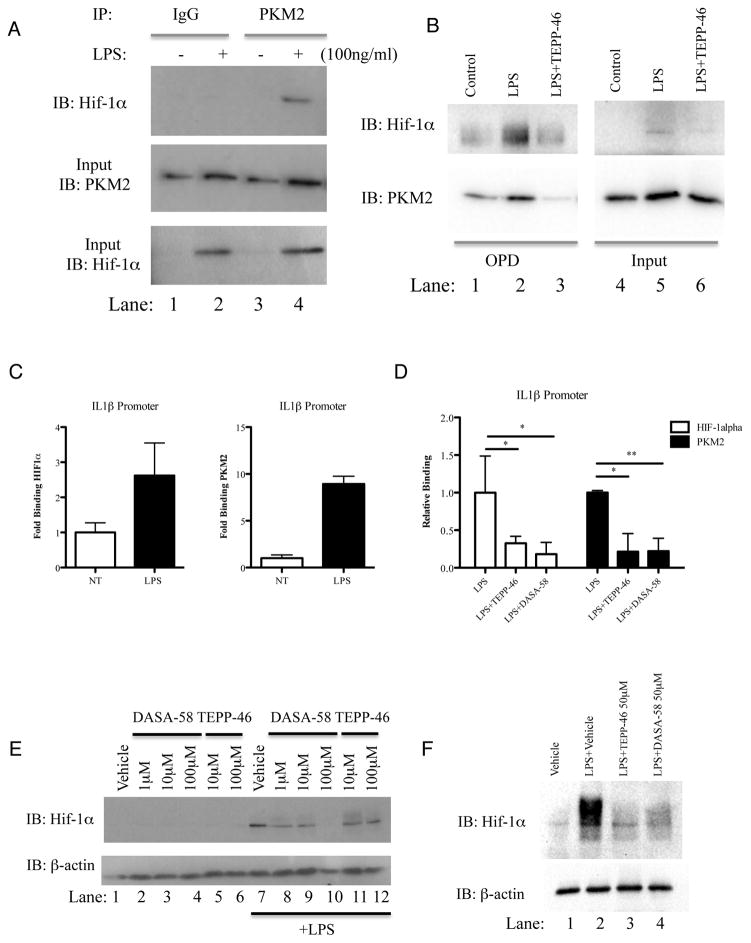
PKM2 has been shown to interact with Hif-1α and stimulate Hif-1α transactivation domain function and recruitment of p300 to the Hif Response Element (HRE) of Hif-1α target genes (Luo et al., 2011). Treatment of BMDMs with LPS for 24 hours increased the association of endogenous PKM2 with endogenous Hif-1α, as shown in Figure 3A.
Figure 3. Dimeric/monomeric PKM2 is required for LPS-induced binding of PKM2 and Hif-1α to the IL-1β promoter.
Immunoprecipitation of PKM2 in BMDMs treated with LPS (24h). Hif-1α measured by immunoblotting (A). (B) BMDMs treated ±TEPP-46 (50μM, 60min) followed by LPS (24h) were lysed and an OPD assay was carried out using oligonucleotide spanning the HIF1α binding site on the IL1β promoter. Samples were probed for Hif-1α (top) and PKM2 (bottom). Representative of n=3. Sequential ChIP–PCR using PKM2 and HIF-1α antibodies and primers specific for −300 position of Il1b in LPS- treated BMDMs (C) and (D) BMDMs treated with TEPP-46/DASA-58 (30 minutes, 50 μM) and LPS (100ng/ml, 24 hrs). ChIP data are calculated as percent of input, represented as fold binding ±SD for one representative experiment (n=3). (E) BMDMs (left panel) or PECs (right panel) (1×106 cells per ml) were pretreated with indicated doses of DASA-58 or TEPP-46 (30 min), followed by LPS (100 ng/ml, 24h). Hif-1α or β-actin expression analyzed by western blotting.
In order to investigate binding of PKM2 to the Hif-1α specific binding site of the IL-1β promoter we next used two approaches. Firstly we employed an oligo pulldown (OPD) assay where an oligonucleotide specific to the Hif-1α binding site of the IL-1β promoter (-408, (Tannahill et al., 2013)) was incubated with lysates from TEPP-46 and LPS treated primary BMDMs as indicated (Figure 3B). These results reveal binding of PKM2 to the IL-1β promoter (lower left panel). In addition, there is an increased binding of both PKM2 and Hif-1α to this promoter site in response to LPS (compare lane 2 to lane 1). Furthermore tetramerization of PKM2 using TEPP-46 inhibits LPS-induced binding of PKM2 and Hif-1α to the IL-1β promoter (compare lane 3 to lane 2).
We extended our binding studies to include Sequential Chromatin Immuno-Precipitation (ChIP) assays. This technique allows us to simultaneously examine the endogenous binding of two transcription factors to chromatin in response to LPS. As seen in Figure 3C, LPS causes increased binding of endogenous Hif-1α to the IL-1β promoter (left panel), and interestingly after this sample was put through a second round of ChIP specific for PKM2 (right panel) we also see an increased binding of endogenous PKM2 to the same copy of DNA in response to LPS. Furthermore, activation of PKM2 using TEPP-46 and DASA-58 causes significant inhibition of LPS induced binding of both Hif-1α and PKM2 to the IL-1β promoter (Figure 3D).
A role for PKM2 in LPS-induced stability of Hif-1α was further verified by directly measuring LPS-induced Hif-1α protein as shown in Figure 3E, where DASA-58 and TEPP-46 inhibited the induction of Hif-1α protein in LPS-activated BMDMs. Both compounds also inhibited LPS- induced Hif-1α protein in peritoneal macrophages (Figure 3F).
PKM2 activators inhibit glycolysis and the accumulation of succinate in LPS-activated macrophages
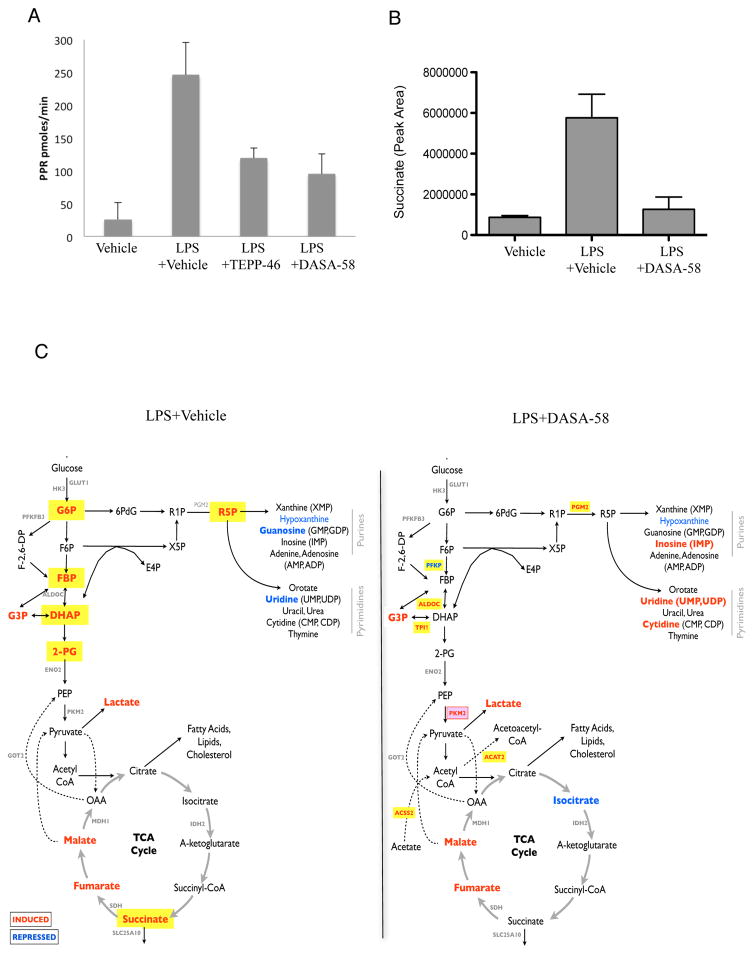
We next examined the metabolic consequences of PKM2 induction by LPS. We first investigated if activation of PKM2 could inhibit LPS- induced glycolysis. BMDMs were treated with TEPP-46 and DASA-58 prior to LPS and extracellular acidification was monitored as shown in Figure 4A. LPS caused up to a ten-fold increase in glycolysis, which is markedly inhibited by both TEPP-46 and DASA-58. Neither TEPP-46 nor DASA-58 had a significant effect on oxidative phosphorylation as measured by oxygen consumption rate (data not shown).
Figure 4. Activation of PKM2 counteracts LPS induced excessive rate of glycolysis and restores cellular levels of succinate.
Rate of glycolysis in BMDMs treated ±TEPP-46 (50μM) or DASA-58 (50μM) ±LPS (A). Succinate levels in LPS-treated BMDMs ±DASA-58 (50μM) represented as relative abundance (B). (C) Schematic map illustrating key metabolites and genes that were significantly enhanced (red) or inhibited (blue) in LPS treated (100ng/ml, 24h) BMDMs ± 50μM DASA-58. All metabolites with significant accumulation (p-value < 0.05) are shown in bold red text. Those in yellow suggest up-regulation specific to LPS stimulation. Statistical analysis performed on 3 separate experiments. Metabolites with p value < 0.05 and fold-change > 10% were deemed to be statistically significant.
One important consequence of LPS-induced metabolic reprogramming in macrophages is a build-up of succinate, which acts as an inflammatory signal contributing to increased Hif-1α dependent IL-1β expression (Tannahill et al., 2013). In agreement with a role for PKM2 in LPS action here, DASA-58 dramatically blocked LPS-induced succinate (Figure 4B).
We carried out a comprehensive metabolomic profiling of LPS-activated BMDMs in the presence or absence of DASA-58. We revealed a clear effect of PKM2 activation by DASA-58 in preventing LPS-induced glycolysis (Figure 4C and Table S6). LPS stimulation led to accumulation of upstream glycolytic intermediates, such as Glucose-6-phosphate (G6P), FBP, Dihydroxyacetone phosphate (DHAP) and Phosphoglyceric acid (2-PG), and TCA cycle intermediates including Malate, Fumarate and Succinate (Figure 4C; Left panel; all metabolites with significant accumulation (p-value < 0.05) are shown in bold red text. Those highlighted in yellow indicate up-regulation following LPS stimulation). Activation of PKM2 prevented the accumulation of any of these glycolytic intermediates. These findings are consistent with the microarray results where we found evidence to suggest depletion of FBP during activation of PKM2. Additionally, the pentose phosphate pathway intermediate Ribose 5-phosphate (R5P), was significantly accumulated in response to LPS (Figure 4C; left panel), however this was alleviated by activation of PKM2 (right panel). Furthermore a significant up-regulation in levels of purines and pyrimidines downstream of the pentose phosphate pathway (such as Cytidine, Inosine, and Uridine) were observed in DASA-58-treated samples (Figure 4C; right panel) suggesting that PKM2 can somehow promote the process of pyridine and pyrimidine biosynthesis downstream of R5P. These metabolomic changes are consistent with DASA-58 activating PKM2, which would lead to a decrease in glycolytic and pentose phosphate pathway intermediates.
Isoform-specific deletion of PKM2 inhibits LPS-mediated activation of Hif-1α and expression of Hif-1α target genes in macrophages
In order to confirm a role for PKM2 in LPS signaling we next used BMDMs generated from mice carrying a conditional PKM2fl/fl allele allowing for Cre-recombinase-mediated deletion (Israelsen et al., 2013) and matched wild type controls. Isolated BMDMs were treated ±600 nM Tamoxifen for 72 hours prior to the experiment in order to excise the PKM2 specific Pkm Exon 10. Pkm2 expression was successfully ablated (Figure 5A), however as expected Pkm1 expression was compensatorily increased (Figure 5B). In addition, PKM2 protein levels were successfully reduced following treatment with Tamoxifen (Figure 5C top panel, compare lanes 5 and 6 to lanes 7 and 8), whereas PKM1 protein was upregulated following Tamoxifen and LPS-treatment compared to control (Figure 5C second panel). LPS-induced Hif-1α protein was dramatically decreased in PKM2 knock-out cells compared to mock treated cells expressing PKM2 (Figure 5C third panel), compare lane 2 to 4 in relation to lane 6 to 8). Furthermore, as a functional consequence of this, BMDMs lacking PKM2 displayed a significantly reduced mRNA expression level of the Hif-1α responsive genes Il1β and Ldha in response to LPS (Figure 5D). In contrast, LPS-induced IL-6 and TNFα expression (Figure 5E) were comparable in Tamoxifen treated versus mock treated PKM2fl/fl BMDMs. This result clearly pointed to the requirement for PKM2 in the activation of HIF1α in response to LPS.
Figure 5. Inhibition of LPS-induced Hif-1α and Hif-1α target genes in PKM2 depleted BMDMs.
BMDMs from mice carrying a PKM2fl/fl allele and relative PKM2+/+ controls were treated ±600 nM Tamoxifen (72h), followed by LPS (24h). Relative mRNA expression levels of Pkm2 (A), Pkm1 (B), Il1b and Ldha (D) were measured by qRT-PCR. PKM2, PKM1, Hif-1α and β-actin protein expression was measured by Western blotting (C). (E) IL-6 and TNFα protein measured by ELISA, depicted as means ± SD of results from triplicate determinations for one representative experiment, n=2. (F) Rate of glycolysis (left) and oxidative phosphorylation (right) in ethanol (PKM2+/+) and Tamoxifen (PKM2−/−) treated LPS-activated BMDMs derived from PKM2fl/fl mice, measured as ECAR and OCR ±SD (n=5). (G) BMDMs derived from PKM2fl/fl mice treated with ethanol (PKM2+/+) or Tamoxifen (PKM2−/−) followed by TEPP-46 or DASA-58 (30 min) and LPS (24h) as indicated. The cells were lysed and expression of il1b mRNA was determined by qRT-PCR.
Using BMDMs generated from PKM2fl/fl mice we measured rate of glycolysis in response to LPS. Macrophages with diminished expression of PKM2 displayed a reduced rate of glycolysis in response to LPS compared to cells with an intact PKM2 gene (depicted in Figure 5F). Furthermore, BMDMs with an impaired PKM2 gene exhibit an increased rate of oxidative phosphorylation as measured by oxygen consumption rate (OCR) (Figure 5F). However, although as expected LPS causes a reduction on OCR in PKM2+/+ cells (Tannahill et al., 2013), the same inhibitory effect was still observed in BMDMs derived from PKM2−/− cells (data not shown). Taken together these results point to a key role for PKM2 in LPS-induced glycolysis.
In order to control for off-target effects of TEPP-46 and DASA-58, Tamoxifen-treated BMDMs derived from conditional PKM2fl/fl deficient mice were treated with TEPP-46 prior to LPS and relative IL-1β expression was measured (Figure 5G). LPS-induced Il1b expression was attenuated in PKM2 ablated cells, with no further inhibitory effect observed following pre-treatment with TEPP-46 or DASA-58.
Activation of PKM2 using TEPP-46 modulates the anti-Mycobacterium tuberculosis response of macrophages
To extend our investigations into PKM2 beyond LPS to an infectious agent that utilizes TLRs other than TLR4 we next examined a bacterial pathogen, Mycobacterium tuberculosis (Mtb). As the main causative agent of tuberculosis, Mtb signals mainly through TLR 2, 6 and 9 (Bafica et al., 2005; Means et al., 1999).
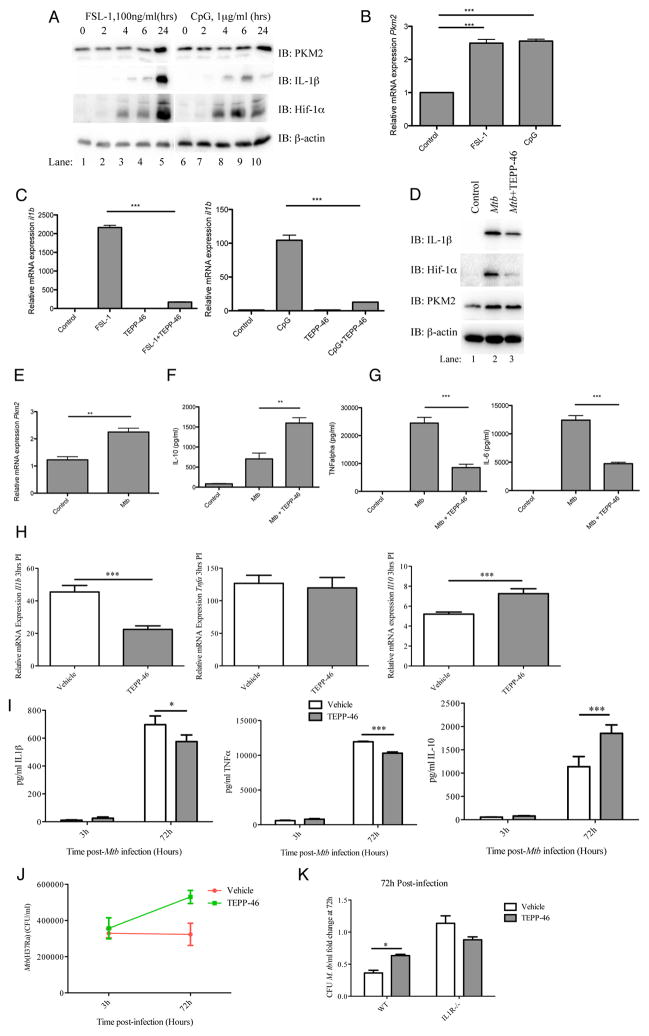
Resting BMDMs were stimulated with FSL-1, a ligand for TLR2 and TLR6 or the TLR9 ligand CpG (Figure 6A) for up to 24 hours. Similarly to LPS these ligands induced expression of PKM2 both at protein level (Figure 6A) and mRNA level (Figure 6B). In addition, both FSL-1 and CpG induced pro-IL-1β and Hif-1α protein expression (Figure 6A). Activation of PKM2 using TEPP-46 significantly inhibited FSL-1 and CpG-induced Il1b mRNA expression (Figure 6C).
Figure 6. Activation of PKM2 modulates anti-mycobacterial macrophage responses.
Cell lysates from FSL-1 and CpG (24hrs) treated BMDMs were analyzed for PKM2, IL-1β, Hif-1α or β-actin expression by western blotting (A), and pkm2 mRNA by qRT-PCR (B). BMDMs pretreated with TEPP-46 (50μM, 30min) were activated using FSL-1 (100ng/ml) and CpG (1μg/ml) for 24 hours. il1b was analyzed by qRT-PCR (C). Expression levels of IL-1β, Hif-1α, PKM2 and β-actin protein (D), pkm2 mRNA (E), IL-10 (F), TNFα and IL-6 protein (G, left and right) were measured in BMDMs ±TEPP-46 (30 min) stimulated using heat inactivated Mtb. (H) BMDMs ±TEPP-46 (25μM) were infected with live Mtb H37Ra (MOI 5 bacteria/cell, 3hrs) and gene expression of Il1b (left), tnf (middle) and Il10 (right) mRNA analysed (qRT-PCR). Data is mean ± SD for triplicate determinations, n=2. (I) BMDMs ±TEPP-46 (25μM) were infected as above (3 and 72hrs). IL-1β (left), TNFα (middle) and IL-10 (right) production were measured in supernatants of infected cells. BMDMs from (I) were lysed and CFU/ml determined (J). (K) BMDMs derived from wild type or IL-1 Type I Receptor knock out cells were infected as for (H) above. Cells were lysed at 72 hours post-infection and CFU/ml determined. Depicted as means ± SD of results from triplicate wells for one representative experiment n=2. *P<0.05, **P<0.01, ***P<0.001 (two-way analysis of variance with post-hoc Bonferroni correction).
We next turned to intact heat-inactivated Mtb as a stimulus. Results from BMDMs treated with Mtb for 24 hours revealed an upregulation of PKM2 protein (Figure 6D, third panel, compare lanes 1 and 2) as well as PKM2 mRNA (Figure 6E). In addition, TEPP-46 inhibited Mtb-induced pro-IL-1β and Hif-1α protein, (Figure 6D), as well as TNFα and IL-6 (Figure 6G), whilst significantly boosting Mtb induced IL-10 (Figure 6F). This result indicates an important role for PKM2 in innate immune host responses originating from Toll-like receptors other than TLR4.
In a model of macrophage infection using live Mtb (strain H37Ra), TEPP-46 inhibited Mtb-induced Il1b mRNA levels, boosted Mtb-induced levels of Il10 mRNA and had no effect on levels of Tnf (Figure 6H). In addition, Mtb-induced IL-1β but also TNFα protein levels were inhibited by TEPP-46, whereas IL-10 protein was significantly increased by activation of PKM2 (Figure 6I).
Whilst no significant differences were observed in bacterial uptake 3 hours post-infection, Figure 6J shows an increased level of intracellular bacteria at 72 hours post-infection in TEPP-46-treated, Mtb-infected BMDMs compared to Mtb–infection alone. These data are consistent with IL-10 promoting pathogen persistence by contributing to Mtb- phagosome maturation (O’Leary et al., 2011).
As illustrated in Figure 6K BMDMs derived from IL-1 Type I Receptor knock out mice when infected with Mtb display an increased bacterial load compared to wild type cells, with no effect of TEPP-46, demonstrating the requirement for intact IL-1 autocrine signaling for efficient bacterial killing.
Activation of PKM2 results in increased bacterial dissemination in a Salmonella typhimurium model of infection
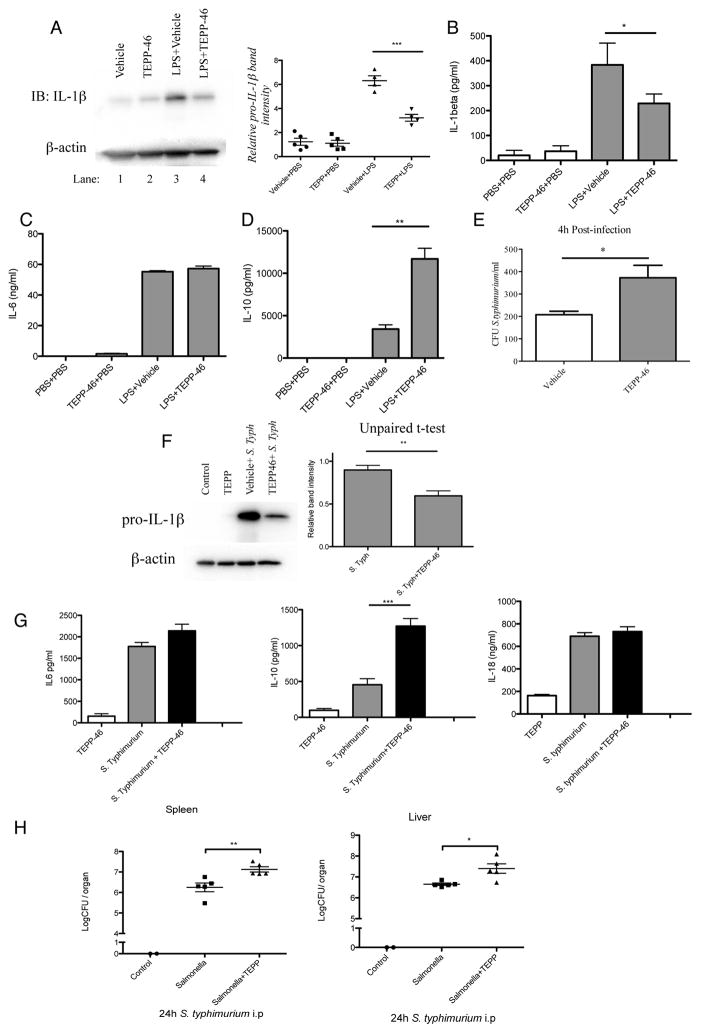
Up-regulation of aerobic glycolysis in macrophages during endotoxemia plays a central part in the pathology of this disease (Tannahill et al., 2013). TEPP-46, but not DASA-58, is pharmacokinetically validated in vivo (Anastasiou et al., 2012). TEPP-46 efficacy as a PKM2 activator had been verified using key in vitro assays as discussed above. To investigate the protective effects of activation of PKM2 in vivo, mice were given TEPP-46 prior to an LPS challenge in a model of sepsis. As shown in Figure 7A this dramatically reduced levels of pro-IL-1β produced by the peritoneal cells of these mice, compared to LPS treatment alone. Furthermore, administration of TEPP-46 prior to LPS resulted in animals expressing less IL-1β in the serum (Figure 7B) whilst IL-6 levels were the same as in the LPS-alone-treated animals (Figure 7C). As observed in BMDMs, serum levels of IL-10 (Figure 7D) were greatly increased in animals given TEPP-46 prior to the LPS challenge, compared to the control group. This suggests that activation of PKM2 in vivo counteracts the inflammatory effects of LPS, most likely through inhibiting Warburg metabolism.
Figure 7. Activation of PKM2 in vivo diminishes the host immune response in LPS-induced sepsis and in an S. typhimurium model of infection.
Pro-IL-1β in PECs isolated from mice injected i.p. with TEPP-46 (50mg/kg) or vehicle control (20% 2-Hydroxypropyl-β-cyclodextrin) for 1 hour, followed by PBS or 15mg/kg LPS for 2 hours (A). Left panel shows one representative sample from each treatment group. Right panel represents densitometry readings of pro-IL-1β western blots from 5 mice per group and treatment, normalized to β-actin. Serum levels of IL-1β (B), IL-6 (C) and IL-10 (D) from mice in (A). n=5 for each group, mean±SEM, **p<0.01. (E) BMDMs were treated ±TEPP-46 (30 min, 25 μM), prior to infection with S. typhimurium UK-1 strain at an MOI of 10 bacteria/cell. Bacterial numbers were assessed at 4h p.i. CFU/ml enumerated 4h after plating. Mice were injected ±TEPP-46 (50mg/kg), 1 hr prior to infection with S. typhimurium (1×106 CFU, 2h). Pro-IL-1β from PECs was measured by western blotting (F), and serum levels of IL-6, IL-10 and IL-18 were measured by ELISA (G). (H) Mice were infected as (F), and sacrificed 24h post infection. Livers and spleens were extracted and Log CFU/organ was determined (n=5 per group, mean ±SEM, two tailed t-test ).
In order to further investigate the effect of TEPP-46 in a model of infection, we used Salmonella typhimurium (strain UK-1) in an in vitro model of infection. BMDMs were treated with TEPP-46 prior to infection with S. typhimurium. 4 hours post infection intracellular bacterial load was counted (Figure 7E). Cells treated with TEPP-46 displayed an increased bacterial load probably due to decreased bacterial killing by the cell.
We next used S. typhimurium in an in vivo model of infection. Similarly to the results obtained from the LPS induced model of sepsis, peritoneal cells from mice treated in vivo with TEPP-46 prior to infection with S. typhimurium display a significant reduction in IL-1β protein expression levels (Figure 7F) 2 hours post infection. Serum levels of IL-6 and IL-18 (Figure 7G) remained unchanged in TEPP-46 treated animals compared to the control group, however once again serum levels of IL-10 were greatly enhanced in animals that were given TEPP-46 prior to infection.
In order to measure bacterial dissemination mice were intraperitoneally infected with S. typhimurium and sacrificed 24h post infection. Livers and spleens were extracted and log colony forming units (CFU)/organ was determined as illustrated in Figure 7H. The bacterial load in the spleens as well as in the livers in animals given TEPP-46 prior to infection were significantly increased. This is likely to be due to decreased bacterial killing due to the ability of TEPP-46 to inhibit IL-1β production whilst simultaneously promoting production of anti- inflammatory IL-10.
DISCUSSION
Inflammatory immune cells, such as ‘M1’ type macrophages, dendritic cells or Th17 cells harbor many of the metabolic changes displayed by tumor cells. This allows the host immune system to support anabolic processes and meet the increased demand for biosynthetic precursors required for mounting an immune response. Central to the metabolic switch observed in tumors is PKM2, which we now demonstrate also plays a key role in the glycolytic switch occurring in LPS-activated macrophages. We demonstrate PKM2 up-regulation and phosphorylation in an activated macrophage, with only a minor change in PKM1 protein expression. As LPS induces formation of a PKM2 and Hif-1α complex that can bind to the IL-1β promoter, this points to a role for PKM2 as an important regulator of Hif-1α activity, and Hif-1α-dependent genes in host defense, as previously described in hypoxia and cancer where nuclear PKM2, with PHD3 acting as a cofactor, regulates the transcriptional activity of Hif-1α (Luo et al., 2011).
In addition, using isoform specific PKM2 conditional knock out cells, we show that induction of PKM2 by LPS is critical for Hif-1α stabilization, in agreement with previous studies demonstrating a PKM2/Hif-1α complex targeting Hif-1α expression (Luo et al., 2011). Hif-1α will also be stabilized by succinate inhibiting PHDs, providing further regulation of Hif-1α.
Current data indicates that PKM2, when highly expressed, exists in an equilibrium of enzymatically inactive pyruvate kinase dimers (or monomers) which can translocate into the nucleus(Christofk et al., 2008; Hitosugi et al., 2009), and enzymatically active tetramers which are retained in the cytosol(Mazurek et al., 2005). The pyruvate kinase activity of the tetramers can be regulated by the concentration of the naturally occurring upstream glycolytic intermediate FBP (Ashizawa et al., 1991; Jurica et al., 1998), serine (Chaneton et al., 2012), as well as by a multitude of post-translational modifications (Wu and Le, 2013) including tyrosine phosphorylation. Activity is controlled by stabilizing or destabilizing the tetramer (Hitosugi et al., 2009). Tyrosine phosphorylation of PKM2 at residue 105 will inhibit the formation of a tetramer by dislodging the binding of FBP. This event has been shown to be important for the Warburg effect in hypoxia and cancer(Hitosugi et al., 2009). We have found that this also occurs in LPS activated macrophages, reflecting the similarities in metabolic changes observed in cancer, and suggesting a role for PKM2 in the Warburg effect in macrophages. This role was confirmed using PKM2 activating compounds DASA-58 and TEPP-46. These well-characterized and highly specific compounds bind PKM2, which then forms a tight tetramer with PKM1-like kinetic properties, an event that is resistant to inhibition by tyrosine phosphorylation. In addition to relieving the build-up of glycolytic intermediates, evident from the higher pyruvate kinase activity of the PKM2 tetramer (Anastasiou et al., 2012), this process also prevents nuclear translocation and therefore decreases the expression of several Hif-1α target genes such as Ldha (Luo et al., 2011). We show that activation of PKM2 by DASA-58 or TEPP-46 forces PKM2 to take on a tetrameric form. This impairs the ability of the PKM2 and Hif-1α complex to bind to the Hif-1α motif in the promoter of the known Hif-1α target gene IL-1β (Peyssonnaux et al., 2007; Zhang et al., 2006), thereby inhibiting the transcriptional activity of Hif-1α and induction of IL-1β and conceivably other Hif-1α-dependent genes in response to LPS. This validates a role for dimeric/monomeric PKM2 in LPS action here. Interestingly, the effects of activating PKM2 also extends into changing the polarizing phenotype of a macrophage, with tetramerization of LPS- induced PKM2 in an M1 macrophage promoting characteristics typical of the M2 phenotype. This further implies the activation of PKM2 as a potential therapeutic target in inflammatory disease.
Activation of PKM2 also inhibited induction of IL-1β and boosted IL-10 production in Mtb-infected macrophages promoting bacterial growth. This extended our observation beyond TLR4 and LPS to a bacterial pathogen that utilizes TLR2, TLR6 and TLR9.
We demonstrate the impact tetramerization of PKM2 has on limiting the host immune response in vivo, by attenuating both LPS and S. typhimurium-induced IL-1β whilst boosting IL-10, leading to impaired clearance of infection evident from increased bacterial dissemination. Our results are in agreement with the study of Yang et al who demonstrated that inhibition of PKM2 with shikonin prevented lethality in a model of septic shock, although the mechanism of shikonin is not fully understood (Yang et al., 2014).
We confirmed that LPS stimulation leads to accumulation of upstream glycolytic intermediates, such as G6P, FBP, DHAP and 2-PG, and TCA cycle intermediates including succinate (Tannahill et al., 2013). Activation of PKM2 using DASA-58 dramatically reverts this glycolytic phenotype. Coupled results of metabolomics and microarray analysis further indicated PKM2 mediated metabolic reprogramming of activated macrophages. The data indicates a combination of transcriptional regulation of key enzymes to increase flux into lactate, divert metabolites into the pentose phosphate pathway and thereby divert pyruvate away from the TCA cycle. These events are all prevented by PKM2 activation by DASA-58 and TEPP-46, which thereby prevent macrophage activation by LPS.
Another consequence of the metabolic changes in LPS-activated macrophages is an increase in succinate. Activation of PKM2 also limits this response and this will lead to Hif-1α destabilization. From our data, the effect of PKM2 activation on LPS-activated macrophages is therefore threefold. Firstly, it will limit the nuclear function of PKM2 and decrease Hif-1α-dependent gene expression. Secondly, the decrease in succinate will further limit Hif-1α through stabilization and degradation. Finally, it will decrease levels of glycolytic and pentose phosphate pathway metabolites, thereby limiting biosynthesis. The increased pyruvate kinase activity of PKM2 will also increase the flux of pyruvate into the TCA cycle. This could explain the increase in IL-10 production, which in the case of M2 macrophage activity may require mitochondrial oxidative metabolism (Krawczyk et al., 2010). Precisely how PKM2 promotes IL-10 production requires further analysis. PKM2 in its dimeric, catalytically less active form, is therefore required for glycolytic reprogramming in response to LPS, most likely because of the role dimeric PKM2 plays in Hif-1α function.
In conclusion, we have revealed a novel role for PKM2 in LPS induced activation of macrophages and in macrophages infected with Mtb, as well as in an in vivo S.typhimurium model of infection providing important insights into the Warburg-like metabolic changes in macrophages, likely to be very important for inflammation and infection. Targeting PKM2 in order to inhibit glycolysis or re-engaging mitochondrial oxidative metabolism in an overactive macrophage may provide novel therapeutic approaches for inflammatory diseases.
EXPERIMENTAL PROCEDURES
Reagents
TEPP-46 and DASA-58 synthesized in accordance with published methods (Boxer et al., 2010; Jiang et al., 2010). LPS used in vitro (100 ng/ml) and in vivo were E. coli, serotype EH100 (Alexis), and 055:B5 (Sigma-Aldrich). 4-hydroxytamoxifen (H7904) from Sigma. Antibodies: anti-PKM2 (3198), antiphospho-PKM2 (Tyr105), β-actin (4267) (all Cell Signaling Technologies), anti-IL-1β (R&D, AF401-NA), anti-HIF-1α (Novus, NB100-449), and anti-PKM1 (Novus, NBP2-14833). Dimethyl sulfoxide (DMSO) used as vehicle control for TEPP/DASA in all in vitro assays.
Mice and cell culture
BMDMs and PECs isolated from C57BL/6 mice from Harlan UK as previously described(Tannahill et al., 2013). All experiments were carried out with prior ethical approval from Trinity College Dublin Animal Research Ethics Committee. Cells were used at 1×106 cells/ml unless otherwise stated. Each ‘n’ represents BMDM/PECs from individual mice.
BMDMs from mice carrying a PKM2fl/fl allele were previously described (Israelsen et al., 2013). Ablation of PKM2 was achieved by adding 600nM 4-hydroxytamoxifen (Sigma H7904) on day 4 of macrophage differentiation, until 24 hrs prior to experiment.
Western blotting
Western blot analysis was carried out as previously described (Fitzgerald et al., 2001). Western blots were developed using autoradiographic film, or using a Gel Doc EZ System gel imaging system alternatively.
RNA Isolation and Gene Expression
RNA was transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For mRNA, 18s ribosomal RNA or RPS18 gene were used as housekeeping controls. Relative quantitation values were calculated using the 2(−Delta Delta C(T)) method (Livak and Schmittgen, 2001).
Nuclear and cytosolic fractionation
Nuclear Extract Kit (Active Motif, 40010) was used according to manufacturers recommendations. 20 μl of each fraction was separated by SDS-PAGE prior to western blotting.
Crosslinking
BMDMs or RAW 264.7 macrophages were treated with 50μM TEPP-46, 20μM DASA-58 or DMSO. Crosslinking was performed using 500μM disuccinimidyl suberate (Thermo Scientific Pierce) for 30 minutes. Lysates were analysed by Western Blot.
Size exclusion chromatography
2–3 mg protein from RAW 264.7 ±10μM TEPP-58 or DMSO (1 hr), followed by 100ng/ml LPS (24hrs) was loaded on a Superdex 200 10/300GL column (GE Healthcare) and eluted with 50mM sodium phosphate, 150mM NaCl, pH7.5. 250μL fractions were taken and analysed by SDS-PAGE and Western Blot.
ELISA
ELISAs were performed according to manufacturers’ instructions. Kits used were IL-1β (DY401), IL-6 (DY406), IL-10 (DY417), TNFα (DY410) and IL-1β (MLB00C, in vivo), IL-10 (M1000B, in vivo) (all from R&D). Results presented as mean±SEM. Two-tailed t-tests were carried out.
Microarray Profiling and Measurement of LC-MS
Microarray and Metabolomics profiling were performed as described in Tannahill et al (Tannahill et al., 2013).
Statistical analysis of microarray data
Statistical analysis of microarray data was carried out in MATLAB. Data was checked for correlation between replicates, (>0.96 on average within groups). Probes without any gene assignments were removed and further filtered on the basis of having a variance, signal or entropy less than 10th percentile of the entire dataset. Remaining probes were analyzed for statistically significant differences (fold-change > 25%) between three groups (two-tailed t-test assuming equal variance). 107 probes (100 genes) were identified as statistically significant across the three groups. p-values were corrected for multiple hypothesis testing by Benjamini- Hochberg’s method to control false discovery rate at 25%.
Co-Immunoprecipitation
BMDMs (±LPS, 24 hrs) were lysed as previously described (Fitzgerald et al., 2001). PKM2 immune complexes were precipitated using anti-PKM2 antibody coupled to protein A/G PLUS agarose beads (Santa Cruz Biotechnology) for 2 h at 4°C. Non-specific Rabbit IgG antibody was used for control. Immunoprecipitated proteins were visualized by western blotting using anti-PKM2 or Hif-1α antibodies respectively. Lysates were blotted for PKM2 and Hif-1α to control for input.
Affinity Purification with Biotinylated Oligonucleotides
Oligonucleotides for the HIF1α binding site on the IL1β promoter were annealed as described (Quinn et al., 2014)(forward, 5′BIO-ggt agg cac gta gat gca cac c-3′; reverse, 5′ggt gtg cat cta cgt gcc tac c-3′). BMDMs (0.5×106 cell/ml) were treated with DASA (50μM) or TEPP (100μM) for 1 hr prior to LPS (24 hours). Oligonuceotide pulldown was performed as previously described (Quinn et al., 2014).
Chromatin Immunoprecipitation
BMDMs (0.5×106 cell/ml) were treated ±DASA-58 or TEPP-46 (1 hr) prior to LPS treatment for 24 h. ChIP was performed as previously described (Quinn et al., 2014; Tannahill et al., 2013). Lysates were incubated with primary antibodies; Anti-HIF-1-α antibody (Abcam, ab2185), negative control anti-IgG (Sigma, I5006), and positive control Pol II Antibody (Santa Cruz Biotechnologies (N20) sc-899). For Sequential ChIP, the precipitated HIF1α sample was reprobed for binding of PKM2 (2 hours incubation, 30μl pre-blocked Protein A/G beads, 30μl PKM2 D78AXP antibody (4053)). qRT-PCR was carried out using primers for the IL1β promoter consensus HIF1α binding site (−408), or the β-actin promoter as a positive control for Pol II binding (data not shown). Data are calculated as percent of input, and represented by one experiment expressed as fold binding (n=3, ±SD).
Extracellular acidification and oxygen consumption rate
XF24 Extracellular Flux analyzer (Seahorse Biosciences) was used. BMDMs were plated at 200,000 cells/well in XF24 plates overnight then ±50μM TEPP-46 or DASA-58 (1hr), followed by LPS for 24 hours. Results were normalized to cell number and are represented as mean ± SEM.
Statistical analysis of metabolomics data
Processed data was log transformed (base 2). Correlation between replicates were >0.94 on average within different treatment groups. Significance was assessed using a two-tailed t-test (assuming equal variance) and metabolites with fold-change > 10% were selected. Metabolites scoring with nominal p-value < 0.05 across any of the four comparisons were selected for further analysis.
Mycobacterium tuberculosis assays
Infections were carried out at an MOI of 5 Mtb H37Ra/macrophage. For RNA analysis, cells were lysed in Trizol Reagent (Invitrogen). ELISA results shown as means +/− SEMs for one representative experiment n=2. *P<0.05, **P<0.01, ***P<0.001 (two-way analysis of variance with post-hoc Bonferroni correction).
In vitro uptake assay
Serial dilutions of lysates from BMDM ±TEPP-46 infected with S. typhimurium strain UK-1 (MOI 10 bacteria/cell) were quantified as CFU/ml. Data shown is mean ± SEM for 3 independent infections.
Endotoxin-induced and S. typhimurium in vivo model of sepsis
Mice were treated ±TEPP-46 (50mg/kg) or vehicle (20% 2- Hydroxypropyl-β-cyclodextrin) i.p. for 1hr. 15mg/kg of LPS, alternatively 1×106 CFU S. typhimurium was administered i.p.. 2 hours post-infection serum was isolated from whole blood and PECs were harvested. To measure bacterial dissemination mice infected as above were sacrificed 24h post infection. Log CFU/liver alt. spleen was determined.
Supplementary Material
Acknowledgments
We thank Science Foundation Ireland, the European Research Council, the Health Research Board, European Union FP7 programme ‘TIMER’, Wellcome Trust, Baggot St Hospital Trust and National Institutes of Health for funding.
Footnotes
AUTHOR CONTRIBUTUTIONS
E.P-McD designed and did experiments, analyzed data and wrote the manuscript, L.A.J.O’N. conceived ideas, oversaw the project and co- wrote the manuscript, A.M.C, M.AR.L, L.E.G., F.J.S., M.W.M.vdB., S.R.Q., R.D-F., and D.J. designed and did experiments and analyzed data, G.G. performed bioinformatics analysis of metabolomic profiling and Illumina micorarray and advised on manuscript, R.J.X., C.C., provided advice, performed metabolomic profiling and Illumina micorarray, J-k.J, W.I., C.T., and M.V-H., provided advice and reagents. L.A.G, F.J.S and J.K performed Mtb experiments.
References
- Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nature chemical biology. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashizawa K, Willingham MC, Liang CM, Cheng SY. In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. The Journal of biological chemistry. 1991;266:16842–16846. [PubMed] [Google Scholar]
- Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. The Journal of experimental medicine. 2005;202:1715–1724. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer MB, Jiang JK, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, et al. Evaluation of substituted N,N′-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. Journal of medicinal chemistry. 2010;53:1048–1055. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, David CJ, Manley JL. Concentration-dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. Nature structural & molecular biology. 2012;19:346–354. doi: 10.1038/nsmb.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies CA, Mansell AS, Brady G, Brint E, Dunne A, Gray P, Harte MT, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Molecular cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Science signaling. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JK, Boxer MB, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, et al. Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorganic & medicinal chemistry letters. 2010;20:3387–3393. doi: 10.1016/j.bmcl.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Mesecar A, Heath PJ, Shi W, Nowak T, Stoddard BL. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure. 1998;6:195–210. doi: 10.1016/s0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- Keller KE, Tan IS, Lee YS. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 2012;338:1069–1072. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Hirsila M, Remes AM, Hassinen IE, Kivirikko KI, Myllyharju J. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. The Journal of biological chemistry. 2007;282:4524–4532. doi: 10.1074/jbc.M610415200. [DOI] [PubMed] [Google Scholar]
- Krawczyk CM, Holowka T, Sun J, Blagih J, Amiel E, DeBerardinis RJ, Cross JR, Jung E, Thompson CB, Jones RG, et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115:4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Seminars in cancer biology. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Means TK, Wang S, Lien E, Yoshimura A, Golenbock DT, Fenton MJ. Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. Journal of immunology. 1999;163:3920–3927. [PubMed] [Google Scholar]
- O’Leary S, O’Sullivan MP, Keane J. IL-10 blocks phagosome maturation in mycobacterium tuberculosis-infected human macrophages. American journal of respiratory cell and molecular biology. 2011;45:172–180. doi: 10.1165/rcmb.2010-0319OC. [DOI] [PubMed] [Google Scholar]
- O’Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C, Cejudo-Martin P, Doedens A, Zinkernagel AS, Johnson RS, Nizet V. Cutting edge: Essential role of hypoxia inducible factor-1alpha in development of lipopolysaccharide-induced sepsis. Journal of immunology. 2007;178:7516–7519. doi: 10.4049/jimmunol.178.12.7516. [DOI] [PubMed] [Google Scholar]
- Quinn SR, Mangan NE, Caffrey BE, Gantier MP, Williams BR, Hertzog PJ, McCoy CE, O’Neill LA. The role of Ets2 transcription factor in the induction of microRNA-155 (miR-155) by lipopolysaccharide and its targeting by interleukin-10. The Journal of biological chemistry. 2014;289:4316–4325. doi: 10.1074/jbc.M113.522730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Noguchi T, Sadahiro S, Hirai H, Yamada K, Matsuda T, Imai E, Tanaka T. Isolation and characterization of the human pyruvate kinase M gene. European journal of biochemistry/FEBS. 1991;198:101–106. doi: 10.1111/j.1432-1033.1991.tb15991.x. [DOI] [PubMed] [Google Scholar]
- Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HJ, Hsieh YJ, Cheng WC, Lin CP, Lin YS, Yang SF, Chen CC, Izumiya Y, Yu JS, Kung HJ, et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1alpha-mediated glucose metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:279–284. doi: 10.1073/pnas.1311249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. Metabolism of tumours. Biochem Z. 1923;142:317–333. [Google Scholar]
- Wu S, Le H. Dual roles of PKM2 in cancer metabolism. Acta biochimica et biophysica Sinica. 2013;45:27–35. doi: 10.1093/abbs/gms106. [DOI] [PubMed] [Google Scholar]
- Yang L, Xie M, Yang M, Yu Y, Zhu S, Hou W, Kang R, Lotze MT, Billiar TR, Wang H, et al. PKM2 regulates the Warburg effect and promotes HMGB1 release in sepsis. Nature communications. 2014;5:4436. doi: 10.1038/ncomms5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Lu Z. Nuclear PKM2 regulates the Warburg effect. Cell cycle. 2013;12:3154–3158. doi: 10.4161/cc.26182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nature cell biology. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Petrovic JM, Callaghan D, Jones A, Cui H, Howlett C, Stanimirovic D. Evidence that hypoxia-inducible factor-1 (HIF-1) mediates transcriptional activation of interleukin-1beta (IL-1beta) in astrocyte cultures. Journal of neuroimmunology. 2006;174:63–73. doi: 10.1016/j.jneuroim.2006.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.