Abstract
Background: Liver kinase B1 (LKB1) is a tumor suppressor in lung adenocarcinoma (LADC). We investigated the proteomic profiles of 45 LADC cell lines with and without LKB1 inactivation. Carbamoyl phosphate synthetase 1 (CPS1), the first rate-limiting mitochondrial enzyme in the urea cycle, was distinctively overexpressed in LKB1-inactivated LADC cell lines. We therefore assessed the role of CPS1 and its clinical relevance in LKB1-inactivated LADC.
Methods: Mass spectrometric profiling of proteome and metabolome and function of CPS1 were analyzed in LADC cell lines. CPS1 and LKB1 expression in tumors from 305 LADC and 160 lung squamous cell carcinoma patients was evaluated by immunohistochemistry. Kaplan-Meier and Cox regression analyses were applied to assess the association between overall survival and CPS1 and LKB1 expression. All statistical tests were two-sided.
Results: CPS1 knockdown reduced cell growth, decreased metabolite levels associated with nucleic acid biosynthesis pathway, and contributed an additive effect when combined with gemcitabine, pemetrexed, or CHK1 inhibitor AZD7762. Tissue microarray analysis revealed that CPS1 was expressed in 65.7% of LKB1-negative LADC, and only 5.0% of LKB1-positive LADC. CPS1 expression showed statistically significant association with poor overall survival in LADC (hazard ratio = 3.03, 95% confidence interval = 1.74 to 5.25, P < .001).
Conclusions: Our findings suggest functional relevance of CPS1 in LKB1-inactivated LADC and association with worse outcome of LADC. CPS1 is a promising therapeutic target in combination with other chemotherapy agents, as well as a prognostic biomarker, enabling a personalized approach to treatment of LADC.
Liver kinase B1 (LKB1) is a serine/threonine kinase, frequently inactivated in some human cancers including lung, breast, and cervical cancer (1). Notably, LKB1 somatic mutations were detected in approximately 20% of lung adenocarcinoma (LADC) (2–5). In addition, DNA methylation, homozygous deletion, and intragenic deletion contribute to LKB1 inactivation (6–8). Aside from the effects of LKB1 inactivation on tumor initiation, previous studies have suggested that LKB1-mutant cancers are biologically distinct from cancer with functional LKB1 and that the loss of LKB1 uniquely confers invasive and metastatic properties (9,10). In addition, coclinical trials with genetically engineered mouse models demonstrated that LKB1-deficient lung tumors impaired the response of Kirsten rat sarcoma viral oncogene homolog (KRAS)–mutant tumors to chemotherapy (9,11). LKB1 is a master regulator of various cellular responses including energy metabolism, cell polarity, and cell growth through direct phosphorylation and activation of AMP-activated protein kinase (AMPK)–related protein kinases (12,13). While various pathways regulated by LKB1 can play a pleiotropic role in cancer initiation and progression, no therapies are currently available for clinical use that specifically target LKB1 inactivation (14). Therefore, elucidation of the functional mechanism(s) associated with LKB1 inactivation has immense translational relevance (15).
In this study, we initially compared the proteomic profiles of 45 LADC cell lines to identify molecular features associated with LKB1 inactivation. Carbamoyl phosphate synthase 1 (CPS1) was identified as a markedly increased protein in LKB1-inactivated LADC cell lines. CPS1 is the first rate-limiting enzyme in the urea cycle, converting ammonium into carbamoyl phosphate, and plays an intricate role in arginine metabolism and pyrimidine metabolism (16–19). We further investigated the effect of CPS1 through knockdown experiments and the consequences of its overexpression on survival in LADC.
Methods
LADC Cell Lines
Proteomic analysis of whole cell lysates (WCE), cell surface proteins, and conditioned media from 45 LADC cell lines was performed using mass spectrometry as previously described (20). Details are provided in the Supplementary Methods (available online). Cell transfection, viral transduction, cell viability assay, cell cycle analysis, ammonia assay, metabolomics profiling, real-time polymerase chain reaction (RT-PCR), and immunoblot analysis are described in the Supplementary Methods (available online).
Tissue Microarrays
The tissue microarrays used in this study comprised 305 surgically resected LADCs and 160 squamous cell carcinoma tumor specimens collected under an institutional review board protocol of The University of Texas MD Anderson Cancer Center and archived as formalin-fixed, paraffin-embedded specimens in The University of Texas Specialized Program of Research Excellence thoracic tissue bank at The University of Texas MD Anderson Cancer Center. All patients gave written informed consent. Details are provided in the Supplementary Methods (available online).
Statistical Analysis
Categorical data were compared by Fisher’s exact test or chi-square test. Continuous variables were compared by Mann-Whitney U test or unpaired t test. Pearson’s correlation was applied to assess the linear association between two variables.
Survival analysis was performed with 262 LADC samples with available survival data from the tissue microarray and an independent gene expression data set of 403 LADCs from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) (2) using Kaplan-Meier survival curves. The log-rank test was used to evaluate the statistical significance of differences of survival curves. Overall survival was defined as the time from the date of the initial surgery (tissue microarray) and the initial pathologic diagnosis (the TCGA data set) to the date of death or last follow-up, at which point the data were censored. Cox proportional hazards model analyses (21) were performed to adjust for covariates of statistical significance in univariate analysis, including sex, age, smoking status, stage, and CPS1 and LKB1 expression, and to estimate the relative hazard of mortality over the follow-up period. Using R statistical software (version 3.2.3), we generated the time-dependent covariates by creating interactions of the predictors and a function of survival time and included the covariates in the model. When any of the time-dependent covariates are significant, then those predictors are not proportional. Details on determination of optimal cutoff values for age and CPS1 and LKB1 mRNA expression levels are provided in the Supplementary Methods (available online).
Statistical analysis was done using R and GraphPad Prism software (version 6.0, GraphPad Software, Inc., La Jolla, CA). We used a two-sided statistical significance level of .05 for all statistical analyses.
Results
Comparison of Proteomic Profiling of LKB1-Inactivated and -Intact LADC Cell Lines
Expression levels of LKB1 protein were determined by immunoblotting across 48 LADC cell lines that encompass the known genomic heterogeneity associated with LADC with respect to known mutations (Supplementary Figure 1, available online). On the basis of the mutational status of LKB1, obtained from the Catalogue of Somatic Mutations in Cancer (COSMIC; http://cancer.sanger.ac.uk/cosmic) (22), and LKB1 protein expression levels, we classified 17 cell lines as LKB1-inactivated and 28 cell lines as LKB1-intact LADC cell lines (Supplementary Table 1, available online). Three cell lines with indeterminate LKB1 status were excluded from further analysis. LKB1 inactivation status was statistically significantly associated with epidermal growth factor receptor (EGFR) and KRAS mutational status (P = .01, chi-square test), supporting previous studies (6), but was not statistically significant related to phenotypic characteristics such as cell morphology or invasion properties (data not shown) (23).
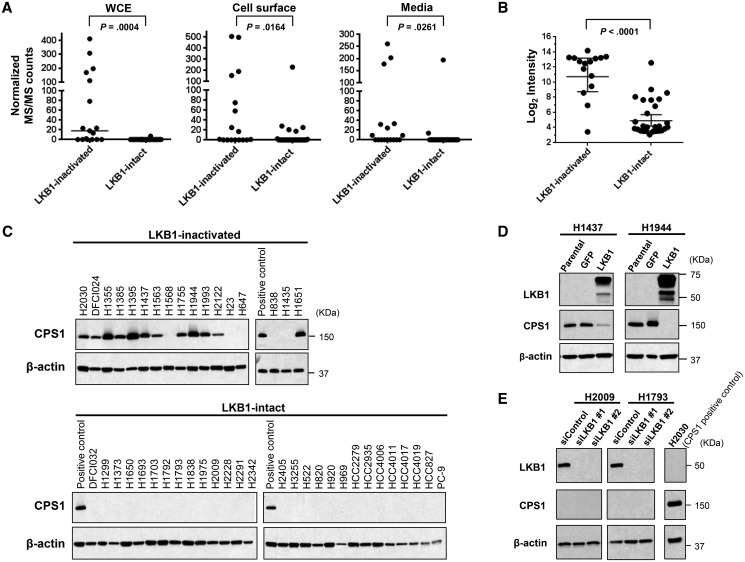
We then performed quantitative proteomic profiling of three cellular protein compartments (whole cell extract [WCE], cell surface proteins, and conditioned media) of the 45 LADC cell lines with known LKB1 status using mass spectrometry as previously described (20,23,24). A total of 213, 325, and 200 proteins were found to be increased in WCE, cell surface proteins, and conditioned media, respectively, from LKB1-inactivated cell lines compared with LKB1-intact cell lines (Supplementary Table 2, available online). Ingenuity Pathway Analysis (IPA: http://www.ingenuity.com/) revealed that the increased proteins in LKB1-inactivated cell lines were highly associated with metabolic pathways, including mitochondrial dysfunction pathways (Supplementary Figure 2, available online), suggesting profound effects of LKB1 on metabolism in LADC. Interestingly, expression levels of CPS1 protein were strikingly higher in all three cellular compartments of LKB1-inactivated cell lines than in LKB1-intact cell lines (P < .001 for WCE, P = .02 for cell surface, and P = .03 for media, Mann-Whitney U test) (Figure 1A; Supplementary Table 2, available online). In addition, mRNA expression levels of CPS1 (23) were statistically significantly increased in LKB1-inactivated cell lines (P < .001, Mann-Whitney U test) (Figure 1B). Increased protein expression of CPS1 with LKB1 inactivation was further confirmed by immunoblotting in LKB1-inactivated and LKB1-intact LADC cell lines (Figure 1C).
Figure 1.
Carbamoyl phosphate synthetase 1 (CPS1) expression in lung adenocarcinoma cell lines. A) CPS1 protein expression levels in whole cell extract (WCE), cell surface proteins, and conditioned media of liver kinase B1 (LKB1)–inactivated (n = 17) and LKB1-intact (n = 28) lung adenocarcinoma (LADC) cell lines based on normalized MS/MS = tandem mass spectral counts. Horizontal lines indicate median. B) mRNA expression of CPS1 in LKB1-inactivated (n = 15) and LKB1-intact (n = 27) LADC cell lines. Horizontal lines indicate median and standard deviation. C) Immunoblot analysis of CPS1 in LKB1-inactivated (n = 17) and LKB1-intact (n = 28) LADC cell lines. D) Immunoblot analysis of LKB1 and CPS1 after overexpression of LKB1 in H1437 and H1944 cell lines. E) Immunoblot analysis of LKB1 and CPS1 after knockdown of LKB1 with siRNA in H2009 and H1793 cells lines. A and B)P values were calculated by two-sided Mann-Whitney U test. C, D, and E) β-actin served as a loading control. C and E) H2030 cell lysates were used as a CPS1-positive control. CPS1 = carbamoyl phosphate synthetase 1; LKB1 = liver kinase B1.
To determine whether CPS1 expression is regulated by LKB1, we induced overexpression and knockdown of LKB1 in LADC cell lines. Overexpression of LKB1 substantially repressed CPS1 protein expression in H1437 and H1944 cell lines (Figure 1D), while knockdown of LKB1 did not increase CPS1 expression in LKB1-intact cell lines H2009 and H1793 (Figure 1E), suggesting that CPS1 is negatively regulated by LKB1 and that additional regulatory mechanisms are required for induction of CPS1. On the other hand, overexpression of CPS1 did not affect LKB1 expression (Supplementary Figure 3, available online).
Analysis of CPS1 Function in LADC Cell Lines
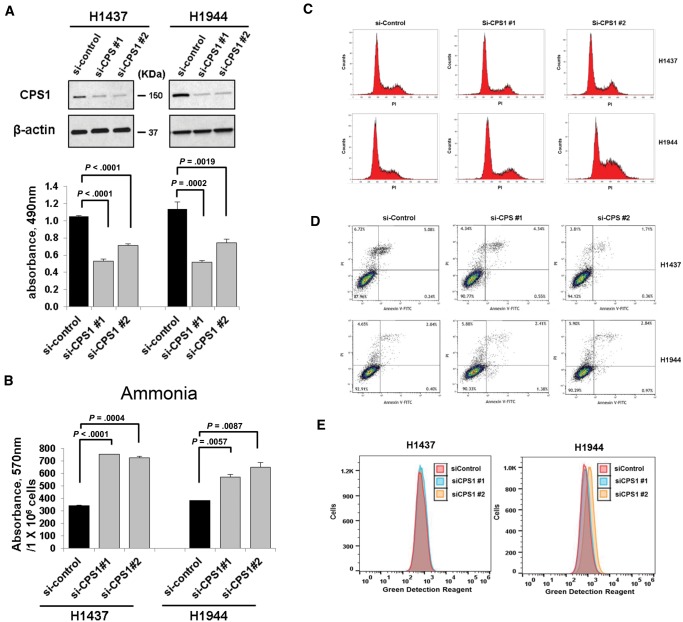
We next explored the functional relevance of CPS1 in LKB1-inactivated LADC. Knockdown of CPS1 statistically significantly reduced cell growth in LKB1-inactivated LADC cell lines but not LKB1-intact LADC cells (Figure 2A;Supplementary Figure 4, available online). On the other hand, CPS1 overexpression did not clearly induce cell growth in either LKB1-inactivated or -intact LADC cell lines without CPS1 expression (Supplementary Figure 5, available online), suggesting that function of CPS1 might be complemented by other genes in CPS1-negative LADC. Growth inhibition in H1944 cells induced by siRNA targeting 3ʹuntranslated region (UTR) of the CPS1 gene was recovered by overexpression of CPS1, which lacks siRNA binding sites in 3ʹUTR (Supplementary Figure 5, available online). Although cell growth was only partially recovered, possibly because of low transfection efficiency (Supplementary Figure 5, available online), the findings further indicate the oncogenic potential of CPS1 in LKB1-inactivated LADC. As expected, CPS1 knockdown statistically significantly increased levels of ammonia in the media (Figure 2B). CPS1 knockdown had no statistically significant effect on G1, G2/M, or sub-G1 population, suggesting that cell cycle arrest and apoptosis were not associated with the reduction of cell proliferation. The findings were further supported by Annexin V assay (Figure 2D). While recent studies indicated that ammonia induces autophagy (25,26), we did not observe the occurrence of autophagy in cells with silenced CPS1 (Figure 2E).
Figure 2.
Effects of carbamoyl phosphate synthetase 1 (CPS1) knockdown in liver kinase B1 (LKB1)–inactivated lung adenocarcinoma cell lines. A) Statistically significant reduction in cell proliferation was observed with MTS = 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay in H1437 and H1944 cells treated with negative control siRNA or CPS1 siRNA. Knockdown of CPS1 with siRNA was confirmed by immunoblotting. β-actin served as a loading control. B) Ammonia was statistically significantly accumulated in the media of H1437 and H1944 cells treated with CPS1 siRNA compared to negative control siRNA. C) Cell cycle analysis was performed using flow cytometry. D) Apoptosis induction was determined with flow cytometric analysis of annexin V and PI = Propidium iodide staining. E) H1437 and H1944 cells were treated with negative control siRNA and CPS1 siRNA for 72 hours. Cationic amphiphilic tracer dye was used to assess the autophagy using flow cytometry. A and B) Columns indicate the average of triplicate samples from a representative experiment, and bars indicate standard deviation. P values were calculated by two-sided unpaired t test. CPS1 = carbamoyl phosphate synthetase 1.
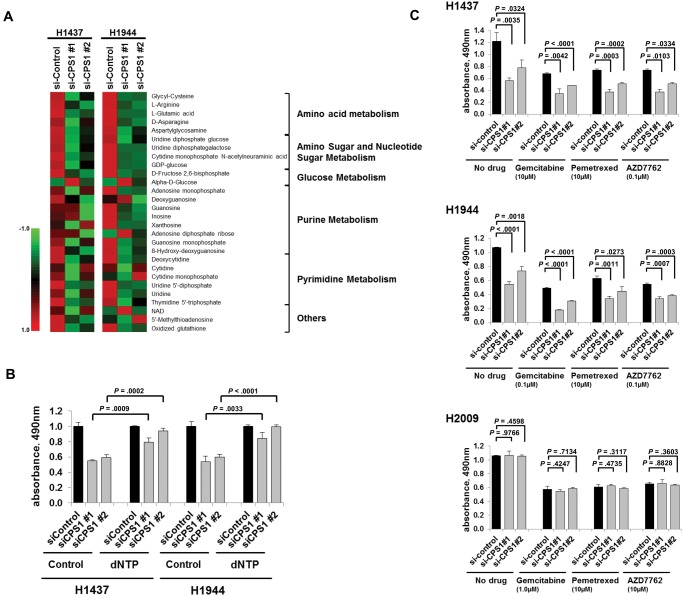
As CPS1 is involved in the arginine and pyrimidine metabolism pathways, we examined metabolites associated with these pathways during CPS1 knockdown using mass spectrometry. Twenty-eight metabolites were identified with high confidence on the basis of NIST MSMS and The Human Metabolome Database (http://www.hmdb.ca/) fragmentation matching. Interestingly, most of the identified metabolites in both purine and pyrimidine metabolism pathways, as well as arginine, were consistently decreased in CPS1 knockdown H1437 and H1944 cells compared with control cells from those cell lines (Figure 3A).
Figure 3.
Effects of carbamoyl phosphate synthetase 1 (CPS1) knockdown on nucleic acid synthesis and on growth inhibition in combination with chemotherapy agents. A) Heatmap of identified metabolites in H1437 and H1944 cells with CPS1 siRNA treatment or negative control. Metabolic pathways were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/). B) MTS assay of H1437 and H1944 cells treated with negative control siRNA or siRNAs against CPS1 in combination with 100 μM dNTP. Columns indicate the average of triplicate samples from a representative experiment, and bars indicate standard deviation. C) MTS assay of two liver kinase B1 (LKB1)–inactivated cell lines (H1437 and H1944) and one LKB1-intact cell line (H2009) treated with negative control siRNA or CPS1 siRNA in combination with gemcitabine, pemetrexed, or AZD7762. Columns indicate the average of triplicate samples from a representative experiment, and bars indicate standard deviation. All P values were calculated by two-sided unpaired t test. CPS1 = carbamoyl phosphate synthetase 1.
While cell cycle arrest at G2/M phase was not observed in siCPS1-treated cells (Figure 2C), supplementation of deoxynucleotide (dNTP) statistically significantly rescued cell growth in cells with siCPS1 treatment (Figure 3B), leading us to speculate that CPS1 knockdown would deplete the pool of metabolites in the nucleic acid synthesis pathways and thus would have additive effects on growth inhibition in combination with other chemotherapy agents. We examined the effects of CPS1 knockdown combined with gemcitabine and pemetrexed, which interfere with DNA synthesis pathways and are frequently used to treat LADC (27), as well as with CHK1 inhibitor AZD7762, to which LKB1-deficient lung cancer was recently shown to be sensitive (28). CPS1 knockdown showed statistically significant additive effects in all drug combinations in the LKB1-inactivated cell lines H1437 and H1944 (Figure 3C) but not in the LKB1-intact cell line H2009, indicating the potential of CPS1 as a therapeutic target alone or combined with these conventional chemotherapy agents or with DNA damage checkpoint inhibitors in LKB1-inactivated LADC.
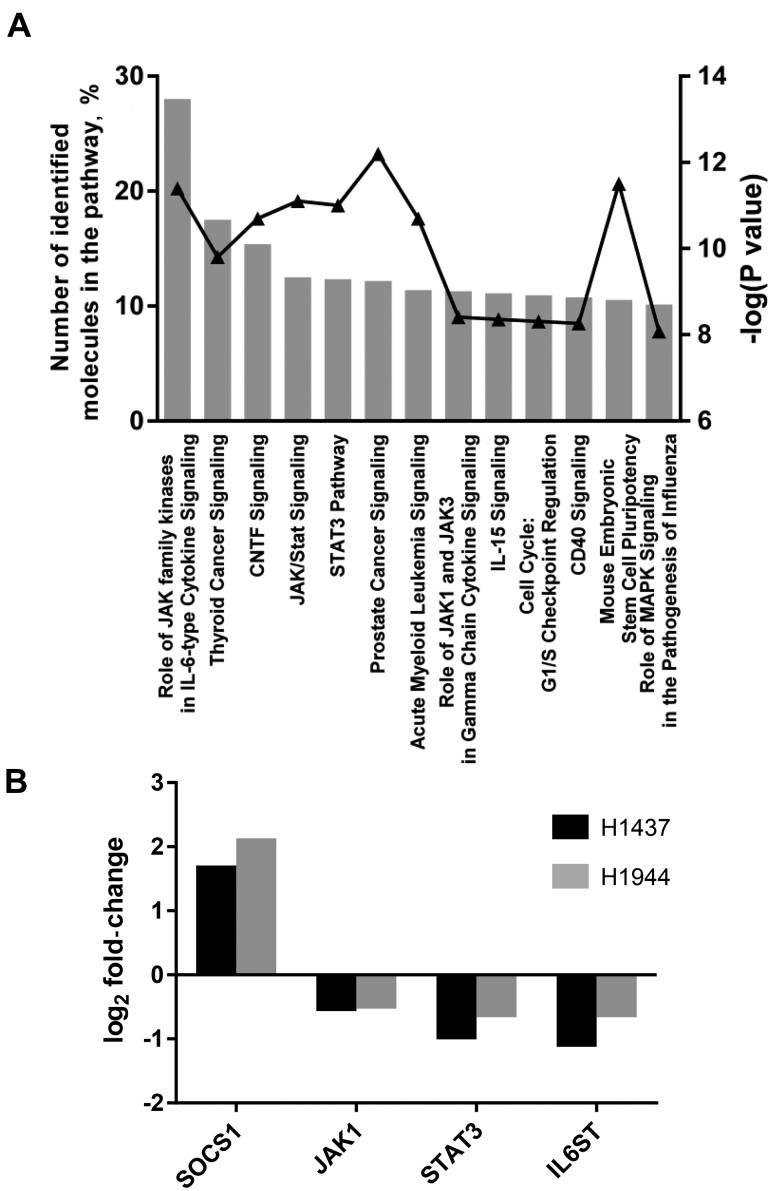
To gain further insights into molecular pathways perturbed by CPS1 knockdown, we performed PrimePCR array (Biorad, Hercules, CA), which consists of 376 cancer-related genes in H1437 and H1944 cells with or without CPS1 knockdown. Eighty-one genes were more than 25% increased or decreased after CPS1 knockdown in both cell lines (Supplementary Table 3, available online). Interestingly, pathway analysis using IPA revealed that among top 13 statistically significant pathways (identification of > 10% genes in the pathway and -log(P value) > 8), eight pathways were associated with Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway: role of JAK family kinases in interleukin-6 (IL-6) cytokine signaling, ciliary neurotrophic factor (CNTF) signaling, JAK/STAT signaling, STAT3 pathway, role of JAK1 and JAK3 in gamma chain cytokine signaling, IL-15 signaling, cluster of differentiation 40 (CD40) signaling, and mouse embryonic stem cell pluripotency (Figure 4A). While mRNA expression of suppressor of cytokine signaling 1 (SOCS1), a negative regulator of the JAK/STAT signaling pathway, was increased by CPS1 knockdown, mRNA expression levels of several key genes in the JAK/STAT signaling pathway, including JAK1, STAT3, and interleukin 6 signal transducer (IL6ST), were consistently decreased (Figure 4B), suggesting that CPS1 knockdown would suppress the JAK/STAT signaling pathway.
Figure 4.
Quantitative real-time polymerase chain reaction array in H1437 and H1944 cell lines. A) Top 13 statistically significant canonical pathways predicted by IPA for 81 genes with altered mRNA expression after carbamoyl phosphate synthetase 1 knockdown. P values were calculated by two-sided Fisher's exact test. B) mRNA expression of SOCS1, JAK1, STAT3, and IL6ST.
CPS1 Expression and Overall Survival in LADC
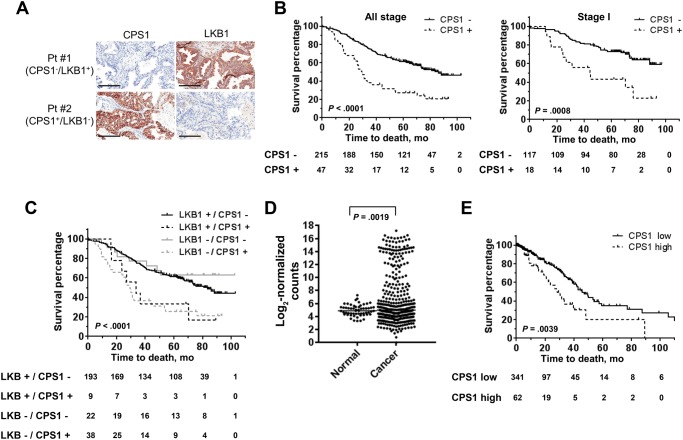
To determine the clinical significance of CPS1 in lung cancer, we examined CPS1 and LKB1 protein expression in LADC and squamous cell carcinoma tissues using a tissue microarray. CPS1 expression was found predominantly in the cytoplasm. Figure 5A depicts representative images of LADC specimens with negative CPS1 and positive LKB1 (Pt #1) and with positive CPS1 and negative LKB1 (Pt #2). In total, 56 (18.4%) of 305 LADC tumors were positively stained with CPS1 (Table 1). CPS1 protein expression was statistically significantly and inversely correlated with LKB1 protein expression: CPS1 was positive in 44 (65.7%) LKB1-negative LADCs but only 12 (5.0%) LKB1-positive LADCs (P < .001, chi-square test), further confirming the findings in LADC cell lines. The CPS1 positivity was statistically significantly associated with smoking status (P = .003, chi-square test), but not with sex, age, or mutational status for EGFR and KRAS (Table 1). On the other hand, only two (1.3%) of 160 squamous cell carcinoma tumors were positively stained with CPS1 (Supplementary Table 4, available online).
Figure 5.
Association of carbamoyl phosphate synthetase 1 (CPS1) expression with lung adenocarcinoma (LADC) patient survival. A) Representative images of immunohistochemical analysis of CPS1 and liver kinase B1 (LKB1). Scale bars = 200 μm. B) Kaplan-Meier survival curves of LADC patients in the tissue microarray stratified according to protein expression of CPS1 (all stage: CPS1-negative n = 215 and CPS1-positive n = 47; stage I: CPS1-negative n = 117 and CPS1-positive n = 18). C) Kaplan-Meier survival curves of LADC patients in the tissue microarray stratified according to protein expression of LKB1 and CPS1 (LKB1-positive/CPS1-negative, n = 193; LKB1-positive/CPS1-positive, n = 9; LKB1-negative/CPS1-positive, n = 38; LKB1-negative/CPS1-negative, n = 22). D) mRNA expression levels of CPS1 in LADC tumors (n = 423) and adjacent normal tissues (n = 56) in The Cancer Genome Atlas (TCGA) data set. P values were calculated by unpaired t test. E) Kaplan-Meier survival curves of LADC patients in the TCGA data set stratified according to mRNA expression of CPS1 (CPS1-low n = 341; CPS1-high n = 62). B, C, and E)P values were calculated by log-rank test. All statistical tests were two-sided. CPS1 = carbamoyl phosphate synthetase 1; LKB1 = liver kinase B1.
Table 1.
CPS1 and LKB1 expression in lung adenocarcinoma tissue microarray
| CPS1 immunostaining |
||||
|---|---|---|---|---|
| Characteristic | Total | Positive, No. (%) | Negative, No. (%) | P* |
| Total | 305 | 56 (18.4) | 249 (81.6) | |
| Sex† | ||||
| Male | 136 | 20 (14.7) | 116 (85.3) | .20 |
| Female | 126 | 27 (21.4) | 99 (78.6) | |
| Age†, y | ||||
| >65 | 132 | 18 (13.6) | 114 (86.4) | .08 |
| ≤65 | 130 | 29 (22.3) | 101 (77.7) | |
| Smoking status† | ||||
| Current smoker | 94 | 22 (23.4) | 72 (76.6) | .003 |
| Former smoker | 124 | 25 (20.2) | 99 (79.8) | |
| Never smoker | 44 | 0 (0.0) | 44 (100.0) | |
| Stage† | ||||
| I | 135 | 18 (13.3) | 117 (86.7) | .22 |
| II | 54 | 13 (24.1) | 41 (75.9) | |
| III | 56 | 13 (23.2) | 43 (76.8) | |
| IV | 17 | 3 (17.6) | 14 (82.4) | |
| Mutation‡ | ||||
| KRAS | 47 | 10 (21.3) | 37 (78.7) | .12 |
| EGFR | 16 | 0 (0.0) | 16 (100.0) | |
| KRAS and EGFR wild | 86 | 13 (15.1) | 73 (84.9) | |
| LKB1 immunostaining | ||||
| Positive | 238 | 12 (5.0) | 226 (95.0) | <.001 |
| Negative | 67 | 44 (65.7) | 23 (34.3) | |
P values were calculated by two-sided Fisher’s exact test. CPS1 = carbamoyl phosphate synthetase 1; EGFR = epidermal growth factor receptor; KRAS = Kirsten rat sarcoma viral oncogene homolog; LKB1 = liver kinase B1.
Information was not available for 43 samples.
Mutations were not investigated in 156 samples.
Kaplan-Meier analysis in 262 LADC patients revealed that positive CPS1 expression was statistically significantly associated with worse overall survival (P < .001, log-rank test) (Figure 5B). Of note, CPS1 positivity was statistically significantly associated with poor overall survival in stage I LADC (P < .001, log-rank test) (Figure 5B). A univariate Cox regression analysis revealed that CPS1 expression, LKB1 expression, stage, and sex were statistically significantly associated with overall survival (Table 2). Multivariable Cox regression analysis indicated that CPS1 expression was the strongest independent predictor of overall survival (P < .001, hazard ratio [HR] = 3.03, 95% confidence interval [CI] = 1.74 to 5.25), while LKB1 expression was not statistically significantly associated with overall survival (Figure 5C and Table 2).
Table 2.
Univariate and multivariable analysis of overall survival in lung adenocarcinoma tissue microarray
| Univariate analysis |
Multivariable analysis |
|||
|---|---|---|---|---|
| Variable | HR (95% CI) | P* | HR (95% CI) | P* |
| Sex | ||||
| Female | Reference | Reference | ||
| Male | 1.73 (1.23 to 2.43) | .002 | 1.68 (1.18 to 2.40) | .004 |
| Age, y | ||||
| >65 | Reference | Reference | ||
| ≤65 | 0.97 (0.69 to 1.36) | .84 | 0.74 (0.51 to 1.07) | .11 |
| Smoking status | ||||
| Current smoker | Reference | Reference | ||
| Former smoker | 0.84 (0.58 to 1.21) | .35 | 0.77 (0.52 to 1.14) | .19 |
| Never smoker | 0.81 (0.49 to 1.34) | .41 | 0.96 (0.56 to 1.65) | .89 |
| Stage | ||||
| I | Reference | Reference | ||
| II | 2.21 (1.44 to 3.41) | <.001 | 1.97 (1.27 to 3.06) | .003 |
| III | 2.45 (1.60 to 3.75) | <.001 | 2.17 (1.39 to 3.38) | <.001 |
| IV | 2.50 (1.33 to 4.70) | .004 | 2.68 (1.42 to 5.08) | .003 |
| CPS1 immunostaining | ||||
| Negative | Reference | Reference | ||
| Positive | 2.57 (1.75 to 3.77) | <.001 | 3.03 (1.74 to 5.25) | <.001 |
| LKB1 immunostaining | ||||
| Negative | Reference | Reference | ||
| Positive | 0.62 (0.42 to 0.91) | .01 | 1.23 (0.72 to 2.11) | .44 |
P values were calculated by two-sided Fisher’s exact test. CI = confidence interval; HR = hazard ratio.
The prognostic potential of CPS1 was further examined in an independent cohort of 403 LADCs in the TCGA data set. mRNA levels of CPS1 were statistically significantly higher in LADC tissues than in normal adjacent tissues (P = .002, unpaired t test) (Figure 5D;Supplementary Figure 6, available online).While LKB1 mRNA expression levels in tumor tissues were not statistically significantly lower than in normal tissues, CPS1 mRNA and LKB1 mRNA expression levels showed a statistically significant inverse correlation (Pearson correlation coefficient = −0.33, P < .001) (Supplementary Figure 6, available online). Concordant with the findings in the tissue microarray, Kaplan-Meier survival curves indicated that high CPS1 mRNA expression was statistically significantly associated with worse overall survival (P = .004, log-rank test) (Figure 5E). In addition, high CPS1 mRNA expression was a statistically significant and independent predictor of overall survival in both univariate and multivariable Cox regression analyses (univariate analysis: HR = 1.96, 95% CI = 1.22 to 3.15, P = .005; multivariable analysis: HR = 2.31, 95% CI = 1.28 to 4.16, P = .005), while LKB1 mRNA expression was not statistically significantly associated with overall survival (Table 3).
Table 3.
Univariate and multivariable analysis of overall survival in lung adenocarcinoma The Cancer Genome Atlas data set
| Univariate analysis |
Multivariable analysis |
|||
|---|---|---|---|---|
| Variable | HR (95% CI) | P* | HR (95% CI) | P* |
| Sex | ||||
| Female | Reference | Reference | ||
| Male | 0.99 (0.66 to 1.48) | .94 | 0.87 (0.55 to 1.36) | .52 |
| Age, y | ||||
| >65 | Reference | Reference | ||
| ≤65 | 0.65 (0.42 to 1.00) | .05 | 0.58 (0.37 to 0.91) | .02 |
| Smoking status | ||||
| Current smoker | Reference | Reference | ||
| Former smoker | 1.43 (0.86 to 2.36) | .16 | 1.68 (0.99 to 2.85) | .05 |
| Never smoker | 1.20 (0.58 to 2.49) | .62 | 1.14 (0.51 to 2.52) | .75 |
| Stage | ||||
| I | Reference | Reference | ||
| II | 1.34 (0.71 to 2.53) | .37 | 1.56 (0.81 to 3.01) | .19 |
| III | 3.86 (1.95 to 7.64) | <.001 | 4.90 (2.41 to 9.96) | <.001 |
| IV | 2.81 (1.18 to 6.68) | .02 | 3.44 (1.38 to 8.61) | .008 |
| CPS1 mRNA expression | ||||
| Low | Reference | Reference | ||
| High | 1.96 (1.22 to 3.15) | .005 | 2.31 (1.28 to 4.16) | .005 |
| LKB1 mRNA expression | ||||
| Low | Reference | Reference | ||
| High | 0.97 (0.61 to 1.54) | .88 | 1.42 (0.79 to 2.54) | .23 |
P values were calculated by two-sided Fisher’s exact test. CI = confidence interval; HR = hazard ratio.
Discussion
In this study we analyzed the proteomic profiles of 45 LADC cell lines to identify molecular features associated with LKB1 inactivation. Protein signatures associated with LKB1 inactivation comprised proteins involved in metabolism, including CPS1, which showed strikingly higher expression levels in LKB1-inactivated cell lines than in LKB1-intact cell lines. CPS1 knockdown reduced cell growth, decreased levels of metabolites associated with nucleic acid biosynthesis, and showed an additive beneficial effect when combined with conventional chemotherapy agents or DNA damage checkpoint inhibitors. In addition, CPS1 expression at both protein and mRNA levels was statistically significantly and independently associated with poor overall survival in LADC. Our findings suggest that CPS1 is a promising therapeutic target and prognostic indicator in LADC.
CPS1 is a multidomain mitochondrial enzyme protein, catalyzing the first committed step of the urea cycle for ammonia detoxification and disposal (16,29). CPS1 is expressed mainly in intestinal epithelial cells and liver cells, as well as several types of cancer including liver, colorectal, stomach, cervical, and pancreatic cancer (29–37). Although expression of CPS1 protein has not been well studied in lung cancer, Kaufman et al. recently characterized gene expression signatures associated with LKB1 loss and demonstrated that 16-gene expression signatures that included CPS1 could accurately predict LKB1 inactivation status (38). In addition, Skoulidis et al. performed an integrative analysis of genomic, transcriptomic, and proteomic profiling of KRAS-mutant LADC and showed CPS1 to be one of the overexpressed genes in a subset of LADCs harboring genetic events in KRAS and LKB1 (39). This previous evidence further supports the CPS1 protein expression in LKB1-inactivated LADC demonstrated in this study through in-depth proteomic profiling of LADC cell lines and tissue microarray analysis.
CPS1 plays a crucial role in removing excessive ammonia by converting ammonium into carbamoyl phosphate in the liver (16), which is a de novo resource of arginine (19) and potential substrate of pyrimidine synthesis (17,18). We demonstrated that knockdown of CPS1 induced accumulation of ammonia, a decrease of metabolites in nucleic acid synthesis pathways, and a decrease of arginine in LKB1-inactivated LADC cell lines. While CPS1 is implicated in pyrimidine synthesis pathways, our metabolomic profiling suggested that metabolites in both pyrimidine and purine synthesis pathways were decreased in cells with siCPS1 treatment. This can be possibly explained by decreased levels of glutamic acid, a prominent intermediate in nitrogen disposal (Figure 3A), because accumulation of ammonia would result in excessive buildup of ammonia and a reduction in metabolites directly associated with nitrogen removal.
Our findings suggest that the decrease of metabolites in the nucleic acid synthesis pathways was associated with siCPS1-induced inhibition of cell growth, and we observed that CPS1 knockdown had a beneficial effect when combined with conventional chemotherapy agents or a DNA damage checkpoint inhibitor. Liu and colleagues have recently indicated that LKB1-mutant lung cancers are vulnerable in nucleotide biosynthesis pathways and sensitive to deoxythymidylate kinase inhibition (28). Therefore, targeting CPS1 in combination with deoxythymidylate kinase also might have promising therapeutic potential in LKB1-inactivated LADC.
Quantitative RT-PCR analyses suggest possible suppression of the JAK/STAT pathway by CPS1 knockdown. Recent studies have reported activation of the JAK/STAT pathway in sex-determining region Y box 2 (SOX2)–driven, LKB1-deficient mouse model of squamous cell lung cancer (47) and increase of IL-6 production and subsequent activation of STAT3 in LKB1-deficient mouse model of LADC (48,49). As IL-6/JAK/STAT signaling axis plays important roles in cell proliferation, survival, invasion, chemoresistance, and immunosuppression, (48,50–53), targeting CPS1 might alter tumor microenvironment and improve efficiency of immunotherapy.
Regarding the clinical relevance of CPS1 in LADC, we demonstrated that increased CPS1 expression at both protein and mRNA levels was statistically significantly associated with poor overall survival. Increased CPS1 expression is also associated with poor prognosis in cervical carcinoma (37) and rectal cancer (33). Of note, positive immunostaining of CPS1 was statistically significantly associated with poorer overall survival in stage I LADC. The survival benefit of adjuvant chemotherapy in stage I non–small cell lung cancer is controversial, while trials have shown that cisplatin-based adjuvant chemotherapy in clinic significantly improved the five-year survival rate in stage II and IIIa non–small cell lung cancer (27). Therefore, CPS1 would be a promising molecular biomarker of poor prognosis in LADC and could enable us to personalize treatment of the disease.
A potential limitation of the study is the lack of CPS1 inhibitor, which hampers immediate application to therapy. However, it has been reported that sirtuin 5 (SIRT5) deacetylates and desuccinylates CPS1 protein and enhances CPS1 activity (43,44). Thus, pharmacologic inhibition of SIRT5 might be a strategy for targeting CPS1. It has been recently demonstrated that the antidiabetic drug metformin, an established AMPK activator, reduced the mouse SIRT5 protein both in cultured hepatocytes and in livers in vivo (45). In addition, phenformin, a mitochondrial inhibitor and analogue of metformin, selectively reduced tumor volume and prolonged survival in a mouse model of lung cancer with KRAS and LKB1 mutations (46), suggesting repurposing metformin and related agents as a promising strategy in the treatment of LKB1-inactivated LADC.
In conclusion, proteomic characterization of LKB1-inactivated LADC cell lines identified CPS1 as a therapeutic target, expanding potential therapeutic options in combination with conventional chemotherapy agents while revealing a potential biomarker for precision medicine in LADC.
Funding
This work was supported by the Canary Foundation, the LUNGevity Foundation, the National Cancer Institute Early Detection Research Network, the Department of Defense (W81XWH-09-LCRP-CTRA and W81XWH-15-1-0127), the National Institutes of Health (U01 CA186150), Lung Cancer Specialized Program of Research Excellence (P50CA70907), the Cancer Center Support Grant from the National Institutes of Health (P30 CA016672), The University of Texas MD Anderson Cancer Center Moon Shots Program, and start-up funds from MD Anderson Cancer Center.
Notes
The funding agencies had no role in design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
The authors have no conflicts of interest to disclose.
Supplementary Material
References
- 1. Sanchez-Cespedes M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 2007;2657:7825–7832. [DOI] [PubMed] [Google Scholar]
- 2. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;5117511:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;4557216:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;1506:1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;1506:1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matsumoto S, Iwakawa R, Takahashi K, et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;2640:5911–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gill RK, Yang SH, Meerzaman D, et al. Frequent homozygous deletion of the LKB1/STK11 gene in non-small cell lung cancer. Oncogene. 2011;3035:3784–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Esteller M, Avizienyte E, Corn PG, et al. Epigenetic inactivation of LKB1 in primary tumors associated with the Peutz-Jeghers syndrome. Oncogene. 2000;191:164–168. [DOI] [PubMed] [Google Scholar]
- 9. Carretero J, Shimamura T, Rikova K, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;176:547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;4487155:807–810. [DOI] [PubMed] [Google Scholar]
- 11. Chen Z, Cheng K, Walton Z, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;4837391:613–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gan RY, Li HB.. Recent progress on liver kinase B1 (LKB1): Expression, regulation, downstream signaling and cancer suppressive function. Int J Mol Sci. 2014;159:16698–16718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alessi DR, Sakamoto K, Bayascas JR.. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. [DOI] [PubMed] [Google Scholar]
- 14. Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat Rev Cancer. 2014;148:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shah U, Sharpless NE, Hayes DN.. LKB1 and lung cancer: More than the usual suspects. Cancer Res. 2008;6810:3562–3565. [DOI] [PubMed] [Google Scholar]
- 16. Summar ML, Dasouki MJ, Schofield PJ, et al. Physical and linkage mapping of human carbamyl phosphate synthetase I (CPS1) and reassignment from 2p to 2q35. Cytogenet Cell Genet. 1995;713:266–267. [DOI] [PubMed] [Google Scholar]
- 17. Pausch J, Rasenack J, Haussinger D, et al. Hepatic carbamoyl phosphate metabolism. Role of cytosolic and mitochondrial carbamoyl phosphate in de novo pyrimidine synthesis. Eur J Biochem. 1985;1501:189–194. [DOI] [PubMed] [Google Scholar]
- 18. Levin B, Oberholzer VG, Sinclair L.. Biochemical investigations of hyperammonaemia. Lancet. 1969;27613:170–174. [DOI] [PubMed] [Google Scholar]
- 19. Morris SM., Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annu Rev Nutr. 2002;22:87–105. [DOI] [PubMed] [Google Scholar]
- 20. Taguchi A, Politi K, Pitteri SJ, et al. Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell. 2011;203:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox DR, Oakes D.. Analysis of survival data. London and New York: Chapman and Hall; 1984. [Google Scholar]
- 22. Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: Exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schliekelman MJ, Taguchi A, Zhu J, et al. Molecular portraits of epithelial, mesenchymal, and hybrid States in lung adenocarcinoma and their relevance to survival. Cancer Res. 2015;759:1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taguchi A, Delgado O, Celiktas M, et al. Proteomic signatures associated with p53 mutational status in lung adenocarcinoma. Proteomics. 2014;14(23–24):2750–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eng CH, Yu K, Lucas J, et al. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010;3119:ra31. [DOI] [PubMed] [Google Scholar]
- 26. Polletta L, Vernucci E, Carnevale I, et al. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy. 2015;112:253–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reck M, Heigener DF, Mok T, et al. Management of non-small-cell lung cancer: Recent developments. Lancet. 2013;3829893:709–719. [DOI] [PubMed] [Google Scholar]
- 28. Liu Y, Marks K, Cowley GS, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov. 2013;38:870–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Butler SL, Dong H, Cardona D, et al. The antigen for Hep Par 1 antibody is the urea cycle enzyme carbamoyl phosphate synthetase 1. Lab Invest. 2008;881:78–88. [DOI] [PubMed] [Google Scholar]
- 30. Fan Z, van de Rijn M, Montgomery K, et al. Hep par 1 antibody stain for the differential diagnosis of hepatocellular carcinoma: 676 tumors tested using tissue microarrays and conventional tissue sections. Mod Pathol. 2003;162:137–144. [DOI] [PubMed] [Google Scholar]
- 31. Liu H, Dong H, Robertson K, et al. DNA methylation suppresses expression of the urea cycle enzyme carbamoyl phosphate synthetase 1 (CPS1) in human hepatocellular carcinoma. Am J Pathol. 2011;1782:652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Siddiqui MT, Saboorian MH, Gokaslan ST, et al. Diagnostic utility of the HepPar1 antibody to differentiate hepatocellular carcinoma from metastatic carcinoma in fine-needle aspiration samples. Cancer. 2002;961:49–52. [PubMed] [Google Scholar]
- 33. Lee YY, Li CF, Lin CY, et al. Overexpression of CPS1 is an independent negative prognosticator in rectal cancers receiving concurrent chemoradiotherapy. Tumour Biol. 2014;3511:11097–11105. [DOI] [PubMed] [Google Scholar]
- 34. Sato T, Kashima K, Gamachi A, et al. Immunohistochemical localization of pyruvate carboxylase and carbamyl-phosphate synthetase I in normal and neoplastic human pancreatic tissues. Pancreas. 2002;252:130–135. [DOI] [PubMed] [Google Scholar]
- 35. Chu PG, Weiss LM.. Immunohistochemical characterization of signet-ring cell carcinomas of the stomach, breast, and colon. Am J Clin Pathol. 2004;1216:884–892. [DOI] [PubMed] [Google Scholar]
- 36. Brentnall TA, Pan S, Bronner MP, et al. Proteins that underlie neoplastic progression of ulcerative colitis. Proteomics Clin Appl. 2009;311:1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thamboo TP, Wee A.. Hep Par 1 expression in carcinoma of the cervix: Implications for diagnosis and prognosis. J Clin Pathol. 2004;571:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaufman JM, Amann JM, Park K, et al. LKB1 loss induces characteristic patterns of gene expression in human tumors associated with NRF2 activation and attenuation of PI3K-AKT. J Thorac Oncol. 2014;96:794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;58:860–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen YR, Sekine K, Nakamura K, et al. Y-box binding protein-1 down-regulates expression of carbamoyl phosphate synthetase-I by suppressing CCAAT enhancer-binding protein-alpha function in mice. Gastroenterology. 2009;1371:330–340. [DOI] [PubMed] [Google Scholar]
- 41. Christoffels VM, van den Hoff MJ, Moorman AF, et al. The far-upstream enhancer of the carbamoyl-phosphate synthetase I gene is responsible for the tissue specificity and hormone inducibility of its expression. J Biol Chem. 1995;27042:24932–24940. [DOI] [PubMed] [Google Scholar]
- 42. Lagace M, Goping IS, Mueller CR, et al. The carbamyl phosphate synthetase promoter contains multiple binding sites for C/EBP-related proteins. Gene. 1992;1182:231–238. [DOI] [PubMed] [Google Scholar]
- 43. Nakagawa T, Lomb DJ, Haigis MC, et al. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;1373:560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Du J, Zhou Y, Su X, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;3346057:806–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buler M, Aatsinki SM, Izzi V, et al. SIRT5 is under the control of PGC-1alpha and AMPK and is involved in regulation of mitochondrial energy metabolism. FASEB J. 2014;287:3225–3237. [DOI] [PubMed] [Google Scholar]
- 46. Shackelford DB, Abt E, Gerken L, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;232:143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mukhopadhyay A, Berrett KC, Kc U, et al. Sox2 cooperates with Lkb1 loss in a mouse model of squamous cell lung cancer. Cell Rep. 2014;81:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Koyama S, Akbay EA, Li YY, et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress t-cell activity in the lung tumor microenvironment. Cancer Res. 2016;765:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Grabner B, Schramek D, Mueller KM, et al. Disruption of STAT3 signalling promotes KRAS-induced lung tumorigenesis. Nat Commun. 2015;6:6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dutta P, Sabri N, Li J, et al. Role of STAT3 in lung cancer. JAKSTAT. 2014;34:e999503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Caetano MS, Zhang H, Cumpian AM, et al. IL6 blockade reprograms the lung tumor microenvironment to limit the development and progression of K-ras-mutant lung cancer. Cancer Res. 2016;7611:3189–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miklossy G, Hilliard TS, Turkson J.. Therapeutic modulators of STAT signalling for human diseases. Nat Rev Drug Discov. 2013;128:611–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu H, Lee H, Herrmann A, et al. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat Rev Cancer. 2014;1411:736–746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.