Abstract
Introduction
We analyzed the effects of the top 20 Alzheimer disease (AD) risk genes on gray-matter density (GMD) and metabolism.
Methods
We ran stepwise linear regression analysis using posterior cingulate hypometabolism and medial temporal GMD as outcomes and all risk variants as predictors while controlling for age, gender, and APOE ε4 genotype. We explored the results in 3D using Statistical Parametric Mapping 8.
Results
Significant predictors of brain GMD were SLC24A4/RIN3 in the pooled and mild cognitive impairment (MCI); ZCWPW1 in the MCI; and ABCA7, EPHA1, and INPP5D in the AD groups. Significant predictors of hypometabolism were EPHA1 in the pooled, and SLC24A4/RIN3, NME8, and CD2AP in the normal control group.
Discussion
Multiple variants showed associations with GMD and brain metabolism. For most genes, the effects were limited to specific stages of the cognitive continuum, indicating that the genetic influences on brain metabolism and GMD in AD are complex and stage dependent.
Keywords: Genome-wide association studies (GWAS), Genetic variation, Imaging genetics, Magnetic resonance imaging (MRI), Fluorodeoxyglucose positron emission tomography (FDG PET), Atrophy, Brain metabolism, Risk genes, ADNI, Brain mapping, Statistical parametric mapping (SPM), Positron emission tomography (PET), Alzheimer disease, AD
1. Background
Alzheimer disease (AD) is a chronic neurodegenerative disease characterized by short-term memory loss in the early disease stages and progressive cognitive and functional deficits as the disease advances. The clinical symptoms result from the deposition of two toxic proteins, β-amyloid (Aβ) and tau, which give rise to neuritic plaques and neurofibrillary tangles, respectively [1]. The clinical appearance of AD is the direct result of neuronal dysfunction and death, which is manifested by brain atrophy and hypometabolism.
Brain imaging is increasingly used to measure AD-associated changes in vivo. Amyloid positron emission tomography (PET), a novel Food and Drug Administration–approved imaging technology, uses selective Aβ tracers to visualize brain amyloidosis and can reliably detect the presence of neuritic plaques in the symptomatic and presymptomatic stages. Brain atrophy is best evaluated with longitudinal studies of magnetic resonance imaging (MRI). The atrophic changes are first noticeable in the medial temporal lobe, eventually spreading through the remainder of the brain as the disease progresses [2]. Contributing to the neuronal death, brain hypometabolism, a decrease in the brain metabolic activity, can be visualized using F18-fluorodeoxyglucose (FDG) PET or single photon emission tomography. The hallmark pattern in AD is early hypometabolism of the posterior cingulate, lateral temporal, and parietal lobes with spread to the frontal lobes as the disease progresses [3].
Seventy to eighty percent of sporadic AD can be attributed to genetic risk [4], [5]. Recent large-scale genome-wide association studies (GWASs) have discovered more than 20 AD gene variants that confer genetic risk [6], [7], [8], [9], [10], [11]. Among these variants is the apolipoprotein E (APOE) gene, which is the most established genetic risk factor for AD. Individuals with a single APOE ε4 allele have a three-fold increase in AD risk, whereas homozygotes have a 12-fold increase [12]. apoE is a major protein component of chylomicrons and is highly expressed in both liver and brain, where it plays a role in lipid metabolism and is thought to be involved in the breakdown of Aβ, both inside and outside of cells. The apoE4 protein is less effective in clearing Aβ, providing a possible explanation for the increased risk of amyloid buildup [13]. With the help of imaging studies, APOE ε4 allele was found to be strongly associated with brain amyloidosis [14], [15], atrophy [16], and hypometabolism [17], [18]. These data indicate that valuable observations related to gene function can be gathered with imaging phenotypes.
Many of the remaining top 20 AD variants have also been implicated in brain metabolism and neurodegeneration. Several SORL1 variants, EPHA1 rs11771145, and CR1 rs6656401 were found to be associated with hippocampal atrophy and cerebrovascular or cardiovascular disease [19], [20]. Additionally, various research groups have shown that ABCA7 rs3764650, MS4A6A rs983392, MS4A6A rs610932 and rs11230161, BIN1 rs6733839 and rs744373, CR1 rs1408077, CR1 rs6656401, CR1 rs3818361, PICALM rs3851179, CLU rs11136000 and rs2279590, CD2AP rs10948363, and CD33 rs3865444 are all associated with MRI-measured brain atrophy on MRI [21], [22], [23], [24], [25], [26], [27]. BIN1 rs7561528 was found to be significantly associated with both hippocampal volume and FDG PET brain metabolism [28]. The studies mentioned have unquestionably contributed to the field of imaging genetics and AD research as a whole, but many of these studies have either analyzed the effect of a single gene variant at a time [19], [20], [21], [22], [24], [26], [27] or investigated the association between a polygenic risk score with the imaging trait, which does not allow us to interpret the individual contribution of genetic variants [23]. The commonly used univariate imaging genetics approach ignores the fact that in any given human subject, many of these risk variants are simultaneously present, and the genetic contribution of each variant should be investigated in the presence of the rest and not in isolation. In addition, these studies have investigated the effects in the pooled samples consisting of asymptomatic individuals, of whom only a portion harbor AD pathology, as well as symptomatic individuals who are in different stages of the disease. Such an approach would miss any stage-specific associations that might occur for genes that influence the timing and course of development of disease traits (e.g., early vs. late neurodegeneration or amyloidosis, early vs. late impairment in a specific cognitive domain) and explain, at least in part, AD heterogeneity.
Using a multivariable approach across the disease spectrum allows for accurate modeling of this complex polygenic disease that is constantly evolving. Here, we report a comprehensive analysis of the associations of all well-validated AD risk variants from recent large-scale GWAS studies with two markers of neurodegeneration—brain gray matter density (GMD) and brain glucose metabolism. Our goal was to establish the relative contribution of the top 20 AD risk genes to changes in GMD and metabolic dysfunction. We hypothesized that we would find gene variants that show a profound effect on these two neurodegenerative phenotypes and that some variants will show associations in a stage-specific manner.
2. Methods
2.1. Subjects
We sourced our study data from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). ADNI is an international longitudinal study with approximately 50 sites across the United States and Canada that was launched in 2003. ADNI's goal is to track the progression of AD using clinical and cognitive tests, MRI, FDG PET, amyloid PET, cerebrospinal fluid, and blood biomarkers (http://adni.loni.usc.edu/study-design).
ADNI has undergone three study cycles: ADNI1, ADNI GO, and ADNI2. Our study population was composed of participants from all three stages [29]. The MRI and FDG PET analyses included all subjects with GWAS and baseline MRI or FDG PET data that were successfully preprocessed. A total of 1564 ADNI subject had baseline MRI and GWAS data. Of those, 65 failed in the MRI preprocessing steps and were excluded from our structural analyses. Our final MRI cohort consisted of 441 cognitively normal (NC) subjects, 764 mild cognitive impairment (MCI) subjects, and 294 dementia subjects (total N = 1499). As not all ADNI1 subjects received FDG PET, our FDG PET cohort was smaller and consisted of 381 NC, 634 MCI, and 243 dementia subjects (total N = 1258). There were 59 subjects with available FDG PET data whose MRI scans failed in the preprocessing steps as described previously. These subjects were included in our FDG PET analyses.
The clinical characteristics of the ADNI cohort were described previously [30]. Diagnosis of AD was based on the National Institute of Neurological and Communicative Disorders and Stroke and the AD and Related Disorders Association criteria [31]. AD subjects were required to have Mini–Mental State Examination (MMSE) [32] scores between 20 and 26 and a Clinical Dementia Rating (CDR) scale score of 0.5–1 at baseline [33]. Qualifying MCI subjects had memory complaints but no significant functional impairment, scored between 24 and 30 on the MMSE, had a global CDR score of 0.5, a CDR memory score of 0.5 or greater, and objective memory impairment on Wechsler Memory Scale–Logical Memory II test [34]. NC subjects had MMSE scores between 24 and 30, a global CDR of 0, and did not meet criteria for MCI and AD. Subjects were excluded if they refused or were unable to undergo MRI; had other neurological disorders, active depression, or history of psychiatric diagnosis, alcohol, or substance dependence within the past 2 years; less than 6 years of education; or were not fluent in English or Spanish. The full list of inclusion/exclusion criteria may be accessed on pages 23–29 of the online ADNI protocol (http://www.adni-info.org/Scientists/ADNIStudyProcedures.html). Written informed consent was obtained from all participants.
2.2. Gene variant selection and imputation
ADNI-1 participants were genotyped using the Illumina Human610-Quad BeadChip array, whereas ADNI-2/GO participants were genotyped using the Illumina HumanOmniExpress BeadChip (Illumina, Inc., San Diego, CA). Our decision to include gene variants was based on the AD GWAS studies that discovered these variants to date [6], [7], [8], [9], [10], [11]. Genes previously associated with the defining AD pathologic hallmark–amyloid pathology were also included in our study [35], [36], [37] (Supplementary Table 1). The total number of variants selected was 36.
ABCA7 rs3752246, BIN1 rs6733839, CASS4 rs7274581, CD2AP rs9349407, CELF1 rs10838725, INPP5D rs35349669, PTK2B rs2883497, SORL1 rs11218343, and SORL1 rs1131497 were not genotyped on either ADNI GWAS array and needed full imputation. The following variants were only genotyped on one of the platforms and needed partial imputation: NME8 rs2718058 in ADNI1 and CLU rs933194, DSG2 rs8093731, MEF2C rs190982, and ZCWPW1 rs1476679 in ADNI-GO/2 (Supplementary Table 2). The imputation procedures have been previously described [38]. Imputation was performed using MACH and minimac methodology and the 1000 Genomes project (www.1000genomes.org) as the reference panel. The accuracy threshold was set at r2 = 0.30.
We assessed Hardy-Weinberg equilibrium (HWE) using the --hardy option in PLINK. In the test, we used a quantitative phenotype (global cortical metabolism) and a case-control phenotype. Our results indicate that all 27 single nucleotide polymorphisms (SNPs) do not show any evidence of deviation from HWE (P-value > .01). The accepted significance threshold for declaring SNPs thought to be in HWE is P-value < .001.
Nine of our 20 genes were represented by more than one SNP. Given that such variants could be in linkage disequilibrium (LD) and introduce colinearity bias, we performed LD analyses followed by Cohen kappa (κ) statistics (Supplementary Table 3 and Supplementary Figure 1). When variants providing identical information (those with high LD and high κ) were detected, we chose the SNP with the smallest amount of missing data. This reduced our variants from 36 to 27.
We assessed the allele frequencies for each gene variant. SNPs were coded by minor allele dosage except for the following: ABCA7 rs3764650 GG/GT versus TT, CASS4 rs7274581 CC/TC versus TT, CLU rs9331949 AG/GG versus AA, DSG2 rs8093731 TT/TC versus CC, FERMT2 rs17125944 CC/TC versus TT, and SORL1 rs112183431 CC/TC versus TT where the minor allele homozygote frequency was less than 2%.
2.3. MRI and FDG PET data acquisition and analyses
The MRI acquisition and preprocessing protocols can be found on www.adni-info.org. ADNI MRI data acquisition and preprocessing have been previously described elsewhere [39], [40], [41]. Briefly, we downloaded preprocessed MRI data from LONI IDA (https://ida.loni.usc.edu) [42]. Seven hundred eighty-five subjects had 3T scans available, and for the remaining 715 subjects, we used 1.5T data. MRI field strength was included as a covariate in all MRI analyses. We analyzed all scans using voxel-based morphometry in Statistical Parametric Mapping (SPM8), as described previously [43], [44]. Scans were downloaded from the ADNI site in NifTI format, coregistered to MNI space, bias corrected, and segmented into gray matter (GM), white matter, and cerebrospinal fluid compartments using SPM templates. GM maps were converted to 1 × 1 × 1 mm voxel resolution and smoothed using 10-mm full-width half maximum Gaussian kernel yielding GM density data. Total intracranial volume (ICV) and baseline mean medial temporal lobe thickness measures were extracted for each subject using FreeSurfer version 5.1, as described previously [45], [46]. The medial temporal region of interest included the entorhinal, fusiform, parahippocampal, and temporopolar cortical areas.
The FDG PET acquisition and preprocessing protocols can be found on www.adni-info.org. PET scanners and related equipment across sites were held to the same qualifications, calibration, and normalization standards, as described in detail [47]. We downloaded preprocessed FDG PET data from LONI IDA (https://ida.loni.usc.edu). These scans were already averaged, aligned to a standard space, resampled to a standard image and voxel size (2 × 2 × 2 mm), smoothed to a uniform resolution as previously described [47]. The downloaded images were aligned to each corresponding MRI image on a subject-by-subject basis in MNI space using SPM8, as previously described [46]. Each scan's intensity was scaled to the pons to create standard uptake value ratio (SUVR) images. Finally, baseline mean FDG SUVR in bilateral posterior cingulate was extracted for each subject [48], [49].
2.4. Statistical analyses
2.4.1. R statistical analyses
The distributions of clinical and demographic characteristics (age, sex, education, MMSE, APOE ε4 allele dosage, diagnosis) for each variant were analyzed using t tests or chi-square tests with two-sided P-values as appropriate.
Our main analyses were done in R. First, we performed stepwise linear regression with all 27 AD risk variants as predictors and age, gender, education, and APOE ε4 allele dosage as covariates in the pooled sample and then in each diagnostic group. We used medial temporal GMD or posterior cingulate SUVR, respectively, as outcome measures. Additional covariates were the diagnosis in the pooled analyses and magnetic field strength and ICV in all MRI analyses. The decision to exclude variables in the stepwise regression models was based on the Akaike information criterion using the critical P-value of .157 [50]. Given that all the risk genes were previously validated (i.e., all were candidate genes) and we used a multivariable model, we set our significance threshold at P < .05. After discovering stage-specific genetic influences, we repeated the pooled sample analyses introducing interaction terms between the genetic variants retained in our models and diagnosis.
2.4.2. Analyses in imaging space
All imaging analyses were done in exploratory fashion. We reproduced the final stepwise regression models using voxelwise regression in SPM8 for visualization purposes to explore, on a whole-brain level, the extent and spatial pattern of these imaging genetic associations established using a region of interest approach. These models included all variants retained in the R stepwise linear regression models and were covaried for age, gender, education, and APOE ε4 allele dosage. Consistent with our original regression model, the pooled analyses also included diagnosis as a covariate, and the MRI analyses were additionally controlled for MRI field strength and ICV. Due to the exploratory nature of our secondary results, we used a less-stringent visualization voxelwise threshold, which was uncorrected P < .01 with a minimum cluster size (k) of 50 voxels. Next, we applied cluster-level family-wise error (FWE) and false discovery rate (FDR) correction and sought out the within-cluster peak effects for all genetic variants identified in our models.
3. Results
Group comparisons of demographic characteristics and distributions of the genotypes that were retained in the regression models are shown in Table 1, Table 2 for the MRI and PET samples, respectively. As expected, AD subjects were the oldest, least educated, had the greatest frequency of APOE ε4 homo- and heterozygotes, and performed the worst on MMSE (Table 1, Table 2). There were no significant differences in age, gender, education, MMSE, and APOE ε4 allele dosage distribution between carriers and noncarriers or by allele dosage for any of the genotypes except the following: DSG2 minor allele carriers were more likely to be male (P = .028 in the FDG sample) and less likely to be APOE ε4 carriers (P = .04 in the MRI sample); EPHA1 rs11767557 minor allele carriers were less likely to be APOE ε4 carriers and had significantly higher MMSE scores in the FDG sample (P = .037 and P = .047, respectively); SORL1 rs11218343 minor allele carriers were less educated and more likely to be male in both samples (MRI: P = .01 and P = .008, and FDG P = .008 and P = .026, respectively), and ZCWPW1 risk allele carriers were significantly less educated and had higher MMSE scores in both samples (MRI P = .0012 and P = .034, and FDG P = .025 and P = .034, respectively). For completeness, the allele dosage for all 27 variants including the ones not retained in our models can be seen in Supplementary Tables 4 and 5.
Table 1.
Descriptive characteristics and distribution of the genotypes of the MRI sample
| Variables | NC (N = 441) | MCI (N = 764) | DEM (N = 294) | P |
|---|---|---|---|---|
| Age, mean years (SD) | 74.1 (5.7) | 72.6 (7.6) | 74.6 (7.9) | <.001 |
| Sex, N male (%) | 222 (50) | 453 (59) | 165 (56) | .01 |
| Education, mean years (SD) | 16.4 (2.6) | 16.0 (2.8) | 15.2 (3.0) | <.001 |
| MMSE, mean (SD) | 29.1 (1.1) | 27.6 (1.8) | 23.3 (2.1) | <.001 |
| APOE ε4, % 0/1/2 | 71/27/2 | 49/40/11 | 34/47/19 | <.001 |
| ABCA7 rs3752246, % 0/1/2 | 69/29/2 | 68/28/4 | 67/30/3 | .58 |
| CELF1 rs10838725, % 0/1/2 | 46/45/9 | 44/46/10 | 45/48/7 | .45 |
| EPHA1 rs11771145, % 0/1/2 | 44/44/12 | 43/45/12 | 45/42/13 | .95 |
| FERMT2 rs17125944, % 0/1 | 85/15 | 84/16 | 78/22 | .02 |
| INPP5D rs35349669, % 0/1/2 | 30/48/22 | 30/46/24 | 26/51/23 | .52 |
| SLC24A4/RIN3 rs10498633, % 0/1/2 | 60/35/5 | 61/34/5 | 60/35/5 | .99 |
| ZCWPW1 rs1476679, % 0/1/2 | 50/41/9 | 51/41/8 | 60/33/7 | .08 |
Abbreviations: MRI, magnetic resonance imaging; NC, normal control; MCI, mild cognitive impairment; DEM, dementia; SD, standard deviation.
NOTE. Bold text indicates a significant P value (P <0.05).
Table 2.
Descriptive characteristics and distribution of the genotypes of the FDG PET sample
| Variables | NC (N = 381) | MCI (N = 634) | DEM (N = 243) | P |
|---|---|---|---|---|
| Age, mean years (SD) | 74.3 (6.2) | 72.6 (7.6) | 75.0 (7.7) | <.001 |
| Sex, N male (%) | 190 (50) | 380 (60) | 148 (61) | .003 |
| Education, mean years (SD) | 16.4 (2.7) | 16.1 (2.7) | 15.4 (2.9) | <.001 |
| MMSE, mean (SD) | 29.0 (1.2) | 27.8 (1.8) | 23.1 (2.8) | <.001 |
| APOE ε4, % 0/1/2 | 73/25/2 | 51/38/11 | 33/50/17 | <.001 |
| CD2AP rs9349407, % 0/1/2 | 49/44/7 | 53/40/7 | 49/40/11 | .22 |
| CELF1 rs10838725, % 0/1/2 | 48/42/10 | 44/46/10 | 43/50/4 | .27 |
| CLU rs11136000, % 0/1/2 | 36/50/14 | 37/50/13 | 43/42/15 | .28 |
| CLU rs9331949, % 0/1 | 95/5 | 97/3 | 95/5 | .31 |
| CR1 rs12034383, % 0/1/2 | 14/49/37 | 15/48/37 | 21/46/33 | .19 |
| DSG2 rs8093731, % 0/1 | 98/2 | 98/2 | 97/3 | .84 |
| EPHA1 rs11771145, % 0/1/2 | 44/44/12 | 45/42/13 | 43/44/13 | .94 |
| EPHA1 rs11767557, % 0/1/2 | 66/30/4 | 70/27/3 | 63/34/3 | .29 |
| MS4A6A rs610932, % 0/1/2 | 31/48/21 | 37/47/16 | 38/45/17 | .25 |
| NME8 rs2718058, % 0/1/2 | 39/47/14 | 40/45/15 | 38/49/13 | .88 |
| PTK2B rs28834970, % 0/1/2 | 42/42/16 | 42/42/16 | 39/49/12 | .39 |
| SLC24A4/RIN3 rs10498633, % 0/1/2 | 61/34/5 | 62/34/4 | 58/37/5 | .87 |
| SORL1 rs11218343, % 0/1 | 91/9 | 92/8 | 93/7 | .55 |
Abbreviations: FDG PET, F18-fluorodeoxyglucose positron emission tomography; NC, normal control; MCI, mild cognitive impairment; DEM, dementia; SD, standard deviation; MMSE, Mini–Mental State Examination.
NOTE. Bold text indicates a significant P value (P <0.05).
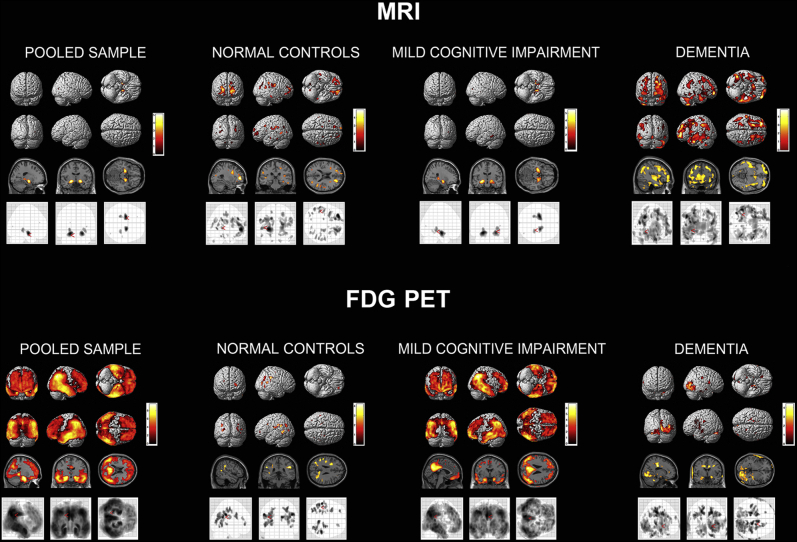
APOE ε4 showed the expected positive association with GMD in the NC and MCI group (Fig. 1 top). In the symptomatic MCI group, we saw a strong hippocampo-centric pattern, indicating that MCI carriers had greater hippocampal loss of GMD compared to MCI noncarriers. In the dementia group, we found the opposite association. APOE ε4-negative dementia subjects had greater cortical loss of GMD than APOE ε4-positive subjects, indicative of greater cortical neurodegeneration. APOE ε4 was associated with widespread hypometabolism in the MCI group (Fig. 1 bottom). APOE ε4 showed a much less pronounced effect in the dementia stage indicating that both carriers and noncarriers experienced significant, widespread hypometabolic changes.
Fig. 1.
APOE4 effect on brain atrophy (top) and hypometabolism (bottom). Results are displayed using P < .01 (uncorrected) and cluster size (k) of 50 voxels. Abbreviations: FDG PET, F18-fluorodeoxyglucose positron emission tomography; MRI, magnetic resonance imaging.
3.1. MRI analyses
3.1.1. Pooled sample
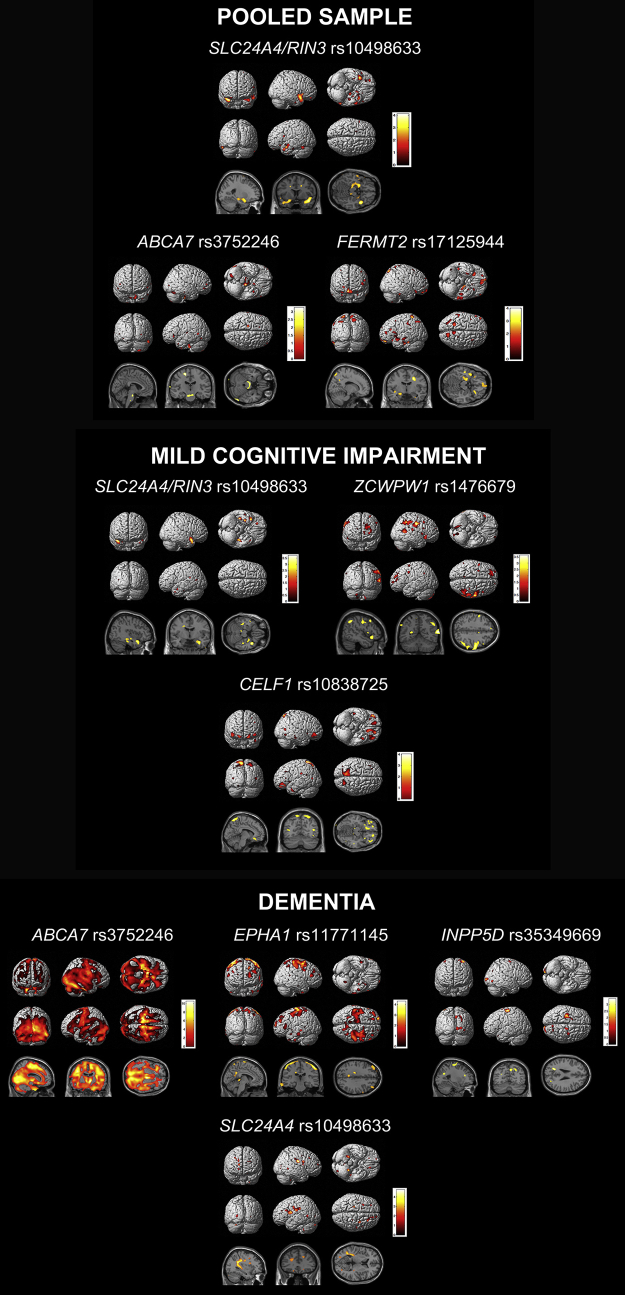
In the pooled sample, the stepwise linear regression model achieved an R2 = 0.4, P < .0001. SLC24A4/RIN3 rs10498633 was the only variant that was significantly associated with mean medial temporal lobe GMD in the pooled sample (χ2 = 11.8, P = .003). ABCA7 rs3752246 and FERMT2 rs17125944 were retained in the regression output based on the selection criteria but were not statistically significant. See Fig. 2 for the exploratory visualization of these associations and Table 3 for FWE- and FDR-corrected cluster-level results and within-cluster peak effects for genetic variants identified in our models.
Fig. 2.
Stepwise linear regression results—MRI analysis. Results are displayed using P < .01 (uncorrected) and cluster size (k) of 50 voxels. Abbreviation: MRI, magnetic resonance imaging.
Table 3.
Family-wise error (FWE) and false discovery rate (FDR)-corrected cluster analyses and within-cluster peak effects for the genetic variants identified in the exploratory MRI analyses
| DX group | Gene variant | Cluster-level |
Peak level |
Talairach coordinates | Brain region (Brodmann area [BA]) | ||||
|---|---|---|---|---|---|---|---|---|---|
| pFWE-corr | qFDR-corr | kE | puncorr | T | puncorr | ||||
| DEM | ABCA7 rs3752246 | <.0001 | <0.0001 | 924757 | <.0001 | 9.72 | <.0001 | −11 −23 63 | L medial frontal gyrus (BA6) |
| EPHA1 rs11771145 | .04 | 0.051 | 13,081 | .001 | 3.86 | <.0001 | −52 −28 61 | L postcentral gyrus (BA2) | |
| .007 | 0.017 | 1889 | <.0001 | 4.01 | <.0001 | 52 −30 59 | R postcentral gyrus (BA2) | ||
| SLC24A4/RIN3 rs10498633 | .007 | 0.012 | 18,621 | <.0001 | 3.78 | <.0001 | −14 1 46 | L cingulate gyrus (BA24) | |
Abbreviations: MRI, magnetic resonance imaging; pFWE-corr, family-wise error-corrected P-value; qFDR-corr, false discovery rate–corrected q-value; kE, cluster size; puncorr, uncorrected P-value; T, T statistic; DEM, dementia.
NOTE. Bold text indicates a significant P value (P <0.05).
3.1.2. Analyses within diagnostic groups
We found neither significant nor trend-level associations in the NC group. In the MCI group, the model achieved an R2 = 0.26. SLC24A4/RIN3 rs10498633 (χ2 = 11.3, P = .004) and ZCWPW1 rs1476679 (χ2 = 7.3, P = .026) reached significance. CELF1 rs10838725 (χ2 = 5.2, P = .073) was trending. In the dementia group, the model achieved an R2 = 0.19, P < .0001. ABCA7 rs3752246 (χ2 = 8.5, P = .014), EPHA1 rs11771145 (χ2 = 11.6, P = .003), and INPP5D rs35349669 (χ2 = 6.4, P = .042) were significantly associated with mean medial temporal lobe GMD. SLC24A4/RIN3 rs10498633 (χ2 = 5.4, P = .068) was trending. See Fig. 2 for the exploratory pattern of these associations.
Next, we repeated the pooled sample analyses introducing interaction terms between the genetic variants retained in our models and diagnosis. The following variants showed significant interaction effect with diagnosis—EPHA1 rs11771145 (F = 3.2, P = .01) and SLC24A4/RIN3 (F = 2.7, P = .03). Please note that the R statistical analyses and the analyses in imaging space might differ. This is because in our R statistics model, we use a circumscribed ROI, in the case of MRI, the medial temporal region, while the whole-brain exploratory results show the effect of the variant across the brain.
3.2. FDG PET analyses
3.2.1. Pooled sample
In the pooled sample, the stepwise linear regression model achieved an R2 = 0.13, P < .0001. EPHA1 rs11767557 showed significant associations with brain metabolism (χ2 = 6.3, P = .042). PTK2B rs28834970 (χ2 = 5.6, P = .059), MS4A6A rs610932 (χ2 = 5.5, P = .065) and SLC24A4/RIN3 rs10498533 (χ2 = 4.6, P = .098) showed trend-level associations. CLU rs11136000 and EPHA1 rs11771145 were included in the model based on the selection criteria but were not statistically significant.
See Fig. 3 for the exploratory visualization of these associations and Table 4 for FWE- and FDR-corrected cluster-level results and within-cluster peak effects for genetic variants identified in our models.
Fig. 3.
Stepwise linear regression results of FDG PET. Results are displayed using P < .01 (uncorrected) and cluster size (k) of 50 voxels. Abbreviation: FDG PET, F18-fluorodeoxyglucose positron emission tomography.
Table 4.
Family-wise error (FWE) and false discovery rate (FDR)-corrected cluster-level analyses and within-cluster peak effects for genetic variants identified in the exploratory FDG PET analyses
| Diagnostic group | Gene variant | Cluster |
Peak effect |
Talairach coordinates | Brain region (Brodmann area [BA]) | ||||
|---|---|---|---|---|---|---|---|---|---|
| pFWE-corr | qFDR-corr | kE | puncorr | T | puncorr | ||||
| Pooled sample | CLU rs1113600 | <.0001 | <0.0001 | 5979 | <.0001 | 4.02 | <.0001 | −48 −4 −32 | L inferior temporal gyrus (BA20) |
| .027 | 0.025 | 1493 | <.0001 | 4.01 | <.0001 | 64 −18 8 | R transverse temporal gyrus (BA42) | ||
| EPHA1 rs11771145 | .049 | 0.102 | 1314 | .0001 | 3.8 | <.0001 | −14 −50 60 | L superior parietal lobule (BA7) | |
| MS4A6A rs610932 | .001 | 0.001 | 2723 | <.0001 | 4.28 | <.0001 | 40 −14 42 | R precentral gyrus (BA4) | |
| PTK2B rs28834970 | .002 | 0.003 | 2426 | <.0001 | 3.96 | <.0001 | −2 −74 −2 | L lingual gyrus (BA18) | |
| NC | CELF1 rs10838725 | .004 | 0.004 | 1960 | <.0001 | 4.19 | <.0001 | 20 −66 44 | R precuneus (BA7) |
| <.0001 | <0.0001 | 2718 | <.0001 | 3.88 | <.0001 | 0 52 −12 | L medial frontal gyrus (BA10) | ||
| .017 | 0.012 | 1514 | <.0001 | 3.5 | <.0001 | 32 −38 −4 | R subgyral, hippocampus | ||
| CR1 rs12034383 | <.0001 | <0.0001 | 37,014 | <.0001 | 4.98 | <.0001 | 18 −36 30 | R cingulate gyrus (BA31) | |
| DSG2 rs8093731 | .017 | 0.008 | 1515 | <.0001 | 4.15 | <.0001 | 6 −4 50 | R medial frontal gyrus (BA6) | |
| PTK2B rs28834970 | <.0001 | <0.0001 | 6279 | <.0001 | 4.24 | <.0001 | 44 −22 6 | L superior temproral gyrus (BA13) | |
| <.0001 | <0.0001 | 6722 | <.0001 | 3.98 | <.0001 | −42 −36 12 | L superior temproral gyrus (BA41) | ||
| SLC24A4/RIN3 rs10498633 | <.0001 | <0.0001 | 20,054 | <.0001 | 4.38 | <.0001 | 22 −4 −30 | R parahippocampal gyrus (BA36) | |
| .007 | 0.004 | 1767 | <.001 | 3.71 | <.0001 | −22 −8 −30 | L parahippocampal gyrus (BA36) | ||
| DEM | NME8 rs2718058 | <.001 | <0.001 | 3551 | <.001 | 4.52 | <.0001 | −14 −88 30 | L cuneus (BA19) |
Abbreviations: FDG PET, F18-fluorodeoxyglucose positron emission tomography; pFWE-corr, family-wise error-corrected P-value; qFDR-corr, false discovery rate–corrected q-value; kE, cluster size; puncorr, uncorrected P-value; T, T statistic; NC, normal control; DEM, dementia.
NOTE. Bold text indicates a significant P value (P <0.05).
3.2.2. Analyses within diagnostic groups
In the NC group, the stepwise linear regression model achieved an R2 = 0.14, P < .0001. SLC24A4/RIN3 rs10498533 (χ2 = 6.3, P = .043), NME8 rs2718058 (χ2 = 6.0, P = .049), and CD2AP rs9349407 (χ2 = 6.1, P = .048) showed significant associations, whereas DSG2 rs8093731 (χ2 = 3.4, P = .064), CR1 rs12034383 (χ2 = 4.9, P = .087), and CELF1 rs10838725 (χ2 = 5.386, P = .068) were trending. CLU rs9331949 and PTK2B rs28834970 were included in the model based on the selection criteria but were not statistically significant. In the MCI group, the stepwise linear regression model achieved an R2 = 0.09, P < .0001. CLU rs9331949 was trending (χ2 = 3.4, P = .065), whereas MS4A6A rs610932 was included based on the selection criteria but was not statistically significant. In the dementia stage, the model achieved an R2 = 0.07, P = .034. SORL1 rs11218343 showed trend-level significance (χ2 = 3.5, P = .063), whereas NME8 rs2718058 and CD2AP rs9349407 were included based on the selection criteria but were not statistically significant. See Fig. 3 for the exploratory visualization of these associations.
Next, we repeated the pooled sample analyses introducing interaction terms between the genetic variants retained in our models and diagnosis. The following variants showed significant interaction effect with diagnosis—CD2AP rs9349407 (F = 2.4, P = .04), CLU rs9331949 (F = 3.5, P = .03), and NME8 rs2718058 (F = 3.1, P = .01), CR1 rs12034383 (F = 2.3, P = .06) and SORL1 rs11218343 (F = 2.9, P = .06) showed trend-level interactions. Please note that the R statistical analyses and the analyses in imaging space might differ. This is because in our R statistics model, we use a circumscribed ROI, in the case of FDG PET the posterior cingulate region, while the whole-brain exploratory results show the effect of the variant across the brain.
4. Discussion
To our knowledge, this is the first comprehensive analysis of the effect of the top 20 AD risk variants on GMD and brain metabolism. In our MRI analyses, we found no genetic influences on GMD in NC. In the MCI stage, SLC24A4/RIN3 rs10498633 and ZCWPW1 rs1476679 showed significant effects, whereas in the dementia stage, ABCA7 rs3752246, EPHA1 rs11771145, and INPP5D rs35349669 were significantly associated with GMD. In our FDG PET analyses, the only significant associations were seen in the NC control group for SLC24A4/RIN3 rs10498533, NME8 rs2718058, and CD2AP rs9349407. The reported associations of ABCA7, ZCWPW1, and INPP5D with GMD, and CD2AP with brain metabolism, are novel.
Many of our variants displayed stage-specific associations, which is likely due to the nature of AD pathological biomarker changes from the presymptomatic stage to dementia. These stage-specific associations are in agreement with the biomarker progression as proposed by Jack et al. [51]. Jack's biomarker progression model postulates that neurodegenerative changes begin in the late asymptomatic stages as NC individuals start to transition to MCI and that FDG PET abnormalities precede brain atrophy [51]. Both neurodegenerative biomarkers become progressively more abnormal over the course of the disease. Our findings agree with this model. Our results indicate neither significant nor trend-level associations with GMD in the NC group, but significant associations were detected in the MCI and dementia stages. We also see a modality effect progression that fits with the Jack model. SLC24A4/RIN3 shows significant association with brain metabolism in the presymptomatic stages and associations with GMD in the MCI group.
Recent analyses from our group on the top 20 AD genes and brain amyloidosis revealed that, after APOE ε4, ABCA7 had the strongest effect on brain amyloidosis, with the effect being most pronounced in the MCI stage [52]. In the present study, we observe a strong association of ABCA7 with GMD in the dementia group. These associations also seem to follow the biomarker progression, as Aβ deposition begins in the presymptomatic to early MCI phase and global decreases in GMD begin in late MCI to dementia phase. Associations of ABCA7 with both brain atrophy [21] and amyloidosis [35], [36], [37] have been previously reported.
In a similar manner, our previous work showed that EPHA1's strongest effect on brain amyloidosis is in the prodromal phase [52]. In the present study, our MRI results show a significant association with GMD in the dementia stages only. To date, two additional studies have described EPHA1 associations with brain atrophy [53], [54] and one also detected an association with brain metabolism as we did here [53].
Next, we found it pertinent to briefly review the literature on the function and central nervous system associations for each of the genes in our models.
ATP-binding cassette subfamily A member 7 (ABCA7) encodes a 2146–amino acid member of the ABC transporter family comprised of proteins involved in lipid transport [55]. ABCA7 is highly expressed in the central nervous system and in microglia [56]. Loss of function of endogenous ABCA7 increases β-secretase cleavage of APP to Aβ in brain lysates and in murine models [57]. Fifteen ABCA7 loci were recently evaluated for associations with cerebrospinal fluid Aβ and tau levels, brain atrophy, and brain hypometabolism. Several ABCA7 variants had significant associations with amyloid deposition, although an association with brain atrophy was not reported there [58]. However, another group has previously found such an association in a different independent imaging genetic cohort [21]. Additionally, one rare missense mutation variant of ABCA7 (rs7297358) was found to be protective for AD [59].
The CD2-associated protein gene (CD2AP) encodes a 639–amino acid scaffolding protein that is named by its association with the T-cell and natural killer cell adhesion molecule cluster of differentiation 2 (CD2). CD2AP is a cytokinetic regulator that might be influencing neuronal survival by reducing the potency of glial cell–derived neurotrophic factor [60]. In cell culture, CD2AP suppression results in lower levels of APP, less Aβ release, and a lower Aβ42/Aβ40 ratio while having little to no effect on Aβ deposition [61]. CD2AP knockdown significantly increases Aβ protein levels, whereas APP remained at a similar level to the wild type [62]. Recent research in CD2AP knockout mice shows that CD2AP's association with LOAD risk may be at least in part due to an effect on the cerebrovascular unit [63]. To our knowledge, there are no reported imaging associations of CD2AP to date.
EPH receptor A1 (EPHA1) belongs to the EPH family of receptor tyrosine kinases. EPHA1 codes for a 976–amino acid protein with a single kinase domain [64]. EPHA1 is highly expressed in cerebral cortex and hippocampus (http://www.proteinatlas.org/ENSG00000146904-EPHA1/tissue) and plays a crucial role in cortical and axonal development [65], [66]. EPHA1 directs contact-dependent bidirectional signaling by binding the membrane-bound ephrin-A family of ligands [67], [68]. As already discussed, EPHA1 has been previously associated with brain atrophy and brain metabolism [53], [54].
Inositol polyphosphate-5-phosphatase D (INPP5D) encodes a 1888–amino acid protein that plays a role in a number of inflammatory pathways and in regulation of cytokine signaling [69], [70]. INPP5E, a closely related gene to INPP5D, is a crucial regulator of autophagy [71]. Autophagy has been shown in many instances to be dysregulated in AD [72], [73], [74]. INPP5D may also play a role in suppression of cytokine release from microglia or astrocytes [75]. INPP5D has been significantly associated with several central nervous system pathologies, including macroscopic and microscopic infarcts, Lewy bodies, and hippocampal sclerosis [76].
NME family member 8 (NME8) encodes a 588–amino acid protein kinase. Until recently, NME8 was associated with nonneurologic diseases such as primary ciliary dyskinesia [77] and osteoarthritis [78], [79]. In a recent ADNI study, however, NME8 rs2718058 was shown to have a neuroprotective effect against hippocampal atrophy and brain hypometabolism [80].
Solute carrier family 24 (sodium/potassium/calcium exchanger) member 4 (SLC24A4)/Ras and Rab Interactor 3 (RIN3) are indicated in AD together because the candidate polymorphism lies between both genes on chromosome 14q32.12. SLC24A4 encodes a 622–amino acid potassium-dependent sodium-calcium exchanger [81]. SLC24A4 appears to take part in lipid metabolism [82]. SLC24A4 CpG methylation sites were also associated with Aβ burden and tau pathology [83]. In a recent study, SLC24A4 knockout mice showed brain glucose hypometabolism [84]. Interestingly, we found a similar effect in NC.
RIN3 encodes a 985–amino acid guanine exchange factor for RAB5B and RAB31 and plays an important role in the transport of early endosomes [85], [86]. In a transgenic model, a mutation in APP was shown to contribute to early endosomal abnormalities and enlargement, which leads to loss of cholinergic neurons [87]. RAB GTPase expression is increased in MCI subjects and in aging brains [88]. RIN3 also interacts with BIN1, which has recently been linked to tau pathology [89]. To our knowledge, there are no reported imaging associations of RIN3 to date.
Zinc finger CW-type and PWWP domain containing 1 (ZCWPW1) codes for a 648–amino acid protein that has recently been identified as a risk variant for late-onset AD [90]. Zinc fingers including ZCWPW1 are crucial components of epigenetic regulation [91], [92], [93]. To our knowledge, there are no reported imaging associations of ZCWPW1 to date.
The APOE ε4 allele dosage effect on cortical GMD warrants some discussion. Structurally, we found the expected negative association of APOE ε4 allele dosage with GMD in NC and MCI with a strong predilection for the medial temporal lobe. In the dementia group, however, APOE ε4-negative individuals showed more profound atrophy than APOE ε4 carriers with a broad neocortical distribution. Given that a far greater proportion of APOE ε4-negative dementia subjects were amyloid negative (94% vs. 28% of the amyloid-positive dementia subjects, P < .0001), one could safely conclude that the profound cognitive decline of many of these subjects is due to other neurodegenerative diseases. Thus, the finding greater atrophy in APOE ε4 noncarrier dementia subjects in extra-hippocampal locations is not surprising.
Several strengths and limitations of our work are worth noting. The major strength of our study is our multivariable approach. This allows us to more accurately model the associations with AD biomarkers as they exist in vivo. Using ADNI is another strength as the ADNI protocol includes a rigorous clinical, biomarker, and genetic characterization for all enrolled subjects. By standardizing their data collection and processing strategies, ADNI minimizes between-site variations as much as possible. ADNI is a multisite study modeled by clinical trials. As such, ADNI uses more stringent inclusion and exclusion criteria typical of clinical trial methodology. Hence all observations made here need to be further replicated in the general population. Another limitation is that our study design is a cross-sectional analysis. From our data alone, it is not possible to reliably draw conclusions about changes in metabolism or atrophy over time. We do, however, intend to address this in future studies by taking a longitudinal approach to our work. Despite this limitation, this study has identified several key genes that may exert their effect in specific stages, which need to be examined in future research.
In conclusion, we found several AD risks and protective loci that may play a key role in GMD and brain metabolism. We also noted stage-specific associations for certain variants, which may follow a specific progression of AD biomarkers throughout the disease. Importantly, many of these stage associations take place in the context of the proposed biomarker timeline.
Research in Context.
-
1.
Systematic review: Our goal was to analyze the effects of the top 20 Alzheimer disease (AD) risk genes on brain gray-matter density (GMD) and metabolism. We assessed relevant literature by searching PubMed and Science Direct for studies describing imaging genetics associations in AD.
-
2.
Interpretation: SLC24A4, ABCA7, and EPHA1 have significant stage-specific associations that followed the predicted biomarker pathway. We discovered novel associations of ZCWPW1, INPP5D, and ABCA7 with brain GMD and CD2AP with brain metabolism.
-
3.
Future directions: Future longitudinal studies of the observed associations will aptly measure defined changes over time. This approach will be critical in accurately modeling the genetic effects on disease progression.
Acknowledgments
The authors thank the members of the ADNI Imaging Core for their contributions to the image preprocessing, the members of the ADNI Biomarker Core for the CSF biomarker analyses, and the investigators at the University of Pittsburgh for the PIB SUVR analyses.
The analyses reported in this article were funded by the NIA R01 AG040770, NIA K02 AG048240, NIA P30 AG010133, NLM K99/R00 LM011384, NIA K01 AG049050, the Alzheimer's Association, the Easton Consortium for Alzheimer's Drug Discovery and Biomarker Development, the Indiana University Strategic Research Initiative, the Indiana University Physician Science Initiative, and the Indiana Clinical and Translational Science Institute.
Data collection and sharing for this project were funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; Euroimmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck, Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Eddie Stage, BS, has nothing to disclose. Tugce Duran, BS, has nothing to disclose. Shannon Risacher, PhD, has nothing to disclose. Naira Goukasian, BS, has nothing to disclose. Triet Do, BS, has nothing to disclose. John West, MS, has nothing to disclose. Holly Wilhalme, MS, has nothing to disclose. Kwangsik Nho, PhD, has nothing to disclose. Meredith Phillips, MS, has nothing to disclose. David Elashoff, PhD, has nothing to disclose. Andrew Saykin, PsyD, has received research support from Eli Lilly and AVID Radiopharmaceuticals. Liana G. Apostolova, MD, serves on an Advisory Board for Eli Lilly and on the Speakers Bureau for Piramal and Eli Lilly. Dr. Apostolova has received research support from GE Healthcare.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.dadm.2016.12.003.
Supplementary data
References
- 1.Tiraboschi P., Hansen L.A., Thal L.J., Corey-Bloom J. The importance of neuritic plaques and tangles to the development and evolution of AD. Neurology. 2004;62:1984–1989. doi: 10.1212/01.wnl.0000129697.01779.0a. [DOI] [PubMed] [Google Scholar]
- 2.Apostolova L.G., Steiner C.A., Akopyan G.G., Dutton R.A., Hayashi K.M., Toga A.W. Three-dimensional gray matter atrophy mapping in mild cognitive impairment and mild Alzheimer disease. Arch Neurol. 2007;64:1489–1495. doi: 10.1001/archneur.64.10.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apostolova L.G., Thompson P.M., Rogers S.A., Dinov I.D., Zoumalan C., Steiner C.A. Surface feature-guided mapping of cerebral metabolic changes in cognitively normal and mildly impaired elderly. Mol Imaging Biol. 2010;12:218–224. doi: 10.1007/s11307-009-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wingo T.S., Lah J.J., Levey A.I., Cutler D.J. Autosomal recessive causes likely in early-onset Alzheimer disease. Arch Neurol. 2012;69:59–64. doi: 10.1001/archneurol.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sleegers K., Lambert J.C., Bertram L., Cruts M., Amouyel P., Van Broeckhoven C. The pursuit of susceptibility genes for Alzheimer's disease: progress and prospects. Trends Genet. 2010;26:84–93. doi: 10.1016/j.tig.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 9.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seshadri S., Fitzpatrick A.L., Ikram M.A., DeStefano A.L., Gudnason V., Boada M. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verghese P.B., Castellano J.M., Holtzman D.M. Apolipoprotein E in Alzheimer's disease and other neurological disorders. Lancet Neurol. 2011;10:241–252. doi: 10.1016/S1474-4422(10)70325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 14.Reiman E.M., Chen K., Liu X., Bandy D., Yu M., Lee W. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer's disease. Proc Natl Acad Sci U S A. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazaris A., Hwang K.S., Goukasian N., Ramirez L.M., Eastman J., Blanken A.E. Alzheimer risk genes modulate the relationship between plasma apoE and cortical PiB binding. Neurol Genet. 2015;1:e22. doi: 10.1212/NXG.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrawis J.P., Hwang K.S., Green A.E., Kotlerman J., Elashoff D., Morra J.H. Effects of ApoE4 and maternal history of dementia on hippocampal atrophy. Neurobiol Aging. 2012;33:856–866. doi: 10.1016/j.neurobiolaging.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiman E.M., Caselli R.J., Yun L.S., Chen K., Bandy D., Minoshima S. Preclinical evidence of Alzheimer's disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- 18.Mosconi L., Nacmias B., Sorbi S., De Cristofaro M.T., Fayazz M., Tedde A. Brain metabolic decreases related to the dose of the ApoE e4 allele in Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2004;75:370–376. doi: 10.1136/jnnp.2003.014993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assareh A.A., Piguet O., Lye T.C., Mather K.A., Broe G.A., Schofield P.R. Association of SORL1 gene variants with hippocampal and cerebral atrophy and Alzheimer's disease. Curr Alzheimer Res. 2014;11:558–563. doi: 10.2174/1567205011666140618101408. [DOI] [PubMed] [Google Scholar]
- 20.Cuenco T., Lunetta K.L., Baldwin C.T., McKee A.C., Guo J., Cupples L.A. Association of distinct variants in SORL1 with cerebrovascular and neurodegenerative changes related to Alzheimer disease. Arch Neurol. 2008;65:1640–1648. doi: 10.1001/archneur.65.12.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez L.M., Goukasian N., Porat S., Hwang K.S., Eastman J.A., Hurtz S. Common variants in ABCA7 and MS4A6A are associated with cortical and hippocampal atrophy. Neurobiol Aging. 2016;39:82–89. doi: 10.1016/j.neurobiolaging.2015.10.037. [DOI] [PubMed] [Google Scholar]
- 22.Ma J., Zhang W., Tan L., Wang H.F., Wan Y., Sun F.R. MS4A6A genotypes are associated with the atrophy rates of Alzheimer's disease related brain structures. Oncotarget. 2016;7:58779–58788. doi: 10.18632/oncotarget.9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohannim O., Hua X., Rajagopalan P., Hibar D.P., Jahanshad N., Grill J.D. Multilocus genetic profiling to empower drug trials and predict brain atrophy. Neuroimage Clin. 2013;2:827–835. doi: 10.1016/j.nicl.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J.Q., Wang H.F., Zhu X.C., Sun F.R., Tan M.S., Tan C.C. GWAS-linked loci and neuroimaging measures in Alzheimer's disease. Mol Neurobiol. 2016 doi: 10.1007/s12035-015-9669-1. [DOI] [PubMed] [Google Scholar]
- 25.Biffi A., Anderson C.D., Desikan R.S., Sabuncu M., Cortellini L., Schmansky N. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chauhan G., Adams H.H., Bis J.C., Weinstein G., Yu L., Toglhofer A.M. Association of Alzheimer's disease GWAS loci with MRI markers of brain aging. Neurobiol Aging. 2015;36:1765.e7–1765.e16. doi: 10.1016/j.neurobiolaging.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furney S.J., Simmons A., Breen G., Pedroso I., Lunnon K., Proitsi P. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer's disease. Mol Psychiatry. 2011;16:1130–1138. doi: 10.1038/mp.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H.F., Wan Y., Hao X.K., Cao L., Zhu X.C., Jiang T. Bridging integrator 1 (BIN1) genotypes mediate Alzheimer's disease risk by altering neuronal degeneration. J Alzheimers Dis. 2016;52:179–190. doi: 10.3233/JAD-150972. [DOI] [PubMed] [Google Scholar]
- 29.Weiner M.W., Veitch D.P., Aisen P.S., Beckett L.A., Cairns N.J., Green R.C., Alzheimer's Disease Neuroimaging Initiative The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2012;8:S1–S68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen R.C., Aisen P.S., Beckett L.A., Donohue M.C., Gamst A.C., Harvey D.J. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74:201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 32.Folstein M.F., Folstein S.E., McHugh P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 33.Morris J.C. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 34.Wechsler D. Psychological Corporation; San Antonio, Texas: 1987. Manual for the Wechsler Memory Scale–revised. [Google Scholar]
- 35.Thambisetty M., An Y., Nalls M., Sojkova J., Swaminathan S., Zhou Y. Effect of complement CR1 on brain amyloid burden during aging and its modification by APOE genotype. Biol Psychiatry. 2013;73:422–428. doi: 10.1016/j.biopsych.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shulman J.M., Imboywa S., Giagtzoglou N., Powers M.P., Hu Y., Devenport D. Functional screening in Drosophila identifies Alzheimer's disease susceptibility genes and implicates Tau-mediated mechanisms. Hum Mol Genet. 2014;23:870–877. doi: 10.1093/hmg/ddt478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes T.M., Lopez O.L., Evans R.W., Kamboh M.I., Williamson J.D., Klunk W.E. Markers of cholesterol transport are associated with amyloid deposition in the brain. Neurobiol Aging. 2014;35:802–807. doi: 10.1016/j.neurobiolaging.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nho K., Shen L., Kim S., Swaminathen S., Risacher S.L., Saykin A.J. The effect of reference panels and software tools on genotype imputation. AMIA Annu Symp Proc. 2011;2011:1013–1018. [PMC free article] [PubMed] [Google Scholar]
- 39.Wyman B.T., Harvey D.J., Crawford K., Bernstein M.A., Carmichael O., Cole P.E. Standardization of analysis sets for reporting results from ADNI MRI data. Alzheimers Dement. 2013;9:332–337. doi: 10.1016/j.jalz.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jack C.R., Jr., Bernstein M.A., Borowski B.J., Gunter J.L., Fox N.C., Thompson P.M. Update on the magnetic resonance imaging core of the Alzheimer's disease neuroimaging initiative. Alzheimers Dement. 2010;6:212–220. doi: 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jack C.R., Jr., Bernstein M.A., Fox N.C., Thompson P., Alexander G., Harvey D. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jack C.R., Jr., Barnes J., Bernstein M.A., Borowski B.J., Brewer J., Clegg S. Magnetic resonance imaging in Alzheimer's Disease Neuroimaging Initiative 2. Alzheimers Dement. 2015;11:740–756. doi: 10.1016/j.jalz.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitwell J.L. Voxel-based morphometry: an automated technique for assessing structural changes in the brain. J Neurosci. 2009;29:9661–9664. doi: 10.1523/JNEUROSCI.2160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Risacher S.L., Kim S., Shen L., Nho K., Foroud T., Green R.C. The role of apolipoprotein E (APOE) genotype in early mild cognitive impairment (E-MCI) Front Aging Neurosci. 2013;5:11. doi: 10.3389/fnagi.2013.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Risacher S.L., Shen L., West J.D., Kim S., McDonald B.C., Beckett L.A. Longitudinal MRI atrophy biomarkers: relationship to conversion in the ADNI cohort. Neurobiol Aging. 2010;31:1401–1418. doi: 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Risacher S.L., Saykin A.J., West J.D., Shen L., Firpi H.A., McDonald B.C. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res. 2009;6:347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jagust W.J., Bandy D., Chen K., Foster N.L., Landau S.M., Mathis C.A. The Alzheimer's Disease Neuroimaging Initiative positron emission tomography core. Alzheimers Dement. 2010;6:221–229. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landau S.M., Harvey D., Madison C.M., Koeppe R.A., Reiman E.M., Foster N.L. Associations between cognitive, functional, and FDG-PET measures of decline in AD and MCI. Neurobiol Aging. 2011;32:1207–1218. doi: 10.1016/j.neurobiolaging.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jagust W.J., Landau S.M., Shaw L.M., Trojanowski J.Q., Koeppe R.A., Reiman E.M. Relationships between biomarkers in aging and dementia. Neurology. 2009;73:1193–1199. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akaike H. Information theory and an extension of the maximum likelihood principle. 2nd International Symposium on Information Theory. 1973:267–281.
- 51.Jack C.R., Jr., Knopman D.S., Jagust W.J., Shaw L.M., Aisen P.S., Weiner M.W. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Apostolova L.G., Risacher S.L., Duran T., Stage E.C., Goukasian N., West J.D. Examining the effect of the top 20 AD risk variants on brain amyloidosis, structural atrophy and metabolism. Alzheimers Dement. 2016;12:P376–P377. [Google Scholar]
- 53.Wang H.F., Tan L., Hao X.K., Jiang T., Tan M.S., Liu Y. Effect of EPHA1 genetic variation on cerebrospinal fluid and neuroimaging biomarkers in healthy, mild cognitive impairment and Alzheimer's disease cohorts. J Alzheimers Dis. 2015;44:115–123. doi: 10.3233/JAD-141488. [DOI] [PubMed] [Google Scholar]
- 54.Ge T., Nichols T.E., Ghosh D., Mormino E.C., Smoller J.W., Sabuncu M.R. A kernel machine method for detecting effects of interaction between multidimensional variable sets: an imaging genetics application. Neuroimage. 2015;109:505–514. doi: 10.1016/j.neuroimage.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaminski W.E., Orso E., Diederich W., Klucken J., Drobnik W., Schmitz G. Identification of a novel human sterol-sensitive ATP-binding cassette transporter (ABCA7) Biochem Biophys Res Commun. 2000;273:532–538. doi: 10.1006/bbrc.2000.2954. [DOI] [PubMed] [Google Scholar]
- 56.Li H., Karl T., Garner B. Understanding the function of ABCA7 in Alzheimer's disease. Biochem Soc Trans. 2015;43:920–923. doi: 10.1042/BST20150105. [DOI] [PubMed] [Google Scholar]
- 57.Satoh K., Abe-Dohmae S., Yokoyama S., St George-Hyslop P., Fraser P.E. ATP-binding cassette transporter A7 (ABCA7) loss of function alters Alzheimer amyloid processing. J Biol Chem. 2015;290:24152–24165. doi: 10.1074/jbc.M115.655076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Q.F., Wan Y., Wang H.F., Sun F.R., Hao X.K., Tan M.S. ABCA7 genotypes confer Alzheimer's disease risk by modulating amyloid-beta pathology. J Alzheimers Dis. 2016;52:693–703. doi: 10.3233/JAD-151005. [DOI] [PubMed] [Google Scholar]
- 59.Sassi C., Nalls M.A., Ridge P.G., Gibbs J.R., Ding J., Lupton M.K. ABCA7 p.G215S as potential protective factor for Alzheimer's disease. Neurobiol Aging. 2016;46 doi: 10.1016/j.neurobiolaging.2016.04.004. 235.e1-e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monzo P., Gauthier N.C., Keslair F., Loubat A., Field C.M., Le Marchand-Brustel Y. Clues to CD2-associated protein involvement in cytokinesis. Mol Biol Cell. 2005;16:2891–2902. doi: 10.1091/mbc.E04-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao F., Jiang H., Srivatsan S., Xiao Q., Lefton K.B., Yamada K. Effects of CD2-associated protein deficiency on amyloid-beta in neuroblastoma cells and in an APP transgenic mouse model. Mol Neurodegener. 2015;10:12. doi: 10.1186/s13024-015-0006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunstan M., Gerrish A., Thomas R.S., Kidd E., Morgan T., Owens H. The effects of CD2AP expression on APP processing. Alzheimers Dement. 2015;11:858. [Google Scholar]
- 63.Cochran J.N., Rush T., Buckingham S.C., Roberson E.D. The Alzheimer's disease risk factor CD2AP maintains blood-brain barrier integrity. Hum Mol Genet. 2015;24:6667–6674. doi: 10.1093/hmg/ddv371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maru Y., Hirai H., Yoshida M.C., Takaku F. Evolution, expression, and chromosomal location of a novel receptor tyrosine kinase gene, eph. Mol Cell Biol. 1988;8:3770–3776. doi: 10.1128/mcb.8.9.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torii M., Hashimoto-Torii K., Levitt P., Rakic P. Integration of neuronal clones in the radial cortical columns by EphA and ephrin-A signalling. Nature. 2009;461:524–528. doi: 10.1038/nature08362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hattori M., Osterfield M., Flanagan J.G. Regulated cleavage of a contact-mediated axon repellent. Science. 2000;289:1360–1365. doi: 10.1126/science.289.5483.1360. [DOI] [PubMed] [Google Scholar]
- 67.Yamazaki T., Masuda J., Omori T., Usui R., Akiyama H., Maru Y. EphA1 interacts with integrin-linked kinase and regulates cell morphology and motility. J Cell Sci. 2009;122:243–255. doi: 10.1242/jcs.036467. [DOI] [PubMed] [Google Scholar]
- 68.Chen G., Wang Y., Zhou M., Shi H., Yu Z., Zhu Y. EphA1 receptor silencing by small interfering RNA has antiangiogenic and antitumor efficacy in hepatocellular carcinoma. Oncol Rep. 2010;23:563–570. [PubMed] [Google Scholar]
- 69.Metzner A., Precht C., Fehse B., Fiedler W., Stocking C., Gunther A. Reduced proliferation of CD34(+) cells from patients with acute myeloid leukemia after gene transfer of INPP5D. Gene Ther. 2009;16:570–573. doi: 10.1038/gt.2008.184. [DOI] [PubMed] [Google Scholar]
- 70.Leung W.H., Tarasenko T., Bolland S. Differential roles for the inositol phosphatase SHIP in the regulation of macrophages and lymphocytes. Immunol Res. 2009;43:243–251. doi: 10.1007/s12026-008-8078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hasegawa J., Iwamoto R., Otomo T., Nezu A., Hamasaki M., Yoshimori T. Autophagosome-lysosome fusion in neurons requires INPP5E, a protein associated with Joubert syndrome. EMBO J. 2016;35:1853–18567. doi: 10.15252/embj.201593148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L., Sheng R., Qin Z. The lysosome and neurodegenerative diseases. Acta Biochim Biophys Sin (Shanghai) 2009;41:437–445. doi: 10.1093/abbs/gmp031. [DOI] [PubMed] [Google Scholar]
- 73.Wolfe D.M., Lee J.H., Kumar A., Lee S., Orenstein S.J., Nixon R.A. Autophagy failure in Alzheimer's disease and the role of defective lysosomal acidification. Eur J Neurosci. 2013;37:1949–1961. doi: 10.1111/ejn.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orr M.E., Oddo S. Autophagic/lysosomal dysfunction in Alzheimer's disease. Alzheimers Res Ther. 2013;5:53. doi: 10.1186/alzrt217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Z.G., Li Y., Ng C.T., Song Y.Q. Inflammation in Alzheimer's disease and molecular genetics: recent update. Arch Immunol Ther Exp (Warsz) 2015;63:333–344. doi: 10.1007/s00005-015-0351-0. [DOI] [PubMed] [Google Scholar]
- 76.Farfel J.M., Yu L., Buchman A.S., Schneider J.A., De Jager P.L., Bennett D.A. Relation of genomic variants for Alzheimer disease dementia to common neuropathologies. Neurology. 2016;87:489–496. doi: 10.1212/WNL.0000000000002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duriez B., Duquesnoy P., Escudier E., Bridoux A.M., Escalier D., Rayet I. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. Proc Natl Acad Sci U S A. 2007;104:3336–3341. doi: 10.1073/pnas.0611405104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mahr S., Burmester G.R., Hilke D., Gobel U., Grutzkau A., Haupl T. Cis- and trans-acting gene regulation is associated with osteoarthritis. Am J Hum Genet. 2006;78:793–803. doi: 10.1086/503849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi D., Nakamura T., Nakajima M., Dai J., Qin J., Ni H. Association of single-nucleotide polymorphisms in RHOB and TXNDC3 with knee osteoarthritis susceptibility: two case-control studies in East Asian populations and a meta-analysis. Arthritis Res Ther. 2008;10:R54. doi: 10.1186/ar2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y., Yu J.T., Wang H.F., Hao X.K., Yang Y.F., Jiang T. Association between NME8 locus polymorphism and cognitive decline, cerebrospinal fluid and neuroimaging biomarkers in Alzheimer's disease. PLoS One. 2014;9:e114777. doi: 10.1371/journal.pone.0114777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kraev A., Quednau B.D., Leach S., Li X.F., Dong H., Winkfein R. Molecular cloning of a third member of the potassium-dependent sodium-calcium exchanger gene family, NCKX3. J Biol Chem. 2001;276:23161–23172. doi: 10.1074/jbc.M102314200. [DOI] [PubMed] [Google Scholar]
- 82.Kraja A.T., Borecki I.B., Tsai M.Y., Ordovas J.M., Hopkins P.N., Lai C.Q. Genetic analysis of 16 NMR-lipoprotein fractions in humans, the GOLDN study. Lipids. 2013;48:155–165. doi: 10.1007/s11745-012-3740-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu L., Chibnik L.B., Srivastava G.P., Pochet N., Yang J., Xu J. Association of brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol. 2015;72:15–24. doi: 10.1001/jamaneurol.2014.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li X.F., Lytton J. An essential role for the K+-dependent Na+/Ca2+-exchanger, NCKX4, in melanocortin-4-receptor-dependent satiety. J Biol Chem. 2014;289:25445–25459. doi: 10.1074/jbc.M114.564450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kajiho H., Saito K., Tsujita K., Kontani K., Araki Y., Kurosu H. RIN3: a novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J Cell Sci. 2003;116:4159–4168. doi: 10.1242/jcs.00718. [DOI] [PubMed] [Google Scholar]
- 86.Kajiho H., Sakurai K., Minoda T., Yoshikawa M., Nakagawa S., Fukushima S. Characterization of RIN3 as a guanine nucleotide exchange factor for the Rab5 subfamily GTPase Rab31. J Biol Chem. 2011;286:24364–24373. doi: 10.1074/jbc.M110.172445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi J.H., Kaur G., Mazzella M.J., Morales-Corraliza J., Levy E., Mathews P.M. Early endosomal abnormalities and cholinergic neuron degeneration in amyloid-beta protein precursor transgenic mice. J Alzheimers Dis. 2013;34:691–700. doi: 10.3233/JAD-122143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li G. Rab GTPases, membrane trafficking and diseases. Curr Drug Targets. 2011;12:1188–1193. doi: 10.2174/138945011795906561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chapuis J., Hansmannel F., Gistelinck M., Mounier A., Van Cauwenberghe C., Kolen K.V. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18:1225–1234. doi: 10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beecham G.W., Hamilton K., Naj A.C., Martin E.R., Huentelman M., Myers A.J. Genome-wide association meta-analysis of neuropathologic features of Alzheimer's disease and related dementias. PLoS Genet. 2014;10:e1004606. doi: 10.1371/journal.pgen.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.He F., Umehara T., Saito K., Harada T., Watanabe S., Yabuki T. Structural insight into the zinc finger CW domain as a histone modification reader. Structure. 2010;18:1127–1139. doi: 10.1016/j.str.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 92.Allen M., Kachadoorian M., Carrasquillo M.M., Karhade A., Manly L., Burgess J.D. Late-onset Alzheimer disease risk variants mark brain regulatory loci. Neurol Genet. 2015;1:e15. doi: 10.1212/NXG.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Karch C.M., Jeng A.T., Nowotny P., Cady J., Cruchaga C., Goate A.M. Expression of novel Alzheimer's disease risk genes in control and Alzheimer's disease brains. PLoS One. 2012;7:e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.