Abstract
This randomized phase II trial compared panitumumab plus fluorouracil, leucovorin, and irinotecan (FOLFIRI) with bevacizumab plus FOLFIRI as second‐line chemotherapy for wild‐type (WT) KRAS exon 2 metastatic colorectal cancer (mCRC) and to explore the values of oncogenes in circulating tumor DNA (ctDNA) and serum proteins as predictive biomarkers. Patients with WT KRAS exon 2 mCRC refractory to first‐line chemotherapy containing oxaliplatin and bevacizumab were randomly assigned to panitumumab plus FOLFIRI or bevacizumab plus FOLFIRI. Of 121 randomly assigned patients, 117 were eligible. Median overall survival (OS) for panitumumab plus FOLFIRI and bevacizumab plus FOLFIRI were 16.2 and 13.4 months [hazard ratio (HR), 1.16; 95% CI, 0.76–1.77], respectively. Progression‐free survival (PFS) was also similar (HR, 1.14; 95% CI, 0.78–1.66). KRAS,NRAS, and BRAF status using ctDNA was successfully examined in 109 patients, and mutations were identified in 19 patients (17.4%). Panitumumab plus FOLFIRI showed favorable survival compared with bevacizumab plus FOLFIRI in WT patients and unfavorable survival in those with mutations (P for interaction = 0.026 in OS and 0.054 in PFS). OS with bevacizumab plus FOLFIRI was better than panitumumab plus FOLFIRI in patients with high serum vascular endothelial growth factor‐A (VEGF‐A) levels and worse in those with low levels (P for interaction = 0.016). Second‐line FOLFIRI plus panitumumab and FOLFIRI plus bevacizumab showed a similar efficacy in patients with WT KRAS exon 2 mCRC. RAS and BRAF mutation in ctDNA could be a negative predictive marker for panitumumab.
Keywords: Bevacizumab, colorectal cancer, liquid biopsy, panitumumab, RAS mutation
Monoclonal antibodies against both vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR) improve overall survival (OS) and progression‐free survival (PFS) in patients with metastatic colorectal cancer (mCRC).1, 2, 3 These antibodies are commonly used as first‐ or second‐line chemotherapy in combination with backbone standard cytotoxic chemotherapy including fluorouracil and leucovorin with either oxaliplatin (FOLFOX) or irinotecan (FOLFIRI).4 Continuous blockade of tumor angiogenesis with bevacizumab, a monoclonal antibody to VEGF‐A, was shown to be superior to chemotherapy alone in patients who failed first‐line treatment containing bevacizumab.5 Meanwhile, panitumumab, a fully human monoclonal antibody was also effective in second‐line as combination with FOLFIRI for patients without mutations in codons 12 and 13 of KRAS exon 2.3 Recent reports indicated that less frequent mutations in KRAS, other than exon 2, and NRAS were also negative predictive markers for efficacy of anti‐EGFR therapies.6, 7, 8, 9
Although several randomized trials compared bevacizumab with anti‐EGFR monoclonal antibodies in combination with FOLFIRI or FOLFOX for wild‐type (WT) KRAS exon 2 mCRC, no definitive results have yet been reported to establish a standard sequence for these treatments.8, 9, 10, 11 Thus, establishment of biomarkers is warranted for optimal selection of patients into treatments and improved overall outcomes. Many of biomarkers clinically available at present require tumor samples such as archival tissues. While it has been reported that biomarker status may change during the treatment course, it is rather difficult to obtain tumor samples repeatedly especially for the second or later line treatment. Nowadays, utility of serum or plasma samples, which can be assessed in a timely manner, has been investigated such as mutational status of tumor oncogenes12, 13, 14 and protein biomarkers including VEGF‐A and human EGFR (HER) family ligands.15, 16, 17, 18 However, there are few reports on oncogenes detected by circulating tumor DNA (ctDNA), so‐called liquid biopsy, and on blood biomarkers to distinguish efficacies of VEGF‐ and EGFR‐targeted therapies in randomized trials of mCRC.
Here, we report the results of a multi‐center, open‐label, randomized phase II study to compare FOLFIRI plus panitumumab with FOLFIRI plus bevacizumab as second‐line chemotherapy in patients with WT KRAS exon 2 mCRC, with comprehensive circulating biomarker analysis.
Materials and Methods
Patients
Prior to enrollment in the study, patients had to fulfill the following criteria: (i) histopathologically proven unresectable distant metastatic or locally advanced colorectal adenocarcinoma; (ii) presence of radiographically confirmed or clinically diagnosed disease progression during or within 3 months after the last dose of first‐line chemotherapy containing fluoropyrimidine, oxaliplatin, and bevacizumab; (iii) confirmation of WT KRAS exon 2 (codon 12 or 13) using paraffin‐embedded tumor tissue by a validated test method. KRAS testing conducted previously or during screening period by local or central laboratory was accepted; (iv) Eastern Cooperative Oncology Group performance status (PS) 0–2; (v) age of 20 years or older; (vi) presence of measurable or non‐measurable disease by Response Evaluation Criteria In Solid Tumor (RECIST) version 1.1; (vii) adequate organ function (supplementary material, available at online). All patients provided written informed consent. This study was approved by the institutional ethics committees of each institution and was registered at the University Hospital Medical Information Network (UMIN) Clinical Trials Registry (protocol ID UMIN000005216).
Treatment schedule and assessment
Patients were randomized in a 1:1 ratio to receive either panitumumab at 6 mg/kg with FOLFIRI once every 2 weeks or bevacizumab at 5 mg/kg with FOLFIRI every 2 weeks in one cycle. Randomization was stratified according to institutions and three groups by Köhne prognostic index.19 Treatment was discontinued upon disease progression, occurrence of unacceptable severe toxicity, or patient request. Radiologic tumor evaluations were repeated every 8 weeks during the first year and every 12 weeks thereafter by each investigator according to RECIST version 1.1. Adverse events (AEs) were graded using the Common Terminology Criteria for Adverse Events version 4.0.
Extended RAS and BRAF mutation analysis using next‐generation sequencing with ctDNA
Patients who provided written informed consent for biomarker research were included in biomarker analysis. Serum samples were collected before the study treatment. Total nucleic acid content was purified from 0.5 to 1.0 mL serum using QIAamp Circulating Nucleic Acid kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The sensitivity assays were performed with genomic DNA extracted from cultured cell lines and healthy human blood samples as described previously.20
Mutant alleles of KRAS and NRAS exon 2 (codons 12 and 13), exon 3 (codons 59, 61,117 and 146), and BRAF exon 15 (codon 600) were assessed.20, 21 Multiplex polymerase chain reaction (PCR) containing pooled primers for KRAS, NRAS, and BRAF was performed with the Complete PCR Reagent set (Sequenom, Japan), followed by the preparation of a barcoded DNA library using the Ion Plus Fragment Library kit (Thermo Fisher Scientific, Japan) and IonXpress barcode adaptors (Thermo Fisher Scientific, Japan). Quantified DNA libraries were pooled and sequenced using the Ion Proton sequencer. DNA sequencing data were accessed through the Torrent Suite v.4.4 (Life Technologies, Carlsbad, CA, USA) software program. The Poisson distribution model was used to determine the presence of mutant alleles. The Poisson coefficient as a cutoff value was set at average error rate plus 7 standard deviations calculated from normal DNA. The significance level of error rate was set at 2 × 10−5.
Serum protein analysis
The following serum proteins were analyzed: heregulin, hepatocyte growth factor (HGF), extracellular domain of HER type 2 (HER2), amphiregulin, betacellulin, EGF, EGFR, epiregulin, heparin‐binding EGF (HB‐EGF), transforming growth factor (TGF)‐α, tenascin C, VEGF‐A, fibroblast growth factor (FGF)‐basic, platelet‐derived growth factor‐BB, and placental growth factor. Enzyme‐linked immunosorbent assay (ELISA) was performed for heregulin and HGF using a commercially available sandwich ELISA kit (DuoSet ELISA development system for human HRG1‐β1 and Quantikine Human HGF Immunoassay; R&D Systems, Minneapolis, MN, USA). Expression of extracellular domain of HER2 was measured by chemiluminescent immunoassay (Siemens Healthcare Diagnostics, Erlangen, Germany). A bead‐based flow cytometric assay using a commercially available WideScreen Human Cancer Panel 2 kit (Merck, Kenilworth, NJ, USA) was performed for the remaining biomarkers.
Statistical analysis
The primary endpoint of this study was OS, which was estimated from the date of trial entry to the date of death from any cause or censored at the date of last follow‐up. Secondary endpoints included PFS, objective response rate (ORR), safety, and biomarker research. PFS was measured from the date of entry into the trial to the time when progression or death without evidence of progression occurred.
This study was initially designed as a randomized, phase II trial to evaluate the superiority of FOLFIRI plus bevacizumab over FOLFIRI plus panitumumab by log‐rank test. However, based on the results of other randomized studies which were reported after initiation of this study,5, 6, 7, 8, 9, 10, 11 the main objective of the study was changed to the estimation of hazard ratio (HR) between the two arms. As such, a revised decision rule regarding the primary endpoint was determined that the two arms had similar efficacy for OS when the point estimate of HR fell in between 0.80 and 1.25. Ensuring 70% or greater probability that the observed HR was between 0.80 and 1.25 when an expected HR was 1.0 required a total of 86 deaths; accordingly this trial included a total of 120 patients with 60 subjects per arm. The study protocol was amended on 14th October 2013.
For biomarker analyses of serum proteins, the predictive and prognostic values were assessed by dichotomizing at the median value (i.e., greater than the median denoted high expression levels and below or equal to the median denoted low expression levels). In the biomarker analyses, a P‐value of < 0.1 was considered as a possible treatment‐by‐marker interaction. All statistical analyses were performed using SAS v. 9.3 (SAS Institute; Cary, NC, USA), and two‐sided P‐values were reported.
Results
Patient population
Between April 2011 and February 2014, a total of 121 patients were enrolled, all of whom received allocated treatment (Fig. S1). Two patients in each treatment arm were excluded from full analysis set (FAS) due to ineligibility; therefore, the FAS included 59 patients in the FOLFIRI plus panitumumab arm and 58 patients in the FOLFIRI plus bevacizumab arm. Patients and disease characteristics were well balanced between the two arms (Table 1). The cutoff date for analysis was August 2015, resulting in a median follow‐up time of 15.4 months in FOLFIRI plus panitumumab arm and 13.4 months in FOLFIRI plus bevacizumab arm. The median treatment cycle was 8 in FOLFIRI plus panitumumab and 10 in FOLFIRI plus bevacizumab. One patient receiving FOLFIRI plus panitumumab continued protocol treatment at the data cut‐off. The most common reason for treatment discontinuation was disease progression in 62.7% of FOLFIRI plus panitumumab and in 72.4% of FOLFIRI plus bevacizumab followed by adverse events.
Table 1.
Baseline patient characteristics
| Characteristics | FOLFIRI plus panitumumab (n = 59) | FOLFIRI plus bevacizumab (n = 58) | ||
|---|---|---|---|---|
| n | % | n | % | |
| Age, years | ||||
| Median, range | 62 | 31–82 | 64 | 26–78 |
| Age, years | ||||
| <65 | 34 | 57.6 | 29 | 50.0 |
| 65 or more | 25 | 42.4 | 29 | 50.0 |
| Gender | ||||
| Male | 34 | 57.6 | 39 | 67.2 |
| Female | 25 | 42.4 | 19 | 32.8 |
| ECOG PS | ||||
| 0 | 47 | 79.7 | 43 | 74.1 |
| 1 | 12 | 20.3 | 15 | 25.9 |
| Köhne index | ||||
| Low | 23 | 39.0 | 25 | 43.1 |
| Intermediate | 14 | 23.7 | 13 | 22.4 |
| High | 22 | 37.3 | 20 | 34.5 |
| Prior surgery | ||||
| Yes | 49 | 83.1 | 47 | 81.0 |
| Prior adjuvant treatment | ||||
| Yes | 16 | 27.1 | 15 | 25.9 |
| Prior first‐line treatment | ||||
| FOLFOX + bevacizumab | 45 | 76.3 | 45 | 77.6 |
| CapeOX + bevacizumab | 14 | 23.7 | 12 | 20.7 |
| SOX + bevacizumab | 0 | 0 | 1 | 1.7 |
| Duration of first‐line | ||||
| 6 months or more | 50 | 84.7 | 49 | 84.5 |
| Measurable lesion | ||||
| Present | 52 | 88.1 | 53 | 91.4 |
| Metastatic sites | ||||
| Liver | 35 | 59.3 | 39 | 67.2 |
| Lung | 31 | 52.5 | 28 | 48.3 |
| Lymph node | 24 | 40.7 | 21 | 36.2 |
| No. of metastatic sites | ||||
| 1 | 22 | 37.3 | 20 | 34.5 |
| 2 or more | 37 | 62.7 | 38 | 65.5 |
CapeOX, capecitabine plus oxaliplatin; ECOG PS, Eastern Cooperative Oncology Group performance status; FOLFIRI, fluorouracil, leucovorin, and irinotecan; FOLFOX, combination of fluorouracil, leucovorin, and oxaliplatin; SOX, S1 plus oxaliplatin.
Efficacy
At the time of analysis, all FAS patients were evaluated for efficacy, and 88 patients (75.2%) died. As the primary analysis, estimating HR of OS between the two arms provided HR of 1.16 [95% confidence interval (CI), 0.76–1.77; Fig. 1a], concluding that the arms showed the similar efficacy based on the decision rule. A median OS was 16.2 months in the FOLFIRI plus panitumumab arm and 13.4 months in the FOLFIRI plus bevacizumab arm. The median PFS was 6.0 months in the FOLFIRI plus panitumumab arm and 5.9 months in the FOLFIRI plus bevacizumab arm (HR, 1.14; 95% CI, 0.78–1.66; Fig. 1b). The ORR in patients with measurable disease was 46.2% in FOLFIRI plus panitumumab arm and 5.7% in FOLFIRI plus bevacizumab (Fisher's exact test, P < 0.001; Table S1). Results from the Cox proportional hazards analysis of OS or PFS according to baseline clinicopathological factors showed possible interactions between history of previous surgery and treatment outcome in OS, while age and duration of first‐line chemotherapy showed interaction in PFS (Tables S2 and S3). No other significant interaction was observed in subgroup analyses.
Figure 1.
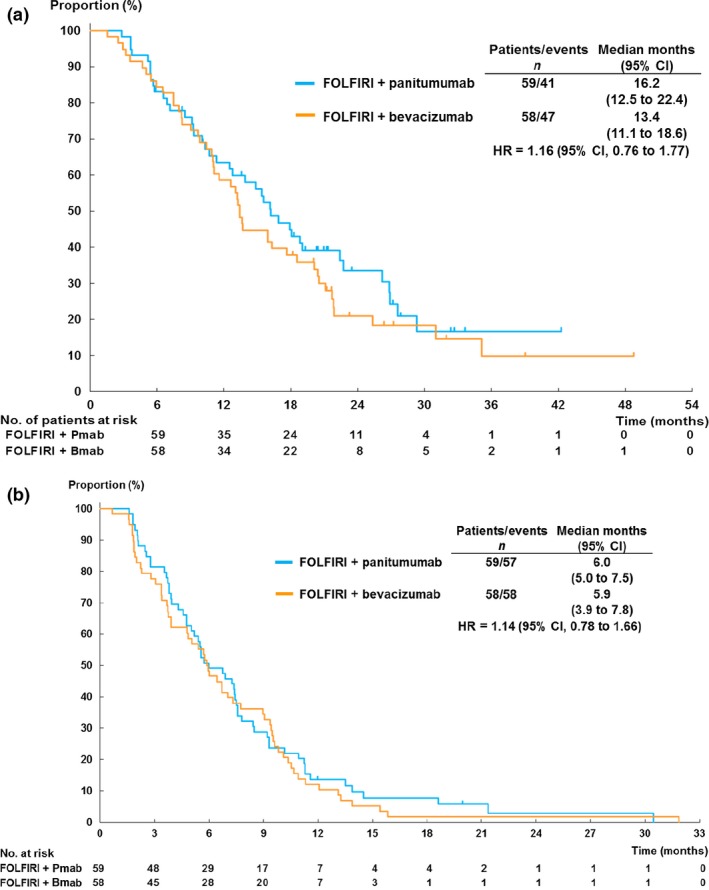
Kaplan–Meier plots of (a) overall survival and (b) progression‐free survival in the full analysis set cohort. Bmab, bevacizumab; CI, confidence interval; HR, hazard ratio; Pmab, panitumumab.
Adverse events
Among the 121 patients who received allocated treatment, 54 patients (88.5%) in the FOLFIRI plus panitumumab arm and 40 patients (66.7%) in the FOLFIRI plus bevacizumab arm experienced AEs with worst grades of 3 or worse (Table 2, P = 0.004). Grade 3 or worse acneiform rash, stomatitis, and hypomagnesemia were more frequent (>10%) in the FOLFIRI plus panitumumab, whereas leucopenia was more frequent in FOLFIRI plus bevacizumab. No treatment‐related deaths occurred.
Table 2.
Grade 3 and 4 adverse events
| Adverse event | FOLFIRI plus panitumumab (n = 61) | FOLFIRI plus bevacizumab (n = 60) | ||
|---|---|---|---|---|
| ≥Grade 3 | ≥Grade 3 | |||
| n | % | n | % | |
| Any adverse events | 54 | 88.5 | 40 | 66.7 |
| Neutropenia | 30 | 49.2 | 28 | 46.7 |
| Stomatitis | 13 | 21.3 | 4 | 6.7 |
| Leucopenia | 11 | 18.0 | 17 | 28.3 |
| Acneiform rash | 10 | 16.4 | 0 | 0.0 |
| Hypomagnesemia | 7 | 11.5 | 0 | 0.0 |
| Anorexia | 6 | 9.8 | 7 | 11.7 |
| Proteinuria | 6 | 9.8 | 2 | 3.3 |
| Non‐neutropenic infection | 5 | 8.2 | 3 | 5.0 |
| Dry skin | 5 | 8.2 | 0 | 0.0 |
| Paronychia | 4 | 6.6 | 0 | 0.0 |
| Thromboembolic events | 4 | 6.6 | 0 | 0.0 |
| Anemia | 3 | 4.9 | 7 | 11.7 |
| Diarrhea | 3 | 4.9 | 4 | 6.7 |
| Nausea | 2 | 3.3 | 3 | 5.0 |
| Vomiting | 2 | 3.3 | 3 | 5.0 |
| Febrile neutropenia | 2 | 3.3 | 3 | 5.0 |
| Fatigue | 1 | 1.6 | 5 | 8.3 |
| Thrombocytopenia | 1 | 1.6 | 3 | 5.0 |
| Any bleeding | 1 | 1.6 | 2 | 3.3 |
| Pneumonitis | 1 | 1.6 | 1 | 1.7 |
| Gastrointestinal perforation | 0 | 0.0 | 1 | 1.7 |
FOLFIRI, fluorouracil, leucovorin, and irinotecan.
Subsequent treatments
The subsequent chemotherapy was administrated in 45 patients (76.3%) in the FOLFIRI plus panitumumab arm and in 46 patients (79.3%) in the FOLFIRI plus bevacizumab arm after the protocol‐specified treatment. Anti‐EGFR monoclonal antibody was used in 72.4% of the patients in FOLFIRI plus bevacizumab arm and 39.0% of those in FOLFIRI plus panitumumab arm (Table S4).
Efficacy and mutation status of KRAS, NRAS and BRAF mutations in serum
Among the 117 patients in FAS cohort, consent for biomarker research was obtained from 109 patients (93.2%; FOLFIRI plus panitumumab arm, 54 patients; FOLFIRI plus bevacizumab arm, 55 patients). Patient characteristics in biomarker population were not significantly different between the two arms (Fig. S1 and Table S5). Efficacy results were also consistent with the FAS results (the median OS, 16.2 months in FOLFIRI plus panitumumab vs 13.4 months in FOLFIRI plus bevacizumab; HR, 1.16; 95% CI, 0.75–1.80). Success rate of mutation analysis was 100%. Any RAS (n = 14, 12.8%) or BRAF mutations (n = 5, 4.6%) in ctDNA were identified in a total of 19 patients (17.4%; Table S6) as mutually exclusive manner. Patients with any RAS or BRAF mutation showed worse OS in FOLFIRI plus panitumumab arm than in FOLFIRI plus bevacizumab arm (median 5.4 vs 8.2 months; HR, 0.42). On the other hand, a trend toward better OS was observed in patients who were WT for all examined genes in FOLFIRI plus panitumumab arm than those in FOLFIRI plus bevacizumab arm (median 18.9 vs 16.1 months; HR 1.21), resulting in significant interaction (P for interaction = 0.026; Fig. 2a). A possible interaction between oncogenic mutations and treatment was also observed for PFS (P = 0.054; Fig. S2A). These observations were consistent after adjustment for stratification variables. For all WT patients, the ORRs were 52.5% in FOLFIRI plus panitumumab and 2.6% in FOLFIRI plus bevacizumab (P < 0.001), whereas it was 0% in FOLFIRI plus panitumumab and 18.2% in FOLFIRI plus bevacizumab (P < 0.01) among those with any RAS or BRAF mutations.
Figure 2.
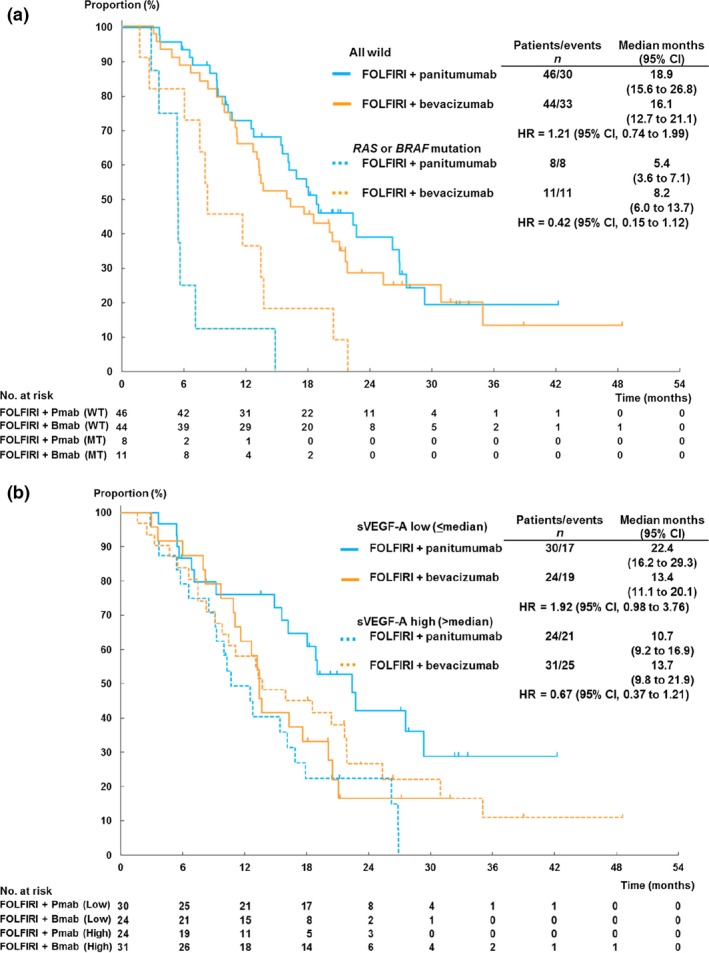
Kaplan–Meier plots of overall survival according to RAS and BRAF mutation status (a) and according to serum VEGF‐A level (b). Bmab, bevacizumab; CI, confidence interval; HR, hazard ratio; Pmab, panitumumab. WT,RAS or BRAF wild type; MT,RAS or BRAF mutant. sVEGF‐A, serum vascular endothelial growth factor‐A.
Efficacy and serum protein levels
No significant differences in serum protein levels were observed between the two arms (Table S7). Panitumumab led to a better OS than FOLFIRI plus bevacizumab in patients with low serum VEGF‐A level (median 22.4 vs 13.2 months; HR, 1.92), whereas FOLFIRI plus bevacizumab showed better OS among those with high serum VEGF‐A level (median 13.7 vs 10.7 months; HR, 0.67), resulting in significant interaction (P for interaction = 0.016; Fig. 2b). These results were consistent when we limited our analysis to all patients with WT RAS or BRAF (HR, 2.29 with low serum VEGF‐A; HR, 0.64 with high serum VEGF‐A; P for interaction = 0.014) or after adjustment for stratification variables. Although no significant interaction between serum VEGF‐A level and PFS was observed (Fig. S2b), the ORR was numerically higher in those patients with low VEGF‐A than those with high VEGF‐A in FOLFIRI plus panitumumab arm (52.0% vs 36.4%) and in FOLFIRI plus bevacizumab arm (9.1% and 3.6%). No other biomarkers were associated with predictive value on treatment outcomes (Figs 3 and S3).
Figure 3.
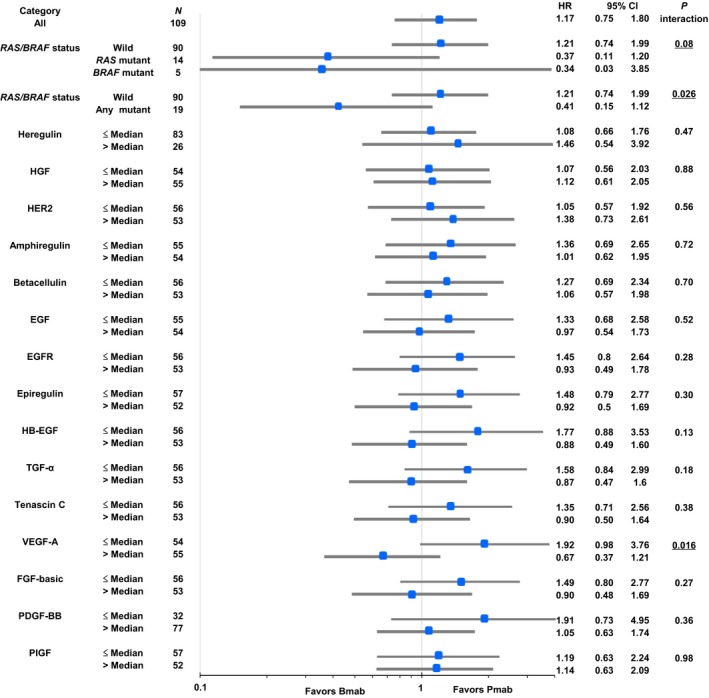
Comparison of overall survival among patient subgroups according to biomarkers. Patient's number of each subgroup was not in half by median value of heregulin or PDGF‐BB because not a few patients were associated with undetectable value or maximum value. CI, confidence interval; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; FGF‐basic, fibroblast growth factor‐basic; HB‐EGF, heparin‐binding EGF; HER2, human epidermal growth factor receptor type 2; HGF, hepatocyte growth factor; HR, hazard ratio; PDGF‐BB, platelet‐derived growth factor‐BB; PIGF, placental growth factor; TGF‐α, transforming growth factor‐α; VEGF‐A, vascular endothelial growth factor‐A.
Discussion
In this randomized study that compared FOLFIRI plus panitumumab with FOLFIRI plus bevacizumab as second‐line chemotherapy in patients with WT KRAS exon 2 mCRC, similar OS and PFS were observed between the two treatments, whereas ORR was higher with panitumumab. These results were in line with a previous study in a similar setting.11
Importantly, this study showed that patients with minor RAS and BRAF mutations, detected in ctDNA from 12.8% and 4.6% of mCRC patients with wild type of KRAS exon 2 in tumor tissue at the diagnosis, showed worse prognosis in FOLFIRI plus panitumumab. These were similar to the results of recent studies indicating that extended mutations in RAS genes and BRAF mutation using archival tumor tissue were negative predictive factors for anti‐EGFR therapy.6, 7, 8 The incidences of RAS mutations other than exon 2 and 3, and BRAF mutation were consistent with those obtained from tumor tissues.6, 7, 8 Although concordance between ctDNA and tumor tissues was not evaluated in this study, it is suggested that gene status detected in ctDNA could reflect the tumor biology. Previous studies indicated that mutational status changed during treatment especially with anti‐EGFR therapy.12, 13 Thus, using ctDNA as non‐invasive procedure to determine patient status immediately just before treatment might be alternative approach in near future.
Fifteen serum proteins, including VEGF‐A, were also evaluated in this study. Of note, the FOLFIRI plus bevacizumab arm was associated with favorable OS in high serum VEGF‐A subgroup, whereas the FOLFIRI plus panitumumab arm showed better OS in low serum VEGF‐A subgroup. These findings suggested that sustained activation of VEGF pathway might predict the benefit of bevacizumab continuation as suggested in previous phase 2 study.18 However, a correlation between VEGF expression and treatment outcome with bevacizumab was arguable in other studies.17, 22 Therefore, validation studies are necessary to conclusively determine whether VEGF‐A can be used to optimize bevacizumab or anti‐EGFR therapy.23
Several studies reported that the mRNA levels of EGFR ligands such as amphiregulin and epiregulin in tumor tissue predicted the efficacy of anti‐EGFR therapy.15, 24, 25 However, in this study, circulating protein levels of none of the EGFR ligands showed significant interaction with treatment efficacy. While upregulation of bypass signaling pathways, such as HER2, HER3, and HGF‐cMET axes, were reported to associate with resistance to anti‐EGFR therapy.16, 26 However, our study showed that the levels of circulating heregulin, extracellular domain of HER2, or HGF did not significantly correlate with resistance to panitumumab therapy. One possible reason for these discrepancies is that circulating ligand levels might not reflect ligand levels in tumor.15
This study has several limitations. First, the number of patients in each treatment arm was relatively small. In addition, we had to set the cut‐off values of biomarkers at median because the sample size was too small to explore several cut‐off values. Thus, larger studies are necessary to confirm and identify optimal cut‐off levels. Second, we did not evaluate biomarkers in tumor tissue. Matched analysis of serum and tumor tissue should be included in future studies to evaluate these concordance and predictive values. Because the translational research is exploratory, statistical correction for multiplicity was not performed. Third, although this study showed higher ORR in FOLFIRI plus panitumumab than FOLFIRI plus bevacizumab, ORR was not conformed by independent radiological reviews.
In conclusion, second‐line FOLFIRI plus panitumumab and FOLFIRI plus bevacizumab showed a similar efficacy in patients with WT KRAS exon 2 mCRC. Additionally, RAS and BRAF mutation in ctDNA could identify could be a negative predictive marker for panitumumab. Finally, serum VEGF‐A might be a candidate biomarker for optimizing anti‐EGFR therapy or bevacizumab.
Disclosure Statement
All authors have no conflicts of interest to declare. All authors had full access to all of the data in the study and had final responsibility for the decision to submit for publication.
Supporting information
Fig. S1. CONSORT flowchart.
Fig. S2. Kaplan–Meier plots of progression‐free survival according to RAS and BRAF mutation status.
Fig. S3. Comparison of progression‐free survival among patient subgroups according to biomarker.
Table S1. Objective response in patients with measurable lesion.
Table S2. Interaction between clinical factors and overall survival.
Table S3. Interaction between clinical factors and progression‐free survival.
Table S4. Subsequent treatments.
Table S5. Baseline patient characteristics of the biomarker population.
Table S6. Frequencies of KRAS, NRAS, and BRAF mutations in circulating tumor DNA.
Table S7. Biomarker measurement results.
Acknowledgments
This study was supported by the West Japan Oncology Group (WJOG), a non‐profit organization. We thank the data managers and other support staff of West Japan Oncology Group, especially Dr. Shinichiro Nakamura, MD, and Kaori Mori.
Cancer Sci 107 (2016) 1843–1850
Clinical Trials Registry (protocol ID UMIN000005216).
Funding Information
West Japan Oncology Group (WJOG).
References
- 1. Hurwitz H, Fehrenbacher L, Novotny W et al Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 2014; 350: 2335–42. [DOI] [PubMed] [Google Scholar]
- 2. Douillard JY, Siena S, Cassidy J et al Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first‐line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 2010; 28: 4697–705. [DOI] [PubMed] [Google Scholar]
- 3. Peeters M, Price TJ, Cervantes A et al Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second‐line treatment in patients with metastatic colorectal cancer. J Clin Oncol 2010; 28: 4706–13. [DOI] [PubMed] [Google Scholar]
- 4. NCCN . NCCN Clinical Practice Guidelines in Oncology. Colon Cancer. NCCN Practice Guidelines in Oncology, version 2. NCCN; USA: 2016.
- 5. Bennouna J, Sastre J, Arnold D et al Continuation of bevacizumab after first progression in metastatic colorectal cancer (ML18147): a randomised phase 3 trial. Lancet Oncol 2013; 14: 29–37. [DOI] [PubMed] [Google Scholar]
- 6. Douillard JY, Oliner KS, Siena S et al Panitumumab‐FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013; 369: 1023–34. [DOI] [PubMed] [Google Scholar]
- 7. Peeters M, Oliner KS, Parker A et al Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase 3 study of metastatic colorectal cancer. Clin Cancer Res 2013; 19: 1902–12. [DOI] [PubMed] [Google Scholar]
- 8. Heinemann V, von Weikersthal LF, Decker T et al FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first‐line treatment for patients with metastatic colorectal cancer (FIRE‐3): a randomised, open‐label, phase 3 trial. Lancet Oncol 2014; 15: 1065–75. [DOI] [PubMed] [Google Scholar]
- 9. Schwartzberg LS, Rivera F, Karthaus M et al PEAK: a randomized, multicenter phase II study of panitumumab plus modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6) or bevacizumab plus mFOLFOX6 in patients with previously untreated, unresectable, wild‐type KRAS exon 2 metastatic colorectal cancer. J Clin Oncol 2014; 32: 2240–7. [DOI] [PubMed] [Google Scholar]
- 10. Venook AP, Niedzwiecki D, Lenz HJ et al CALGB/SWOG 80405: Phase III trial of irinotecan/5‐FU/leucovorin (FOLFIRI) or oxaliplatin/5‐FU/leucovorin (mFOLFOX6) with bevacizumab (BV) or cetuximab (CET) for patients (pts) with KRAS wild‐type (wt) untreated metastatic adenocarcinoma of the colon or rectum (MCRC). J Clin Oncol 2014; 32: 5s (suppl; abstr LBA3). [Google Scholar]
- 11. Hecht JR, Cohn A, Dakhil S et al SPIRITT: a randomized, multicenter, phase ii study of panitumumab with FOLFIRI and bevacizumab with FOLFIRI as second‐line treatment in patients with unresectable wild type KRAS metastatic colorectal cancer. Clin Colorectal Cancer 2015; 14: 72–80. [DOI] [PubMed] [Google Scholar]
- 12. Misale S, Yaeger R, Hobor S et al Emergence of KRAS mutations and acquired resistance to anti‐EGFR therapy in colorectal cancer. Nature 2012; 486: 532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diaz LA Jr, Williams RT, Wu J et al The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012; 486: 537–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tabernero J, Lenz HJ, Siena S et al Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol 2012; 16: 937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khambata‐Ford S, Garrett CR, Meropol NJ et al Expression of epiregulin and amphiregulin and K‐ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 2007; 25: 3230–7. [DOI] [PubMed] [Google Scholar]
- 16. Yonesaka K, Zejnullahu K, Okamoto I et al Activation of ERBB2 signaling causes resistance to the EGFR‐directed therapeutic antibody cetuximab. Sci Transl Med 2011; 3: 99ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jayson GC, deHaas S , Delmar P et al Evaluation of plasma VEGF‐A as a potential predictive pan‐tumor biomarker for bevacizumab. Proceedings of the 2011 European Multidisciplinary Cancer Congress, Stockholm, Sweden, September 23–27, 2011 (abstr 804).
- 18. Cremolini C, Loupakis F, Bocci G et al Circulating angiogenic factors as predictors of benefit from bevacizumab (bev) beyond progression in metastatic colorectal cancer (mCRC): translational analyses from the phase III BEBYP trial. J Clin Oncol 2013; 31: (Suppl. 4; abstr 382). [Google Scholar]
- 19. Köhne CH, Cunningham D, Di CF et al Clinical determinants of survival in patients with 5‐fluorouracil based treatment for metastatic colorectal cancer: results of a multivariate analysis of 3825 patients. Ann Oncol 2002; 13: 308–17. [DOI] [PubMed] [Google Scholar]
- 20. Sakai K, Tsurutani J, Yamanaka T et al Extended RAS and BRAF Mutation analysis using next‐generation sequencing. PLoS ONE 2015; 10: e0121891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kukita Y, Uchida J, Oba S et al Quantitative identification of mutant alleles derived from lung cancer in plasma cell‐free DNA via anomaly detection using deep sequencing data. PLoS ONE 2013; 8: e81468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lambrechts D, Lenz HJ, de Haas S et al Markers of response for the antiangiogenic agent bevacizumab. J Clin Oncol 2013; 31: 1219–30. [DOI] [PubMed] [Google Scholar]
- 23. Vallböhmer D, Zhang W, Gordon M et al Molecular determinants of cetuximab efficacy. J Clin Oncol 2005; 23: 3536–44. [DOI] [PubMed] [Google Scholar]
- 24. Seligmann JF, Elliott F, Richman S et al Combined epiregulin (EREG) and amphiregulin (AREG) expression levels as a biomarker of prognosis and panitumumab benefit in RAS‐wt advanced colorectal cancer (aCRC). J Clin Oncol 2014; 32: 5s (Suppl; abstr 3520). [Google Scholar]
- 25. Yonesaka K, Takegawa N, Satoh T et al Combined analysis of plasma amphiregulin and heregulin predicts response to cetuximab in metastatic colorectal Cancer. PLoS ONE 2015; 10: e0143132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cushman SM, Jiang C, Hatch AJ et al Gene expression markers of efficacy and resistance to cetuximab treatment in metastatic colorectal cancer: results from CALGB 80203 (Alliance). Clin Cancer Res 2015; 21: 1078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. CONSORT flowchart.
Fig. S2. Kaplan–Meier plots of progression‐free survival according to RAS and BRAF mutation status.
Fig. S3. Comparison of progression‐free survival among patient subgroups according to biomarker.
Table S1. Objective response in patients with measurable lesion.
Table S2. Interaction between clinical factors and overall survival.
Table S3. Interaction between clinical factors and progression‐free survival.
Table S4. Subsequent treatments.
Table S5. Baseline patient characteristics of the biomarker population.
Table S6. Frequencies of KRAS, NRAS, and BRAF mutations in circulating tumor DNA.
Table S7. Biomarker measurement results.


