Abstract
Dissemination of carbapenem resistance via Enterobacteriaceae, particularly among Klebsiella pneumoniae and Escherichia coli, is a major public health concern. Rapid methods for determining antimicrobial susceptibility are important to ensure adequate and appropriate use of antimicrobial agents and to limit the spread of these bacteria. In the current study, we compared the rapidity, sensitivity and specificity of traditional methods and molecular-based Xpert Carba-R PCR assay to identify sixty isolates, (26 E. coli and 34 K. pneumoniae). The specificity of MicroScan was 100% while sensitivity to ertapenem (ERT), imipenem (IMI), and meropenem (MER) was 93%, 68.9%, and 55.17%, respectively. For the modified Hodge test, the specificity was 96.77% and sensitivity was 89.65%. Although some results of phenotypic assays matched with the definite PCR identification, some results were misleading. Out of the 29 positive PCR samples, three samples of K. pneumoniae were negative for the MHT and one E. coli sample was MHT positive but negative for the PCR. Nine samples were positive for the PCR but were determined as carbapenem sensitive by MicroScan. While MicroScan and MHT requires several hours and multi-steps to obtain results, Xpert Carba-R PCR assay takes less than an hour. Therefore, we recommend using Gene xpert Carba-R assay for the optimal carbapenemnase detection with reducing material, manpower and cost. Also it is important to know the type of carbapenemase is present.
Keywords: Klebsiella pneumonia, E. coli, Carbapenemase, Modified Hodge test, MicroScan, Gene Xpert Carba-R assay, Antibiotic resistance, Phenotypic, PCR
1. Introduction
Enterobacteriaceae comprises a group of gram negative bacteria that are known to cause several important infections such as urinary tract infections, cystitis, wound sepsis, pneumonia, bacteremia, and meningitis (Mitchell and Yehuda, 2008). Multiple antibiotic resistance, particularly resistance to carbapenem, among Enterobacteriaceae is a major public health concern. Carbapenem-resistant Enterobacteriaceae, reported mostly in hospital-associated infections, is associated with more than 50% mortalities which poses a great challenge in patient care units (Poirel et al., 2011, Brink et al., 2011, Munoz-Price et al., 2014, McLaughlin et al., 2014, Nordmann, 2014). Resistance to carbapenems is attributed to the ability to produce three different categories of carbapenemases; (1) class A (serine carbapenemases, such as Klebsiella pneumoniae carbapenemase (KPC)), (2) class B, metallo-β-lactamases, such as IMP, VIM, and NDM, and (3) class D (OXA carbapenemases, such as OXA-23 and OXA-48) (Queenan and Bush, 2007, Miriagou et al., 2010).
The rapid identification of carbapenem resistance organisms is critical not only for the initiation of the proper antimicrobial regimen but also for stopping their spread. Several phenotypic methods are available for detection of carbapenemase enzymes such as; modified Hodge test (MHT), combined disk test (CDT) and inhibitor based E-test. However, none of the current phenotypic tests offer optimal sensitivities and specificities. Modified Hodge test (MHT) has been extensively used as a specific method to detect un-typed carbapenemase activity and is currently the only method recommended by the Clinical and Laboratory Standards Institute (CLSI) (Orstavik and Odegaard, 1971). Although MHT is useful for detection of carbapenemases, it has low sensitivity (Girlich et al., 2012) and low specificity for metallo-enzymes (Cuzon et al., 2011, Girlich et al., 2012). In addition, MHT may produce false positive results mainly due to CTX-M-producing strains with reduced outer membrane permeability and high-level AmpC-producers (Girlich et al., 2008, Pasteran et al., 2009). Similarly for the inhibitor-based E-test, a test for detection of K. pneumoniae carbapenemases (KPC), false-positive test results may occur if AmpC β lactamases are coproduced (Pasteran et al., 2009, Pournaras et al., 2010, van Dijk et al., 2014).
Thus, phenotypic methods are growth dependent and time-consuming as it takes 18–24 h to completion, not clinically useful and results are also subjective (Cuzon et al., 2011, Girlich et al., 2012). Therefore, identification by molecular methods such as real time-PCR have been shown to be sensitive and more accurate for identification of carbapenemase genes. Moreover, diagnosis by molecular methods is comparatively faster than phenotypic assays as the extra overnight incubation step is superfluous. (Manchanda et al., 2011, Liu et al., 2012, Rachana et al., 2014). The current study was designed to evaluate and compare the sensitivity and specificity of the phenotypic assays, MHT, MicroScan, and E-test, with Gene Expert Carba-R assay, as a rapid, low-cost real-time PCR method, for the detection of carbapenemase-producing K. pneumoniae and E. coli isolates in Riyadh region.
2. Materials and methods
2.1. Bacterial isolates and culture conditions
Sixty non-duplicated isolates were collected from patients admitted to Prince Sultan Military Medical City in Riyadh, Saudi Arabia from 2014 to 2015. Both outpatients and inpatients were included and samples were obtained from urine. Isolates were maintained by routinely culturing on blood agar (BA) plates and MacConkey agar plates for 18–24 h at 37 °C. Identification of the isolates were performed using several assays such as colonial morphology, Oxidase test, API 20E (bioMerieux, Inc) and MicroScan walkaway 96 plus (SIEMENS Healthcare diagnostics). Of the sixty isolates, twenty six were E. coli and thirty four were K. pneumoniae.
2.2. Phenotypic screening of carbapenemases
2.2.1. Antimicrobial susceptibility tests by MicroScan system
Resistance to and MIC values of 14 antibiotics including imipenem, meropenem, ertapenem were determined by MicroScan walkaway 96 plus (SIEMENS Healthcare diagnostics) as described by manufacturer’s instructions. Briefly, 0.5 McFarland bacterial suspension from each isolate was prepared and transferred to the gram negative bacteria panel (Neg panel Type 49). Following 18–24 h incubation, MIC values were obtained and analyzed according to the standard Clinical and Laboratory Standards Institute, 2014 guidelines (CLSI). Isolates that were resistant to imipenem, meropenem or ertapenem were considered carbapenem-resistant.
2.2.2. Modified Hodge test (MHT)
The ability of carbapenem-resistant isolates to produce carbapenemases was investigated by MHT technique. Briefly, 0.5 McFarland suspension of carbapenem susceptible strain E. coli ATCC 25922 was streaked as a lawn over Mueller Hinton agar (MHA) plates and disks loaded with meropenem (10 μg) or ertapenem were placed in the center of the test area. Isolates to be tested along with a positive control (K. pneumoniae ATCC1705) and negative control (K. pneumoniae ATCC1706) were streaked as straight lines from the edge of the meropenem disk to the edge of the plate. Inoculated plates were allowed to dry for 15 min at room temperature and incubated overnight at 35 ± 2 °C for 16–24 h. The test was considered positive when a clover leaf-like shape was observed.
2.3. Molecular Xpert® Carba-R assay
Xpert® Carba-R (Cepheid, Sunnyvale, USA) is a fully automated and integrated system for sample preparation, DNA extraction, amplification and qualitative detection of target genes using multiplex real time PCR assays. This new assay involves a single-use disposable cartridge that holds PCR reagents and hosts the PCR processes. The cartridge contains primers that target and amplify a number of carbapenemase resistance genes; blaIMP, blaVIM, blaNDM, blaKPC, and blaOXA-48. Xpert Carba-R assay kit contains sufficient reagents to process 10 samples. In addition, it contains a positive control which carries all the 5 beta-lactamase gene sequences detected and negative control with no beta-lactamase gene sequences. Isolates to be tested were inoculated into the sample reagent and loaded into the cartridge as described by manufacturers’ instructions. Results were analyzed using GeneXpert Software Version 4.3.
3. Results
3.1. Antibiotic susceptibility of E. coli and K. pneumoniae
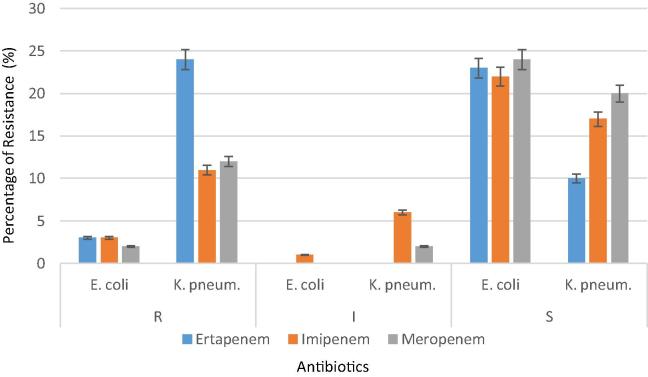
MicroScan antibiotic susceptibility profile of the 60 clinical isolates revealed that E. coli and K. pneumoniae have different susceptibilities where 24 (40%) isolates of K. pneumoniae and only 3 (5%) isolates of E. coli were resistant to imipenem, meropenem and ertapenem (Table 1). Of the 26 E. coli isolates, 3 (11.5%), 2 (7.69%), and 3 (12%) were resistant to imipenem, meropenem and ertapenem, respectively, while 22 (84.6%), 24 (92.3%) and 23 (88%) were sensitive to imipenem, meropenem and ertapenem, respectively. Only one (4%) isolate exhibited intermediate susceptibility to imipenem. For the 34 K. pneumoniae isolates, 11 (32%), 12 (35%), and 24 (70.58%) were resistant to imipenem, meropenem and ertapenem, respectively, while 17 (50%), 20 (59%) and 10 (29.4%) were sensitive to imipenem, meropenem and ertapenem, respectively (Fig. 1). Only 2 (6%) and 6 (18%) isolates exhibited intermediate susceptibility to meropenem and imipenem, respectively. Interestingly, these isolates have high resistance values against other antibiotics as indicated from MICs. Five (19%) out of 26 E. coli isolates and 19 (52.7) out of 36 K. pneumoniae were defined as multi drug resistant isolates. Of the 60 isolates only 2 (3.3%) were resistant to all the tested antibiotics except Amikacin and only two were sensitive to all antibiotics. Five (14.7%) of K. pneumoniae isolates were resistant to all tested antibiotics but exhibited intermediate susceptibility to Amikacin. Only one E. coli isolate was sensitive to all antibiotics except for Ampicillin.
Table 1.
Overall rates of antibiotics susceptibility test of E. coli (n = 26) and K. pneumoniae isolates (n = 34) as determined by MicroScan.
| Antibiotics | R⁎n (%) |
I⁎n (%) |
S⁎n (%) |
ESBL⁎n (%) |
||||
|---|---|---|---|---|---|---|---|---|
| E. coli | K. pneum. | E. coli | K. pneum. | E. coli | K. pneum. | E. coli | K. pneum. | |
| Ertapenem | 3 (11.5) | 24 (70.5) | 0 | 0 | 23 (88.4) | 10 (29.4) | 0 | 0 |
| Imipenem | 3 (11.5) | 11 (32.3) | 1 (3.8) | 6 (17.6) | 22 (84.6) | 17 (50) | 0 | 0 |
| Meropenem | 2 (7.6) | 12 (35.2) | 0 | 2 (5.8) | 24 (92.3) | 20 (58.8) | 0 | 0 |
| Amikacin | 2 (7.6) | 12 (35.2) | 0 | 10 (29.4) | 24 (92.3) | 12 (35.2) | 0 | 0 |
| Amox/kclav | 9 (3.6) | 29 (85.8) | 5 (19) | 2 (5.8) | 12 (46.1) | 3 (8.8) | 0 | 0 |
| Ampicillin | 25 (96) | 33 (97) | 0 | 0 | 1 (3.8) | 1 (2.9) | 0 | 0 |
| Cefepime | 25 (96) | 27 (79) | 0 | 1 (2.9) | 1 (3.8) | 6 (17.6) | 0 | 0 |
| Cefotaxime | 4 (15.3) | 17 (50) | 0 | 0 | 1 (3.8) | 6 (17.6) | 21 (80.7) | 11 (32.3) |
| Ceftazidime | 4 (15.3) | 16 (47) | 0 | 0 | 1 (3.8) | 6 (17.6) | 21 (80.7) | 12 (35.2) |
| Cefuroxime | 25 (96) | 32 (94) | 0 | 0 | 1 (3.8) | 2 (5.8) | 0 | 0 |
| Ciprofloxacin | 15 (57.6) | 29 (85) | 0 | 0 | 11 (42.3) | 5 (14.7) | 0 | 0 |
| Gentamicin | 15 (57.6) | 23 (67.6) | 1 (3.8) | 0 | 10 (38.4) | 11 (32.3) | 0 | 0 |
| PipTzo | 6 (23) | 27 (79) | 1 (3.8) | 1 (2.9) | 19 (73) | 6 (17.6) | 0 | 0 |
| TrimethSulfa | 16 (61.5) | 18 (53.9) | 0 | 0 | 10 (41.2) | 16 (47) | 0 | 0 |
(S = sensitive, R = resistant, I = intermediate, ESBL = Extended spectrum beta-lactamase).
Figure 1.
Antibiotic resistance profile of the 60 isolates of E. coli and K. pneumoniae as determined by MicroScan. (R = resistant, I = intermediate, S = sensitive).
3.2. Carbapenem resistant isolates produce carbapenemase
The ability of E. coli and K. pneumoniae isolates to produce carbapenemases was tested using MHT. Out of the 60 isolates, 26 (43%) were positive for carbapenemase production. Of these, 23 (67.6%) out of 34 isolates were positive for K. pneumoniae and only 4 out of 26 were positive for E. coli (Table 2 and Fig. 2).
Table 2.
Modified Hodge test results of isolates (n = 60).
| Number of isolates (%) |
||
|---|---|---|
| MHT positive | MHT negative | |
| Carbapenemase producer | Non-carbapenemase producer | |
| K. pneumoniae (n = 34) | 23 (38.3%) | 11 (18.3%) |
| E. coli (n = 26) | 4 (6.6%) | 22 (36.6%) |
| Total (n = 60) | 27 (45%) | 33 (55%) |
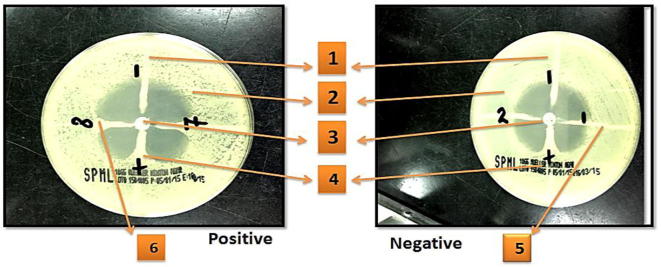
Figure 2.
Example of E. coli isolates showing a positive and negative results with MHT on Muller Hinton Agar. Symbols; (1) K. pneumoniae negative control ATCC 1706. (2) E. coli 25922. (3) Meropenem disk. (4) K. pneumoniae positive control ATCC1705. (5) Negative MHT result. (6) Positive MHT result.
3.3. Rapid identification of carbapenem genes using Gene Xpert Carba-R assay
A total of 60 isolates were tested by the Xpert Carba-R Assay for the target carbapenem-resistant gene sequences (blaIMP, blaVIM, blaNDM, blaKPC, and blaOXA-48). The results were interpreted by the Gene-Xpert software from measured fluorescent signals and embedded calculation algorithms (Fig. 3). PCR results indicated that 29 (48.3%) out of 60 isolates were positive for carbapenem resistance determine genes. As shown in Table 5, OXA-48 gene was the most frequent where it was detected in 23 (38.33%) isolates, (1 (4%) isolate of E. coli, and 22 (64.7%) isolates of K. pneumoniae), followed by NDM in 4 (6.6%) isolates (2 (8%) in E. coli and 2 (5.8%) in K. pneumoniae isolates), VIM was detected only in one isolate of K. pneumoniae (2.9%). KPC and IMP-1 genes were not detected in any of the tested isolates.
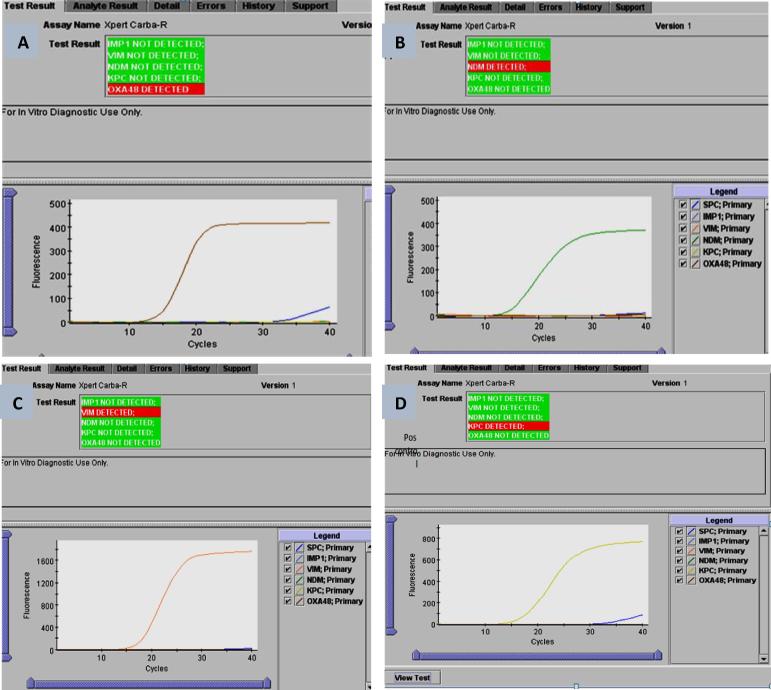
Figure 3.
Gene Xperts Carba-R assay analysis. (A) OXA-48 gene detected in K. pneumoniae isolates, (B) NDM gene detected in E. coli isolates, (C) VIM gene detected in K. pneumoniae isolates, and (D) positive control KPC of K. pneumoniae.
Table 5.
Comparison between phenotypic assays (MicroScan and MHT) and Xpert Carba-R molecular assay.
| Strain No. | Species | MHT | MicroScan |
Xpert Carba-R | |||||
|---|---|---|---|---|---|---|---|---|---|
| ERT | MIC | IMI | MIC | MER | MIC | ||||
| 1 | K. pneumoniae | −ve | S | ⩽0.25 | S | ⩽1 | S | ⩽1 | OXA-48 |
| 3 | K. pneumoniae | +ve | R | 2 | S | ⩽1 | R | ⩽1 | OXA-48 |
| 5 | K. pneumoniae | +ve | R | 2 | S | ⩽1 | S | ⩽1 | OXA-48 |
| 6 | K. pneumoniae | +ve | R | 2 | S | ⩽1 | S | ⩽1 | OXA-48 |
| 7 | K. pneumoniae | −ve | S | 0.5 | S | ⩽1 | S | ⩽1 | OXA-48 |
| 8 | K. pneumoniae | −ve | R | >1 | R | >8 | R | >8 | NDM |
| 9 | K. pneumoniae | +ve | R | >1 | I | 2 | S | ⩽1 | OXA-48 |
| 15 | K. pneumoniae | +ve | R | >1 | S | ⩽1 | S | <1 | OXA-48 |
| 30 | E. coli | +ve | S | ⩽0.25 | S | ⩽1 | S | ⩽1 | -ve |
| 41 | E. coli | +ve | R | 2 | S | 2 | S | ⩽1 | OXA-48 |
| 47 | K. pneumoniae | +ve | R | >1 | S | ⩽1 | S | ⩽1 | OXA-48 |
| 48 | K. pneumoniae | +ve | R | 4 | S | ⩽1 | S | ⩽1 | OXA-48 |
| 56 | K. pneumoniae | +ve | R | >1 | S | <1 | S | <1 | OXA-48 |
3.4. Rapidity, sensitivity and specificity of Xpert Carba-R assay in comparison with phenotypic tests
The results showed a notable variation between phenotypic assays, MicroScan and MHT, and molecular-based Xpert Carba-R PCR assay in terms of rapidity, sensitivity and specificity. The specificity of MicroScan was 100% while its sensitivity to ETP, IMP, and MER was 93%, 68.9%, and 55.17%, respectively. For the modified Hodge test, the specificity was 96.77% and sensitivity was 89.65%. Although some results of phenotypic assays matched with the definite PCR identification, some results were misleading. Out of the 29 positive PCR samples (i.e. carbapenem resistant), three samples of K. pneumoniae (samples; 1, 7, and 8) were negative for the MHT and one E. coli sample was MHT positive but negative for the PCR. For the MicroScan results, 9 samples (8 for K. pneumoniae and 1 for E. coli) were determined as carbapenem sensitive for (Imipenem and Meropenem). These 9 samples were determined as carbapenem resistance by PCR (Table 5). Another important item that should be considered is the time factor. In the phenotypic assays where MicroScan takes 18–24 h and MHT 16-24 h, PCR takes less than an hour. From the above findings, PCR is rapid, accurate and sensitive assay for detection of carbapenem-resistant Enterobacteriaceae.
4. Discussion
Enterobacteriaceae members such as E. coli and K. pneumoniae are among the most important causative agents of hospital-acquired and community-onset infections of human beings (Paterson, 2006). Carbapenemases production by these bacteria is associated with therapy failures, dissemination, and increased mortality rates (Nordmann et al., 2011). Therefore, reliable detection methods with rapidity, high sensitivity and specificity is required (Nordmann, 2014). The rapid identification of carbapenem resistance bacteria allows physicians not only to describe the proper antimicrobial regimen but also to restrict their spreading. The preliminary screening for carbapenemase producers in clinical specimens is based first on phenotypic tests, whereas, confirmation tests are mainly based on molecular assays. However, traditional phenotypic methods have some disadvantages such as; are time-consuming, difficult to interpret, and the sensitivity/specificity vary between different species (Nordmann et al., 2011, Miriagou et al., 2010, Cheng-Yen and Jiunn-Jong, 2015, Walsh et al., 2011).
The phenotypic assays, MHT has been suggested as the gold standard technique to detect carbapenemase producing bacteria in the past years (Nordmann et al., 2012). Of the 60 isolates tested in the current study, 27 (45%) were MHT positive (Table 2, Fig. 2). Our results showed that MHT failed to detect one isolate of K. pneumoniae which was PCR positive for NDM gene. As well, two K. pneumoniae isolates were OXA-48 producers but were negative for MHT (Table 5). The overall sensitivity and specificity of the MHT were 89.6% and 96.77%, respectively. Doyle et al., found that the sensitivity and specificity for MHT was 61% and 93%, respectively (Doyle et al., 2012). MHT is less reliable to detect NDMs, VIMs, and IMPs producing bacteria; however, it may be useful for detecting KPC and OXA-48 producers (Nordmann et al., 2011, Castanheira et al., 2011, Doyle et al., 2012).
Resistance to carbapenem agents is due to carbapenemase and presence of other resistance mechanisms, such as ESBLs, porin mutations and/or presence of efflux pumps (Chambers, 2010). In our study, carbapenem resistance rates among K. pneumoniae isolates were 24 (70.5%), and 5 (19.2%) among E. coli isolates, while we found 36 isolates sensitive suggesting resistance mechanism other than carbapenemase production (Rachana et al., 2014).
Most carbapenem resistant isolates had high resistance rates against other antibiotics including amikacin, augmentin, ampicillin, cefepime, cefotaxime, ceftazidime, ciprofloxacin, cefuroxime, gentamicin, piperacillin/tazobactam, and trimethoprim/ sulfamethoxazole (Table 1). The resistance rates for carbapenem resistant K. pneumoniae isolates were 45.8%, 100%, 100%, 83.3%, 87.7%, 87.7%, 95.8%, 75%, 95.8% and 62.5% respectively, while resistance rates for 5 isolates of carbapenem resistant E. coli were 0%, 60%, 100%, 100%, 100%, 100%, 100%, 100%, 40%, 40%, 40% respectively.
Current study found that two isolates of K. pneumoniae harbored OXA-48 gene exhibited sensitivity to IMI, MER and ERT antibiotics by Microscan (Table 1, Fig. 1). As well, seven isolates of K. pneumoniae produced OXA-48 gene and seemed resistant only to ertapenem. For E. coli isolates, one OXA-48 producer isolate exhibited resistance to ertapenem but not for IMI or MER (Table 1, Table 5). Several studies had reported that automated susceptibility systems incorrectly labeled up to 87% of carbapenemase-producing K. pneumoniae isolates as susceptible to imipenem (Tenover et al., 2006). Ertapenem resistance seems to be a marker for carbapenemase production when automated testing methods are used (Tenover et al., 2006, Anderson et al., 2007). This necessitates further testing by phenotypic and genotypic methods. If resources are limited, an elevated MIC for ertapenem could be used as a screening method to determine which isolates need further testing (Tenover et al., 2006, Anderson et al., 2007).
Xpert Carba-R PCR assay can differentiate the most prevalent carbapenemase genes including KPC, NDM, VIM, OXA-48 and IMP-1 in less than an hour. Tenover et al., 2013; showed that the sensitivity and specificity of Xpert MDRO for detecting KPC or VIM were 100% and 99.0%, respectively (Tenover et al., 2013). Other study evaluated the performance of three commercial molecular assays for detecting major families of carbapenemases; they found that the sensitivity and specificity of Xpert Carba-R kit is 71–100% and 100% for different carbapenemase families (Findlay et al., 2015).
However, our study revealed high prevalence of OXA-48 type carbapenemase-producing in 24 (38.33%) isolates, followed by NDM type in 5 isolates, VIM was found less detected only in one isolate of K. pneumoniae (Table 3 and Fig. 3). The absence of VIM-1, KPC and IMP genes suggest that these isolates are not major sources of carbapenemases in the Kingdom (Table 4). Zowawi et al., 2014; found that the most common carbapenemases were of the OXA-48 and NDM respectively. Other studies showed that the OXA-type carbapenemases is a growing problem and the most important reason for resistance to IMI and MER Acinetobacter’s worldwide (Gur et al., 2008, Carvalho et al., 2009, Kuo et al., 2010). Other studies performed in Saudi Arabia indicated that OXA-48 and NDM-1 types are the dominant carbapenemases among the Enterobacteriaceae (Memish et al., 2014). The emergence of OXA-48 and NDM carbapenemases among the Enterobacteriaceae in Saudi Arabia may reflect the extensive population flows between the Kingdom, other part of the Middle East, and North Africa where OXA-48 is widespread and the Indian subcontinent where NDM are common (Nordmann et al., 2011).
Table 3.
Gene Xperts Carba-R results in all isolates (n = 60).
| No. | KPC | NDM | VIM | OXA-48 | IMP-1 | No. | KPC | NDM | VIM | OXA-48 | IMP-1 | NO. | KPC | NDM | VIM | OXA-48 | IMP-1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | −ve | +ve | −ve | −ve | −ve | 21 | −ve | −ve | −ve | −ve | −ve | 41 | −ve | −ve | −ve | −ve | −ve |
| 2 | −ve | −ve | −ve | +ve | −ve | 22 | −ve | −ve | −ve | −ve | −ve | 42 | −ve | −ve | −ve | −ve | −ve |
| 3 | −ve | −ve | −ve | +ve | −ve | 23 | −ve | −ve | −ve | −ve | −ve | 43 | −ve | −ve | −ve | +ve | −ve |
| 4 | −ve | −ve | −ve | +ve | −ve | 24 | −ve | −ve | −ve | −ve | −ve | 44 | −ve | −ve | −ve | −ve | −ve |
| 5 | −ve | +ve | −ve | −ve | −ve | 25 | −ve | −ve | −ve | −ve | −ve | 45 | −ve | −ve | −ve | −ve | −ve |
| 6 | −ve | −ve | −ve | −ve | −ve | 26 | −ve | −ve | −ve | −ve | −ve | 46 | −ve | −ve | −ve | −ve | −ve |
| 7 | −ve | −ve | −ve | +ve | −ve | 27 | −ve | −ve | −ve | −ve | −ve | 47 | −ve | −ve | −ve | −ve | −ve |
| 8 | −ve | −ve | −ve | +ve | −ve | 28 | −ve | −ve | −ve | −ve | −ve | 48 | −ve | −ve | −ve | +ve | −ve |
| 9 | −ve | −ve | −ve | +ve | −ve | 29 | −ve | −ve | −ve | −ve | −ve | 49 | −ve | −ve | −ve | +ve | −ve |
| 10 | −ve | +ve | −ve | −ve | −ve | 30 | −ve | −ve | −ve | −ve | −ve | 50 | −ve | −ve | −ve | +ve | −ve |
| 11 | −ve | −ve | −ve | +ve | −ve | 31 | −ve | −ve | −ve | −ve | −ve | 51 | −ve | −ve | −ve | +ve | −ve |
| 12 | −ve | −ve | −ve | +ve | −ve | 32 | −ve | −ve | −ve | −ve | −ve | 52 | −ve | −ve | −ve | +ve | −ve |
| 13 | −ve | −ve | +ve | −ve | −ve | 33 | −ve | −ve | −ve | −ve | −ve | 53 | −ve | −ve | −ve | +ve | −ve |
| 14 | −ve | −ve | −ve | +ve | −ve | 34 | −ve | −ve | −ve | −ve | −ve | 54 | −ve | −ve | −ve | −ve | −ve |
| 15 | −ve | −ve | −ve | +ve | −ve | 35 | −ve | +ve | −ve | −ve | −ve | 55 | −ve | −ve | −ve | −ve | −ve |
| 16 | −ve | −ve | −ve | +ve | −ve | 36 | −ve | +ve | −ve | −ve | −ve | 56 | −ve | −ve | −ve | +ve | −ve |
| 17 | −ve | −ve | −ve | +ve | −ve | 37 | −ve | −ve | −ve | +ve | −ve | 57 | −ve | −ve | −ve | +ve | −ve |
| 18 | −ve | −ve | −ve | +ve | −ve | 38 | −ve | −ve | −ve | −ve | −ve | 58 | −ve | −ve | −ve | +ve | −ve |
| 19 | −ve | −ve | −ve | −ve | −ve | 39 | −ve | −ve | −ve | −ve | −ve | 59 | −ve | −ve | −ve | −ve | −ve |
| 20 | −ve | −ve | −ve | −ve | −ve | 40 | −ve | −ve | −ve | −ve | −ve | 60 | −ve | −ve | −ve | −ve | −ve |
Samples number; 1–16, 18, 21, 26, 31, 36–38, 40, 45–51, 54–56 are for K. pneumoniae and samples number; 17, 19, 20, 22, 23–25, 27–30, 32–35, 39, 41–44, 52, 53, 57–60 are for E. coli.
Table 4.
Prevalence of carbapenem resistance determining genes among all isolates (n = 60).
| Carbapenem resistance genes | No. of total isolates | Percentage (%) |
|---|---|---|
| OXA-48 | 23 | 38.33% |
| NDM | 4 | 6.66% |
| VIM | 1 | 1.66% |
| KPC | – | – |
| IMP | – | – |
Thus awareness of carpbapenemase producing Enterobacteriaceae in Gulf cooperation council countries has important implications for controlling the spread of carpbapenemase producing Enterobacteriaceae in the Middle East and in hospitals accommodating patients transferred from the region (Zowawi et al., 2014). Thus after we evaluated phenotypic and molecular tests, we recommend using molecular tests such Gene Expert R assay for optimal carbapenemnase detection and it is important to know the type of carbapenemase gene present, reducing material, manpower and cost (Doyle et al., 2012), besides recommendation for an initial screening for Enterobacteriaceae by using ertapenem in addition to meropenem and imipenem.
Acknowledgements
This research project was supported by a grant from the “Research Center of the Female Scientific and Medical Colleges”, Deanship of Scientific Research, King Saud University.
Footnotes
Peer review under responsibility of King Saud University.
References
- Anderson K. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J. Clin. Microbiol. 2007;45:2723–2725. doi: 10.1128/JCM.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink A.J., Coetzee J., Clay C.G., Sithole S., Richards G.A., Poirel L., Nordmann P. Emergence of New Delhi metallo-beta-lactamase (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC-2) in South Africa. J. Clin. Microbiol. 2011;50(2):525–527. doi: 10.1128/JCM.05956-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho K., Carvalho-Assef A., Peirano G., Santos L., Pereira M., Asensi M. Dissemination of multidrug-resistant Acinetobacter baumannii genotypes carrying bla (OXA-23) collected from hospitals in Rio de Janeiro, Brazil. Int. J. Antimicrob. Agents. 2009;34(1) doi: 10.1016/j.ijantimicag.2008.12.009. 25-8. [DOI] [PubMed] [Google Scholar]
- Castanheira M., Deshpande L., Mathai D., Bell J., Jones R., Mendes R. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob. Agents Chemother. 2011;55:1274–1278. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H. Carbapenems and monobactams. In: Gerald L., editor. Mandlle, Dougals and Bennett’s Principles and Practice of Infectious Diseases. seventh ed. Elsevier Publishers; USA: 2010. pp. 341–346. [Google Scholar]
- Cheng-Yen K., Jiunn-Jong W. Carbapenem resistance: epidemiology, mechanisms and screening methods. J. Biomed. Lab. Sci. 2015;27:1–9. [Google Scholar]
- Cuzon G., Ouanich J., Gondret R., Naas T., Nordmann P. Outbreak of OXA- 48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob. Agents Chemother. 2011;55:2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D., Peirano G., Lascols C., Lloyd T., Church D., Pitout J. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J. Clin. Microbiol. 2012;50:3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay J., Hopkins K.L., Meunier D., Woodford N. Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J. Antimicrob. Chemother. 2015;70(5):1338–1342. doi: 10.1093/jac/dku571. [DOI] [PubMed] [Google Scholar]
- Girlich D., Poirel L., Nordmann P. Do CTX-M beta-lactamases hydrolyse ertapenem? J. Antimicrob. Chemother. 2008;62(5):1155–1156. doi: 10.1093/jac/dkn317. [DOI] [PubMed] [Google Scholar]
- Girlich D., Poirel L., Nordmann P. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 2012;50:477–479. doi: 10.1128/JCM.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur D., Korten V., Unal S., Deshpande L.M., Castanheira M. Increasing carbapenem resistance due to the clonal dissemination of oxacillinase (OXA-23 and OXA-58)-producing Acinetobacter baumannii: report from the Turkish SENTRY Program sites. J. Med. Microbiol. 2008;57:1529–1532. doi: 10.1099/jmm.0.2008/002469-0. [DOI] [PubMed] [Google Scholar]
- Kuo H., Yang C., Lin M., Cheng W., Tien N., Liou M. Distribution of blaOXA-carrying imipenem-resistant Acinetobacter spp. in 3 hospitals in Taiwan. Diagn. Microbiol. Infect. Dis. 2010;66(2):195–199. doi: 10.1016/j.diagmicrobio.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Liu W. Sensitive and rapid diagnostic method for detection of New Delhi metallo-beta-lactamase gene by loop-mediated isothermal amplification. J. Clin. Microbiol. 2012;50(5):1580–1585. doi: 10.1128/JCM.06647-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchanda V., Rai S., Gupta S., Rautela R.S., Chopra R., Rawat D.S., Verma N., Singh N.P., Kaur I.R., Bhalla P. Development of Taq Man real-time polymerase chain reaction for the detection of the newly emerging form of carbapenem resistance gene in clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii. Indian J. Med. Microbiol. 2011;29(3):249–253. doi: 10.4103/0255-0857.83907. [DOI] [PubMed] [Google Scholar]
- McLaughlin M., Advincula M., Malczynski M., Qi C., Bolon M., Scheetz M.H. Correlations of antibiotic use and carbapenem resistance in Enterobacteriaceae. Antimicrob. Agents Chemother. 2014;57:5131–5133. doi: 10.1128/AAC.00607-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z., Assiri M., ALMasri M., Roshdy H., Hathout H., Kaasy M., Gatermann S.G., Yezli S. Molecular characterization of carbapenemase production among gram-negative bacteria in Saudi Arabia. Microb. Drug Resist. 2014;21(3):307–314. doi: 10.1089/mdr.2014.0121. [DOI] [PubMed] [Google Scholar]
- Miriagou V., Cornaglia G., Edelstein M., Galani I., Giske C., Gniadkowski M. Acquired carbapenemases in gram-negative bacterial pathogens: detection and surveillance issues. Clin. Microbiol. Infect. 2010;16(2):112–122. doi: 10.1111/j.1469-0691.2009.03116.x. [DOI] [PubMed] [Google Scholar]
- Mitchell J., Yehuda C. Carbapenem-resistant Enterobacteriaceae. A potential threat. JAMA. 2008;300(24):2911–2913. doi: 10.1001/jama.2008.896. [DOI] [PubMed] [Google Scholar]
- Munoz-Price L. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2014;13(9):785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Poirel L., Toleman M., Walsh T.R. Does broad spectrum β-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gramnegative bacteria? J. Antimicrob. Chemother. 2011;66:689–692. doi: 10.1093/jac/dkq520. [DOI] [PubMed] [Google Scholar]
- Nordmann P. Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Med. Mal. Infect. 2014;44:51–56. doi: 10.1016/j.medmal.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Nordmann P., Gniadkowski M., Giske C., Poirel L., Woodford N., Miriagou V. Identification and screening of carbapenemase producing Enterobacteriaceae. Clin. Microbiol. Infect. 2012;18(5):432–438. doi: 10.1111/j.1469-0691.2012.03815.x. [DOI] [PubMed] [Google Scholar]
- Orstavik I., Odegaard K. Simple test for penicillinase production in Staphylococcus aureus. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 1971;79(6):855–856. [PubMed] [Google Scholar]
- Pasteran F., Mendez T., Guerriero L., Rapoport M., Corso A. Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J. Clin. Microbiol. 2009;47(6):1631–1639. doi: 10.1128/JCM.00130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson D.L. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control. 2006;34:20–28. doi: 10.1016/j.ajic.2006.05.238. [DOI] [PubMed] [Google Scholar]
- Poirel L., Revathi G., Bernabeu S., Nordmann P., Naas T. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob. Agents Chemother. 2011;55(2):934–936. doi: 10.1128/AAC.01247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournaras S., Poulou A., Tsakris A. Inhibitor-based methods for the detection of KPC carbapenemase-producing Enterobacteriaceae in clinical practice by using boronic acid compounds. J. Antimicrob. Chemother. 2010;65:1319–1321. doi: 10.1093/jac/dkq124. [DOI] [PubMed] [Google Scholar]
- Queenan A., Bush M.K. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachana S., Lavanya V., Sreevidya S., Aparna B., Nagapriyanka E., Vemu L. Comparative evaluation of multiplex PCR and routine laboratory phenotypic. J. Clin. Diagn. Res. 2014;8(12):23–26. doi: 10.7860/JCDR/2014/10794.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F., Canton R., Kop J., Chan R., Ryan J., Weir F., Ruiz-Garbajosa P., LaBombardi V., Persing D.H. Detection of colonization by carbapenemase-producing gram-negative Bacilli in patients by use of the Xpert MDRO assay. J. Clin. Microbiol. 2013;51:3780–3787. doi: 10.1128/JCM.01092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F., Kalsi R., Williams P., Carey R., Stocker S., Lonsway D. Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg. Infect. Dis. 2006;12:1209–1213. doi: 10.3201/eid1208.060291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk K., Voets G.M., Scharringa J., Voskuil S., Fluit A.C., Rottier W.C., Leverstein-Van Hall M.A., Cohen Stuart J.W. A disc diffusion assay for detection of class A, B and OXA-48 carbapenemases in Enterobacteriaceae using phenyl boronic acid, dipicolinic acid and temocillin. Clin. Microb. Infect. 2014;20(4):345–349. doi: 10.1111/1469-0691.12322. [DOI] [PubMed] [Google Scholar]
- Walsh T., Weeks J., Livermore D., Toleman M. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 2011;11(5):355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- Zowawi H. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf cooperation council: dominance of OXA-48 and NDM producers. Antimicrob. Agents Chemother. 2014;58(6):3085–3090. doi: 10.1128/AAC.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]