Abstract
In eukaryotes, the Cu/Zn superoxide dismutase (SOD1) is a major cytosolic cuproprotein with a small fraction residing in the mitochondrial intermembrane space (IMS) to protect against respiratory superoxide. Curiously, the opportunistic human fungal pathogen Candida albicans is predicted to express two cytosolic SODs including Cu/Zn containing SOD1 and manganese containing SOD3. As part of a copper starvation response, C. albicans represses SOD1 and induces the non-copper alternative SOD3. While both SOD1 and SOD3 are predicted to exist in the same cytosolic compartment, their potential role in mitochondrial oxidative stress had yet to be investigated. We show here that under copper replete conditions, a fraction of the Cu/Zn containing SOD1 localizes to the mitochondrial IMS to guard against mitochondrial superoxide. However in copper starved cells, localization of the manganese containing SOD3 is restricted to the cytosol leaving the mitochondrial IMS devoid of SOD. We observe that during copper starvation, an alternative oxidase (AOX) form of respiration is induced that is not coupled to ATP synthesis but maintains mitochondrial superoxide at low levels even in the absence of IMS SOD. Surprisingly, the copper-dependent cytochrome c oxidase (COX) form of respiration remains high with copper starvation. We provide evidence that repression of SOD1 during copper limitation serves to spare copper for COX and maintain COX respiration. Overall, the complex copper starvation response of C. albicans involving SOD1, SOD3 and AOX minimizes mitochondrial oxidative damage whilst maximizing COX respiration essential for fungal pathogenesis.
Introduction
Superoxide dismutases (SOD) play vital roles in the biology of reactive oxygen species (ROS) by disproportionating superoxide anion free radicals into hydrogen peroxide and molecular oxygen [1, 2]. There are three major SOD families including copper and zinc (Cu/Zn) SODs that use copper as the catalytic co-factor, a separate SOD family with either manganese or iron [3], and a rare family of nickel containing SODs [4]. Gram-negative bacteria restrict copper containing SODs to the periplasmic/extracellular space while manganese and iron SODs are intracellular/cytosolic [5, 6]. An analogous partitioning occurs in eukaryotic mitochondria where a Cu/Zn SOD (known as SOD1) lies in the intermembrane space (IMS) and a manganese SOD (known as SOD2) resides in the mitochondrial matrix [7–15]. SOD1 is also cytosolic in eukaryotes and its distribution between the cytosol and mitochondrial IMS involves a disulfide relay system and the copper chaperone for SOD1, CCS [16–18].
In mitochondria, superoxide anion is formed as a byproduct of respiration and is released into both the matrix and IMS where it reacts with SOD2 and SOD1 respectively [19, 20]. With conventional respiration involving the full electron transport chain and cytochrome c oxidase (COX), superoxide is released to the matrix by complex I [21], while complex III releases superoxide to both the IMS and matrix [19, 22]. COX is not the only form of respiration and certain fungi, plants, and protists express an alternative oxidase (AOX) that accepts electrons directly from coenzyme Q [23–29]. Unlike COX, AOX respiration is not coupled to ATP production [28]. One example of an organism that utilizes both COX and AOX respiration is the polymorphic fungus Candida albicans [30–33]. C. albicans is an opportunistic human pathogen that exists as a commensal of the human flora, but can become pathogenic in immune compromised individuals. Unlike fermenting yeasts such as Saccharomyces cerevisiae, C. albicans relies heavily on COX respiration for ATP [34, 35]. Thus, the rationale for retaining AOX respiration is not clear, particularly since AOX could potentially compete with COX for coenzyme Q electrons.
Aside from dual modes of respiration, C. albicans has an unusually large collection of six SOD enzymes including the Cu/Zn containing SOD1 [36, 37], the manganese containing SOD2 in the mitochondrial matrix [38], three extracellular copper-only SODs (SOD4,5,6) [39–42] and a second manganese containing SOD3 predicted to be cytosolic, as is the case with SOD1 [43]. We have recently shown that SOD1 and SOD3 are reciprocally expressed according to copper status: copper replete yeast only express SOD1 while copper starved cells repress SOD1 and induce SOD3 [44]. This switch in SOD enzymes is mediated by the copper sensing regulator MAC1 and is set into motion during fungal invasion of the kidney [44]. While both SOD1 and SOD3 are predicted to be cytosolic, it is unknown whether either can enter the mitochondrial IMS to deal with superoxide release in this compartment.
Herein we investigate the role of C. albicans SOD1 versus SOD3 in mitochondrial oxidative stress protection. We demonstrate that under copper replete conditions, the Cu/Zn containing SOD1 partitions between the cytosol and mitochondrial IMS and protects against mitochondrial superoxide. However, during copper starvation, the mitochondrial IMS becomes devoid of a SOD enzyme since the manganese containing SOD3 localizes exclusively to the cytosol. In spite of no IMS SOD, mitochondrial superoxide is not elevated. We find that during copper limitation, C. albicans induces AOX respiration, which suppresses mitochondrial superoxide and bypasses the need for an IMS SOD. In spite of extreme copper starvation, copper dependent COX respiration remains high and we provide evidence for a copper sparing mechanism whereby repression of SOD1 helps maintain high COX respiration in the face of copper starvation.
Materials and Methods
Yeast strains and culture conditions
C. albicans strains used in this study were derived from SC5314 including CA-IF100 (arg4Δ/arg4Δ, leu2Δ/leu2Δ::cmLEU2, his1Δ/his1Δ::cdHIS1, URA3/ura3Δ) and isogenic strains CA-IF001 (sod1Δ::cmLEU2/sod1Δ::cdHIS1) and CA-IF011 (sod3Δ::cmLEU2/sod3 Δ::cdHIS1) [41]. The SN152 strain (his1Δ/his1Δ, leu2Δ/leu2Δ, arg4Δ/arg4Δ, URA3/ura3Δ::imm434, IRO1/iro1Δ::imm434) and isogenic mac1Δ::LEU2/mac1Δ::HIS1 strain were obtained from the Fungal Genetics Stock Center [45, 46]. The ccs1Δ/Δ strain (ccs1Δ::cmLEU2/ccs1::cdHIS1) was derived from SN78 (ura3Δ::imm434/ura3Δ::imm434, leu2Δ/leu2Δ, his1Δ/his1Δ) as previously described [47]. The strain constitutively expressing SOD1 was derived from CA-IF001 (sod1Δ::cmLEU2/SOD1) as previously described in [44]. In the so-called SOD1con strain, the single allele of SOD1 was engineered with a mutation in the MAC1 binding site at position +148 by gene replacement. A similar gene replacement was used to engineer the control SODrep with an intact MAC1 binding site [44].
C. albicans cells were cultured at 30°C in either enriched media (YPD; BD Difco) containing 1% yeast extract, 2% peptone, and 2% dextrose (w/v) or in a synthetic complete (SC) media with 0.67% yeast nitrogen base (US Biologicals) and 2% dextrose (w/v). Where indicated, cells were starved for copper by supplementing the growth media with 800 μM of the extracellular Cu(I) chelator BCS.
Cell fractionation and analysis of SOD proteins and enzymatic activity
Mitochondria and post-mitochondrial supernatant (PMS)/largely cytosolic fractions were obtained from C. albicans essentially as described [31]. Briefly, cells were grown in YPD to an OD600 between 1.0 and 4.0, were harvested and successively washed in MilliQ deionized water and 0.1 M Tris-SO4 pH 9.4, 10 mM DTT, followed by incubation in the same buffer for 30 minutes at 30°C shaking at 100 RPM. Cells were harvested, and spheroplasts formed by incubating cells in 1.2 M sorbitol, 20 mM KH2PO4/K2HPO4 pH 7.4 with 0.3 mg/L Lyticase (Sigma). Spheroplasts were isolated by centrifugation at 1000 x g for 5 minutes and resuspended in 0.6 M sorbitol 20 mM K+HEPES pH 7.4 and lysed by dounce homogenization to create whole cell lysates. The lysate was spun at 12,000 x g for 10 minutes at 4°C to resolve PMS and crude mitochondria fractions. Mitochondria were washed twice in 1.2 M sorbitol 20 mM KH2PO4/K2HPO4 pH 7.4 buffer and where indicated, were resolved further into IMS and matrix fractions by resuspending in 20 mM K+ HEPES pH 7.4 and briefly vortexing for 5 seconds to rupture the outer membrane. The mitochondrial fraction was centrifuged at 12,000 x g for 10 minutes at 4°C to separate the matrix (pellet) from the soluble IMS. Prior to analysis by gel electrophoresis, crude whole mitochondria or matrix fractions were resuspended in a 10 mM sodium phosphate buffer 7.4 containing 0.1% Triton X-100 5 mM EDTA, 5 mM EGTA, 50 mM NaCl, 10% glycerol, a procedure that solubilizes the matrix and allows for visualization of matrix SOD2 [48].
SOD protein expression and localization was analyzed by immunoblot as previously described [44]. 70 μg of whole cell spheroplast lysates and the same cell equivalents of PMS and mitochondrial fractions were loaded onto 4–12% Bis-Tris gels (Thermo Fisher). Where indicated whole cell lysates of C. albicans were prepared by glass bead homogenization [44] and 30 μg of lysate protein was used. Blots were probed with anti-SOD1 and SOD3 primary antibodies as previously described [44]. Anti-SOD1 is a polyclonal antibody directed against C. elegans Sod-1 (JH766) that reacts well with copper and zinc containing SOD molecules across eukaryotes [48]. The anti-SOD3 polyclonal antibody is directed against a peptide unique to C. albicans SOD3 that is absent in mitochondrial SOD2 [47]. Anti-PGK1 at a 1:5000 dilution (Acris Antibodies, #AP21371AF-N) was used as a cytosolic marker. The polyclonal anti-SOD2 antibody is directed against S. cerevisiae SOD2 and was prepared as described [49]. This anti-SOD2 cross-reacts well with C. albicans SOD2 and was used at a 1:5000 dilution as a mitochondrial marker. On immunoblots, all four antibodies are highly specific. SOD enzymatic activity was analyzed by native gel electrophoresis and nitroblue tetrazolium staining as previously described [44] using 30 μg total cell lysate.
RNA analysis
C. albicans cells were grown in SC medium in the absence or presence of 800 μM BCS to an OD600 between 1.0 and 2.0. RNA was isolated from 20 OD600 cell units using an RNeasy mini kit (Qiagen), and then subsequently converted to cDNA using Superscript IV Reverse Transcriptase kit (Thermo Fisher Scientific). Real time PCR was performed as previously described [44]. cDNA was diluted 20-fold before PCR amplification with iQ SYBR Green Supermix (Bio-Rad) and values were normalized to TUB2 transcripts in each sample. The SOD3 and TUB2 primers were used as described [44]; AOX2 primers are GTTGGTCAAGGGGTTTTCACTAATG and ACTGCCACTTCAGGGATTTTCATGG.
Measurements of mitochondrial superoxide, oxygen consumption and cellular copper
Mitochondrial superoxide was measured in C. albicans cells initially cultured in YPD media to early stationary phase (OD600 6.0–10.0), then diluted back to OD600 ≈0.1 in SC medium and grown to OD600 of 1.0–2.0. Background fluorescence is minimized in cells from SC medium. 5 μM of MitoSOX Red (Life Technologies) in DMSO or the equivalent volume of DMSO control was added to 500 μl of culture and incubated for 45 minutes in round bottom tubes at 250 rpm in the dark at 30°C. Where indicated, cells were co-incubated with MitoSOX red and 500 nM MitoTracker Green FM (Thermo Scientific). To monitor effects of AOX inhibition, cultures were incubated for 1 hour with 5 mM SHAM prior to MitoSOX addition. Cells were washed three times in phosphate buffered saline (PBS), resuspended in PBS and subjected to either microscopic visualization of MitoSOX Red fluorescence using a Zeiss Observer Z1 fluorescence microscope with a Zeiss Plan-Apochromat 63x objective, or to fluorescence quantification on a Biotek Synergy HT microplate reader with excitation and emission at 510 nm and 580 nm respectively. Fluorescence quantification used 2 x 107 of cells in 200 μl PBS.
Oxygen consumption measurements were conducted on whole cells essentially as described [35] using cells grown in YPD to early log (OD600 of 1.0–2.0). Ten OD600 units of cells were harvested, washed once in PBS and resuspended in 2 ml of YPD. Oxygen consumption was measured polarographically using a Clark-type oxygen electrode in a magnetically stirred, thermostatically controlled 1.5-ml chamber at 25°C (Oxytherm; Hansatech). Following 1 min, 5mM salicylhydroxamic acid (SHAM) was added to the oxygen chamber to inhibit AOX respiration followed by addition of 1mM potassium cyanide (KCN) to inhibit COX respiration. The change in oxygen saturation over time was used to calculate rates of oxygen consumption. The degree to which the oxygen consumption rate is inhibited by SHAM or KCN was used to derive the percent oxygen consumption by AOX or COX, respectively.
Copper content of whole cells was measured on the same cultures used for oxygen consumption. 20 OD600 units of cells were washed twice in 10 mM Tris pH 8.0 1 mM EDTA, and twice in deionized water. Cells were resuspended in 500 μl MilliQ deionized water before analysis of copper by atomic absorption spectroscopy (AAS) on a PerkinElmer Life Sciences AAnalyst 600 graphite furnace instrument as described [44].
Results
A fraction of Candida albicans SOD1, but not SOD3, localizes to the mitochondrial intermembrane space.
In standard enriched medium, C. albicans cells are copper replete and only express SOD1, not SOD3 (Fig 1A). Supplementing cultures with the extracellular Cu(I) chelator BCS (bathocuproinedisulfonic acid) results in a ten-fold reduction in intracellular copper (Fig 1B), and cells respond to this copper starvation stress by repressing SOD1 and inducing the manganese containing SOD3 (Fig 1A) [44]. Using these differential copper conditions we examined the localization of SOD1 versus SOD3.
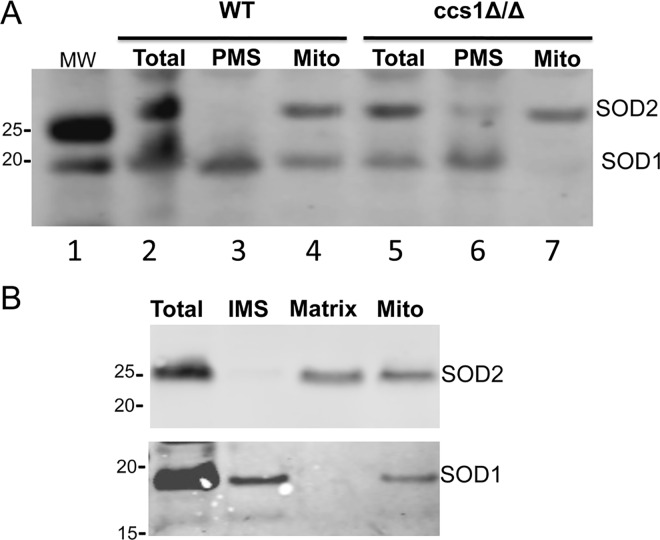
Fig 1. A fraction of C. albicans SOD1 but not SOD3 localizes to the mitochondria IMS.
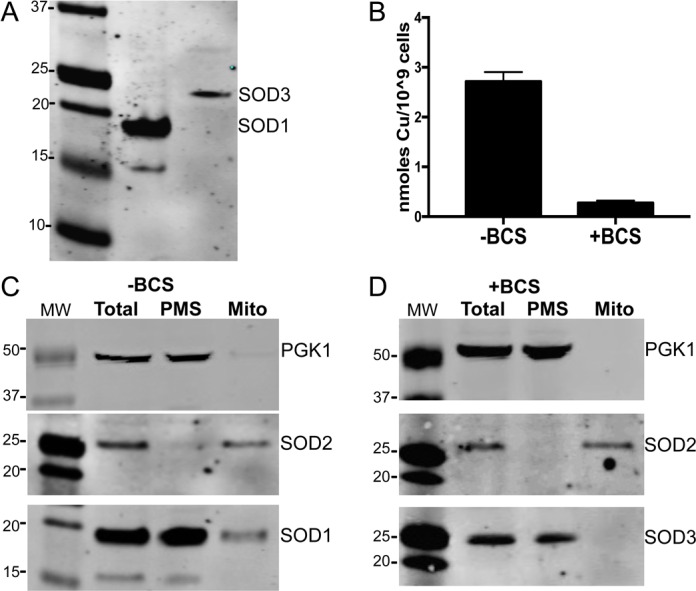
C. albicans strain CAIF-100 was grown in enriched medium in either the presence or absence of 800 μM of the extracellular Cu(I) chelator BCS as indicated. (A) Lysates were prepared from spheroplasts and were analyzed for SOD1 and SOD3 by immunoblot. (B) Total cellular copper was measured by atomic absorption spectroscopy. (C-D) Spheroplast cell lysates (Total) were fractionated into the largely cytosolic post-mitochondrial supernatant (PMS) and crude mitochondria (mito) by differential centrifugation. 70 μg of total lysate protein and the same cell equivalents of PMS and mitochondria were analyzed by immunoblot for SOD2 (marker of mitochondrial matrix), PGK1 (marker of cytosol) and either SOD1 (C) or SOD3 (D). MW = molecular weight markers. SOD1 often appears as two bands, both of which are down-regulated by copper starvation.
We first addressed whether the Cu/Zn containing SOD1 partitions between the cytosol and mitochondria as was shown for S. cerevisiae and mammalian cells [8, 9, 50]. Whole cell lysates were subjected to differential centrifugation to resolve crude mitochondria from the largely cytosolic or post mitochondrial supernatant (PMS) fraction. Identical cell equivalents of each fraction were analyzed by western blot. As seen in Fig 1C, the majority of SOD1 fractionates with the cytosolic maker PGK1, consistent with a largely cytosolic localization of the protein. We also observed a small fraction of SOD1 partitioning with the mitochondria together with the mitochondrial marker, SOD2. By quantification of the results from five experimental trials, we find that mitochondrial SOD1 represents 5% of total cellular SOD1 (standard deviation = 0.0247). This value is quite comparable to the ≈1–5% reported for human and S. cerevisiae mitochondrial SOD1 [8, 14]. As was shown for human and S. cerevisiae SOD1 [16, 51], C. albicans SOD1 appears to require its copper chaperone CCS for mitochondrial uptake, as SOD1 was not detected in mitochondria from ccs1Δ/Δ cell (Fig 2A, compare lanes 4 and 7). Additionally, upon fractionation of IMS and matrix components, we observed that SOD1 localizes to the IMS while the manganese containing SOD2 partitions to the mitochondrial matrix (Fig 2B), precisely as was shown for S. cerevisiae and mammalian SODs [8–11, 13–15]. Altogether, the studies of Figs 1C and 2 demonstrate that under copper replete conditions, C. albicans parallels other eukaryotes by importing a small fraction of SOD1 into the mitochondrial IMS.
Fig 2. C. albicans SOD1 localizes to the mitochondrial IMS and requires its copper chaperone CCS1 for mitochondrial localization.
(A) Cell lysates from wild type (CAIF-100) and the isogenic ccs1Δ/Δ strain were fractionated into PMS and mitochondria and analyzed by immunoblot for SOD1 and SOD2. (B) The mitochondrial fraction derived from 70 μg total lysate protein was further fractionated into IMS and matrix as described in Materials and Methods and analyzed for SOD1 and SOD2 by immunoblot. Numbers on left indicate molecular weight markers.
We next addressed the status of C. albicans mitochondria when cells are starved for copper and switch from Cu/Zn containing SOD1 to the manganese containing SOD3. In the experiment of Fig 1D, the mitochondrial and PMS fractions of copper starved cells were probed for SOD3. We observed that unlike SOD1, SOD3 was only visible in the PMS, and over three experimental trials there was no detectable SOD3 in the mitochondrial fraction (representative result shown in Fig 1D).
Mitochondrial superoxide under copper replete versus copper starvation conditions
We sought to understand the consequences of no IMS SOD during copper starvation. To monitor mitochondrial superoxide we utilized the superoxide specific fluorescent probe MitoSOX Red, a derivative of dihydroethidium designed to target mitochondria. As seen in Fig 3A, MitoSOX yields a punctate pattern of fluorescence in C. albicans wild type cells mirroring that of MitoTracker Green FM, validating its use as a specific marker of mitochondrial ROS in C. albicans. Under copper replete conditions, sod1Δ/Δ mutants also display a punctate pattern of fluorescence (Fig 3C) that appeared enhanced compared to the signal obtained with wild type cells analyzed in parallel (Fig 3B). To quantitate any changes between wild type and sod1Δ/Δ cells, MitoSOX fluorescence was measured spectrophotometrically at 580 nm. Over four experimental trials with eight independent samples, sod1Δ/Δ cells consistently exhibited a ≈2- fold increase in MitoSOX detectable fluorescence (Fig 4A). The MitoSOX signal was also increased in sod2Δ/Δ cells lacking the mitochondrial matrix SOD2 (Fig 4B). Thus, MitoSOX Red is an effective indicator of mitochondrial ROS in C. albicans, sensitive to both IMS (SOD1-relevant) and mitochondrial matrix (SOD2-relevant) superoxide.
Fig 3. MitoSOX Red as a probe for mitochondrial superoxide in C. albicans.
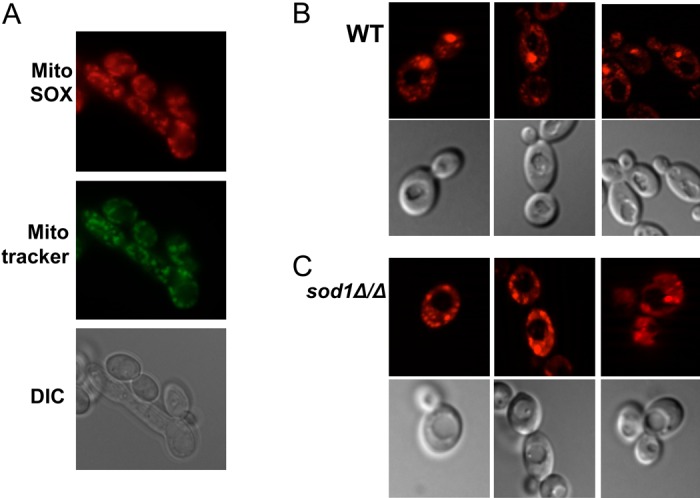
Log phase CA-1F100 C. albicans wild type cells (A, B) or the isogenic sod1Δ/Δ strain (C) grown under non-stress conditions were incubated with 5 mM MitoSOX Red as a marker for mitochondrial superoxide and imaged by fluorescence microscopy at 63X. (A) “DIC” = light microscopy images and “MitoTracker” = cells treated with both MitoSox Red and 500 μM MitoTracker Green as a marker for mitochondria. (B,C) Wild type and sod1Δ/Δ cells were examined in parallel where the top and bottom rows show representative MitoSox Red fluorescence and DIC images, respectively.
Fig 4. Effects of copper starvation and sod gene mutations on mitochondrial superoxide and AOX2 expression.
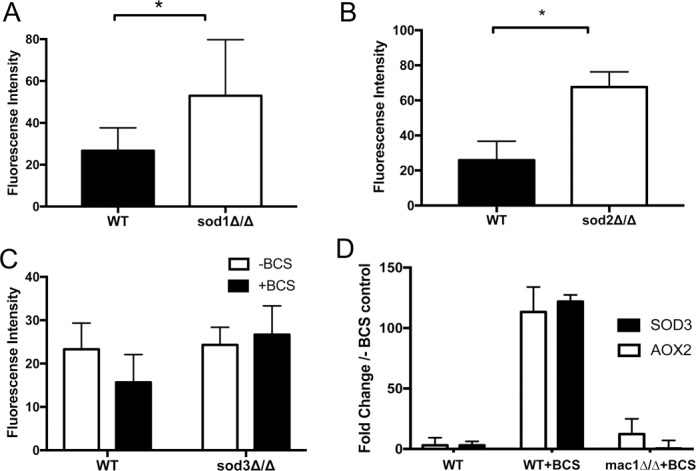
(A-C) MitoSOX Red fluorescence was measured spectrophotometrically at 580 nm where fluorescence intensity is the signal obtained with 2x107 cells as described in Materials and Methods. (A-B) A comparison of the effects of sod1Δ/Δ versus sod2Δ/Δ mutations on MitoSOX Red fluorescence where results represent the averages of eight and four independent samples respectively, and statistical significance was determined using a paired two-tailed Student’s t-test. Asterisks indicate statistical significance with P values < 0.05. Error bars represent standard deviation. The baseline MitoSOX Red fluorescence of non-stressed wild type cells typically varies between 20–40 fluorescent units, but regardless of this baseline, sod1Δ/Δ and sod2Δ/Δ mutants exhibited enhanced fluorescence. (C) The indicated strains were grown in the presence or absence of 800 μM BCS as designated. Results represent the averages of three independent cultures where error bars are standard deviation. The difference between minus and plus BCS with the wild type strain is not statistically significant (p = 0.2063). (D) AOX2 and SOD3 mRNA were quantified by qRT-PCR from wild type (SN152) and the isogenic mac1Δ/Δ strain as described in Materials and Methods. Values are normalized to that obtained with wild type SN152 C. albicans cells without BCS. Shown are averages of two biological replicates where error bars are standard deviation.
Using MitoSOX Red, we probed the mitochondrial ROS status of copper deficient cells. We anticipated an elevation in mitochondrial superoxide, since the mitochondrial IMS appears devoid of a SOD enzyme under these conditions. However, MitoSOX Red fluorescence was not elevated in either wild type cells or sod3Δ/Δ strains treated with BCS (Fig 4C) consistent with the notion that SOD3 does not enter the mitochondria. It was therefore possible that copper starvation stress induces a non-SOD antioxidant to minimize mitochondrial ROS.
Modes of mitochondrial respiration under copper replete versus copper starvation conditions
As mentioned above, C. albicans uses two forms of respiration: COX respiration that employs the full electron transport chain and is a major source of cellular ATP, and AOX respiration that accepts electrons directly from coenzyme Q and is not coupled to ATP production [23, 30, 31]. C. albicans expresses dual AOX isoforms, AOX1 and AOX2 [32, 33], and AOX2 was previously shown to be a target of the same MAC1 regulator that controls SOD1 and SOD3 expression [52]. Copper regulation of AOX2 had not been previously reported and we find using quantitative real-time PCR a very strong induction of AOX2 mRNA in cells starved for copper (Fig 4D). This pronounced induction of AOX2 mimics that of SOD3 mRNA and is abolished by mac1Δ/Δ mutations (Fig 4D).
We tested whether the induction of AOX2 mRNA correlates with utilization of the AOX pathway for respiration. Fig 5A and 5B shows representative oxygen consumption experiments where cells were successively treated with salicylhydroxamic acid (SHAM) to inhibit AOX respiration followed by treatment with potassium cyanide KCN to inhibit COX [31]. By calculating the rates of oxygen consumption in the presence and absence of these inhibitors, the fraction of respiration due to AOX versus COX can be ascertained. We find that during copper replete conditions, COX respiration accounts for essentially all of the oxygen consumption while AOX respiration is undetectable (Fig 5C and 5D). However, during copper starvation, there was a pronounced induction of AOX respiration, accounting for nearly 25% of total oxygen consumption; COX respiration proportionately declined (Fig 5C and 5D). AOX respiration is clearly induced during copper starvation.
Fig 5. AOX respiration contributes to oxygen consumption during copper starvation.
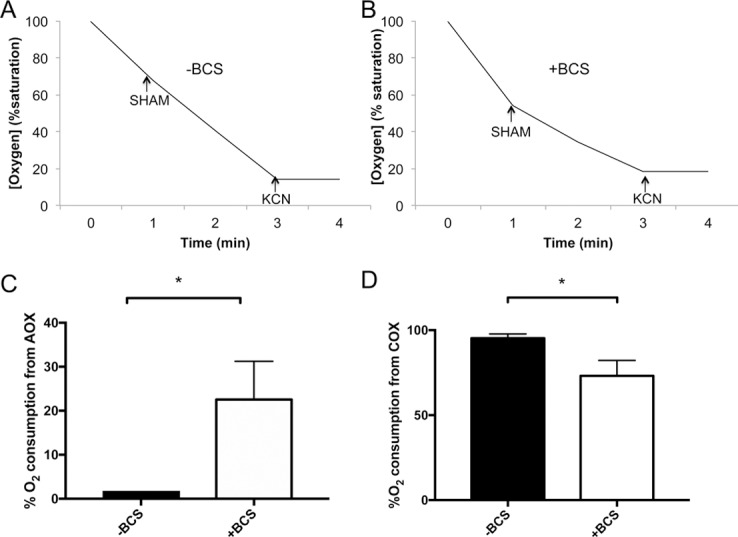
C. albicans strain CA-IF100 grown in the presence or absence of BCS was subjected to measurements of oxygen consumption using a Clark electrode. (A,B) Shown are representative profiles of oxygen consumption as monitored by percent oxygen saturation in solution. At the indicated time points, cells were treated with 5 mM SHAM to inhibit AOX respiration followed by 1 mM KCN to inhibit COX respiration. (C,D) The rates of oxygen consumption were derived from oxygen saturation curves. Plotted are the percentages of oxygen consumption inhibited by SHAM and attributed to AOX (C) and the oxygen consumption inhibited by KCN and attributed to COX (D). Results represent the averages of three independent samples with error bars representing standard deviation. Statistical significance was determined using a paired two-tailed Student’s t-test. Asterisks indicate statistical significance with P values < 0.05.
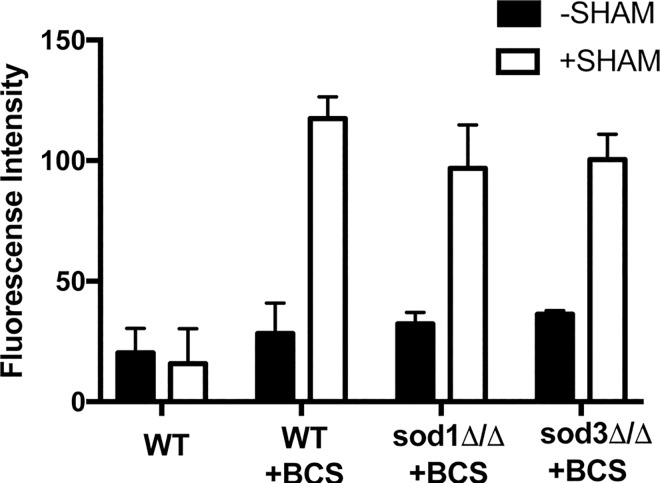
AOX respiration bypasses complex III of the electron transport chain, and since complex III is a source of IMS superoxide [19], AOX induction could conceivably minimize mitochondrial ROS. To test the impact of AOX on mitochondrial superoxide, MitoSOX Red fluorescence was measured in cells in which AOX was inhibited by SHAM. As seen in Fig 6, SHAM had no impact on mitochondrial ROS in copper replete wild type cells, consistent with the observation of no AOX respiration under these conditions (Fig 5B). However, SHAM treatment had a marked effect on mitochondrial ROS under copper starvation conditions. In wild type cells, MitoSOX Red fluorescence was enhanced 3–4 fold when AOX was inhibited with SHAM (Fig 6). The same is true of sod1Δ/Δ and sod3Δ/Δ strains starved for copper, demonstrating that the change in superoxide occurs independent of these SOD enzymes (Fig 6). Together, the studies of Figs 5 and 6 demonstrate that during copper starvation, C. albicans induces AOX respiration to reduce mitochondrial superoxide.
Fig 6. Effect of inhibiting AOX respiration on mitochondrial superoxide under copper starvation.
MitoSOX Red fluorescence was measured in the designated strains. Where indicated, cells were treated with 5 mM SHAM for 1 hour to inhibit AOX respiration prior to MitoSOX Red incubation. Results represent the averages of two independent cultures with error bars representing standard deviation.
Evidence for copper sparing with SOD1
Since cytochrome c oxidase is a copper dependent enzyme, it was curious that COX respiration falls no more than 30% (Fig 5D) when intracellular copper levels drop by ≥90% (Fig 1B). Under these same copper starvation conditions, activity of copper dependent SOD1 is undetectable, largely due to SOD1 gene repression. Does this repression of SOD1 serve to spare copper for COX? To address this, we used a C. albicans strain engineered to constitutively express SOD1.
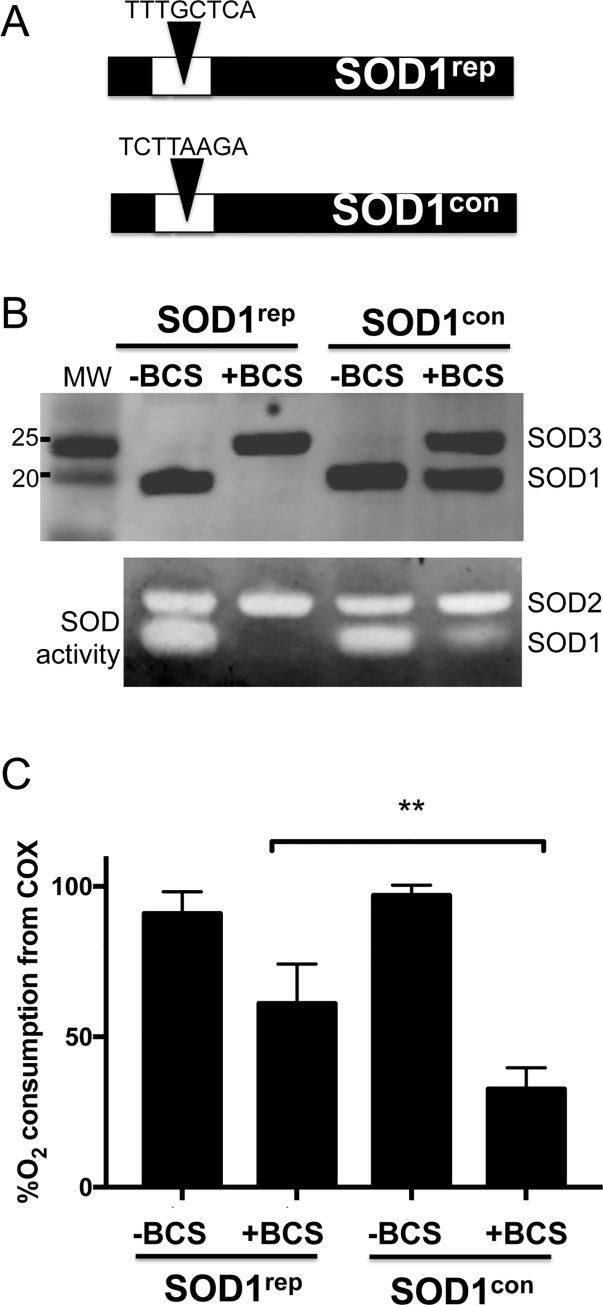
C. albicans SOD1 contains a single intronic MAC1 site that mediates SOD1 repression during copper starvation [44] (illustrated in Fig 7A). As seen in Fig 7B top, mutating this MAC1 site in chromosomal SOD1 results in constitutive SOD1 expression and cells express both SOD1 and SOD3 with copper starvation, consistent with previous findings [44]. We observe that constitutively expressed SOD1 is not fully active with copper starvation due to limitation of its copper co-factor; nevertheless it can secure sufficient copper for 30–40% of the activity seen with copper replete conditions (Fig 7B bottom and legend). We tested whether this limited copper activation of SOD1 can impact COX respiration. As seen in Fig 7C, the control strain expressing repressible SOD1 with intact MAC1 site exhibited the anticipated ≈30% decrease in COX respiration with copper starvation. By comparison, COX respiration drops ≈60% with copper starvation in the strain that constitutively expresses SOD1. Thus, even a small retention in SOD1 activity during copper starvation conditions is sufficient to inhibit COX respiration. We conclude that the repression of SOD1 during copper starvation helps maximize COX respiration.
Fig 7. Constitutive expression of SOD1 inhibits COX repression during copper starvation.
(A) Schematic showing the coding region (black) and intron (white) of the SOD1 gene with sequence of the wild type and mutated MAC1 sites in SOD1rep (SOD1 repressed by copper) and SOD1con (SOD1 constitutively expressed). (B) Strains expressing a single copy of either SOD1rep or SOD1con were grown in the presence or absence of BCS and whole cell lysates were (top) immunoblotted for SOD1 and SOD3 as in Fig 1A and (bottom) subjected to SOD enzymatic activity analysis by the native gel assay. Numbers on left of immunoblot represent molecular weight markers and the positions of SOD1 and SOD2 migration on the native gel are indicated on the right. Densitometric tracings show that the intensity of SOD1 activity in SOD1con +BCS is 38% that of the corresponding–BCS sample. (C) Cytochrome c oxidase respiration was measured as in Fig 5D as a function of KCN inhibitable oxygen consumption. Results represent the averages of two independent experiments with a total of four biological replicates with error bars representing standard deviation. Statistical significance was determined using a paired two-tailed Student’s t-test. Asterisks indicate statistical significance with P values < 0.005.
Discussion
Copper is an essential nutrient of restricted availability and when cellular copper declines, priorities must be established to spare the nutrient. In the case of C. albicans, the response to low copper involves down-regulating the cuproprotein SOD1. Like other eukaryotes, SOD1 resides in both the C. albicans cytosol and mitochondrial IMS, and when copper declines, C. albicans maintains ROS homeostasis by replacing SOD1 with two copper-independent enzymes including SOD3 in the cytosol and AOX in the mitochondria (see model, Fig 8). We show here that AOX serves to minimize mitochondrial ROS in the absence of an IMS SOD enzyme. The role of SOD3 in the cytosol is less clear, but may act to substitute for SOD1 in cell signaling. Previously, we have shown that in S. cerevisiae, cytosolic SOD1 functions in glucose sensing and signaling [53], and the laboratories of Thiele and Zheng have shown a role for nuclear SOD1 in gene regulation and the response to DNA damage [54, 55]. C. albicans SOD3 could very well replace SOD1 in one or more of these extra-mitochondrial roles in signaling through ROS.
Fig 8. Adaption to copper in Candida albicans involving SOD enzymes and the mode of mitochondrial respiration.
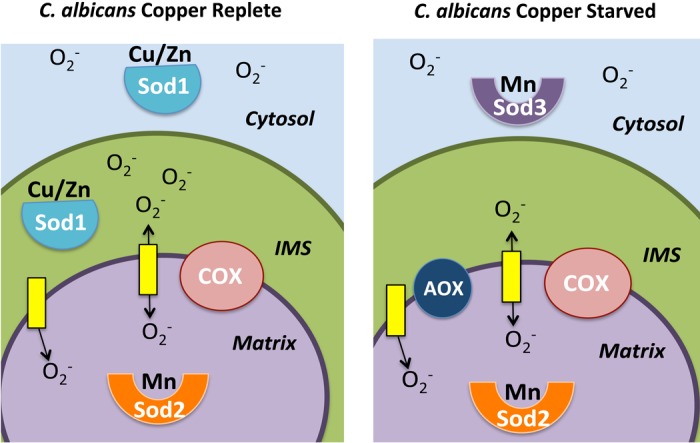
Cartoon to illustrate C. albicans adaptation to low copper by alternating SOD enzymes and inducing AOX respiration. When copper is abundant (left), C. albicans utilizes COX respiration and SOD1 is active in both the IMS and the cytosol. During copper starvation (right), cells switch to expressing SOD3 in the cytosol, but the IMS becomes devoid of a SOD enzyme. AOX respiration is induced which helps to lower IMS superoxide. Regardless of copper conditions, COX respiration remains active and mitochondrial matrix superoxide is managed by SOD2.
We propose that the down-regulation of C. albicans SOD1 during copper starvation serves to free up substantial metal for other processes. We find that copper-dependent COX respiration remains high in C. albicans during copper starvation; however this activity can be compromised when even a small level of copper is distributed to SOD1, as was found using a constitutively expressed allele of SOD1. By down-regulating SOD1 during copper starvation, C. albicans can spare this metal for COX respiration, crucial for this opportunistic fungal pathogen that relies heavily on COX respiration for ATP. The notion of sparing copper for respiration is reminiscent of what has been reported by Merchant and colleagues for the photosynthetic algae, Chlamydomonas reinharti [56]. C. reinhardtii does not express a copper requiring SOD1 [57], rather a major source of cellular copper is plastocyanin used for photosynthesis. In this organism when copper levels are low, plastocyanin is degraded and allows for copper to be allocated for the synthesis of COX [56]. To compensate for loss of plastocyanin, C. reinhardtii induces iron requiring Cytochrome c6 [58, 59], analogous to the induction of the non-copper alternatives SOD3 and AOX that substitute for SOD1 in C. albicans.
AOX respiration is not coupled to ATP synthesis, and the rationale for retaining this apparently futile mode of oxygen consumption has been the subject of much investigation. Many previous studies support a role for AOX in offsetting certain defects associated with inhibition of COX respiration. By directly accepting electrons from coenzyme Q, AOX can help maintain the NAD/NADH balance through complex I activity [60] or prevent superoxide formation when downstream portions of the electron transport chain are interrupted [61–63]. Consistent with its role in correcting COX deficiencies, AOX gene expression is induced by inhibitors of respiration in several organisms [64–66]. Here we provide a new layer to AOX functionality, specifically under copper limiting conditions. In C. albicans, AOX not only offsets deficiencies in COX respiration but compensates for loss of IMS SOD1 by minimizing mitochondrial ROS. Inside the animal host, C. albicans is subject to great fluctuations in copper availability and experiences copper starvation stress during invasion of the kidney [44]. The adaptation described here involving SOD1, SOD3, AOX and COX is expected to minimize mitochondrial oxidative damage while maximizing COX respiration required for pathogenesis [34, 35].
Lastly, why has C. albicans retained the copper containing SOD1 when the fungus is susceptible to copper starvation stress in vivo and the requirement for SOD1 appears to be bypassed by cytosolic Mn SOD3 and mitochondrial AOX? Our previous work in S. cerevisiae has shown that in addition to scavenging superoxide, SOD1 serves as a copper buffer and can minimize copper toxicity in cases of copper excess [67]. C. albicans SOD1 may have a similar role in copper detoxification. In the case of Chlamydomonas, it has been suggested that Cu-plastocyanin acts as a storage depot for copper that can be rapidly allocated to COX when copper becomes limiting [56]. By the same token, SOD1 may temporarily hold copper that is re-distributed to COX or other sites as needed.
Acknowledgments
We thank Professor S. Claypool and O. Ogunbona for invaluable assistance with oxygen consumption measurements, Christine Lee for assistance with microscopy, Cissy Li and Julie Gleason for helpful discussions and Angelique Besold of careful review of this manuscript.
This work was supported by NIH grants RO1 AI119949, T32 CA009110, and F31 GM113637.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by NIH grants RO1 AI119949, T32 CA009110, and F31 GM1136. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Benov L, Chang LY, Day B, Fridovich I. Copper, zinc superoxide dismutase in Escherichia coli: periplasmic localization. Arch Biochem Biophys. 1995;319:508–11. 10.1006/abbi.1995.1324 [DOI] [PubMed] [Google Scholar]
- 2.McCord JM, Fridovich I. Superoxide dismutase: An enzymic function for erythrocuprein (hemocuprein). J Biol Chem. 1969;244:6049–55. [PubMed] [Google Scholar]
- 3.Abreu IA, Cabelli DE. Superoxide dismutases-a review of the metal-associated mechanistic variations. Biochim Biophys Acta. 2010;1804(2):263–74. 10.1016/j.bbapap.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 4.Dupont CL, Neupane K, Shearer J, Palenik B. Diversity, function and evolution of genes coding for putative Ni-containing superoxide dismutases. Environ Microbiol. 2008;10(7):1831–43. 10.1111/j.1462-2920.2008.01604.x [DOI] [PubMed] [Google Scholar]
- 5.Enriquez RP, Do RN. Bioavailbility of metal ions and evolutionary adapatations. Life. 2012;2:274–85. 10.3390/life2040274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broxton CN, Culotta VC. SOD Enzymes and Microbial Pathogens: Surviving the Oxidative Storm of Infection. PLoS Pathog. 2016;12(1):e1005295 10.1371/journal.ppat.1005295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weisiger RA, Fridovich I. Mitochondrial superoxide dismutase. J Biol Chem. 1973;248:4793–6. [PubMed] [Google Scholar]
- 8.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A Fraction of Yeast Cu,Zn-Superoxide Dismutase and Its Metallochaperone, CCS, Localize to the Intermembrane Space of Mitochondria. J Biol Chem. 2001;276:38084–9. 10.1074/jbc.M105296200 [DOI] [PubMed] [Google Scholar]
- 9.Mattiazzi M, D'Aurelio M, Gajewski CD, Martushova K, Kiaei M, Beal MF, et al. Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J Biol Chem. 2002;277(33):29626–33. 10.1074/jbc.M203065200 [DOI] [PubMed] [Google Scholar]
- 10.Jaarsma D, Rognoni F, Duijn Wv, Verspaget HW, Hassdijk ED, Haasdijk ED, et al. CuZn superoxide dismutase (SOD1) accumulates in vacuolated mitochondria in transgenic mice expressing amyotrophic lateral sclerosis-linked SOD1 mutations. Acta Neuropathol. 2001;102:293–305. [DOI] [PubMed] [Google Scholar]
- 11.Higgins CM, Jung C, Ding H, Xu Z. Cu, Zn superoxide dismutase that causes motoneuron degeneration is present in mitochondria in the CNS. J Neurosci. 2002;22:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inarrea P, Moini H, Han D, Rettori D, Aguilo I, Alava MA, et al. Mitochondrial respiratory chain and thioredoxin reductase regulate intermembrane Cu,Zn-superoxide dismutase activity: implications for mitochondrial energy metabolism and apoptosis. Biochem J. 2007;405(1):173–9. 10.1042/BJ20061809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inarrea P, Moini H, Rettori D, Han D, Martinez J, Garcia I, et al. Redox activation of mitochondrial intermembrane space Cu,Zn-superoxide dismutase. Biochem J. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldsteins G, Keksa-Goldsteine V, Ahtoniemi T, Jaronen M, Arens E, Akerman K, et al. Deleterious role of superoxide dismutase in the mitochondrial intermembrane space. J Biol Chem. 2008. [DOI] [PubMed] [Google Scholar]
- 15.Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutase (SOD) in rat liver. Cu/Zn SOD in mitochondria. J Biol Chem. 2001;276:38388–93. 10.1074/jbc.M105395200 [DOI] [PubMed] [Google Scholar]
- 16.Kawamata H, Manfredi G. Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space. Antioxid Redox Signal. 2010;13(9):1375–84. 10.1089/ars.2010.3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddehase S, Grumbt B, Neupert W, Hell K. The disulfide relay system of mitochondria is required for the biogenesis of mitochondrial Ccs1 and Sod1. J Mol Biol. 2009;385(2):331–8. 10.1016/j.jmb.2008.10.088 [DOI] [PubMed] [Google Scholar]
- 18.Kloppel C, Suzuki Y, Kojer K, Petrungaro C, Longen S, Fiedler S, et al. Mia40-dependent oxidation of cysteines in domain I of Ccs1 controls its distribution between mitochondria and the cytosol. Mol Biol Cell. 2011;22(20):3749–57. 10.1091/mbc.E11-04-0293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller FL, Liu Y, Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem. 2004. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Yu L, Yu C-A. Generation of Superoxide Anion by Succinate-Cytochrome c Reductase from Bovine Heart Mitochondria. J Biol Chem. 1998;273(51):33972–6. [DOI] [PubMed] [Google Scholar]
- 21.Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turrens JF, Alexandre A, Lehninger AL. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch Biochem Biophys. 1985;237:408–14. [DOI] [PubMed] [Google Scholar]
- 23.Joesph-Horne TH, D.W.; Wood P.M. Fungal respiration: A fusion of standard and alternative components Biochemica et Biophysica Acta 2001(1504):179–95. [DOI] [PubMed] [Google Scholar]
- 24.Hanqing F, Kun S, Mingquan L, Hongyu L, Xin L, Yan L, et al. The expression, function and regulation of mitochondrial alternative oxidase under biotic stresses. Mol Plant Pathol. 2010;11(3):429–40. 10.1111/j.1364-3703.2010.00615.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore AL, Umbach AL, Siedow JN. Structure-function relationships of the alternative oxidase of plant mitochondria: a model of the active site. J Bioenerg Biomembr. 1995;27(4):367–77. [DOI] [PubMed] [Google Scholar]
- 26.Chaudhuri M, Ott RD, Hill GC. Trypanosome alternative oxidase: from molecule to function. Trends Parasitol. 2006;22(10):484–91. 10.1016/j.pt.2006.08.007 [DOI] [PubMed] [Google Scholar]
- 27.Krab K. Kinetic and regulatory aspects of the function of the alternative oxidase in plant respiration. J Bioenerg Biomembr. 1995;27(4):387–96. [DOI] [PubMed] [Google Scholar]
- 28.Sluse FE, Jarmuszkiewicz W. Alternative oxidase in the branched mitochondrial respiratory network: an overview on structure, function, regulation, and role. Braz J Med Biol Res. 1998;31(6):733–47. [DOI] [PubMed] [Google Scholar]
- 29.Vanlerberghe GC, McIntosh L. ALTERNATIVE OXIDASE: From Gene to Function. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:703–34. 10.1146/annurev.arplant.48.1.703 [DOI] [PubMed] [Google Scholar]
- 30.Helmerhorst EJ, Murphy MP, Troxler RF, Oppenheim FG. Characterization of the mitochondrial respiratory pathways in Candida albicans. Biochim Biophys Acta. 2002;1556(1):73–80. [DOI] [PubMed] [Google Scholar]
- 31.Helmerhorst EJ, Stan M, Murphy MP, Sherman F, Oppenheim FG. The concomitant expression and availability of conventional and alternative, cyanide-insensitive, respiratory pathways in Candida albicans. Mitochondrion. 2005;5(3):200–11. 10.1016/j.mito.2005.04.001 [DOI] [PubMed] [Google Scholar]
- 32.Huh WKK S.O. Molecular Cloning and Functional Expression of Alternative Oxidase from Candida albicans. Journal of Bacteriology. 1999;181(13):4098–41001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huh WK, Kang SO. Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem J. 2001;356(Pt 2):595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker JM, Kauffman SJ, Hauser M, Huang L, Lin M, Sillaots S, et al. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc Natl Acad Sci U S A. 2010;107(51):22044–9. 10.1073/pnas.1009845107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Chen H, Florentino A, Alex D, Sikorski P, Fonzi WA, et al. Enzymatic dysfunction of mitochondrial complex I of the Candida albicans goa1 mutant is associated with increased reactive oxidants and cell death. Eukaryot Cell. 2011;10(5):672–82. 10.1128/EC.00303-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang CS, Rhie G, Kim ST, Kim YR, Huh WK, Baek YU, et al. Copper- and zinc-containing superoxide dismutase and its gene from Candida albicans. Biochim Biophys Acta. 1999;1427(2):245–55. [DOI] [PubMed] [Google Scholar]
- 37.Hwang CS, Rhie GE, Oh JH, Huh WK, Yim HS, Kang SO. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology. 2002;148(Pt 11):3705–13. 10.1099/00221287-148-11-3705 [DOI] [PubMed] [Google Scholar]
- 38.Hwang CS, Baek YU, Yim HS, Kang SO. Protective roles of mitochondrial manganese-containing superoxide dismutase against various stresses in Candida albicans. Yeast. 2003;20(11):929–41. 10.1002/yea.1004 [DOI] [PubMed] [Google Scholar]
- 39.Gleason JE GA, Peterson RL, Taylor AB, Holloway SP, Waninger-Saroni J, Cormack B, Cabelli DE, Hart PJ and Culotta VC. Candida albicans SOD5 represents the prototype of a new class of copper only superoxide dismutase enzymes required for pathogen defense. 2014 [DOI] [PMC free article] [PubMed]
- 40.Martchenko M, Alarco AM, Harcus D, Whiteway M. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol Biol Cell. 2004;15(2):456–67. 10.1091/mbc.E03-03-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol Microbiol. 2009;71(1):240–52. 10.1111/j.1365-2958.2008.06528.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson RL, Galaleldeen A, Villarreal J, Taylor AB, Cabelli DE, Hart PJ, et al. The Phylogeny and Active Site Design of Eukaryotic Cu-only Superoxide Dismutases. J Biol Chem. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamarre C, LeMay JD, Deslauriers N, Bourbonnais Y. Candida albicans expresses an unusual cytoplasmic manganese-containing superoxide dismutase (SOD3 gene product) upon the entry and during the stationary phase. J Biol Chem. 2001;276(47):43784–91. 10.1074/jbc.M108095200 [DOI] [PubMed] [Google Scholar]
- 44.Li CX, Gleason JE, Zhang SX, Bruno VM, Cormack BP, Culotta VC. Candida albicans adapts to host copper during infection by swapping metal cofactors for superoxide dismutase. Proc Natl Acad Sci U S A. 2015;112(38):E5336–42. 10.1073/pnas.1513447112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4(2):298–309. 10.1128/EC.4.2.298-309.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCluskey K, Wiest A, Plamann M. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J Biosci. 2010;35(1):119–26. [DOI] [PubMed] [Google Scholar]
- 47.Gleason JE, Li CX, Odeh HM, Culotta VC. Species-specific activation of Cu/Zn SOD by its CCS copper chaperone in the pathogenic yeast Candida albicans. J Biol Inorg Chem. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen LT, Culotta VC. Activation of Cu/Zn superoxide dismutases from C. elegans does not require the copper chaperone CCS. J Biol Chem. 2005;280:41373–9. 10.1074/jbc.M509142200 [DOI] [PubMed] [Google Scholar]
- 49.Luk E, Yang M, Jensen LT, Bourbonnais Y, Culotta VC. Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J Biol Chem. 2005;280(24):22715–20. 10.1074/jbc.M504257200 [DOI] [PubMed] [Google Scholar]
- 50.Higgins CM, Jung C, Xu Z. ALS-associated mutant SOD1G93A causes mitochondrial vacuolation by expansion of the intermembrane space and by involvement of SOD1 aggregation and peroxisomes. BMC Neurosci. 2003;4(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Field LS. The impact of copper,zinc superoxide dismutase and the mitochondria on oxidative stress and life span in Saccharomyces cerevisiae [PhD]. Baltimore: Johns Hopkins University; 2003.
- 52.Woodacre A, Mason RP, Jeeves RE, Cashmore AM. Copper-dependent transcriptional regulation by Candida albicans Mac1p. Microbiology. 2008;154(Pt 5):1502–12. 10.1099/mic.0.2007/013441-0 [DOI] [PubMed] [Google Scholar]
- 53.Reddi AR, Culotta VC. SOD1 Integrates Signals from Oxygen and Glucose to Repress Respiration. Cell. 2013;152(1–2):224–35. 10.1016/j.cell.2012.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsang CK, Liu Y, Thomas J, Zhang YJ, Zheng XFS. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun. 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wood LK, Thiele DJ. Transcriptional activation in yeast in response to copper deficiency involves copper-zinc superoxide dismutase. J Biol Chem. 2009;284(1):404–13. 10.1074/jbc.M807027200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kropat J, Gallaher SD, Urzica EI, Nakamoto SS, Strenkert D, Tottey S, et al. Copper economy in Chlamydomonas: prioritized allocation and reallocation of copper to respiration vs. photosynthesis. Proc Natl Acad Sci U S A. 2015;112(9):2644–51. 10.1073/pnas.1422492112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allen MD, Kropat J, Tottey S, Del Campo JA, Merchant SS. Manganese Deficiency in Chlamydomonas Results in Loss of PSII and MnSOD Function, Sensitivity to Peroxides, and Secondary Phosphorus- and Iron-deficiency. Plant Physiol. 2007;143:263–77. 10.1104/pp.106.088609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merchant S, Hill K, Howe G. Dynamic interplay between two copper-titrating components in the transcriptional regulation of cyt c6. EMBO J. 1991;10(6):1383–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merchant S, Bogorad L. Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardi. Mol Cell Biol. 1986;6(2):462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vemuri GN, Eiteman MA, McEwen JE, Olsson L, Nielsen J. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104(7):2402–7. 10.1073/pnas.0607469104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao YW, Shi K, Fu LJ, Zhang S, Li X, Dong DK, et al. The reduction of reactive oxygen species formation by mitochondrial alternative respiration in tomato basal defense against TMV infection. Planta. 2012;235(2):225–38. 10.1007/s00425-011-1483-z [DOI] [PubMed] [Google Scholar]
- 62.Cvetkovska M, Vanlerberghe GC. Alternative oxidase modulates leaf mitochondrial concentrations of superoxide and nitric oxide. New Phytol. 2012;195(1):32–9. 10.1111/j.1469-8137.2012.04166.x [DOI] [PubMed] [Google Scholar]
- 63.Grahl N, Dinamarco TM, Willger SD, Goldman GH, Cramer RA. Aspergillus fumigatus mitochondrial electron transport chain mediates oxidative stress homeostasis, hypoxia responses and fungal pathogenesis. Mol Microbiol. 2012;84(2):383–99. 10.1111/j.1365-2958.2012.08034.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Minagawa N, Koga S, Nakano M, Sakajo S, Yoshimoto A. Possible involvement of superoxide anion in the induction of cyanide-resistant respiration in Hansenula anomala. FEBS Lett. 1992;302(3):217–9. [DOI] [PubMed] [Google Scholar]
- 65.Li Q, Ritzel RG, McLean LL, McIntosh L, Ko T, Bertrand H, et al. Cloning and analysis of the alternative oxidase gene of Neurospora crassa. Genetics. 1996;142(1):129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan L, Li M, Cao Y, Gao P, Cao Y, Wang Y, et al. The alternative oxidase of Candida albicans causes reduced fluconazole susceptibility. J Antimicrob Chemother. 2009;64(4):764–73. 10.1093/jac/dkp273 [DOI] [PubMed] [Google Scholar]
- 67.Culotta VC, Joh HD, Lin SJ, Slekar KH, Strain J. A physiological role for Saccharomyces cerevisiae copper/zinc superoxide dismutase in copper buffering. J Biol Chem. 1995;270:29991–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.