Abstract
This study was carried out to identify drought-responsive genes in a drought tolerant faba bean variety (Hassawi 2) using a suppressive subtraction hybridization approach (SSH). A total of 913 differentially expressed clones were sequenced from a differential cDNA library that resulted in a total of 225 differentially expressed ESTs. The genes of mitochondrial and chloroplast origin were removed, and the remaining 137 EST sequences were submitted to the gene bank EST database (LIBEST_028448). A sequence analysis identified 35 potentially drought stress-related ESTs that regulate ion channels, kinases, and energy production and utilization and transcription factors. Quantitative PCR on Hassawi 2 genotype confirmed that more than 65% of selected drought-responsive genes were drought-related. Among these induced genes, the expression levels of eight highly up-regulated unigenes were further analyzed across 38 selected faba bean genotypes that differ in their drought tolerance levels. These unigenes included ribulose 1,5-bisphosphate carboxylase (rbcL) gene, non-LTR retroelement reverse related, probable cyclic nucleotide-gated ion channel, polyubiquitin, potassium channel, calcium-dependent protein kinase and putative respiratory burst oxidase-like protein C and a novel unigene. The expression patterns of these unigenes were variable across 38 genotypes however, it was found to be very high in tolerant genotype. The up-regulation of these unigenes in majority of tolerant genotypes suggests their possible role in drought tolerance. The identification of possible drought responsive candidate genes in Vicia faba reported here is an important step toward the development of drought-tolerant genotypes that can cope with arid environments.
Keywords: Vicia faba, Hassawi 2, Drought, Suppressive subtraction hybridization, Drought-responsive genes
1. Introduction
There is an urgent need to increase food production in order to address worldwide population increase. Water is the key limiting factor in agriculture and food production in vast tracts of arid and semi-arid climates. Climate change is anticipated to further decrease rainfall in many of these arid areas. The faba bean (Vicia faba L.), is known for its high protein concentration in its seeds. It ranks fourth among the most important legume crops in the world, after dry beans, dry peas and chickpeas (Toker et al., 2007). The crop is a staple food that provides adequate nutrition to many people in the Middle East. The Kingdom of Saudi Arabia imports approximately 100 million tons of seed legumes annually to fill the gap between local production and consumption; this import costs approximately 145 million SAR each year (Ministry of Agriculture, 2012). The demand for seed legumes has significantly increased in recent years. The per capita consumption of legumes increased from 3.6 kg in 1999–2001 to 4.4 kg in 2002–2004 and continues to increase to date (Ministry of Agriculture, 2012). The faba bean trade is showing a high market growth in KSA, and farmers have expressed an increasing interest in the incorporation of genes for agronomic importance in this crop (growth traits, absence of tannins, and resistance to stresses).
Despite its importance, the faba bean subjects to drought stress throughout its lifetime due to water scarcity, particularly in the central part of the Kingdom, where rainfall is low. Increasing the drought tolerance and crop water use continue to be growing issues of concern because of the increasing demand for water and improved environmental quality by the human population (Hatfield et al., 2001). Therefore, it is highly desirable to investigate means to improve drought tolerance in faba beans, especially for cultivation in a dry country such as Saudi Arabia. Such studies will help to develop faba bean genotypes that produce better yields in water-limited environments.
The faba bean genome is large, approximately 13,000 Mb (Raina and Rees, 1983), which makes it one of the largest genomes among legumes. Molecular knowledge of legume genes and the identification of genetic diversity and genetic linkages with RFLP and PCR markers (Van de Ven et al., 1991, Torres et al., 1993, Link et al., 1995) have established the first steps in the application of molecular markers to V. faba breeding. Nevertheless, limited genetic tools are available for the faba bean due to its genome complexity, not much progress has been made in understanding drought tolerance and relevant genes in this crop.
The development of molecular technology and bioinformatics has facilitated a large-scale screening for responsive genes. Both microarrays and Affymetrix gene chips have been employed to detect and analyze genes involved in various regulatory pathways in Arabidopsis thaliana (Wu et al., 2003, Misson et al., 2005, Morcuende et al., 2007, Muller et al., 2007). Similarly, transcriptome analyses were carried out using cDNA microarrays to identify genes regulated by phosphorus deficiency in rice (Wasaki et al., 2003). For other plant species, particularly field legumes, the study of responsive genes remain a challenge due to limited genome sequence information (Drame et al., 2007). A Suppression Subtractive Hybridization cDNA library was constructed to identify genes in the common bean that respond to phosphorus deficiency, and 82 candidate clones were obtained and sequenced (Tian et al., 2007). The Suppression Subtractive Hybridization (SSH) technique is based on the selective amplification of differentially expressed sequences, and this approach overcomes the technical limitations of traditional subtraction methods (Diatchenko et al., 1996). Differentially expressed genes, whose expression levels range from highly abundant to rare, can be cloned with equal probability using SSH. SSH has been used widely to study cell differentiation and development in animals and human cancer. This technique was introduced to the study of rice development (Liu et al., 2001) to identify genes involved in cold acclimation and associated stresses in wheat (Houde et al., 2006). Previous studies of gene expression in the common bean, Glycine max and faba bean have also been reported (Drame et al., 2007, Guo et al., 2008, Barros de Carvalho et al., 2013, Abid et al., 2015). SSH needs relatively smaller amounts of the initial materials, with lower costs, and fewer false positives present within the results (Sahebi et al., 2015).
The present study reports the identification of drought tolerance candidate genes in drought tolerant faba bean variety, Hassawi 2 via Suppression Subtractive Hybridization as well as subsequent sequence analysis and real-time PCR-based validation using potential candidate genes. This approach will allow trait manipulation and eventually lead to the development of drought-tolerant faba bean genotypes.
2. Materials and methods
2.1. Drought stress simulation
The faba bean variety Hassawi 2, which is highly drought tolerant (Ammar et al., 2014), was used in this study. Seeds of this genotype were germinated in pots filled with loamy sandy soil and grown under controlled conditions in the growth chamber for 15 days until the seedlings were fully developed. The osmotic pressure (0.1 bar and 15 bars) was measured to determine the field capacity and permanent wilting point for the soil used. The soil field capacity was identified at 12% water content. The plants were exposed to stress by withholding water for 4 weeks, at which point drought symptoms seriously affect stressed plants. The leaf tissues from control and stressed plants were excised when stressed plants were about to reach the wilting point, dipped directly in liquid N2, and stored properly at −86 °C until RNA isolation.
2.2. RNA isolation and SSH library construction
The total RNA was isolated from control and stressed leaves using the RNeasy mini kit (Qiagen, USA) according to manufacturer’s protocol. The RNA concentration was determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA), and the RNA integrity was assessed on a 1.2% agarose gel. An Oligotex mRNA mini kit (Qiagen, USA) was used to isolate the mRNA from the total RNA. Differentially responsive genes were identified using the Suppressive Subtractive Hybridization strategy from Clontech™ (Clontech, Palo Alto, CA) PCR Select cDNA subtraction kit in duplicate. Only forward subtractions were performed to compare the gene expression of the faba bean under control and stress conditions. The specific transcripts identified were found in the stressed sample that was used as a tester while the control sample (without stress) used as driver. The secondary PCR products of SSH were inserted into a pGEM T-easy (Promega) cloning vector according to manufacturer’s protocol. The ligated mixtures were transformed into Escherichia coli DH5α competent cells and cultured on solid LB medium containing ampicillin, X-Gal and IPTG. Selected positive clones were grown in liquid LB medium containing ampicillin. The plasmids were then purified using mini-prep and sequenced.
2.3. Sequencing and bioinformatics
The sequencing reactions were performed in a thermal cycler using an Applied Biosystems BigDye® Terminator v3.1 kit according to the manufacturer’s protocol. The sequencing reaction was precipitated with EtOH and analyzed in an ABI 3130xl 16 capillary sequencer (Applied Biosystems). The sequence trace files were base-called using the Phred program (Ewing and Green, 1998, Ewing et al., 1998) and all low-quality bases (<Q20, 99% <accuracy) were eliminated from the sequence ends, followed by the application of SeqClean to shorten the Poly-A/T. The vector and other contaminating microbial sequences were removed using VecScreen (http://www.ncbi.nlm.nih.gov/VecScreen/). After trimming, all EST sequences shorter than 100 bp were discarded. The high quality sequences were used to assemble contigs using the Codon Code Aligner software (CodonCode Corporation, USA). BLAST searches were carried out after clustering and assembly to identify similarities between ESTs and other database sequences. All unisequences were compared to the SwissProt and GenBank non-redundant protein and nucleotide databases using either the blastx (E-value ⩽10−6) or blastn (Evalue#1 ⩽10−6) programs (Altschul et al., 1990). Gene Ontology (GO) (Harris et al., 2006) annotation was performed with BLAST2GO (Conesa et al., 2005, Gotz et al., 2008) based on the sequence similarity. The default settings were applied for the annotation (blastx against NCBI non-redundant (nr) protein database, E-value filter ⩽10−3, HSP length cutoff of 33, maximum10 BLAST hits per sequence to sequence description tool and annotation cutoff of 55). Furthermore, InterProScan was performed, and the InterProScan results were merged with the GO annotation to improve annotability. Finally, the biological pathways were analyzed using the KEGG (Kyoto Encyclopedia of Genes and Genomes) (Ogata et al., 1999).
2.4. Quantitative real-time polymerase chain reaction (RT-qPCR)
Thirty-five primers were designed based on the potential candidate identified by the gene sequence analysis using the batch primers3 software and real-time PCR was performed to identify real candidate genes for drought tolerance. The relative expression profiles of the potential genes were assessed using the Hassawi 2 genotypes. Further validation was carried out using 38 faba bean genotypes with eight selected primers. The total RNA was extracted from the leaves of 38 V. faba genotypes using plant RNA reagents (Invitrogen). First strand cDNA synthesis was performed using the ProtoScript first strand synthesis kit (New England BioLabs) according to the manufacturer’s protocol. The PCR was tested for each primer combination for 35 potential unigenes, and the amplicons were analyzed on an agarose gel to confirm the presence of a single band of expected size. All reactions were run in triplicate on an ABI machine 7500 (Applied Biosystems, USA). Faba bean indigenous ribosomal gene (forward primer 5′-CTTGCAGTCAAGCTCCCTTC-3′ and reverse primer 5’-CCTTGTCCCAAGACAGACCA-3′) from faba bean was selected as a reference gene because it was consistently and reproducibly expressed compared with Actin and ElFIAF reference genes. Each reaction consisted of 1 μl of cDNA (100 ng), 10 μl of SYBR Green PCR master mix (Applied Biosystems), 0.7 μl of forward and reverse primer (10 μM) each and a balance of water for a final volume of 20 μl. The following thermocycling protocol was used: 50 °C for two minutes, an initial denaturation at 95 °C for 10 min, 40 cycles that each consisted of denaturation at 95 °C for 0.15 min and annealing/extension at 58 °C for 1 min. A melting curve protocol was carried out to ensure the absence of any multiple amplicon and primer dimers. The cycle threshold (Ct) values were automatically calculated and exported to MS Excel for further analysis. The comparative Ct method was used to evaluate the expression differences. The Ct values of the control and stress samples were normalized with and internal standard and the ΔΔCt were calculated using following formula:
ΔΔCt = ΔCttarget (mCttarget − mCtendogenous control)–ΔCtcontrol (mCtcontrol − mCtendogenous control). The fold differences were determined using power formula: 2−ΔΔCt. One hundred and thirty seven ESTs sequences were submitted to the gene bank EST database accessions number (LIBEST_028448).
3. Results
3.1. Construction of suppression-subtracted cDNA library
The suppressive subtraction hybridization (SSH) approach was used to identify the drought-responsive genes in the Hassawi 2 faba bean genotype after this crop was subjected to drought stress. cDNA was synthesized from the mRNA of stressed and controlled samples using the PCR select™ – cDNA subtraction kit (Clontech, USA). The cDNA of drought-stressed samples was used as a tester, while that of the control served as a driver. Both cDNAs were digested with Rsa1, which yields blunt ends, followed by phenol/chloroform-extraction, ethanol-precipitation and resuspension in water. The digested tester cDNA was subdivided into two equal portions and ligated separately with two different adaptors (adapter 1 and adapters 2R). Because specific genes to judge the ligation efficiency are not known, the kit human skeletal muscle control was simultaneously processed to assess the ligation efficiency. The ligation efficiency confirmation by the control DNA (human skeletal muscle) yielded ideal results based on the kit expected results (Supplementary Fig. 1). Once adaptor ligation has been confirmed, two rounds of hybridization were carried out to normalize and enrich the differentially expressed cDNAs according to the manufacturer’s protocol with the following changes: an excess of driver cDNA was added to each tester cDNA and heat denatured to generate cDNA that can be exponentially amplified using the two new adaptors. First, subtraction was performed by hybridizing the ligated tester and driver adapter-ligated cDNA. A second hybridization was then performed to equalize the high and low abundance cDNA in the tester by adding excess driver cDNA. The secondary differentially amplified PCR products were visualized on a 1.5% agarose gel run in 1X TAE buffer and stained in EtBr (Supplementary Fig. 2). Each subtraction was enriched for differentially expressed cDNA sequences. The subtraction results obtained were matched to the control subtracted sample as well as the kit expected results.
3.2. Sequencing and functional characterization of SSH cDNA sequences
The secondary PCR products were cloned into the pGEM easy vector, positive clones were identified, the plasmid was purified with mini-prep, and the DNA was sequenced. A total of 913 positive clones from two forward subtracted libraries were sequenced since they represent up-regulated genes in response to drought stress. The cleaned 789 ESTs were assembled into 222 unigenes (78 contigs and 144 singletons) after removing and trimming low quality as well as vector and adapter sequences using the codoncode aligner software (CodonCode Corporation, Dedham, MA, USA). The homology of these sequences with the gene bank nucleotide (Blastn) and protein (blastx) databases was assessed against nr/nt databases.
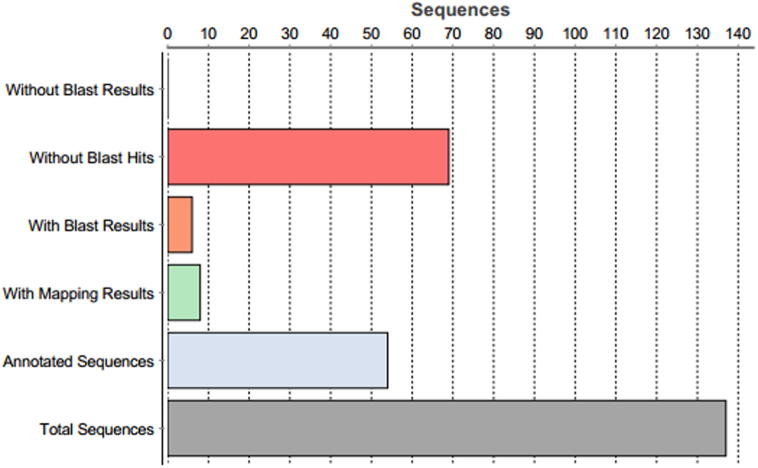
After an initial analysis, unigenes showing homology with mitochondrial and rRNA sequences were removed, and the remaining 137 unigenes were obtained with an average length of 450 bp and deposited in the dbEST division of gene bank (LIBEST_028448). These unigenes were further analyzed with Blast2Go (http://blast2go.org/). The Blast2go analysis of 137 unigenes revealed 68 unigenes with blast hits, 69 without blast hits, 8 with mapping results and 54 with annotated sequences (Fig. 1). The InterProScan results showed 84 sequences without InterProScan and 53 with InterProScan (Supplementary Fig. 3). A majority of blast match hits belonged to proteins from legumes species, with maximum hits from G. max, Cicer arietinum, Medicago truncatula, and Pisum sativum, (Supplementary Fig. 4). The annotated transcripts or unigenes were also mapped to nine different predicted KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (Supplementary Table 1). The predicted KEGG pathways included carbon fixation in photosynthetic organisms (five unigenes), glyoxylate and dicarboxylate metabolism (four unigenes), phenylalanine metabolism (one unigenes), glycolysis/gluconeogenesis (one unigene), starch, sucrose metabolism (one unigene), oxidative phosphorylation (one unigene) phosphatidylinositol signaling pathway (one unigene) phenylpropanoid biosynthesis (one unigene) and pentose and glucuronate interconversions (one unigene).
Figure 1.
Blastx statistics of 137 Vicia faba sequences analyzed by Blast2go.
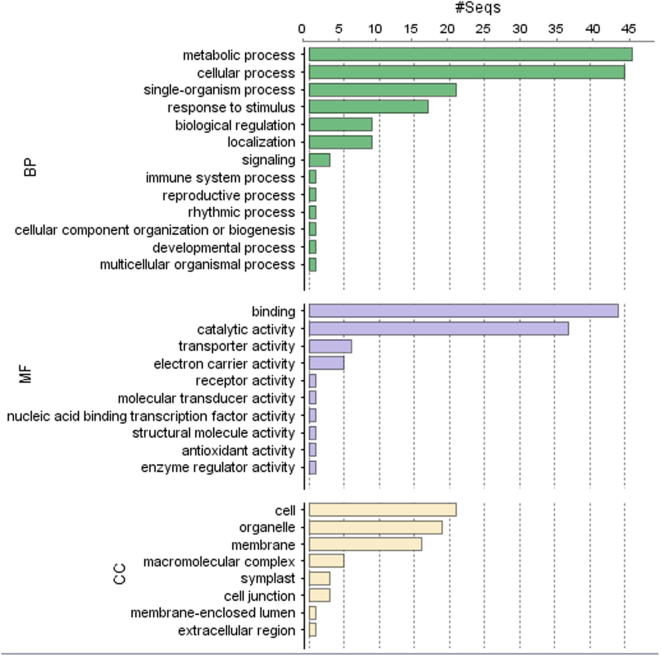
At the second GO level, the sequences were divided into three principal GO categories: biological process (BP), molecular function (MF) and cellular location (Fig. 2). Many of the ESTs in the biological process category (>30%) were linked to metabolic and cellular process, whereas the remaining ESTs were involved in biological regulation, cellular component organization, development process, localization, multicellular organismal process, signaling, single organism process and response to stimulus (Fig. 2 BP). Based on the molecular functional category, the majority of these ESTs were involved in binding and catalytic activities. Other ESTs were linked to antioxidants, transporters, receptors, nucleic acid binding transcription factors, molecular transducers, enzyme regulatory, and electron carrier activities (Fig. 2MF). ESTs were also grouped based on the cellular compartment (Fig. 2 CC), such as the cell (30%), membrane (23%), and organelle (28%) and macromolecular components (7%).
Figure 2.
Functional classifications of the 137 unigenes that were assigned to GO terms (second level GO terms). The three GO categories, biological process (BP), molecular function (MF), and cellular component (CC) are presented.
A number of potential drought responsive genes were detected based on the BLAST results (Table 1). Some of these genes included sequences with homology to the ribulose 1,5-bisphosphate carboxylase (rbcL) gene (KSU-FB-SSH-C18), zinc finger CCCH domain-containing protein (KSU-FB-SSH-C35), probable cyclic nucleotide-gated ion channel (KSU-FB-SSH-S53), polyubiquitin (KSU-FB-SSH-S10), H+-ATPase (KSU-FB-SSH-S79), calcium-dependent protein kinase (KSU-FB-SSH-S80) and putative respiratory burst oxidase-like protein C (KSU-FB-SSH-S83). The detailed descriptions of 137 unigenes obtained by Blast2Go are shown in Supplementary Table 2. The unidentified genes that did not yield a BLAST hit could be of particular interest as new genes and require further bioinformatic analysis to reveal their function.
Table 1.
Important drought-related unigenes identified in Vicia faba L. (Hassawi 2) under stress conditions.
| Unigene name | Putative function | Accession No. | Min. eValue |
|---|---|---|---|
| KSU-FB-SSH-S31 | ATP synthase beta subunit | JZ705344 | 4.08E−82 |
| KSU-FB-SSH-S80 | Calcium-dependent protein kinase sk5-like | JZ705302 | 2.10E−60 |
| KSU-FB-SSH-C42 | Cellular nucleic acid-binding protein | JZ705269 | 3.35E−24 |
| KSU-FB-SSH-S23 | Chlorophyll a-b binding protein | JZ705349 | 7.79E−52 |
| KSU-FB-SSH-S86 | Dehydration-responsive element binding protein 2 | JZ705296 | 3.16E−54 |
| KSU-FB-SSH-C11 | Gag pol polyprotein | JZ705292 | 4.72E−27 |
| KSU-FB-SSH-C34 | Gypsy retrotransposon integrase-like protein 1-like | JZ705275 | 1.76E−30 |
| KSU-FB-SSH-S50 | Hydroxypyruvate reductase isoform partial | JZ705329 | 1.48E−49 |
| KSU-FB-SSH-S84 | kda class ii heat shock | JZ705298 | 5.85E−38 |
| KSU-FB-SSH-S85 | Maturation-associated partial | JZ705297 | 1.20E−61 |
| KSU-FB-SSH-S58 | Non-ltr retroelement reverse related | JZ705323 | 5.33E−04 |
| KSU-FB-SSH-C13 | Photosystem ii 32 kda partial | JZ705291 | 2.59E−100 |
| KSU-FB-SSH-S79 | Plasma membrane H+ ATPase | JZ705303 | 1.26E−77 |
| KSU-FB-SSH-S81 | Plasma membrane intrinsic protein | JZ705301 | 8.74E−59 |
| KSU-FB-SSH-S10 | Polyubiquitin | JZ705361 | 2.62E−31 |
| KSU-FB-SSH-S82 | Potassium channel | JZ705300 | 3.80E−99 |
| KSU-FB-SSH-S53 | Probable cyclic nucleotide-gated ion channel 5-like isoform x1 | JZ705328 | 6.75E−97 |
| KSU-FB-SSH-S83 | Respiratory burst oxidase homolog protein a-like | JZ705299 | 2.70E−101 |
| KSU-FB-SSH-S44 | Retrotransposon unclassified | JZ705333 | 2.23E−27 |
| KSU-FB-SSH-S18 | Ribonuclease uk114-like | JZ705354 | 1.08E−51 |
| KSU-FB-SSH-C18 | Ribulose-bisphosphate carboxylase oxygenase large partial | JZ705289 | 7.65E−53 |
| KSU-FB-SSH-S87 | Transcription factor tga5-like | JZ705295 | 3.42E−68 |
| KSU-FB-SSH-C35 | Zinc finger c-x8-c-x5-c-x3-h type protein | JZ705274 | 9.15E−15 |
3.3. Expression profiling of selected potential candidate genes by RT-qPCR
Thirty-five potential genes were selected as potential candidate genes for faba bean drought tolerance based on the sequence analyses. Specific primers were designed (Supplementary Table 3) using BatchPrimer3 v1.0 (You et al., 2008). Conventional PCR was carried out to confirm the specificity of primers, and the primers showed single band of expected size on an agarose gel. To investigate whether these genes were linked to drought tolerance, RT-qPCR was carried out on the cDNA from the drought-tolerant Hassawi 2 genotype grown under drought stress. A melting curve analysis showed a single peak for each amplicon and that most of these selected ESTs were differentially expressed, proving their potential as drought tolerance candidates. The genomic DNA contamination of all RNA samples was tested by omitting the reverse transcriptase from the RT-qPCR reaction. These control reactions did not yield amplification products. No primer dimers or secondary PCR products were detected.
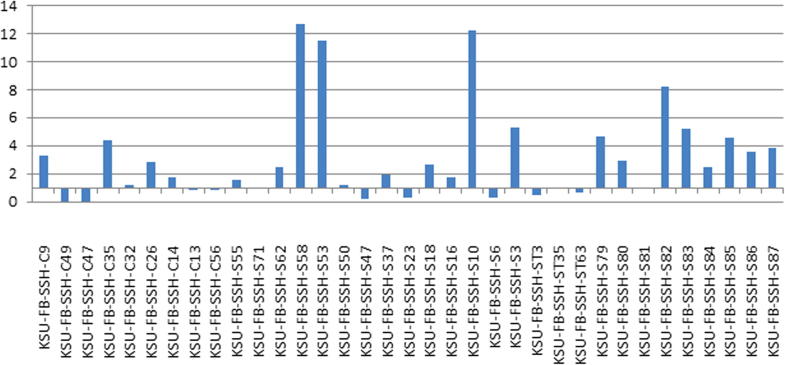
The results of the real-time PCR showed a wide range of expression levels of 35 selected unigenes according to their 2−ΔΔCt values (Fig. 3). The results indicated that the expression levels of 17 unigenes were higher in the Hassawi 2 genotype after drought stress, while the expression levels of 6 unigenes were decreased in the Hassawi 2 genotype after drought stress. However, the expression levels of the remaining genes (out of 35 selected ESTs) were slightly up-regulated, down-regulated or unchanged compared to the control. The unigenes KSU-FB-SSH-C9, KSU-FB-SSH-S79, KSU-FB-SSH-C35, KSU-FB-SSH-S58, KSU-FB-SSH-S53, KSU-FB-SSH-S10, KSU-FB-SSH-S3, KSU-FB-SSH-S80, KSU-FB-SSH-S82, KSU-FB-SSH-S83, KSU-FB-SSH-S85, KSU-FB-SSH-S86 and KSU-FB-SSH-S87 were highly up-regulated, and KSU-FB-SSH-C49, KSU-FB-SSH-C47, KSU-FB-SSH-S47, KSU-FB-SSH-S23, KSU-FB-SSH-S6, and KSU-FB-SSH-ST3 were down-regulated compared to the control. Based on the initial RT-qPCR screening of potential genes, 8 up-regulated unigenes were selected for further analysis to assess the responses of these genes across 38 selected faba bean genotypes. These unigenes were chosen because they are highly up-regulated compared to other unigenes in RT-qPCR in Hassawi 2 genotype and furthermore these unigenes were possibly associated with drought response. These unigenes were KSU-FB-SSH-C9, KSU-FB-SSH-S58, KSU-FB-SSH-S53, KSU-FB-SSH-S10, KSU-FB-SSH-S3, KSU-FB-SSH-S80, KSU-FB-SSH-S82, and KSU-FB-SSH-83. The expression pattern varied among the 38 genotypes (Table 2). A variable expression profile was generated for KSU-FB-SSH-C9. This gene was highly up-regulated in 10 genotypes and moderately down-regulated in 6 genotypes. KSU-FB-SSH-C9 was most up-regulated (fold difference >3.5) in the Hassawi 2 genotype, followed by the H5 genotype. This gene was moderately up-regulated (fold difference >2.5) in the Goff1, ILB4347 and X943, genotypes. However, in sensitive genotypes, such as Pakistani, Giza461, Misr3 and Giza 40, this gene was down-regulated (<0.5). The up-regulation of this unigene in majority of tolerant genotypes suggests that this gene may play a role in drought tolerance.
Figure 3.
Expression pattern of 35 selected unigenes against Hassawi 2 genotype of faba bean grown under stress conditions. The upper bars above 1 show the level of up-regulation in genotypes, while lower bars below 1 show the level of down-regulation. The bar at 1 indicates no differential expression.
Table 2.
Fold difference in expression level of target unigenes in different genotypes.
| Genotype/primers | Origin | KSU-FB-SSH-S53 | KSU-FB-SSH-S58 | KSU-FB-SSH-S3 | KSU-FB-SSH-S80 | KSU-FB-SSH-S83 | KSU-FB-SSH-S10 | KSU-FB-SSH-S82 | KSU-FB-SSH-C9 |
|---|---|---|---|---|---|---|---|---|---|
| Hassawi 1 | KSA | 5.22 | 11.35 | 5.22 | 1.57 | 5.2 | 6.53 | 4.3 | 2.5 |
| Hassawi 2 | KSA | 11.23 | 16.74 | 6.23 | 4.21 | 7.28 | 11.33 | 10.5 | 4 |
| Hassawi 3 | KSA | 3.22 | 5.21 | 4.69 | 1.5 | 2.11 | 7.22 | 4.11 | 2 |
| Luz | Spain | 3.08 | 2.78 | 2.56 | 0.66 | 4.94 | 5.59 | 0.76 | 0.53 |
| Goff1 | KSA | 3.12 | 21.09 | 10.17 | 2.13 | 4.1 | 6.23 | 4.21 | 3.11 |
| Pakistani | Pakistan | 0.28 | 11.91 | 1.1 | 1.94 | 0.26 | 1.35 | 4.33 | 0.22 |
| Gazira1 | Sudan | 3.19 | 17.9 | 8.55 | 1.97 | 2.38 | 0.18 | 1.72 | 0.53 |
| T.W. | Denmark | 6.61 | 3.04 | 5.23 | 3.54 | 2.58 | 10.19 | 6.49 | 2.11 |
| Yamani | Yamen | 1.11 | 3.73 | 1.6 | 3.11 | 0.44 | 4.84 | 3.12 | 1.5 |
| Aquadolce | Spain | 4.84 | 2.96 | 2.36 | 1.47 | 2.1 | 6.48 | 3.55 | 0.95 |
| Kamline | Spain | 1.97 | 5.72 | 6.57 | 1.41 | 1.59 | 0.04 | 2.55 | 1.34 |
| Gazira2 | Sudan | 3.53 | 7.88 | 1.17 | 4.32 | 0.46 | 3.12 | 3.07 | 2.23 |
| Nubaria2 | Egypt | 1.57 | 10.37 | 3.24 | 1.55 | 1.83 | 6.36 | 3.25 | 0.83 |
| Nubaria3 | Egypt | 3.16 | 3.1 | 4.99 | 0.75 | 4.88 | 8.15 | 10.91 | 1.33 |
| Sakha1 | Egypt | 1.43 | 15.84 | 6.57 | 1.78 | 2.31 | 9.49 | 0.94 | 1.15 |
| Sakha2 | Egypt | 1.69 | 12.22 | 0.39 | 3.82 | 1.13 | 0.01 | 9.32 | 0.72 |
| Giza3 | Egypt | 1.69 | 6.83 | 1.32 | 0.71 | 0.53 | 1.77 | 12.14 | 0.62 |
| Giza461 | Egypt | 0.86 | 0.98 | 0.45 | 0.24 | 0.49 | 1.02 | 16.5 | 0.09 |
| X957 | Egypt | 0.18 | 10.5 | 0.24 | 0.98 | 1.2 | 0.53 | 8.21 | 0.94 |
| Misr1 | Egypt | 3.31 | 12.03 | 2.53 | 2.01 | 5.24 | 5.42 | 9.33 | 0.43 |
| Misr3 | Egypt | 3.3 | 0.52 | 1.91 | 0.33 | 0.4 | 3.71 | 15.63 | 0.2 |
| 985/252/95 | Egypt | 3.3 | 8.29 | 3.87 | 2.1 | 2.99 | 14.77 | 5.73 | 1.25 |
| L.56 | KSA | 3.44 | 18.27 | 7.99 | 1.94 | 4.12 | 0.02 | 11.33 | 1.13 |
| Giza492 | Egypt | 2.69 | 20.41 | 6.91 | 3.19 | 6.73 | 13.28 | 10.09 | 0.86 |
| Pop.3 | Egypt | 7.21 | 11.96 | 4.13 | 0.82 | 4.03 | 22.26 | 6.04 | 2.33 |
| ILB4358 | ICARDA | 7.21 | 1.35 | 3.8 | 0.33 | 0.1 | 9.63 | 11.38 | 1.5 |
| ILB4347 | ICARDA | 3.41 | 11.33 | 1.87 | 0.51 | 1.64 | 5.45 | 3.87 | 2.59 |
| 1016/752/95 | Egypt | 2.69 | 15.35 | 8.76 | 0.76 | 9.23 | 6..23 | 7.45 | 2.34 |
| Sudan | Sudan | 10.62 | 1.07 | 1.65 | 0.15 | 0.7 | 0.05 | 11.4 | 0.54 |
| Giza402 | Egypt | 2.83 | 10.86 | 3.36 | 0.46 | 3.33 | 6.27 | 8.14 | 0.59 |
| Giza Blanka | Egypt | 1.99 | 4.08 | 2.43 | 0.45 | 1.32 | 3.11 | 8.31 | 1.53 |
| X1931 | Egypt | 2.8 | 0.39 | 4.75 | 0.33 | 3.49 | 9.03 | 6.03 | 0.99 |
| L.4 | KSA | 9.88 | 0.55 | 1.28 | 0.31 | 7.95 | 14.16 | 3.11 | 1.26 |
| Pop.4 | Egypt | 6.44 | 2.07 | 4.06 | 0.5 | 2.98 | 12.63 | 5.98 | 1.7 |
| Giza40 | Egypt | 0.9 | 0.05 | 0.75 | 0.04 | 0.1 | 1.38 | 4.98 | 0.4 |
| H5 | KSA | 4.8 | 8.42 | 3.27 | 1.12 | 0.8 | 6.23 | 3.5 | 3.58 |
| Sakha4 | Egypt | 8.04 | 6.94 | 4.61 | 1.44 | 1.87 | 8.8 | 1.32 | 1.7 |
| X943 | Egypt | 9.23 | 11.7 | 1.9 | 2 | 5.23 | 1.08 | 3.49 | 2.6 |
KSU-FB-SSH-S58, KSU-FB-SSH-S53, KSU-FB-SSH-S10 and KSU-FB-SSH-S82 (Table 2) were up-regulated in the majority of studied genotypes. However, the expression levels of these genes were up-regulated by more than 10-fold in the Hassawi 2 genotype. The high fold difference indicated that these genes may be involved in drought resistance. The expression of KSU-FB-SSH-S3 varied among the studied genotypes. Its expression was the highest (>10-fold compared to the control) in the Goff1 genotype. The other genotypes, Hassawi 2, Hassawi 1, Gazira 1, T.W., Kamline, Sakhai1, L.56, Giza 492, and 1016/752/95, expressed significant levels of this gene (>5-fold), while this gene was either slightly up-regulated or down-regulated in the remaining genotypes. The expression of KSU-FB-SSH-S80 was variable; it was up-regulated in 19 genotypes and down-regulated in most of the remaining genotypes. This gene was up-regulated more than 4-fold in the Hassawi 2 and Gazira 2 genotypes compared with Hassawi 2 control, while it was down-regulated in the following genotypes: Pop.3, ILB4358, ILB4347, 1016/752/95, Sudani, Giza 402, Giza Blanka, X1931, L.4, Pop.4, Giza 40, Sakha4 and X943. The expression profile of KSU-FB-SSH-S83 was similar to that of most other genes, except in a few genotypes in which it was down-regulated. It was highly expressed (>7-fold) in Hassawi 2, L.4 and 1016/752/95, and moderately expressed (<5-fold) in Hassawi 1, Giza 492, Misr 1, and X943. It was slightly down-regulated in only a few genotypes.
4. Discussion
Changes in gene expression are associated with a wide range of biological and pathological processes. The identification of differentially expressed genes has been crucial to better understand the molecular and cellular mechanisms underlying stress tolerance (Ghorbel and Murphy, 2011). Suppression Subtractive Hybridization (SSH) has been developed to identify differentially expressed genes in many legume crops including pigeon pea under water stress (Kumar et al., 2015). The results demonstrate the power of the SSH strategy to identify differentially expressed cDNAs. A high proportion of identified genes were novel with no significant homology to any genes in the gene bank databases, suggesting that novel genes/mechanisms are active in the stress tolerance of the faba bean. These genes are of a special interest and require further analysis to predict the protein sequence architecture and their possible functions. However, a large number of subtracted genes were drought-responsive, and some were potential real candidate genes. These genes were involved in the transporter, molecular transducer, antioxidant, binding, catalytic, and electron carrier activities that are believed to be induced by drought.
KSU-FB-SSH-S79 belongs to the p-type H+-ATPase family, which plays an important role in generating electrochemical gradients that drive solute transport and water flux. The expression of H+ATPase is crucial for signal transduction in response to external environmental stimuli (Hentzen et al., 1996). Multiple ATPase gene families are reportedly also expressed in A. thaliana (Harper et al., 1989, Pardo and Serrano, 1989, Houlne and Boutry, 1994, Nakajima et al., 1995). Therefore, the differential expression of this gene could suggest that it plays a strong role in attenuating drought stress. Another potential candidate gene is the calcium-dependent protein kinase 1 (KSU-FB-SSH-S80), which is expressed in guard cells as a result of excess cellular calcium. This protein plays an important role in abscisic acid (ABA)-mediated stomatal closure (Schroeder et al., 2001, Ramanjulu and Bartels, 2002). The modulation of the Ca2+ level initiates a protein phosphorylation cascade that ultimately targets proteins involved in the transcriptional activation of stress-regulated genes (Xiong et al., 2002). KSU-FB-SSH-ST50 and KSU-FB-SSH-S53 are probable cyclic nucleotide-gated ion channel 5-like isoforms with voltage-gated potassium channel activity.
Another identified EST was KSU-FB-SSH-S81, a homolog of aquaporin, which is part of the stress-protecting protein family that facilitates water uptake by forming pores. Thus, this gene plays an important role in cell growth and photosynthesis activity after dehydration (Oono et al., 2003). This protein is also reportedly differentially accumulated in drought-resistant plants (Montalvo-Hernandez et al., 2008). Potassium channel proteins are also important to maintain osmolality in response to drought. The homolog to the potassium channel identified in this study was KSU-FB-SSH-S82. A heat shock protein (KSU-FB-SSH-S84) was also differentially accumulated in this study. These proteins are widely distributed in nature and involved in protein re-folding and assembly; drought and salinity stresses reportedly induce their expression (Alamillo et al., 1995, Campalans et al., 2001), and they also assist plant recovery after abiotic stress. Another potential drought resistance gene identified in this study was putative respiratory burst oxidase-like protein C (KSU-FB-SSH-S83), which encodes NADPH oxidase. Drought or dehydration leads to the production of reactive oxygen species (ROS), such as singlet oxygen, superoxide, hydrogen peroxide, and hydroxyl radicals. Thus, the photosynthetic activity is reduced because ROS are mainly produced in chloroplast. The induction of enzymes that lead to antioxidant activity helps protect plants from ROS-induced damage. Therefore, putative respiratory burst oxidase-like protein C plays an important role in neutralizing ROS.
Dehydrin-like protein homolog (KSU-FB-SSH-S85), which belongs to the late embryogenesis abundant (LEA) protein class, was also identified. Various studies showed that LEA accumulates in vegetative plants during drought periods prevent plant desiccation (Moons et al., 1997). These proteins act as molecular chaperones for protein structure maintenance. Dehydrin proteins have already been identified as water stress-induced proteins in many studies (Close, 1997, Cellier et al., 1998). The two transcription factor homologs identified in this study were KSU-FB-SSH-S87 (bZIP 132) and drought-responsive element binding protein 2 (DREB2), (KSU-FB-SSH-S86). These ESTs may play a role in the activation of downstream genes via a signal transduction cascade following stress exposure. Several transcription factors are reportedly induced by stress (Bartels and Sunkars, 2005, Abe et al., 1997, Ashraf and Foolad, 2007, Ashraf, 2010). Some other stress-induced genes in Hassawi 2 included KSU-FB-SSH-S10, homologous to ubiquitin 11, which is involved in the cellular protein modification process. Some identified unigenes represent genes involved in photosynthesis, such as ribulose 1,5-bisphosphate carboxylase/oxygenase larger chain, chlorophyll a/b binding protein, and photosystem reaction proteins. Quantitative PCR also confirmed their abundance under drought stress conditions.
KSU-FB-SSH-S9 and KSU-FB-SSH-ST59 were homologous to ribulose bisphosphate carboxylase oxygenase. KSU-FB-SSH-C35 is the zinc finger CCCH domain-containing protein. KSU-FB-SSH-C14 is the ATP-dependent zinc metalloprotease chloroplastic-like protein. The results clearly suggest the differential expression of a potential signal transduction cascade that triggers the activation of drought tolerance genes in the faba bean.
The identified gene classes include the principal components of signal transduction; transmembrane proteins that transmit stimuli inside the cell (KSU-FB-SSH-S79, KSU-FB-SSH-C81), kinases that activate proteins inside the plant cell (KSU-FB-SSH-S80), transcription factor/activators that increase the transcription of drought genes (KSU-FB-SSH-S87, KSU-FB-SSH-C42, and KSU-FB-SSH-S86) and some stress genes, such as KSU-FB-SSH-S82, KSU-FB-SSH-S53 and KSU-FB-SSH-C18 (Table S2). This finding provides an overview of a possible signal transduction cascade in response to drought stress in faba plants. Further analysis is required to confirm these results.
Recently, some studies reportedly utilized the SSH strategy to identify differentially expressed genes in some legume species, including drought-responsive genes in the pigeon pea, with similar results (Qiao et al., 2012). The BLAST analysis yielded a total of 182 uniquely expressed sequence tags (ESTs), among which 142 (78%) exhibited high homology to previously identified or putative proteins; however, 40 (22%) were not homologous to any genes in the database. These studies further confirmed that 35 differentially expressed genes were involved in drought stress tolerance. The differential transcript accumulation in the chickpea during the early phases of compatible interaction with a necrotrophic fungus, Ascochyta rabie, has also been studied (Jaiswal et al., 2012). Differentially expressed genes during seed development in Phaseolus vulgaris (Abid et al., 2012) included 72 unique ESTs, 12 of which were identified based on their redundancy.
The expression patterns of thirty-five selected unigenes in response to drought conditions were analyzed by RT-qPCR. The RT-qPCR results confirmed that more than 65% of the selected drought-responsive genes (24 of the 35) were drought-inducible across faba bean genotypes. Eight of these highly up-regulation unigenes were selected, and their relative expression was analyzed across 38 selected faba bean genotypes. The expression patterns of these genes were variable across the 38 genotypes (Table 2). Overall KSU-FB-SSH-C9, KSU-FB-SSH-S58, KSU-FB-SSH-S53, KSU-FB-SSH-S10, KSU-FB-SSH-S3, KSU-FB-SSH-S82, KSU-FB-SSH-S80 and KSU-FB-SSH-83 were up-regulated; these genes were significantly up-regulated in most genotypes, although they were down-regulated in some genotypes. In general, the results obtained here are coherent with results obtained by Kumar et al. (2015) on pigeon pea and Abid et al. (2015) on faba bean under drought induced conditions. Furthermore, current study reported many novel genes giving no significant hits to nucleotide databases, indicating their possible significance in a new/modified tolerance mechanism(s) that needs to be further analyzed. In addition, many of the reported genes also exhibited drought related functions and could be further utilized in breeding for drought tolerance in faba bean.
5. Conclusion
Results showed that 24 potential genes in the faba bean were potentially associated with drought stress. The predicted function of some genes highlights their significance in drought tolerance. The differential expression of some drought-responsive genes was confirmed by real-time RT-qPCR. Furthermore, several novel genes with unknown functions that may play a potential role in drought tolerance were identified, and these roles need to be further analyzed. These novel genes may reveal a new mechanism(s) that may also be active in faba beans. Therefore, this study sets the landmark for drought-related genes in the faba bean for further functional analyses and may be useful for improving drought tolerance in faba bean.
Acknowledgment
This work was supported by NSTIP strategic technologies program number (BIO680-02) in the Kingdom of Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.sjbs.2016.05.011.
Appendix A. Supplementary data
References
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosakawa D., Shinozaki K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abid G., Sassi K., Muhovski Y., Jacquemin J.M., Mingeot D., Tarchoun N., Baudoin J.P. Identification and analysis of differentially expressed genes during seed development using suppression subtractive hybridization (SSH) in Phaseolus vulgaris. Plant Mol. Biol. Rep. 2012;30:719–730. [Google Scholar]
- Abid G., Muhovski Y., Mingeot D., Watillon B., Toussaint A., Mergeai G., M’hamdi M., Sassi K., Jebara M. Identification and characterization of drought stress responsive genes in faba bean (Vicia faba L.) by suppression subtractive hybridization. Plant Cell Tissue Org. 2015;121:367–379. [Google Scholar]
- Alamillo J., Almoguera C., Bartels D., Jordano J. Constitutive expression of small heat shock proteins in vegetative tissues of the resurrection plant Craterostigma plantagineum. Plant Mol. Biol. 1995;29:1093–1099. doi: 10.1007/BF00014981. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ammar M.H., Anwar F., El-Harty E.H., Migdadi H.M., Abdelkhalik S.M., Alghamdi S.S. Physiological and yield responses of faba bean (Vicia faba L.) to drought stress. J. Agron. Crop Sci. 2014;201:280–287. [Google Scholar]
- Ashraf M. Inducing drought tolerance in plants, recent advances. Biotechnol. Adv. 2010;28:169–183. doi: 10.1016/j.biotechadv.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Ashraf M., Foolad M.R. Role of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007;59:206–216. [Google Scholar]
- Barros de Carvalho G.A., Batista J.S., Marcelino-Guimarães F.C., Nascimento L.C., Hungria M. Transcriptional analysis of genes involved in nodulation in soybean roots inoculated with Bradyrhizobium japonicum strain CPAC15. BMC Genomics. 2013;14:153–163. doi: 10.1186/1471-2164-14-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D., Sunkars R. Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 2005;24:23–58. [Google Scholar]
- Campalans A., Pages M., Messeguer R. Identification of differentially expressed genes by the cDNA-AFLP technique during dehydration of almond. Tree Physiol. 2001;21:633–643. doi: 10.1093/treephys/21.10.633. [DOI] [PubMed] [Google Scholar]
- Cellier F., Conéjéro G., Breitler J.C., Casse F. Molecular and physiological responses to water deficit in drought tolerant and drought sensitive sunflower lines Helianthus annuus L., accumulation of dehydrin transcripts correlates with tolerance. Plant Physiol. 1998;116:319–328. doi: 10.1104/pp.116.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T.J. Dehydrins, a commonalty in the response of plants to dehydration and low temperature. Physiol. Plant. 1997;100:291–296. [Google Scholar]
- Conesa A., Gotz S., Garcia-Gome J.M., Terol J., Talon M. Blast2GO, a universal tool for annotation. Visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Diatchenko L., Lau Y.F.C., Campbell A.P., Chenchik A., Moqadam F., Huang B., Lukyanov S., Lukyanov K., NadyaGurskaya N., Eugene D.S., Siebert P.D. Suppression subtractive hybridization, a method for generating differentially regulated or tissue specific cDNA probes and libraries. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drame K.N., Clavel D., Repillin A., Passaquet C., Fodel Y.Z. Water deficit induces variation in expression of stress-responsive genes in two peanut Arachis hypogaea L. cultivars with different tolerance to drought. Plant Physiol. Biochem. 2007;45:236–243. doi: 10.1016/j.plaphy.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Ghorbel M.T., Murphy D. Suppression subtractive hybridization. In: Adalberto Merighi., editor. Neuropeptides, Methods and Protocols. 2011. pp. 237–259. (Methods Mol. Biol.). [DOI] [PubMed] [Google Scholar]
- Gotz S., Garcia-Gomez J.M., Terol J., Williams T.D., Nagaraj S.H. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Zhang L., Zhao J., Liao H., Zhuang C., Yan X. Identification of temporally and spatially phosphate-starvation responsive genes in Glycine max. Plant Sci. 2008;175:574–584. [Google Scholar]
- Harper J.F., Surowy T.K., Sussman M.R. Molecular cloning and sequence of cDNA encoding the plasma membrane proton pump H+-ATPase of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1234–1238. doi: 10.1073/pnas.86.4.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M.A., Clark J.I., Ireland A., Lomax J., Ashburner M. The gene ontology GO project. Nucleic Acids Res. 2006;34:D322–326. [Google Scholar]
- Hatfield J.L., Sauer T.J., Prueger J.H. Managing soils to achieve greater water use efficiency, a review. Agron. J. 2001;93:271–280. [Google Scholar]
- Hentzen A.E., Smart L.B., Wimmers L.E., Fang H.H., Schroeder J.I., Bennett A.B. Two plasma membrane H+-ATPase genes expressed in guard cells of Vicia faba are also expressed throughout the plant. Plant Cell Physiol. 1996;37:650–659. doi: 10.1093/oxfordjournals.pcp.a028994. [DOI] [PubMed] [Google Scholar]
- Houde M., Belcaid M., Ouellet F., Danyluk J., Monroy A.F., Dryanova A., Gulick P., Bergeron A., Laroche A., Links M.G., MacCarthy L., Crosby W.L., Sarhan F. Wheat EST resources for functional genomics of abiotic stress. BMC Genomics. 2006;7:149–170. doi: 10.1186/1471-2164-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlne G., Boutry M. Identification of an Arabidopsis thaliana gene encoding a plasma membrane H+-ATPase whose expression is restricted to anther tissues. Plant J. 1994;5:311–317. doi: 10.1111/j.1365-313x.1994.00311.x. [DOI] [PubMed] [Google Scholar]
- Jaiswal P., Cheruku J.R., Kumar K., Yadav S., Singh A., Kumari P., Dube S.C., Upadhyaya K.C., Verma P.K. Differential transcript accumulation in chickpea during early phases of compatible interaction with a necrotrophic fungus Ascochyta rabiei. Mol. Biol. Rep. 2012;39:4635–4646. doi: 10.1007/s11033-011-1255-7. [DOI] [PubMed] [Google Scholar]
- Kumar R.R., Yadav S., Shrinivas D., Kumar Srivastava A., Shitole V., Naik G.R. Transcriptome of pigeonpea roots under water deficit analyzed by suppression subtractive hybridization. J. Agric. Sci. Tech. 2015;17:1333–1345. [Google Scholar]
- Link W., Dixkens C., Singh M., Schwall M., Melchinger A.E. Genetic diversity in European and Mediterranean Faba bean germplasm revealed by RAPD markers. Theor. Appl. Genet. 1995;90:27–32. doi: 10.1007/BF00220992. [DOI] [PubMed] [Google Scholar]
- Liu J., Liu J., Yuan Z., Qian X., Qian M., Yang Isolation and identification of genes expressed differentially in rice inflorescence meristem with suppression subtractive hybridization. Chin. Sci. Bull. 2001;46:98–101. [Google Scholar]
- Ministry of Agriculture . Ministry of Agriculture KSA; 2012. Annual Statistical Book. [Google Scholar]
- Misson J., Raghothama K.G., Jain A., Jouhet J., Block M.A., Bligny R., Ortet P., Creff A., Somerville S., Rolland N., Doumas P., Nacry P., Herrerra-estrella L., Nussaume L., Thibaud M.C. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvo-Hernandez L., Piedra-Ibarra E., Gomez-Silva L., Lira-Carmona R., Acosta-Gallegos J.A., Vazquez-Medrano J., Xoconostle-CazaRes B., Ruiz-Medrano R. Differential accumulation of mRNAs in drought-tolerant and susceptible common bean cultivars in response to water deficit. New Phytol. 2008;177:102–113. doi: 10.1111/j.1469-8137.2007.02247.x. [DOI] [PubMed] [Google Scholar]
- Moons A., De Keyser A., Van Montagu M. A group 3 LEA cDNA of rice responsive to abscisic acid but not to jasmonic acid shows variety-specifc differences in salt stress response. Gene. 1997;191:197–204. doi: 10.1016/s0378-1119(97)00059-0. [DOI] [PubMed] [Google Scholar]
- Morcuende R., Bari R., Gibon Y., Zheng W.M., Pant B.D., Blasing O., Usadel B., Czechowski T., Udvardi M.K., Stitt M., Scheible W.R. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007;30:85–112. doi: 10.1111/j.1365-3040.2006.01608.x. [DOI] [PubMed] [Google Scholar]
- Muller R., Morant M., Jarmer H., Nilsson L., Nielsen T.H. Genome wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 2007;143:156–171. doi: 10.1104/pp.106.090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima N., Saji H., Aono M., Kondo N. Isolation of cDNA for a plasma membrane H+-ATPase from guard cells of Vicia faba L. Plant Cell Physiol. 1995;36:919–924. doi: 10.1093/oxfordjournals.pcp.a078839. [DOI] [PubMed] [Google Scholar]
- Ogata H., Goto S., Sato K., Fujibuchi W., Bono H. KEGG, Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y., Seki M., Nanjo T., Narusaka M., Fujita M., Satoh R., Satou M., Sakurai T., Ishida J., Akiyama K. Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca 7000 full-length cDNA microarray. Plant J. 2003;34:868–887. doi: 10.1046/j.1365-313x.2003.01774.x. [DOI] [PubMed] [Google Scholar]
- Pardo J.M., Serrano R. Structure of a plasma membrane H+-ATPase gene from the plant Arabidopsis thaliana. J. Biol. Chem. 1989;264:8551–8562. [PubMed] [Google Scholar]
- Qiao G., Wen X., Yu L., Ji X. Identification of differentially expressed genes preferably related to drought response in pigeon pea Cajanus cajan inoculated by arbuscular mycorrhizae fungi AMF. Acta Physiol. Plant. 2012;34:1711–1721. [Google Scholar]
- Raina S.N., Rees H. DNA variation between and within chromosome complements of Vicia species. Heredity. 1983;51:335–346. [Google Scholar]
- Ramanjulu S., Bartels D. Drought- and desiccation-induced modulation of gene expression in plants. Plant Cell Environ. 2002;25:141–151. doi: 10.1046/j.0016-8025.2001.00764.x. [DOI] [PubMed] [Google Scholar]
- Sahebi M., Hanafi M.M., Azizi P., Hakim A., Ashkani S., Abiri R. Suppression subtractive hybridization versus next-generation sequencing in plant genetic engineering: challenges and perspectives. Mol. Biotechnol. 2015;57(10):880–903. doi: 10.1007/s12033-015-9884-z. [DOI] [PubMed] [Google Scholar]
- Schroeder J.I., Kwak J.M., Allen G.J. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature. 2001;410:327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- Tian J., Venkatachalam P., Liao H., Yan X.L., Raghothama K.G. Molecular cloning and characterization of phosphorus starvation responsive genes in common bean Phaseolus vulgaris L. Planta. 2007;227:151–165. doi: 10.1007/s00425-007-0603-2. [DOI] [PubMed] [Google Scholar]
- Toker C., Canci H., Yildirim T. Evaluation of perennial wild Cicer species for drought resistance. Genet. Res. Crop Evol. 2007;54:1781–1786. [Google Scholar]
- Torres A.M., Weeden N.F., Martın A. Linkage among isozyme, RFLP and RAPD markers in Vicia faba. Theor. Appl. Genet. 1993;85:937–945. doi: 10.1007/BF00215032. [DOI] [PubMed] [Google Scholar]
- Van de Ven W.T.G., Waugh R., Duncan N., Ramsay G., Dow N., Powell W. Development of a genetic linkage map in Vicia faba using molecular and biochemical techniques. Aspects Appl. Biol. 1991;27:49–54. [Google Scholar]
- Wasaki J., Yonetani R., Kuroda S., Shinano T., Yasaki J., Fujii F., Shimbo K., Yamamoto K., Sakata K., Sasaki T., Kishimoto N., Kikuchi S., Yamagishi M., Osaki M. Transcriptome analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ. 2003;26:1515–1523. [Google Scholar]
- Wu P.L., Ma X., Hou M., Wang Y., Wu F., Liu X., Deng W. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 2003;132:1260–1271. doi: 10.1104/pp.103.021022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Schumaker K.S., Zhu J.K. Cell signaling during cold, drought and salt stress. Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You F.M., Huo N., Yong Q.G., Luo M.C., Ma Q., Hane D., Lazo G.R., Dvorak J., Anderson O.D. BatchPrimer3, a high throughput web application for PCR and sequencing primer designing. BMC Bioinf. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.