Abstract
In biology field, the ontology application relates to a large amount of genetic information and chemical information of molecular structure, which makes knowledge of ontology concepts convey much information. Therefore, in mathematical notation, the dimension of vector which corresponds to the ontology concept is often very large, and thus improves the higher requirements of ontology algorithm. Under this background, we consider the designing of ontology sparse vector algorithm and application in biology. In this paper, using knowledge of marginal likelihood and marginal distribution, the optimized strategy of marginal based ontology sparse vector learning algorithm is presented. Finally, the new algorithm is applied to gene ontology and plant ontology to verify its efficiency.
Keywords: Ontology, Similarity measure, Sparse vector, Margin
1. Introduction
The term “Ontology” refers to a knowledge representation and conceptual shared model. It is widely used in gene computing, knowledge management and information retrieval, which also witnesses its effectiveness in the various applications. Besides, the concept semantic model was welcomed and borrowed by scholars in social science, medical science, biology science, pharmacology science and geography science (for instance, see Gregor et al., 2016, Kaminski et al., 2016, Forsati and Shamsfard, 2016, Pesaranghader et al., 2016, Huntley et al., 2016, Brown et al., 2016, Palmer et al., 2016, Terblanche and Wongthongtham, 2016, Farid et al., 2016, Carmen Suarez-Figuero et al., 2016).
Traditionally, we take ontology model as a graph , where each vertex v in the ontology graph G represents a concept and each edge of it represents a relationship between concepts and . A few years ago, ontology similarity-based technologies were quite popular among researchers due to its wide range of applications. For instance, GO-WAR algorithm was raised by Agapito et al. (2016) to explore cross-ontology association rules in which GO terms present in its sub-ontologies of GO. What’s more, after mining publicly available GO annotated datasets which show how GO-WAR outperforms current state of the art approaches, a deep performance evaluation of GO-WAR was discovered. Chicco and Masseroli (2016) put forward a computational pipeline which can predict novel ontology-based gene functional annotations by means of various semantic and machine learning methods. Then, in order to categorize the predicted annotations by their likelihood of being correct, a new semantic prioritization rule was achieved in their papers. The definition of GO ontological terms, molecular function, biological process and cellular components were given in detail by Umadevi et al. (2016), and he also found the relations to that of the disease genes with p-value < 0.05. Bajenaru et al. (2016) raised the constituent parts and architecture of the proposed ontology-based e-learning system and a framework for its application in reality. However, based on the OWL2 rules and the reasoning process of the OntoDiabetic system, Sherimon and Krishnan (2016) shifted his attention to the modeling and implementation of clinical guidelines. In terms of fuzzy logic, Bobillo and Straccia (2016) extended it to classical ontologies. With the help of pre-existing information about ontologies, such as terminology and ontology structure, Trokanas and Cecelja (2016) worked out a framework for evaluation of ontology for reuse to calculate a compatibility metric of ontology suitability for reuse and hence integration. To illustrate, the framework was explained in a Chemical and Process Engineering perspective. With the aim to allow users to quickly compute, manipulate and explore Gene Ontology (GO) semantic similarity measures, Mazandu et al. (2016) proposed A-DaGO-Fun. Auffeves and Grangier (2016) raised a new quantum ontology to make usual quantum mechanics fully compatible with physical realism. Hence, the physical properties in the ontology are attributed jointly to the system. In addition, Hoyle and Brass (2016) defined a statistical mechanical theory which expresses the process of annotating an object with terms selected from an ontology.
With the consideration of ontology similarity measure and ontology mapping, some effective learning tricks turn out to work well. With the harmonic analysis and diffusion regularization on hypergraph, Gao et al. (2013) proposed a new ontology mapping algorithm. Gao and Shi (2013) raised a novel ontology similarity computation technology considering operational cost in the real applications. Using ADAL trick, an ontology sparse vector learning algorithm was worked out by Gao et al. (2015) to make contributions to the ontology similarity measuring and ontology mapping. Then, Gao et al. (2016) proposed an ontology optimization tactics using distance calculating and learning. Several theoretical analysis of ontology algorithm mentioned but not defined in detail in this paper can refer to Gao et al., 2012, Gao and Xu, 2013, and Gao and Zhu (2014).
The marginal based ontology algorithm for ontology similarity computation and ontology mapping are given in the paper. By means of the sparse vector, the ontology graph is mapped into a real line and vertices into real numbers. Then, based on the difference between their corresponding real numbers, the similarity between vertices is measured. The rest of the paper is structured like this: the notations and setting are presented in Section 2; the ontology sparse vector optimization algorithm is raised in Section 3, in addition, the technologies to tackle the details in algorithm are also included here; and, the experiments on gene science and plant science are taken to show the efficiency of the algorithm in the last section.
2. Setting
Let V represent an instance space. Concerning each vertex in ontology graph, a p dimension vector represents the information: its name, instance, attribute and structure, and semantic information of the concept. All the information is related to the vertex and that is contained in name and attributes components of its vector. Let v = {v1, … , vp} be a vector that corresponds to a vertex v. To make the representation, clearer and further, we take a try to confuse the notations. So we consider using v to denote both the ontology vertex and its corresponding vector. The ontology learning algorithms are set to obtain an optimal ontology (score) function f: , and the similarity between two vertices is determined by the difference between two corresponding real numbers. The core of this algorithm is dimensionality reduction, i.e., choosing one dimension vector to express p dimension vector. Specifically, an ontology function f is a dimensionality reduction function f: .
In the real application, one sparse ontology function is expressed by
| (1) |
Here is a sparse vector and is a noise term. The sparse vector is used to decrease the components that is not necessary to zero. Then, we learn the sparse vector , so that we can determine the ontology function f.
The general versions for learning is learned in the paper. Let be a sample set with n vertex, be the matrix of n samples such that each sample vertex lies in a p dimension space, and be the vector of outputs of the these n sample vertex. Hence, the regression function Eq. (1) can be expressed as the linear model:
| (2) |
where is the n dimension vector for noise which are normally distributed, with variance .
An estimate of the sparse vector is obtained from the general regression obtains after solving the optimization problem below:
| (3) |
where is the loss term, is the -norm balance term that measures the sparseness of vector , and is the balance parameter which controls the sparsity level. On the selection of the balance parameter , readers can refer to Mancinelli et al., 2013, Mukhopadhyay and Bhattacharya, 2013, Ishibuchi and Nojima, 2013, and Varmuza et al. (2014) for more details about the method of cross-validation.
3. Ontology algorithm describing
In our paper, we consider the special case of Eq. (3), and it can be stated as
| (4) |
where .
We suppose the normal likelihood have independent priors about the ontology coefficients of the expression , , and a representative conjugate prior have the error precision . It’s clear to those who fully grasp the criterion Bayesian patterns in which the exponential power class prior can’t be a conjugate prior with the normal likelihood for .
Let be a normal probability distribution function with mean and variance , where is denoted as the density of stable distribution of index . Then, the above mentioned class of distributions can be formulated as
| (5) |
Let be the standard Gamma function. In terms of placing the independent normal priors on the ontology coefficients and considering as the hyperprior on , we infer
| (6) |
Our ontology framework will be presented as follows and then approximate marginal distributions for ontology parameters can be obtained, too. The marginal likelihood of the given ontology data in Eq. (4) or in many other non-trivial patterns can’t be yielded analytically. But the integral can easily be approximated in which the marginal likelihood conditional on and can be decomposed. Set . For fixed and , we deduce
| (7) |
where denote the lower bound on the marginal likelihood and is the Kullback–Leibler divergence between two distributions. Since is a strict non-negative function which equals to zero if and only if , the first term in Eq. (7) forms the lower bond of . Assume be the approximation of the posterior density . We have .
Let be the subvector of and be the expectation with respect to distributions with . We consider the factorized expression
| (8) |
In terms of maximizing the lower bound with respect to , we infer
| (9) |
By virtue of the above mentioned normal mixture expression of the exponential power distribution and the solution obtained from Eq. (9), the error variance and the approximate marginal posterior distributions of ontology coefficients can be determined. However, since the mixing distribution is unknown, the explicit expression for can’t be calculated. Fortunately, using the value of , the expression of can be deduced.
Let
and
The approximate marginal posterior distributions of the ontology coefficients and the error precision can be stated as follows:
| (10) |
| (11) |
According to the fact that we don’t get an available explicit form for its approximate distribution, these above presented moments are clear except for . We infer , , and . In view of (9), we can express the approximate marginal distribution of as
| (12) |
In terms of Eq. (6), evaluated at is the normalizing constant for this term. Moreover, we deduce the following fact by derivation of both sides of Eq. (6) with respect to and evaluate it again at ,
| (13) |
Then, by normalization operation, we have
| (14) |
From what we discussed above, we conclude the following facts:
Based on simplifications, we have
| (15) |
Finally, the iterative produce for ontology coefficients can be stated as
| (16) |
where denotes the expectation determined at the k-th iteration.
4. Simulation studies
In this section, two simulation experiments related to ontology similarity measure are presented. In order to be close to the setting of ontology algorithm, we choose a vector with p dimension to express each vertex’s information. All the information of name, instance, attribute and structure of vertex is contained in the vector. Here the instance of vertex means the set of its reachable vertex in the directed (or, undirected) ontology graph.
In order to make comparisons more accurate, the main algorithm runs in C++, in view of available LAPACK and BLAS libraries for linear algebra computations. All experiments are taken on a double-core CPU with memory of 8 GB.
4.1. Experiment on biology data
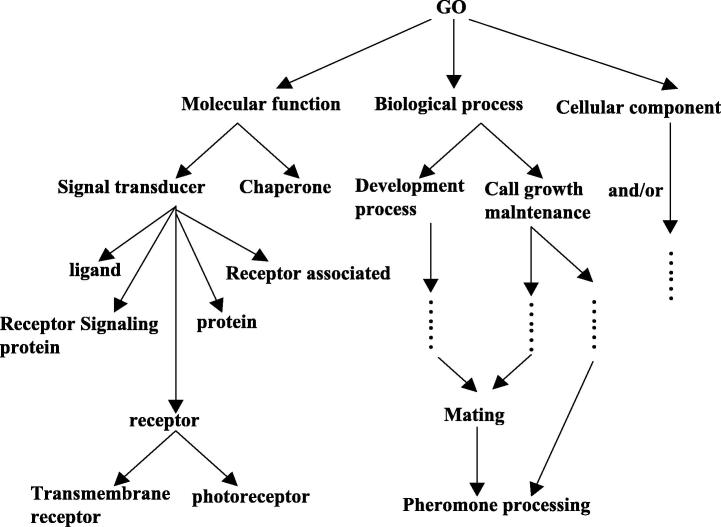
In the first experiment, we choose “Go” ontology O1 whose basic structure can be shown in http://www.geneontology.org (Fig. 1 shows the basic structure of O1). P@N (Precision Ratio, see Craswell and Hawking (2003) for more detail) is traditionally used to judge the equality of the experiment, and we also choose it for its efficiency. At first, the experts give the closest N concepts for every vertex on the ontology graph. Then using the algorithm, we compute the precision ratio, so that we can get the first N concepts for every vertex on ontology graph. Ontology algorithms in Gao et al., 2013, Gao et al., 2016, Gao and Shi, 2013 are also applied into “Go” ontology. At last, the precision ratio obtained from the four methods is gotten and given in Table 1.
Figure 1.
“Go” ontology.
Table 1.
Experiment data for ontology similarity measure.
| P@3 average precision ratio | P@5 average precision ratio | P@10 average precision ratio | P@20 average precision ratio | |
|---|---|---|---|---|
| Our Algorithm | 56.49% | 68.27% | 81.24% | 93.71% |
| Algorithm in Gao et al. (2013) | 56.46% | 67.72% | 78.38% | 79.39% |
| Algorithm in Gao and Shi (2013) | 56.44% | 65.73% | 78.39% | 89.72% |
| Algorithm in Gao et al. (2016) | 49.87% | 63.64% | 76.02% | 85.46% |
From the data in Table 1, we can find that when N = 3, 5, 10 or 20, the precision ratio obtained from our algorithm is higher than that obtained by algorithms proposed in Gao et al., 2013, Gao et al., 2016, Gao and Shi, 2013. Particularly, such precision ratios are increasing apparently with N increasing. Thus, our algorithm is better than the method presented by Gao et al., 2013, Gao et al., 2016, Gao and Shi, 2013.
4.2. Experiment on plant data
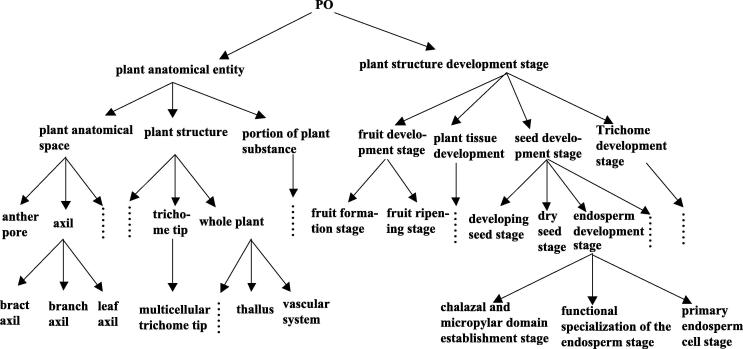
In this subsection, we use “PO” ontology O2, whose structure is presented in http://www.plantontology.org. (Fig. 2 shows the basic structure of O2), to check the efficiency of our new algorithm in ontology similarity measuring. Similarly, we use the P@N again for this experiment. Moreover, the ontology methods in Gao et al., 2013, Gao et al., 2016, Gao and Shi, 2013 are applied to the “PO” ontology. We calculate the data using the three algorithms, and then we compare the results with that gotten from the new algorithm. Part of the data can be referred to Table 2.
Figure 2.
“PO” ontology O2.
Table 2.
Experiment data for ontology similarity measure.
| P@3 average precision ratio | P@5 average precision ratio | P@10 average precision ratio | P@20 average precision ratio | |
|---|---|---|---|---|
| Our Algorithm | 53.60% | 66.64% | 90.04% | 96.73% |
| Algorithm in Gao et al. (2013) | 36.63% | 44.60% | 58.45% | 70.06% |
| Algorithm in Gao and Shi (2013) | 36.96% | 45.08% | 60.17% | 73.99% |
| Algorithm in Gao et al. (2016) | 53.58% | 65.17% | 88.21% | 93.85% |
From the data in Table 2, we can find that when N = 3, 5, 10 or 20, the precision ratio gotten from our algorithm is higher than that from algorithms proposed in Gao et al., 2013, Gao et al., 2016, Gao and Shi, 2013. Particularly, such precision ratios are increasing apparently with N increasing. Thus, our algorithm is better and more effective than the method presented by Gao et al., 2013, Gao et al., 2016, Gao and Shi, 2013.
5. Conclusions
Borrowed the marginal technology for ontology sparse vector computation in this paper, we proposed a new computation algorithm on the basis of the marginal distribution and the analysis of the convergence criterion problem. The simulation data obtained from the experiments shows the high efficiency of our newly proposed algorithm in biology and plant science. Hence, the ontology sparse algorithm sees the promising application prospects for biology science.
Acknowledgments
We thank the reviewers for their constructive comments in improving the quality of this paper. This work was supported in part by NSFC (Nos. 11401519 and 61262070).
Footnotes
Peer review under responsibility of King Saud University.
References
- Auffeves A., Grangier P. Contexts, systems and modalities: a new ontology for quantum mechanics. Found. Phys. 2016;46:121–137. [Google Scholar]
- Agapito G., Milano M., Guzzi P.H., Cannataro M. Extracting cross-ontology weighted association rules from gene ontology annotations. IEEE-ACM Trans. Comput. Biol. 2016;13:197–208. doi: 10.1109/TCBB.2015.2462348. [DOI] [PubMed] [Google Scholar]
- Bajenaru L., Smeureanu I., Balog A. An ontology-based e-learning framework for healthcare human resource management. Stud. Inform. Control. 2016;25:99–108. [Google Scholar]
- Bobillo F., Straccia U. The fuzzy ontology reasoner fuzzyDL. Knowl.-Based Syst. 2016;95:12–34. [Google Scholar]
- Brown R.B.K., Beydoun G., Low G., Tibben W., Zamani R., Garcia-Sanchez F., Martinez-Bejar R. Computationally efficient ontology selection in software requirement planning. Inform. Syst. Front. 2016;18:349–358. [Google Scholar]
- Carmen Suarez-Figuero M., Gomez-Perez A., Fernandez-Lopez M. Scheduling ontology development projects. Data Knowl. Eng. 2016;102:1–21. [Google Scholar]
- Chicco D., Masseroli M. Ontology-based prediction and prioritization of gene functional annotations. IEEE-ACM Trans. Comput. Biol. 2016;13:248–260. doi: 10.1109/TCBB.2015.2459694. [DOI] [PubMed] [Google Scholar]
- Craswell, N., Hawking, D., 2003. Overview of the TREC 2003 web track. In: Proceeding of the Twelfth Text Retrieval Conference, Gaithersburg, Maryland, NIST Special Publication 78–92.
- Farid H., Khan S., Javed M.Y. DSont: DSpace to ontology transformation. J. Inf. Sci. 2016;42:179–199. [Google Scholar]
- Forsati R., Shamsfard M. Symbiosis of evolutionary and combinatorial ontology mapping approaches. Inform. Sci. 2016;342:53–80. [Google Scholar]
- Gao W., Gao Y., Liang L. Diffusion and harmonic analysis on hypergraph and application in ontology similarity measure and ontology mapping. J. Chem. Pharm. Res. 2013;5:592–598. [Google Scholar]
- Gao W., Gao Y., Zhang Y. Strong and weak stability of k-partite ranking algorithm. Information. 2012;15:4585–4590. [Google Scholar]
- Gao W., Shi L. Ontology similarity measure algorithm with operational cost and application in biology science. BioTechnol. An Indian J. 2013;8:1572–1577. [Google Scholar]
- Gao, W., Xu, T. W., 2013. Stability analysis of learning algorithms for ontology similarity computation. Abstr. Appl. Anal. http://dx.doi.org/10.1155/2013/174802.
- Gao W., Zhu L.L., Wang K.Y. Ontology sparse vector learning algorithm for ontology similarity measuring and ontology mapping via ADAL technology. Int. J. Bifurcat. Chaos. 2015;25 [Google Scholar]
- Gao, W., Zhu, L. L., 2014. Gradient learning algorithms for ontology computing. Comput. Intell. Neurosci. http://dx.doi.org/10.1155/2014/438291. [DOI] [PMC free article] [PubMed]
- Gao Y., Farahani M.R., Gao W. Ontology optimization tactics via distance calculating. Appl. Math. Nonlin. Sci. 2016;1:159–174. [Google Scholar]
- Gregor D., Toral S., Ariza T., Barrero F., Gregor R., Rodas J., Arzamendia M. A methodology for structured ontology construction applied to intelligent transportation systems. Comput. Stand. Interf. 2016;47:108–119. [Google Scholar]
- Hoyle D.C., Brass A. Statistical mechanics of ontology based annotations. Physica A. 2016;442:284–299. [Google Scholar]
- Huntley R.P., Sitnikov D., Orlic-Milacic M. Guidelines for the functional annotation of microRNAs using the Gene Ontology. RNA. 2016;22:667–676. doi: 10.1261/rna.055301.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibuchi H., Nojima Y. Repeated double cross-validation for choosing a single solution in evolutionary multi-objective fuzzy classifier design. Knowl.-based Syst. 2013;54:22–31. [Google Scholar]
- Kaminski M., Nenov Y., Grau B.C. Datalog rewritability of disjunctive datalog programs and non-horn ontologies. Artif. Intell. 2016;236:90–118. [Google Scholar]
- Mancinelli G., Vizzini S., Mazzola A., Maci S., Basset A. Cross-validation of delta N-15 and FishBase estimates of fish trophic position in a Mediterranean lagoon: the importance of the isotopic baseline. Estuar. Coast. Shelf Sci. 2013;135:77–85. [Google Scholar]
- Mazandu G.K., Chimusa E.R., Mbiyavanga M., Mulder N.J. A-DaGO-Fun: an adaptable gene ontology semantic similarity-based functional analysis tool. Bioinformatics. 2016;32:477–479. doi: 10.1093/bioinformatics/btv590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Bhattacharya S. Cross-validation based assessment of a new Bayesian palaeoclimate model. Environmetrics. 2013;24:550–568. [Google Scholar]
- Palmer C., Urwin E.N., Pinazo-Sanchez J.M., Cid F.S., Rodriguez E.P., Pajkovska-Goceva S., Young R.I.M. Reference ontologies to support the development of global production network systems. Comput. Ind. 2016;77:48–60. [Google Scholar]
- Pesaranghader A., Matwin S., Sokolova M., Beiko R.G. SimDEF: definition-based semantic similarity measure of gene ontology terms for functional similarity analysis of genes. Bioinformatics. 2016;32:1380–1387. doi: 10.1093/bioinformatics/btv755. [DOI] [PubMed] [Google Scholar]
- Sherimon P.C., Krishnan R. OntoDiabetic: an ontology-based clinical decision support system for diabetic patients. Arab. J. Sci. Eng. 2016;41:1145–1160. [Google Scholar]
- Terblanche C., Wongthongtham P. Ontology-based employer demand management. Software Pract. Exper. 2016;46:469–492. [Google Scholar]
- Trokanas N., Cecelja F. Ontology evaluation for reuse in the domain of process systems engineering. Comput. Chem. Eng. 2016;85:177–187. [Google Scholar]
- Umadevi S., Premkumar K., Valarmathi S., Ayyasamy P.M., Rajakumar S. Identification of novel genes related to diabetic retinopathy using protein-protein interaction network and gene ontologies. J. Biol. Syst. 2016;24:117–127. [Google Scholar]
- Varmuza K., Filzmoser P., Hilchenbach M., Krüger H., Silén J. KNN classification-evaluated by repeated double cross validation: Recognition of minerals relevant for comet dust. Chemometr. Intell. Lab. Syst. 2014;138:64–71. [Google Scholar]