Abstract
Cellular genomic DNA is replicated by a multiprotein replisome machine. The replisome contains numerous essential factors that unwind, prime and synthesize each of the two strands of duplex DNA. The antiparallel structure of DNA, and unidirectional activity of DNA polymerases, requires the two strands of DNA to be extended in opposite directions, and this structural feature requires distinctive processes for synthesis of the two strands. Genome duplication is of central importance to all cell types, and one may expect the replisome apparatus to be conserved from bacteria to human, as is the case with RNA polymerase driven transcription and ribosome mediated translation. However, it is known that the replication factors of bacteria are not homologous to those of archaea and eukaryotes, indicating that the replication process evolved twice, independently, rather than from a common ancestor cell. Thus, the different domains of life may exhibit significant differences in their mechanistic strategy of replication. In this review, we compare and contrast the different structures and mechanistic features of the cellular replication machinery in the three domains of life.
Keywords: DNA helicase, CMG, Replisome, DNA polymerase, Primase
INTRODUCTION
All cells must replicate the genetic instructions for life, held in the vast nucleotide sequence array of large DNA genomes. The replication process requires numerous protein factors that work together, somewhat like a sewing machine, referred to as a “replisome”. The replisome machine must not only duplicate the cellular genome, but must do so with extraordinary precision to preserve the species.
Cells from all three domains of life, bacteria, archaea, and eukaryota utilize duplex DNA as the genetic material and one might expect that this central life process would have evolved from the last universal common ancestor cell (i.e. LUCA). Indeed, the replisome apparatus of archaea and eukaryotes appear to have evolved from a common ancestor [1–3]. However, this does not appear to be the case for bacteria. Most components of the replisome machinery in bacteria are non-homologus in sequence and have different structures from those of archaea and eukaryotes, indicating that the replisome machinery evolved independently for bacteria and archaea/eukaryotes [4,5]. Two different ancestor lineages for the replication apparatus also suggests that LUCA may have replicated by a different means than modern- day cells.
The mechanics of separating duplex DNA, and of replicating antiparallel strands, imposes certain restrictions on how distinct the replication process can be in different cell types. For example, the two strands of the duplex must be separated for replication to occur, and accordingly all cell types employ ATP driven helicases to drive strand separation. Indeed, helicases are employed in a wide variety of DNA and RNA metabolic processes and their sequences and structures assort into several different classes [6]. The nucleotide precursors are 5’ activated in all cell types and this imposes a 5’-3’ unidirectional process for DNA chain growth. Therefore, only one strand of duplex DNA can be replicated continuously, and the antiparallel strand must be synthesized as a series of discontinuous fragments (i.e. semi-discontinuous replication). Furthermore, DNA polymerases cannot start their own chains, probably due to low intracellular dNTP concentrations, and thus did not evolve to bind two dNTPs at once to form the initial phosphodiester bond. Hence, a “primase” activity is present in all cells that use the more abundant rNTPs to synthesize a short RNA primer for the initiation of DNA synthesis. In addition, the very large size of cellular genomes, and finite accuracy of DNA polymerases result in an inevitable low frequency of misinsertions. To counteract this inherent imprecision, the replicative DNA polymerases in all cell types contain a proofreading 3’-5’ exonuclease activity that removes, most insertion errors made by the DNA polymerase. Hence, cells of all three domains of life share the basic enzymatic functions of helicase, primase, DNA polymerase, and 3’-5’ exonuclease. However, the evolutionary relationships of the enzymes that carry out these analogous functions in bacteria compared to archaea/eukaryotes are surprisingly distinct. There are also many other proteins in addition to these core enzymes that are required for genome duplication, and the way that these enzymes work together in different cell types is also quite distinct.
There have been many outstanding advances in our knowledge of eukaryotic replication in the past 5–10 years. While knowledge about eukaryotic replication is not as advanced as in the bacterial field, a clearer picture of the similarity and differences of the replisome apparatus in the major cell types has come into focus. This review uses these new advances to compare and contrast the major replisome operations of cells from the three domains of life.
Clamps and clamp loaders
The DNA polymerase, primase and helicase are the major enzymatic factors at a replication fork and were discovered long ago. However, two additional factors that are required for replication in all cells were discovered more recently. These two factors are the sliding DNA clamp and the clamp loader [7]. Interestingly, the sequences and crystal structures of the clamp and clamp loader from different cell types have revealed that the clamp and clamp loaders of all modern-day cells share a common cellular ancestor, unlike the other core enzymatic factors of DNA replication [8]. The bacterial clamp is referred to as the beta subunit and is a homodimer in all bacterial cells examined thus far (Figure 1a) [9]. The archaeal and eukaryotic clamps are a homotrimer referred to as PCNA (Figure 1b) [9]. While the oligomeric structures of beta and PCNA are different (dimer vs. trimer), the overall structures are nearly super imposable and share an identical chain-folding pattern. Both beta and PCNA are constructed from the 6-fold repetition of a globular domain. In bacteria, three domains are spliced together to form one subunit, which dimerizes to form the six-domain ring. In archaeal and eukaryotic PCNA, two domains are spliced together to form one subunit, which trimerizes to form the six-domain ring. A primary function of these clamps is to encircle duplex DNA and to bind directly to the DNA polymerase, thereby acting as a mobile tether that holds the polymerase to DNA for processive DNA synthesis. Hence, as the DNA polymerase moves forward during DNA synthesis, it pulls the clamp along behind it. Following the discovery of sliding clamps in DNA replication, it has been discovered that the same clamps are utilized by a wide variety of DNA metabolic proteins. Hence, sliding clamps are used by mismatch repair proteins, ligase, translesion DNA polymerases, cell cycle kinases and many other factors [10,11].
Figure 1.
Clamps and clamp loaders. a) The beta clamp of E. coli is a homodimer that consists of 6 globlular domains (pdb 2POL). b) The eukaryotic PCNA clamp is a homotrimer that consists of 6 domains (1AXC). Structure (panel c) and illustration (panel d) of the T4 clamp loader bound to an open clamp (grey) and DNA (yellow) (3U60). Panels c and d are reproduced with permission from Figure (2a) of [12].
Sliding clamps do not assemble onto DNA by themselves; they require a multi-subunit clamp loader that uses ATP to open and close the ring around DNA [7]. Clamp loaders of all cells are composed of 5 homologous subunits, and each subunit is a member of the AAA+ family of ATPases [8,12]. Clamp loaders from many cell types, and the classic T4 phage system, have been studied both biochemically and structurally (Figure 1c,1d). The five subunits are arranged in a circle held tightly by the C-terminal domains, and there is a gap between the N-terminal AAA+ domains of two of the subunits. The gap functions to allow DNA to pass into the inside the clamp loader, which forms a composite DNA binding site that encircles the DNA and positions it through an opened clamp that is held beneath the AAA+ domains (Figure 1c,1d). After DNA is positioned in the clamp loader and through the clamp, ATP is hydrolyzed which enables closure of the clamp around DNA and ejection of the clamp loader, leaving the clamp on DNA for function with other proteins.
DNA helicase
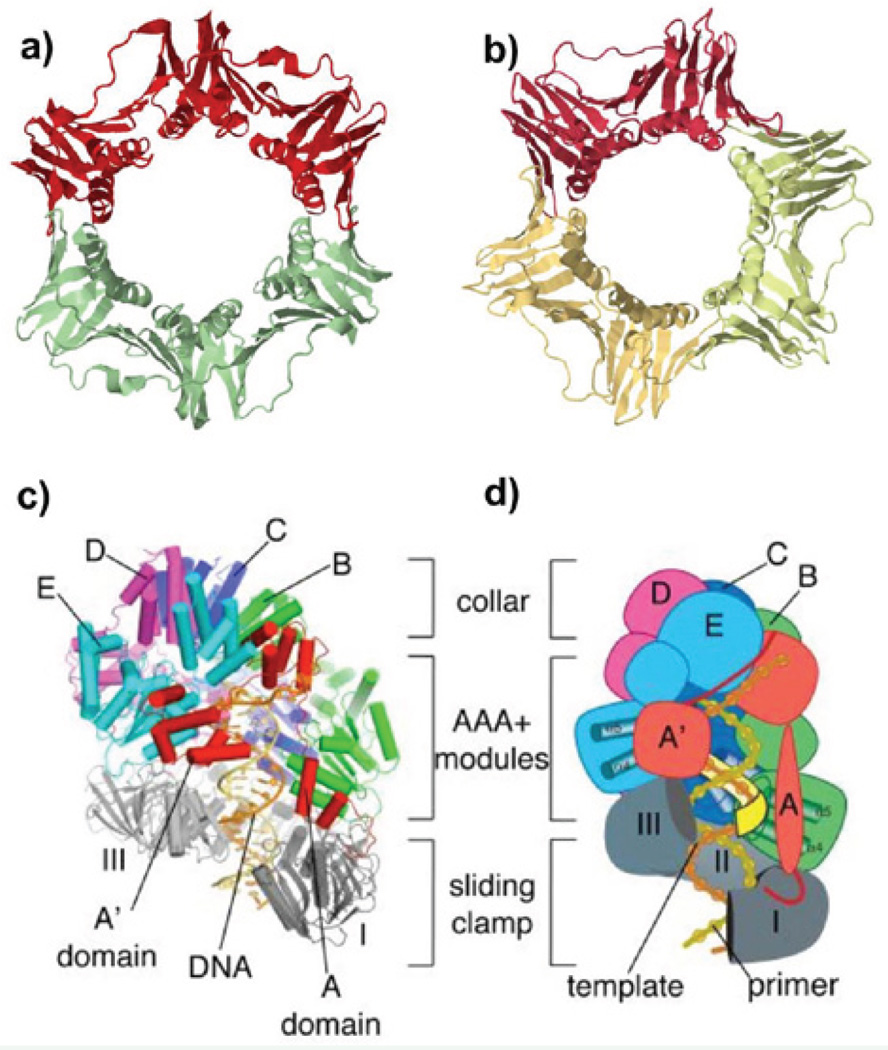
DNA helicases are ubiquitous in nature and they function to unwind DNA for a variety of DNA metabolic transactions. The unwound strands are coated and protected by a single-strand binding protein in all cell types, referred to as SSB (Single-Strand Binding protein) in bacterial cells and RPA (Replication Protein A) in archaeal and eukaryotic cells. The bacterial and archaeal/eukaryotic replicative helicases are arranged as a hexamer ring that encircles DNA, illustrated schematically in Figure (2a) [13–15]. Each subunit of the helicase consists of two major domains, an N-terminal domain (NTD) and a C- terminal domain (CTD), resulting in a hexameric N-tier ring and C-tier ring. The ATP sites are located in the C-tier. The bacterial hexameric helicase motor domains are fashioned from the RecA fold, while the archaeal and eukaryotic helicase motor domains are sculpted from the AAA+ fold [13–15]. The bacterial replicative helicase, exemplified by E. coli DnaB, translocates 5’-3’ along single-strand DNA, placing it on the lagging strand. In contrast, the archaeal and eukaryotic helicases, referred to as MCM (Mini Chromosome Maintenance), translocate 3’-5’ placing them on the leading strand for replication fork advance [13–15]. As the helicase translocates on one strand of DNA, the other strand is excluded from the inner channel and thus the helicase acts as a moving wedge to melt the duplex (Figure 2a,2b). This mechanism of unwinding is referred to as “steric exclusion” [13–15].
Figure 2.
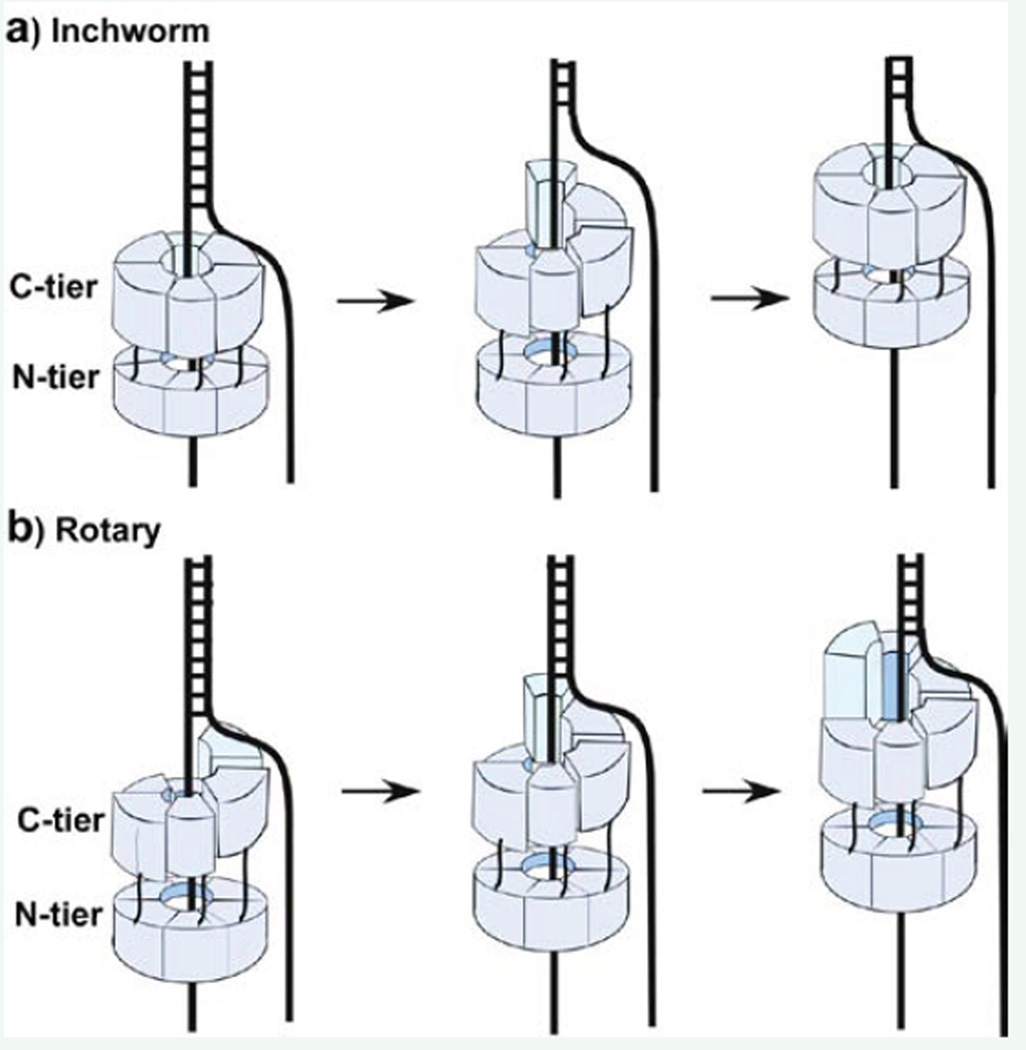
Proposed translocation mechanisms of hexameric helicases. Replicative helicases are hexameric rings composed of an N-tier and C-tier; the motor domains are in the C-tier. a) Inchworm mechanism. Left: the C- and N-tiers are parallel and compact. Middle: The C-tier opens at one interface and expands into a spiral structure, melting DNA and leaving the N-tier a flat uninterrupted ring. Right: The C-tier reassumes the compact shape and translocates the N-tier on DNA. b) Rotary model. Left: The C-tier is shaped as a spiral lock washer and connects to a flat N-tier ring. Middle: ATP hydrolysis translates the lock washer shape by one protomer, resulting in DNA translocation. Right: ATP hydrolysis translates the lock washer shape by one protomer, resulting in DNA translocation.
Unlike the homohexamer DnaB helicase of bacteria, and the MCM homohexamer helicase of archaea, the eukaryotic MCM subunits are encoded by separate genes, forming the MCM2-7 heterohexamer [16]. While each MCM gene is essential for ongoing synthesis in the cell [17], the helicase activity does not require that each ATP site be competent for hydrolysis [18,19]. Thus, each subunit in the eukaryotic MCM2-7 complex may play an individual role. Purification of the eukaryotic helicase was initially performed in the Drosophila system, and characterization showed it to be composed of MCM2-7 in complex with one copy of Cdc45 and one GINS heterotetramer [20]. The eukaryotic helicase is referred to as CMG, an acronym for Cdc45-MCM2-7-GINS [20]. Recombinant CMG has been produced from S. cerevisiae, Drosophila and human [19,21,22]. In each case, the 11-subunit complex is an active helicase. The accessory factors, Cdc45-GINS, lack ATP sites and it is proposed that they function to hold the MCM2-7 motor ring into a proper configuration for helicase activity [19]. The archaeal cell contains homologues to GINS and Cdc45, and thus archaea may also contain a CMG complex, although an archaeal CMG complex has yet to be identified [23].
Structural studies of eukaryotic CMG by EM single-particle 3D reconstruction techniques have elucidated the structure of CMG from Drosophila and from budding yeast [24,25]. The studies show that the Cdc45-GINS accessory factors are attached to one side of the MCM2-7 ring. Interestingly, the accessory factors span the MCM2-5 subunit interface, the interface that opens and closes for DNA entry at the origin [26]. Hence, the accessory factors may function to hold the MCM2-7 ring closed during processive helicase translocation on DNA. Recent high resolution cryoEM studies of CMG indicate that it translocates along DNA by passing DNA from the C-tier to the N-tier [27–29]. In fact, two conformers of CMG indicate a maximum distance change between the N- and C-tiers of 20 angstroms, suggesting that CMG may inchworm along DNA during ATP hydrolysis, illustrated in Figure (2a) [28,29]. An inchworm mechanism of translocation on DNA is also employed certain monomeric helicases [30,31]. In contrast, the homohexameric helicases are proposed to function by a rotary stair-casing mechanism, in which each ATP hydrolysis step results in the motion of one protomer along DNA, and that translation of this motion around the ring results in translocation along DNA, illustrated in Figure (2b) [13,32,33]. Further studies are needed to firm up the mechanism of hexameric DNA helicases.
Primase
The antiparallel structure of duplex DNA, coupled with the unidirectional action of DNA polymerases requires that one strand is duplicated in the opposite direction of fork propagation. Furthermore, DNA polymerases can only extend a preexisting primed site and cannot initiate synthesis de novo like RNA polymerases. Thus, all cells contain a primase that makes short RNA primers to initiate DNA synthesis. In bacteria, the monomeric DnaG protein forms short RNA primers of about a dozen nucleotides upon which a clamp is assembled for extension by the replicative DNA polymerase III [34]. The structure of the active site region of bacterial DnaG primase is evolutionarily related to topoisomerase [35]. In sharp contrast, eukaryotic cells contain a heterodimeric complex required for RNA primer synthesis with homology to X family DNA polymerases instead of topoisomerase [36]. Archaeal cells contain a heterodimeric primase with homology to the eukaryotic heterodimer [36]. Unlike archaea, the eukaryotic primase subunits are harbored within a larger 4-subunit complex that contains a DNA polymerase, referred to as DNA polymerase (Pol) alpha-primase. Pol alpha-primase generates a 25–35 nucleotide hybrid RNA-DNA primer in which a 7–10 nucleotide RNA is handed internally to the DNA polymerase for extension by dNTPs [37].
The eukaryotic Pol alpha-primase has been demonstrated to adhere to other components of the replisome machinery, while in bacterial systems the DnaG primase functions as an independent enzyme with only transient interaction with other replisome components [38]. Cell pullouts from budding yeast using a tag specific for CMG reveals a large assembly, referred to as the replisome promoting complex (RPC) [38,39]. The RPC contains, in addition to CMG, Pol alpha-primase as well as MCM10, Ctf4, FACT, Mrc1, Tof1, Csm3 and Topo I [38,39]. Hence, the eukaryotic Pol alpha-primase is an integral component of a moving replisome, unlike the distributive action of bacterial DnaB primase.
DNA polymerases and replisome organization
Eukaryotic cells utilize three distinct B family DNA polymerases, Pol epsilon, Pol delta and Pol alpha-primase [40,41]. The current view is that Pol epsilon functions on the leading strand; Pol delta replicates the lagging strand, and Pol alpha-primase acts to prime both strands [42]. These assignments are suggested by genetic studies [42–46], strand specific polymerase-DNA cross linking studies [47], and biochemical replication studies using pure proteins [21,25,28,48,49]. However, there remains uncertainty about these assignments and further work is needed to clarify this issue [50,51]. Unlike eukaryotes, bacterial cells use multiple copies of an identical DNA polymerase of the C-family [52,53]. C- family DNA polymerases are only found in bacteria and they share no sequence homology to the eukaryotic and archaeal B-family DNA polymerases, indicating a distinct evolutionary lineage of the B- and C-family DNA polymerases [54]. However, all DNA polymerases examined thus far take the shape of a right hand, with palm, fingers and thumb sub domains [55]. The palm sub domain contains the active site and is the most conserved element among different polymerase families. The structure of the palm sub domain of C-family polymerases shares similarity to the X-family of nucleotidyl transferases, while the folding pattern of the palm in B-family polymerases is similar to that of A- and Y-family DNA polymerases [54–56]. Thus, the bacterial and eukaryotic DNA polymerases, like the primase and helicase, appear to have a distinct evolutionary heritage.
The absence of a common ancestor cell for the fundamental enzymes of DNA replication stands in contrast to the other major nucleic acid processes of transcription and translation. The RNA polymerase of bacteria and eukaryotes share homology and structure, and therefore evolved from a common cellular ancestor. The same is true for the ribosomes of bacteria, archaea and eukaryotes, which use a universal genetic, code, and thus derive from the last universal cellular ancestor (i.e. LUCA). The implication of distinctive evolution of replisome components in the different domains of life is that the replication process in LUCA was different from modern-day cells. Perhaps the ancient genomic material was RNA, which would explain why bacterial and archaeal/eukaryotic cells evolved their own replication machinery to duplicate DNA. Regardless of the reason, evolution would appear to have arrived at two different solutions to cellular replication.
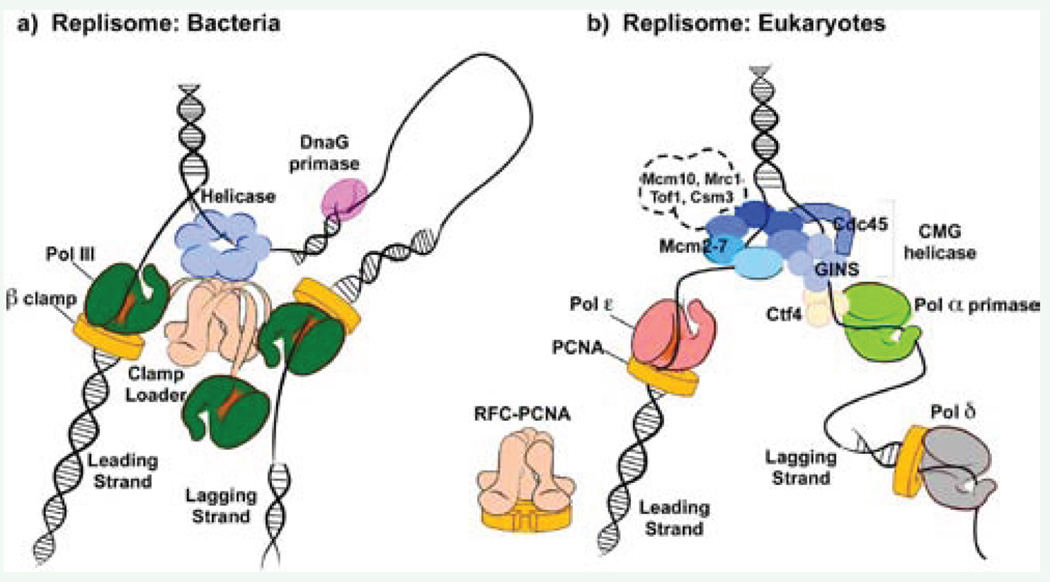
The bacterial replisome is organized into three copies of Pol III, connected to a single clamp loader, as illustrated in Figure (3a) [57]. The clamp loader contains three copies of the tau subunit which has a C-terminal region that is not required for the clamp loading activity [58]. This C-terminal region of each tau subunit contains a domain(s) attached to a flexible tether, each of which binds a molecule of Pol III and also connects to the hexameric DnaB helicase (Figure 3a). This three Pol-clamp loader-DnaB helicase complex constitutes a stabile replisome machine that remains bound to the replication fork for about 100 kb of synthesis in single-molecule studies without dissociating from the DNA [10,59,60]. During duplex DNA replication, one Pol III-clamp complex extends the leading stand, while the lagging strand is copied by the other two Pol III molecules which likely take turns extending RNA primers into Okazaki fragments [61]. The primase transiently interacts with DnaB to form RNA primers about once every 1–2 kb [62]. Thus, the lagging strand is synthesized by a repetitive cycle involving priming, clamp loading, polymerase binding to the clamp, polymerase extension of the Okazaki fragment, and polymerase recycling to a new RNA primed site. In each Okazaki fragment synthesis cycle, the lagging strand DNA polymerase hops from a completed DNA fragment to a new RNA primer, leaving the “used” clamp behind on the daughter duplex. The leftover clamps are used by proteins that replace the RNA with DNA and seal fragments together. The Okazaki fragment cycle produces transient DNA loops on the lagging strand because the polymerase remains tightly adhered to the replisome complex at the forked junction.
Figure 3.
Comparison of replisomes from bacteria and eukaryotes. a) The bacterial replisome is organized by a central clamp loading machine (beige) that has C-terminal extension, which extrude from the top and bind the hexameric helicase (blue) that encircles the lagging strand. The clamp loader extensions also bind three copies of Pol III (green), the replicative DNA polymerases. One Pol III functions on the continuous leading strand, tethered to DNA by the beta clamp (yellow). The other two Pol III molecules extend lagging strand fragments that are initiated by short RNA primers synthesized by primase (pink). b) The eukaryotic replisome is organized by the CMG helicase (blue), which functions with Pol epsilon (red) on the leading strand and bind Pol alpha-primase (green) through a Ctf4 trimer (light yellow) on the lagging strand. Lagging strand primers are extended by Pol delta (grey). Both Pol epsilon and Pol delta function with a PCNA clamp (yellow). Whether the RFC clamp loader and Pol delta directly connect to other replisome proteins is not known. Other factors that move with replisomes are shown within the dashed outline; their exact connection points are not yet known.
The eukaryotic replisome is only recently coming into focus, illustrated in Figure (3b). Biochemical and structural studies show that Pol epsilon forms a tight complex with the CMG helicase, localizing Pol epsilon to the leading strand to which the CMG helicase is attached [25,49]. Note that the eukaryotic CMG helicase encircles the leading strand while the bacterial DnaB helicase encircles the lagging strand. The observation that Pol epsilon forms a tight complex with CMG helicase contrasts with bacterial systems in which the leading polymerase interacts either weakly, or indirectly with the helicase [34,58]. Furthermore, priming of the lagging strand in eukaryotes is performed by Pol alpha-primase which is a component of the RPC. Thus, unlike bacterial DnaG primase, the eukaryotic primase is an integral part of the replisome machinery. Another striking difference between eukaryotic and bacterial replisome mechanics is the absence of direct connections between the eukaryotic RFC clamp loader and other components of the replisome. Likewise, the lagging strand Pol delta has no known strong contacts to the replisome, and thus there is no existing evidence for DNA looping during lagging strand synthesis. In addition, Okazaki fragments in eukaryotes are only 100–200 nucleotides long, much shorter than the 1–2 kb in bacteria [34,58].
In a recent development, reconstitution of a eukaryotic replisome that utilizes all three DNA polymerases to replicate the leading and lagging strands has been accomplished using pure proteins [48]. Replication from an origin has also been reconstituted, although Pol delta, RFC and PCNA were not included in the origin studies [63]. Replication of the leading strand has also been reconstituted in a human system [22]. The in vitro systems have revealed that Pol epsilon is stabilized for processive leading strand synthesis, probably due to the direct interaction of Pol epsilon with CMG helicase. In contrast, Pol delta only forms short DNA products on the leading strand [21,22,48,49]. However, on the lagging strand Pol delta is functional in Okazaki fragment extension while Pol epsilon is inactive [48]. Hence, asymmetric action of the eukaryotic DNA polymerases at a replication fork is recapitulated in vitro. In addition to the helicase, primase and DNA polymerases, elucidation of the RPC has identified numerous other proteins that travel with the eukaryotic replisome [39]. Among these factors is the MCM10 protein, which is essential for cell viability, yet the exact function of MCM10 is not well understood [64]. The eukaryotic replisome also contains Ctf4, a homotrimeric scaffolding protein that cross-links Pol alpha-primase to CMG [38]. The RPC also contains Mrc1, Tof1 and Csm3, factors that function in checkpoint signaling and in programmed fork arrest [65–67]. The RPC also contains the FACT complex (Facilitates Chromatin Transcription) [39]. FACT is a nucleosome handling factor that was discovered for its ability to help RNA polymerase surmount barriers imposed by nucleosomes [68]. The presence of FACT in the RPC suggests that the eukaryotic replisome carries this factor to help deal with nucleosomes that package the genome. Interestingly, the MCM2 subunit has recently been shown to bind histones, suggesting that the eukaryotic replisome may also deal with nucleosomes directly [69,70].
CONCLUSIONS
Biochemical and structural analysis of bacterial replication has illuminated the structure and function of the bacterial replisome machine. Yet several questions persist regarding details of lagging strand replication, the mechanism by which hexameric helicases function, and processes by which the replisome interweaves its actions with repair processes. In contrast, development of an in vitro system for eukaryotic replication has lagged far behind, mostly due to the increased complexity and numerous proteins required for this process. In vitro reconstitution of a eukaryotic replisome system that duplicates both the leading and lagging strands has recently been developed [48]. This breakthrough sets the foundation for future studies to understand the detailed mechanism of the eukaryotic replisome machinery. Obvious questions include: What mechanisms underlie the placement of different DNA polymerases on the leading and lagging strands? The eukaryotic genome is packaged into nucleosomes and they must be displaced to replicate the DNA. How does the replisome deal with nucleosomes? Does it recruit particular chromatin remodelers, or is the handling of nucleosomes intrinsic to the replisome machine? Some regions of the chromosome are highly condensed into heterochromatin. Is the heterochromatin a more difficult challenge to the replisome? DNA damage by spontaneous endogenous reactions, such as hydrolysis and oxidation, require that DNA repair be a continual process. The replisome is expected to periodically encounter some DNA lesions before they can be repaired. How does the replisome deal with DNA lesions? Do specialized DNA polymerases gain access to the replisome for by pass of DNA lesions? Do the enzymes of recombinational repair interface and coordinate with the DNA replication machinery? The eukaryotic replisome is known to be posttranslationally modified in response to DNA damage [71,72]. To what end do these modifications serve? Furthermore, the PCNA clamp is ubiquinylated upon DNA damage. Does this modification recruit specialized translesion DNA polymerases as proposed [73]? Detailed answers to these and many other questions are certain to result in new and exciting discoveries in the future.
Acknowledgments
The authors are grateful for funding from the US National Institutes of Health (GM115809) and the Howard Hughes Medical Institute.
ABBREVIATIONS
- MCM
Minichromosome Maintenance
- GINS
Go-Ichi-Ni-San
- CMG
Cdc45-MCM2-7-GINS
- Pol
DNA Polymerase
- RPC
Replisome Progression Complex
- RPA
Replication Protein A
- SSB
Single-Strand Binding Protein
- PCNA
Proliferating Cell Nuclear Antigen
- RFC
Replication Factor C
- FACT
Facilitates Chromatin Transcription
Footnotes
Conflict of Interest
The authors declare that neither financial interest nor conflict of interest exists.
REFERENCES
- 1.Beattie TR, Bell SD. Molecular machines in archaeal DNA replication. Curr Opin Chem Biol. 2011;15:614–619. doi: 10.1016/j.cbpa.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 2.Kelman LM, Kelman Z. Archaeal DNA replication. Annu Rev Genet. 2014;48:71–97. doi: 10.1146/annurev-genet-120213-092148. [DOI] [PubMed] [Google Scholar]
- 3.Makarova KS, Koonin EV. Archaeology of eukaryotic DNA replication. Cold Spring Harb Perspect Med. 2013;3:12963. doi: 10.1101/cshperspect.a012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forterre P. Why are there so many diverse replication machineries? J Mol Biol. 2013;425:4714–4726. doi: 10.1016/j.jmb.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 5.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 7.Jeruzalmi D, O’Donnell M, Kuriyan J. Clamp loaders and sliding clamps. Curr Opin Struct Biol. 2002;12:217–224. doi: 10.1016/s0959-440x(02)00313-5. [DOI] [PubMed] [Google Scholar]
- 8.Kelch BA, Makino DL, O’Donnell M, Kuriyan J. Clamp loader ATPases and the evolution of DNA replication machinery. BMC Biol. 2012;10:34. doi: 10.1186/1741-7007-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong XP, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E.coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 10.Georgescu R, Langston L, O’Donnell M. A proposal: Evolution of PCNA’s role as a marker of newly replicated DNA. DNA Repair (Amst) 2015;29:4–15. doi: 10.1016/j.dnarep.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan M, Kelman LM, Kelman Z. The archaeal PCNA proteins. Biochem Soc Trans. 2011;39:20–24. doi: 10.1042/BST0390020. [DOI] [PubMed] [Google Scholar]
- 12.Kelch BA, Makino DL, O’Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 16.Bochman ML, Schwacha A. The MCM complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 18.Bochman ML, Bell SP, Schwacha A. Subunit organization of MCM2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol Cell Biol. 2008;28:5865–5873. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/MCM2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgescu RE, Langston L, Yao NY, Yurieva O, Zhang D, Finkelstein J, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/MCM2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci USA. 2012;109:6042–6047. doi: 10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell SD. DNA replication: archaeal oriGINS. BMC Biol. 2011;9:36. doi: 10.1186/1741-7007-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J, Shi Y, Georgescu RE, Yuan Z. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samel SA, Fernández-Cid A, Sun J, Riera A, Tognetti S, Herrera MC, et al. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 2014;28:1653–1666. doi: 10.1101/gad.242404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abid Ali F, Renault L, Gannon J, Gahlon HL, Kotecha A, Zhou JC. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donnell M, Li H. The Eukaryotic Replisome Goes Under the Microscope. Curr Biol. 2016;26:247–256. doi: 10.1016/j.cub.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Z, Bai L, Sun J, Georgescu R, Liu J. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016;23:217–224. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 32.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 33.Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151:267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Podobnik M, McInerney P, O’Donnell M, Kuriyan J. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J Mol Biol. 2000;300:353–362. doi: 10.1006/jmbi.2000.3844. [DOI] [PubMed] [Google Scholar]
- 36.Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta. 2010;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perera RL, Torella R, Klinge S, Kilkenny ML, Maman JD, Pellegrini L. Mechanism for priming DNA synthesis by yeast DNA polymerase α. Elife. 2013;2:482. doi: 10.7554/eLife.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 40.Méndez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 2003;25:1158–1167. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- 41.Stillman B. DNA polymerases at the replication fork in eukaryotes. Mol Cell. 2008;30:259–260. doi: 10.1016/j.molcel.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clausen AR, Lujan SA, Burkholder AB, Orebaugh CD, Williams JS, Clausen MF, et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol. 2015;22:185–191. doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nick Mc Elhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu C, Gan H, Han J, Zhou ZX, Jia S, Chabes A, et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 2014;56:551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Georgescu RE, Schauer GD, Yao NY, Langston LD, Yurieva O, Zhang D, et al. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. Elife. 2015;4:4988. doi: 10.7554/eLife.04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, Yao NY, et al. CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci U S A. 2014;111:15390–15395. doi: 10.1073/pnas.1418334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson RE, Klassen R, Prakash L, Prakash S. A Major Role of DNA Polymerases δ in Replication of Both the Leading and Lagging DNA Strands. Mol Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stillman B. Reconsidering DNA Polymerases at the Replication Fork in Eukaryotes. Mol Cell. 2015;59:139–141. doi: 10.1016/j.molcel.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mc Inerney P, Johnson A, Katz F, O’Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 53.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lamers MH, Georgescu RE, Lee SG, O’Donnell M, Kuriyan J. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 55.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 56.Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 57.O’Donnell M, Jeruzalmi D, Kuriyan J. Clamp loader structure predicts the architecture of DNA polymerase III holoenzyme and RFC. Curr Biol. 2001;11:935–946. doi: 10.1016/s0960-9822(01)00559-0. [DOI] [PubMed] [Google Scholar]
- 58.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 59.Georgescu RE, Yao NY, O’Donnell M. Single-molecule analysis of the Escherichia coli replisome and use of clamps to bypass replication barriers. FEBS Lett. 2010;584:2596–2605. doi: 10.1016/j.febslet.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao NY, Georgescu RE, Finkelstein J, O’Donnell ME. Single-molecule analysis reveals that the lagging strand increases replisome processivity but slows replication fork progression. Proc Natl Acad Sci USA. 2009;106:13236–13241. doi: 10.1073/pnas.0906157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Georgescu RE, Kurth I, O’Donnell ME. Single-molecule studies reveal the function of a third polymerase in the replisome. Nat Struct Mol Biol. 2011;19:113–116. doi: 10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tougu K, Marians KJ. The interaction between helicase and primase sets the replication fork clock. J Biol Chem. 1996;271:21398–21405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- 63.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thu YM, Bielinsky AK. Enigmatic roles of MCM10 in DNA replication. Trends Biochem Sci. 2013;38:184–194. doi: 10.1016/j.tibs.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bairwa NK, Mohanty BK, Stamenova R, Curcio MJ, Bastia D. The intra-S phase checkpoint protein Tof1 collaborates with the helicase Rrm3 and the F-box protein Dia2 to maintain genome stability in Saccharomyces cerevisiae. J Biol Chem. 2011;286:2445–2454. doi: 10.1074/jbc.M110.189456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bando M, Katou Y, Komata M, Tanaka H, Itoh T, Sutani T, et al. Csm3, Tof, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J Biol Chem. 2009;284:34355–34365. doi: 10.1074/jbc.M109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, et al. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell. 2008;32:106–117. doi: 10.1016/j.molcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 69.Huang H, Strømme CB, Saredi G, Hödl M, Strandsby A, González-Aguilera C, et al. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat Struct Mol Biol. 2015;22:618–626. doi: 10.1038/nsmb.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H, Wang M, Yang N, Xu RM. Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2. Protein Cell. 2015;6:693–697. doi: 10.1007/s13238-015-0190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ilves I, Tamberg N, Botchan MR. Checkpoint kinase 2 (Chk2) inhibits the activity of the Cdc45/MCM2-7/GINS (CMG) replicative helicase complex. Proc Natl Acad Sci U S A. 2012;109:13163–13170. doi: 10.1073/pnas.1211525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hedglin M, Benkovic SJ. Regulation of Rad6/Rad18 Activity During DNA Damage Tolerance. Annu Rev Biophys. 2015;44:207–228. doi: 10.1146/annurev-biophys-060414-033841. [DOI] [PMC free article] [PubMed] [Google Scholar]