Abstract
Circular nucleic acid molecules can have chemical and biological properties very different from those of the corresponding linear nucleic acid polymers. Described here are methods used recently for construction of such circular molecules, and some of the properties that can arise from making this topological change. Among the unusual properties found for circular nucleic acids are: strong resistance to degradation in biological media; high affinity of binding to other nucleic acids; high sequence selectivity in nucleic acid binding; topological linkage to biomolecules; and the ability to template the synthesis of specific repeating nucleic acid and protein polymers. These properties may be useful in biochemical, medical diagnostic and therapeutic applications.
The nucleic acids constitute a family of polymers that have a number of properties that make them both interesting to humans and also relatively simple to synthesize, modify and characterize. Not only do DNAs and RNAs encode our genetic information and thus make life possible, but they also form predictable structures with relatively predictable properties. Noncovalent forces can bring about organization of the polymer coils to form double, triple and quadruple helices, and the covalent architecture can be assembled both by harnessing nature’s enzymes and by chemical synthesis. Nucleic acids are commonly manipulated by scientists to modify the properties of living organisms, and they are also being used outside living systems as biochemical tools and in nanoscale molecular architectures.
Topological modification of the linear chains of DNAs and RNAs can result in large changes in their properties. Evolution has shown us that circular DNAs and RNAs can have large advantages in survival over linear counterparts: most of the DN A in nature is in fact circular rather than linear (i.e. bacterial and viral DNAs are usually circular)1. The reasons for this probably include the fact that circular DNAs evade common degradation mechanisms in most organisms, and circular molecules have particularly simple mechanisms for being copied.
Recently, scientists have developed good methods for constructing synthetic DNAs of desired sizes and sequences, and in the past few years we have found ways to make topological modifications as well. Here I describe how such modifications to the polymer form can be effected, and some of the resulting properties that arise on making the change from linearity to circularity.
Construction of DNA and RNA circles
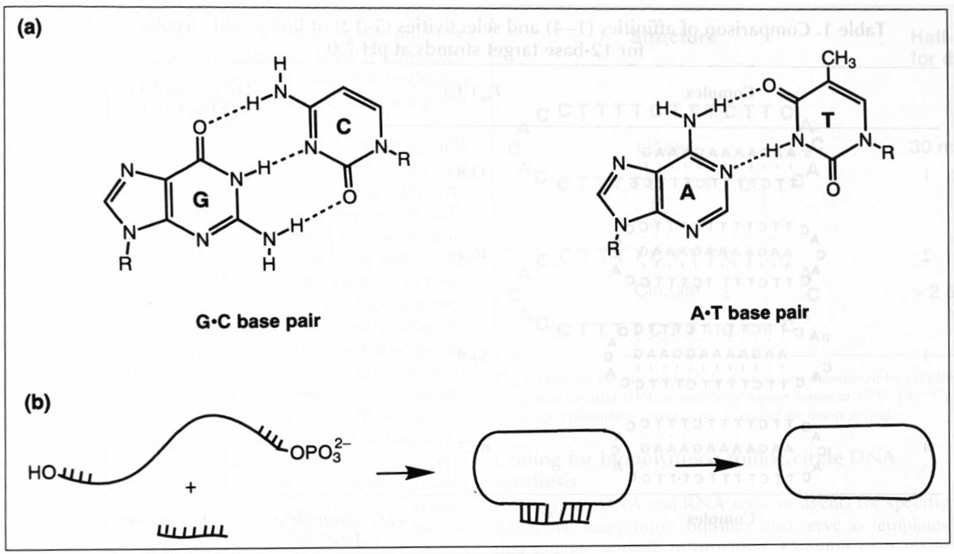
The predictable base pairing of DNA strands to form a double helix (Fig. 1a) makes the cyclization of a nucleic acid strand much more efficient than cyclizations of other long non-associating polymers. The most successful methods for joining opposite ends of a single strand of DNA or RNA involve the use of a short synthetic template or splint of DNA that is complementary to the two remote ends (see Fig. 1b)2,3
Fig. 1.
(a) The structures of the two classical Watson–Crick base pairs in DNA. G and Care guanine and cytosine; A and T are adenine and thymine. (b) Illustration of how circles are formed from linear DNA/RNA strands using a Watson–Crick complementary ‘template’ or ‘splint’ (a short oligonucleotide) to bring the ends together.
The specific reaction that joins the ends to form a circle is formation of a phosphodiester from a phosphate monoester on one end of the strand and a hydroxyl group on the other end. This reaction can be carried out with an enzyme (commonly a DNA ligase or RNA ligase), or chemical (nonenzymatic) methods can now be used, which often utilize BrCN (Refs 4 and 5) or carbodiimides6 to form the same bond.
The linear DNA or RNA precursor of the circle can either be derived from nature using molecular biological methods, or it can be synthesized chemically using an automated DNA synthesizer. The only requirements, in principle, are the presence of a phosphate on one end and a hydroxyl on the other. We have chemically synthesized single-stranded circles as short as 26 nucleotides and as long as 246 nucleotides; in addition, biological methods can generate strands up to thousands of nucleotides in length, and these can be cyclized using related methods7. It should be noted that I am discussing single-stranded nucleic acid circles here; most natural DNA circles are composed of double-stranded helices, which have considerably different topological and biological properties1.
Recent studies have also addressed the synthesis of DNA and RNA circles even smaller than those discussed above8–10. DNA single strands are quite flexible, and even cyclic mono- and dinucleotides are known. Methods for formation of circles ~2–20 nucleotides in length are discussed in the chemical literature8–10. Those smaller molecules cannot rely on the use of templates to join the ends, because the binding to a template is too weak for such small molecules.
Circular DNAs or RNAs can be characterized by showing that they are resistant to degradation by end-modifying enzymes such as exonucleases2; circles have no ends and so cannot be cleaved by these enzymes. In addition, partial cleavage of a circle by enzymatic or chemical methods results in initial formation of a population of molecules having a single size (that of the linear precursor)11, while linear molecules will be cleaved to generate a ladder of different-sized products. Finally, circular nucleic acids have different mobility by gel electrophoresis than do their linear counterparts, and such differences can be used to identify specific structures.
Small circles as ligands for DNA and RNA targets
Very small circular DNAs can be designed and constructed to act as ligands for binding specific sequences of single-stranded RNA and DNA2,11–15 DNA- or RNA-binding molecules are currently being examined in many laboratories for potential uses as medical diagnostic agents (for checking genetic sequences)16 and as possible therapeutic agents meant to bind and inhibit specific disease-related genes17.
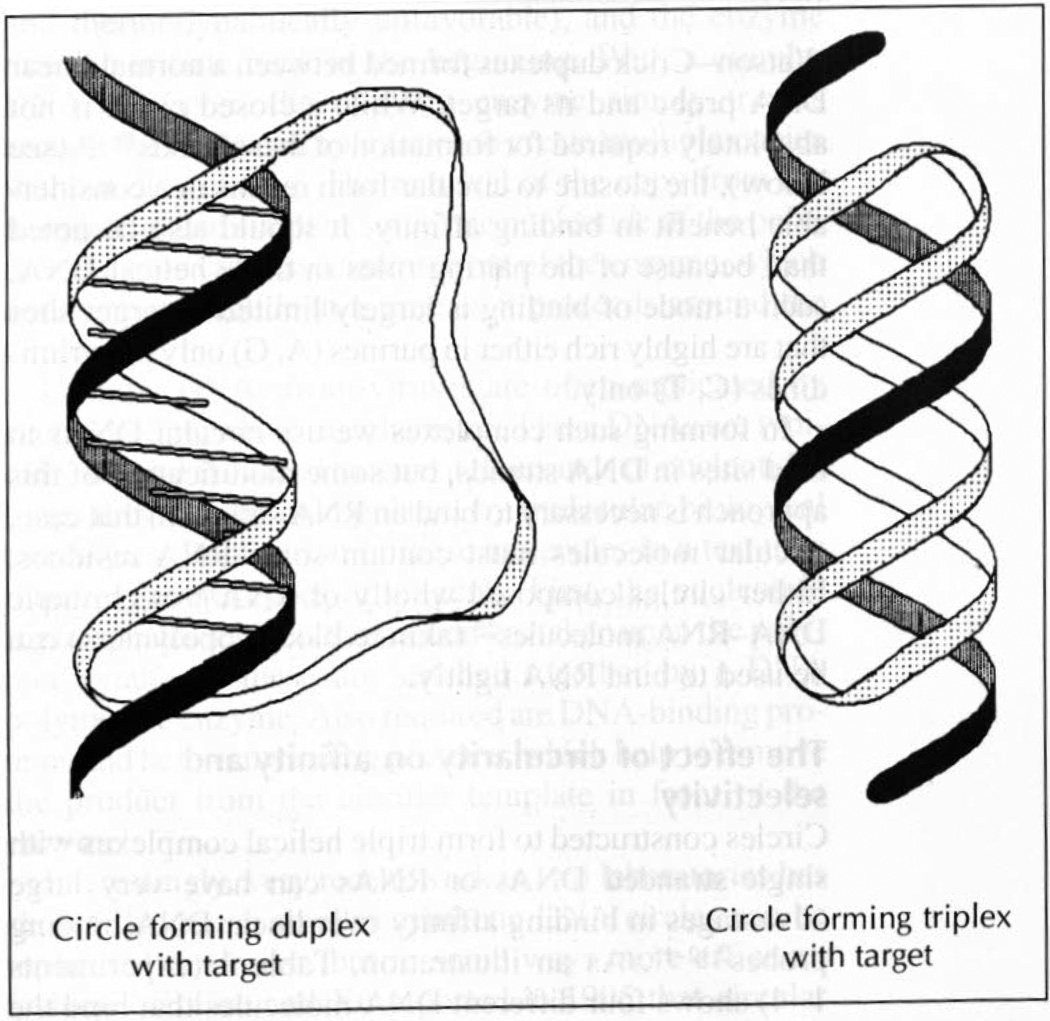
We have shown that circular DNAs 28–74 nucleotides in length can form very tight complexes at pH 7.0 in aqueous solution with a complementary DNA strand2,12–15,18. These circles were designed to form a three-stranded (triple helical) complex with the single-stranded target (Fig. 2). The design of such a circle includes two binding domains that contact the target strand. The orientation of one domain, complementary in the Watson–Crick fashion, is antiparallel to the target (DNA strand orientation being defined by 5′-to-3′ ribose orientation). The opposite binding domain in the circle is complementary in a parallel (called Hoogsteen) fashion to the other side of the same target. These two binding domains are bridged by loops of five nucleotides or so on either side.
Fig. 2.
Illustration of how circular oligonucleotides {DNA or RNA) can form double helical or Triple helical complexes with a single target strand of DNA or RNA.
In this novel mode of binding, a circle sandwiches or clamps its target between the opposing sides of the circle. This results in formation of four or five hydrogen bonds between the circular ligand and each base of the target, which means a total of about 50 H-bonds for a 12-base target site (as well as more than 30 important π-stacking interactions throughout the complex). This can result in complexes that are much more stable than standard Watson–Crick duplexes formed between a normal linear DNA probe and its target. While a closed circle is not absolutely required for formation of these bonds19–21 (see below), the closure to circular form results in a considerable benefit in binding affinity. It should also be noted that, because of the pairing rules in triple helical DNA. such a mode of binding is largely limited to target sites that are highly rich either in purines (A, G) only or pyrimidines (C, T) only.
In forming such complexes we use circular DNAs to bind sites in DNA strands, but some modification of this approach is necessary to bind an RNA strand. In that case, circular molecules must contain some RNA residues. Either circles composed wholly of RNA11 or chimeric DNA–RNA molecules22 (akin to block copolymers) can be used to bind RNA tightly.
The effect of circularity on affinity and selectivity
Circles constructed to form triple helical complexes with single-stranded DNAs or RNAs can have very large advantages in binding affinity over linear DNA-binding probes2,12–15. As an illustration. Table 1 (experiments 1–4) shows four different DNA molecules that bind the same 12-base target sequence at neutral pH and physiological ionic strength in water. The comparison shows that a standard linear complementary probe, which forms a duplex with its target, binds with a thermal melting temperature (Tm) of 43.8°C (corresponding to a free energy, ΔG, of c. −43.3 kJ mol−1) under these conditions.
Table 1.
Comparison of affinities (1–4) and selectivities (5–12) of linear and circular DNAs for 12-base target strands at pH 7.0
| Complex | Tm (°C) | −ΔG°37(complex) (kJ mol−1) |
|
|---|---|---|---|
| 1 | 43.8 | 43.3 | |
| 2 | 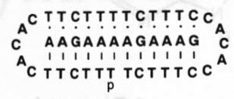 |
46.4 | 45.4 |
| 3 | 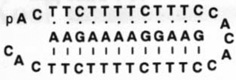 |
54.9 | ~57 |
| 4 | 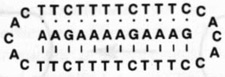 |
62.3 | 68.4 |
| Complex | X | −ΔG°37(complex) (kJ mol−1) |
Kd(complex) (M) | |
|---|---|---|---|---|
| 5 | 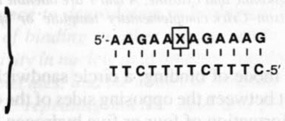 |
A | 43.3 | 3 × 10−8 |
| 6 | T | 26.9 | 2 × 10−5 | |
| 7 | G | 29.8 | 6 × 10−6 | |
| 8 | C | 24.8 | 5 × 10−5 | |
| 9 |  |
A | 68.9 | 9 × 10−13 |
| 10 | T | 38.2 | 2 × 10−7 | |
| 11 | G | 42.8 | 3 × 10−8 | |
| 12 | C | 37.0 | 4 × 10−7 |
Molecules designed to wrap around and form a triple helical complex are, by contrast, considerably better at binding, with free energies that are more favorable by an additional 4–25 kJ mol−1 (and Tm values 3–18°C higher) (compare 2 and 3 with 1). Importantly, the closure to circular form leads to a significantly larger jump in binding affinity (compare 2 and 3 with 4). The circular ligand binds the target with an equilibrium association constant that is four orders of magnitude higher than the standard linear ligand. Thus, by altering the topology of the DNA strand we can greatly alter its ability to act as a binding agent.
The reasons for this difference appear to be twofold. First, it is evident that formation of the extra noncovalent interactions in the triplex relative to the duplex gives a significant binding advantage. Second, the topological modification also gives an added benefit; this is probably due to entropic effects13. Because a circular molecule is somewhat more rigid than a linear one, it loses less entropy on forming a complex, and so free energy is more favorable for binding.
In addition to binding affinity, there is the important issue of selectivity. A compound that is to be used for a diagnostic probe would ideally bind selectively to the correct target sequence as opposed to targets that might be different by as few as one nucleotide. There are three billion bases of DNA sequence in a human cell, leading to many accidental partial sequence similarities for a given target sequence.
We have found that circular triplex-forming ligands can have very large selectivities against even single mismatches in a desired target sequence14,23. This selectivity is illustrated in Table 1 (5–12), which compares the abilities of linear and circular molecules to discriminate against slightly incorrect target strands of DNA. We define selectivity as the difference in free energy of binding the right sequence versus a mismatched sequence under approximately physiological solution conditions. The data show that a linear strand binding the sequence 5′-AAGAAXAGAAAG (where X is varied) prefers X = A by 14–19 kJ mol−1. The circular DNA, however, prefers X = A by 26–32 kJ mol−1 at 37°C. Thus, we have shown that a circular ligand can discriminate against a single mismatch out of 12 nucleotides by a difference of 4–6 orders of magnitude in equilibrium binding constant (= 1/Kd where Kd is the equilibrium dissociation constant, shown in the table), while the linear probe is selective by only 1–2 orders of magnitude.
Resistance to degradation
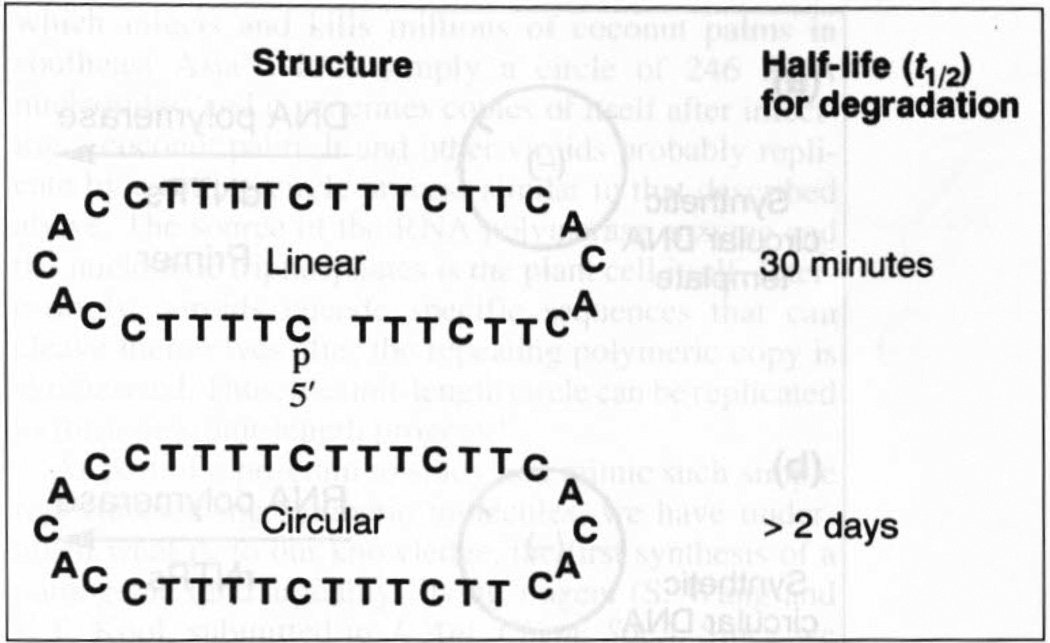
A highly desirable property of molecules to be used as tools or therapeutic agents in biological fluids is the ability to withstand degradation by the various enzymes present. To examine this question for circular DNAs we tested the degradation of linear and circular forms of the same short DNA sequence in fresh undiluted human serum at 37°C (Ref. 24). We found (Fig. 3) that the circular DNA was highly resistant to degradation (actually, no degradation was observed), while the linear molecule had a half-life of c. 30 minutes. The reason for this appears to be that the primary DNA-degrading activity in blood comes from enzymes called exonucleases. These enzymes hydrolyze a DNA strand base by base from one end. Because they lack ends, circles are completely resistant. It remains to be seen whether this resistance will also be evident inside cells or in whole animals. Interestingly, early experiments in our laboratory indicate that circular RNAs in human blood serum are not nearly so resistant to degradation (S. Wang and E.T. Kool, unpublished).
Fig. 3.
Data for the susceptibility to degradation (monitored by gel electrophoresis) of linear and circular DNAs in undiluted human serum at 37°C. The ‘5′ p’ symbol indicates a phosphate group at the 5′ end of the linear strand.
Topological linking
Circular DNAs can also bind single strands without triplex formation if desired, instead forming only standard Watson–Crick bonds11,25. While such binding does not occur with high affinity, it leads to another interesting possibility, which results from the right-handed twist of the duplex. Circular DNAs formed with a template or splint that is longer than the circle itself form a complex in which the template passes through the circle (see Fig. 2). If the template itself is circular (such as a single-stranded virus) then the small circle becomes topologically linked to the target, forming a catenated structure. Such catenated DNA ligands (termed ‘padlock probes’)25 cannot be washed away from their target, and so may serve as efficient diagnostic probes for single-stranded genetic sequences.
Coding for biopolymers: rolling circle DNA synthesis
Not only do DNA and RNA serve as agents for specific molecular recognition, but they also serve as templates that encode genetic information. Copying of a given genetic sequence results in the formation of multiple progeny. This replication is the basis of life on earth.
Circles have some unique advantages in replication1. First, the entire sequence can be copied by an enzyme that simply travels around the circle more than one complete turn, regardless of where it starts. In linear DNAs it is crucial that the copying start at one extreme end and continue completely to the far end, or some genetic information is lost. Moreover, to get a growing population, copying must occur more than once. For a linear DNA this requires that after one copy is made the product strand must dissociate from the template (which is kinetically and thermodynamically unfavorable), and the enzyme must re-initiate back at the beginning. With a circular DNA, however, the copying enzyme simply travels around the circle multiple times to create multiple copies without dissociating. The removal of the copy from the template is also simpler, since one nucleotide of the polymer at a time can dissociate in front of the enzyme, which adds one nucleotide at a time as it proceeds around the circle.
Circular DNAs from viruses are often replicated by such a ‘rolling circle’ mechanism1. These DNAs are typically several thousand to tens of thousands of nucleotides long; they can be manipulated by molecular biological techniques. Replication of these molecules in a test tube requires several types of reagents. First, the nucleotide triphosphates serve as the monomers taken up in the polymer synthesis; these are stitched together by a DNA polymerase enzyme. Also required are DNA-binding proteins and helix-unwinding proteins, which help to remove the product from the circular template in front of the enzyme1.
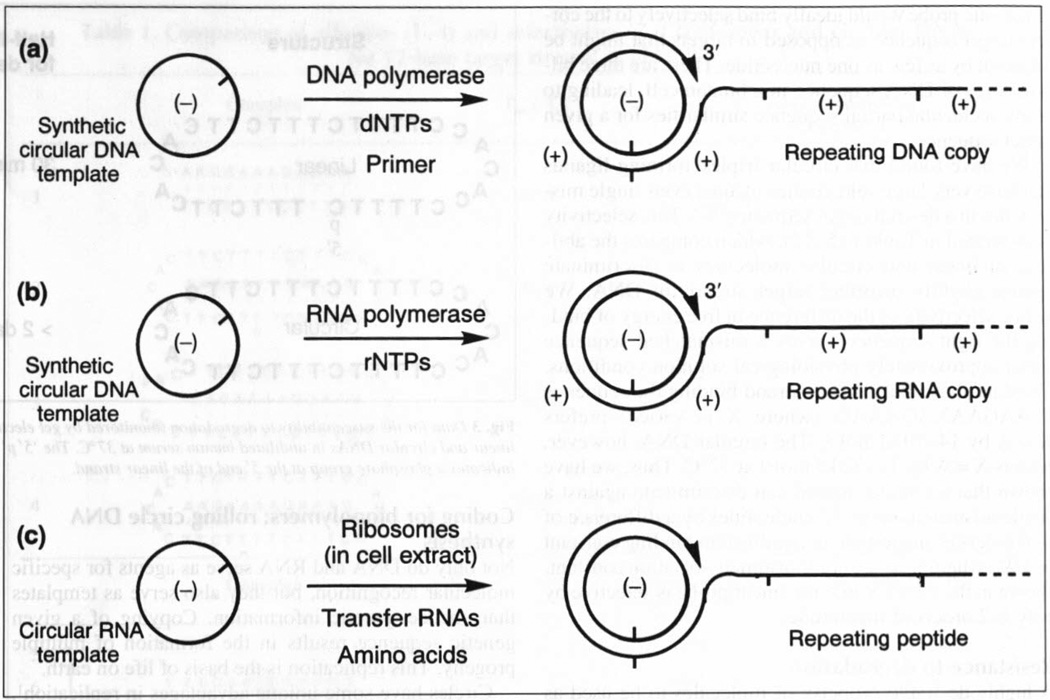
Interestingly, very recent work in two laboratories has shown that much smaller synthetic DNA circles can be replicated in similar, but in some ways more efficient, fashion26,27. Fire and Xu reported in 1995 that circular DNAs as small as 34 nucleotides can be copied simply by supplying a DNA primer (a short DNA strand required by the enzyme to begin synthesis), the four monomer triphosphates (deoxyadenosine triphosphate, deoxycytosine triphosphate, deoxythymidine triphosphate, deoxyguanosine triphosphate), and a DNA polymerase enzyme (Fig. 4a)26. The enzyme extends the primer by inserting the bases encoded by the circle; as it travels around the circle multiple times it produces a long repeating polymer strand of DNA, which consists of complementary copies of the circle joined end-to-end. Treatment of this product with a restriction endonuclease enzyme (which cleaves at a specific short sequence of nucleotides) cleaves it into monomer-length oligonucleotides of specific sequence.
Fig. 4.
Illustrations of rolling circle reactions for synthesis of repairing polymers, (a) Synthesis of DNA, using a DNA circle, a DNA polymerase and dNTP (deoxynucleoside triphosphate) monomers. The end where active synthesis occurs is marked ‘3′’. (b) Synthesis of RNA using a DNA circle, an RNA polymerase and rNTP (ribonucleoside triphosphate) monomers, (c) Synthesis of protein using an RNA circle, ribosomes. tRNAs and amino acid monomers. The ‘notch’ on the circle in each case indicates the initiation site; the (+) and (−) signs are shown to indicate that a copied strand is not identical, but is complementary to the circle in the Watson–Crick sense.
Our laboratory has also been working on this problem. In 1992 we began copying small circular DNAs using this rolling circle strategy (E.T. Kool, US Patent Application, March 1993); in our case the circles originally had been designed to act as ligands, and we decided to test their replication properties as well. We have now shown that DNA circles as small as 26 nucleotides can serve as efficient substrates for a number of commercially available DNA polymerase enzymes (D. Liu, S.L. Daubendiek. M. A. Zillmann and E.T. Kool, submitted to J. Am. Chem. Soc.). The products are repeating DNA polymers and are often longer than 12 000 nucleotides, which means that the enzyme has traveled at least 280 times around the circle before dissociating. We have also demonstrated that the repeating polymers can be cut, using a restriction enzyme, into unit-length oligonucleotides. It should be noted that this is a novel way to amplify a DNA sequence, using the enzyme, the primer and the circle in sub-stoichiometric amounts; only the nucleotide triphosphate monomers are consumed stoichiometrically.
This rolling DNA polymer synthesis from synthetic circles is unusual for at least two reasons. First, replication of larger (viral) circles requires extra proteins (discussed above) to aid in unwinding the product from the circular template1. With very small circles this is unnecessary, possibly because the circles are so small that a double helix cannot be formed all the way around. Thus, unwinding may occur spontaneously. The second unusual aspect is the remarkably small size of the circle: a 26-mer circle has a diameter of ~30 Å, or half that of the polymerase enzyme itself! Clearly, there will be a DNA circle that is too small to be replicated, but we have not yet reached that lower limit.
Rolling circle RNA synthesis
Enzymatic RNA synthesis differs from DNA synthesis in that RNA polymerase enzymes are known to require promoters, rather than primers, for initiation of synthesis28. Specifically, RNA polymerases do not require a short piece of RNA to ‘prime’ the strand but, rather, they read the DNA template sequence and initiate RNA strand synthesis specifically at a certain string of c. 20 nucleotides. This string, called a promoter, serves to signal the enzyme to start RNA synthesis immediately after the promoter.
When considering whether it might be possible to make RNA by a rolling process we therefore expected that it would be necessary to place a promoter sequence within the DNA circle. This is not the case, however. At least in some cases, very small DNA circles can encode long repeating RNA strands even when the circles contain no promoters. We have examined circles ranging from 28 to 74 nucleotides in size, and all give repeating RNA polymers (Fig. 4b) (S.L. Daubendiek and E.T. Kool, unpublished). All that is required is RNA nucleotide triphosphates, the circle and an RNA polymerase enzyme. As was seen for DNA synthesis, we found that long polymers (c. 9000 nucleotides in length) were produced.
Thus, it appears rather general that very small synthetic DNA circles can encode repeating nucleic acid polymers of DNA or of RNA. Potential uses of such repeating polymers might be as probes for biological hybridization experiments, and as encoders of repeating peptides (see below).
Rolling circle protein synthesis
The natural transmission of genetic information leads from double-stranded DNA (the chromosome) to single-stranded RNA (as messenger RNA: mRNA) to protein (the enzymes). DNA is transcribed by RNA polymerase enzymes to give mRNA, and mRNA is read and translated by ribosomes, which assemble proteins of the encoded sequence of amino acids. Above, I have described how circular DNAs can encode repeating polymers of DNA (using DNA polymerases), and how they can encode repeating polymers of RNA (using RNA polymerases). A third, and very interesting, possibility is that circular RNAs might encode repeating polymers of protein, using ribosomes to carry out the translation. Repeating protein polymers have been finding increasing application recently29.
Although translation of RNA to protein is normally carried out starting at one end of linear RNA strands, recently it has been shown that there are some special sequences of RNA, a few hundred nucleotides long, that can signal a ribosome to begin translation in the middle of an RNA strand30. Very recently, Chen and Sarnow showed that it is possible enzymatically to construct a circular RNA that contains such a start signal, and that ribosomes can, in fact, use the circle as a template for translation31. This results in the production of a repeating polypeptide as the ribosome travels multiple times around the circle (Fig. 4c). These authors very cleverly encoded a specific segment of amino acids that can be cleaved with the peptidase enzyme thrombin, and so the repeating polymeric protein could be cleaved predictably into shorter, sequence-defined peptide units.
Natural circular replicators
As was noted in the introduction, circular forms of DNA and RNA in nature are more common than linear forms of the polymers. This is because the organisms that survive today, which use circular DNA topology, in large part outcompeted others that had linear DNAs. Some of the most primitive replicating ‘organisms’ in nature are the viruses (the DNAs of which are mostly circular), which act as parasites on bacteria and on higher organisms. However, an even more primitive type of replicating agent is known: the molecules known as viroids are the smallest known disease-causing agents32. They usually act as parasites in various plant species.
Most relevant to this discussion is the fact that viroids arc small single-stranded circular RNAs. The smallest known viroid is the Coconut Cadang Cadang Viroid, which infects and kills millions of coconut palms in southeast Asia33. It is simply a circle of 246 RNA nucleotides, and it generates copies of itself after infecting a coconut palm. It and other viroids probably replicate by a rolling circle process similar to that described above. The source of the RNA polymerase enzyme and the nucleotide triphosphates is the plant cell itself. Interestingly, viroids encode specific sequences that can cleave themselves after the repeating polymeric copy is synthesized. Thus, the unit-length circle can be replicated to form new unit-length progeny32.
As part of a program to study and mimic such simple replicators using synthetic molecules, we have undertaken what is, to our knowledge, the first synthesis of a pathogenic (and arguably ‘living’) agent (S. Wang and E.T. Kool, submitted to J. Am. Chem. Soc.). Since we have found that small circular DNAs can encode repeating RNA polymers, we hypothesize that a circular DNA version of a viroid might be used to generate the viroid itself by a rolling mechanism, either in a test tube or in a plant cell. We have therefore undertaken the chemical synthesis of a circular 246-nucleotide DNA copy of the Coconut Cadang Cadang Viroid. The strategy we are using is to synthesize four separate quadrants of the molecule ranging in size from 45 to 74 nucleotides long. These are then ligated together using four separate 30-base splints and a DNA ligase enzyme. The ability to construct such natural replicating molecules synthetically would be useful, allowing us to easily make altered versions to study the mechanism of infection and replication.
Prospects for the future
Future studies will undoubtedly uncover additional unusual and useful properties of circular DNAs and RNAs. Circular synthetic DNAs might find use as medical diagnostic agents, where their high sequence selectivity would be beneficial. Synthetic DNAs are also being placed in spatial arrays on solid supports for use in sequencing34, and it is possible that circular structures might find some application in this type of approach. It is even conceivable that cyclic DNAs or their modified analogues might be directly useful as therapeutic agents against viral infections and cancers35,36. Cyclic structures will undoubtedly be combined into larger, more complex molecules that have even better binding properties for DNA and RNA targets37. Beyond the biological properties such as those discussed here, molecules composed of circular DNAs are already being used as architectures for nanometer-scale construction38,39. This will continue in the future as techniques for synthesis and handling of DNAs become yet more sophisticated.
Acknowledgments
We thank the National Institutes of Health (CM46625), the Office of Naval Research, and the Army Research Office for generous support. I also thank my co-workers, whose names appear in the references.
References
- 1.Kornberg A, Baker TA, editors. DNA Replication. 2nd. W.H. Freeman; 1992. p. 113. [Google Scholar]
- 2.Prakash G, Kool ET. J. Am. Chem. Soc. 1992;114:3523. doi: 10.1021/ja00035a056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolinnaya NC, et al. Nucleic Acids Res. 1993;21:5403. doi: 10.1093/nar/21.23.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanaya E, Yanagawa H. Biochemistry. 1986;25:7423. doi: 10.1021/bi00371a026. [DOI] [PubMed] [Google Scholar]
- 5.Dolinnaya NC, Sokolova NI, Gryaznova OI, Shabarova ZA. Nucleic Acids Res. 1988;16:3721. doi: 10.1093/nar/16.9.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashley GW, Kushlan DM. Biochemistry. 1991;30:2927. doi: 10.1021/bi00225a028. [DOI] [PubMed] [Google Scholar]
- 7.Pan T, Gutell RR, Uhlenbeck OC. Science. 1991;254:1361. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- 8.Barbato S, DeNapoli L, Picciali C, Santacroce C. Tetrahedron Lett. 1987;289:5727. [Google Scholar]
- 9.de Vroom E, Broxterman HJG, Sliedregt LAJM, van der Marel CA, van Boom JH. Nucleic Acids Res. 1988;16:4607. doi: 10.1093/nar/16.10.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capobianco ML, Carcuro A, Tondelli L, Garbesi A, Bonora GM. Nucleic Acids Res. 1990;18:2661. doi: 10.1093/nar/18.9.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Kool ET. Nucleic Acids Res. 1994;22:2326. doi: 10.1093/nar/22.12.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kool ET. J. Am. Chem. Soc. 1991;113:6265. [Google Scholar]
- 13.D’Souza DJ, Kool ET. Bioorg. Med. Chem. Lett. 1994;4:965. doi: 10.1016/S0960-894X(01)80664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prakash G, Kool ET. J. Chem. Soc., Chem. Commun. 1991:1161. doi: 10.1039/C39910001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Kool ET. J. Am. Chem. Soc. 1994;116:8857. doi: 10.1021/ja00098a075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strobel SA, Doucette-Stamm LA, Riba L, Housman DE, Dervan PB. Science. 1991;254:1639. doi: 10.1126/science.1836279. [DOI] [PubMed] [Google Scholar]
- 17.Uhlmann E, Peyman A. Chem. Rev. 1990;90:543. [Google Scholar]
- 18.Rubin E, Rumney S, Kool ET. Nucleic Acids Res. 23 doi: 10.1093/nar/23.17.3547. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xodo LE, Manzini G, Quadrifoglio F. Nucleic Acids Res. 1990;18:3557. doi: 10.1093/nar/18.12.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giovannangeli C, Montenay-Garestier T, Rougée M, Chassignol M, Thuong NT, Hétène C. J. Am. Chem. Soc. 1991;113:7775. [Google Scholar]
- 21.D’Souza DJ, Kool ET. J. Biomol. Struct. Dyn. 1992;10:141. doi: 10.1080/07391102.1992.10508634. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Kool ET. Nucleic Acids Res. 1995;23:1157. doi: 10.1093/nar/23.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Kool ET. Biochemistry. 1995;34:9774. doi: 10.1021/bi00030a015. [DOI] [PubMed] [Google Scholar]
- 24.Rumney S, Kool ET. Angew. Chem., Int. Ed. Engl. 1992;31:1617. doi: 10.1002/anie.199216171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BC, Landegren U. Science. 1994;265:2085. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 26.Fire A, Xu S-Q. Proc. Natl Acad. Sci. USA. 1995;92:4641. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daubendiek SL, Ryan K, Kool ET. J. Am. Chem. Soc. 1995;117:7818. doi: 10.1021/ja00134a032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chamberlin M, Ring J. J. Biol. Chem. 1973;248:2235. [PubMed] [Google Scholar]
- 29.Urry DW, Gowda DC, Peng S, Parker TM, ling N, Harris RD. Biopolymers. 1994;34:889. doi: 10.1002/bip.360340708. [DOI] [PubMed] [Google Scholar]
- 30.Le SY, Chen J-H, Sonenberg N, Maizel JV., Jr Nucleic Acids Res. 1993;21:2445. doi: 10.1093/nar/21.10.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C-Y, Sarnow P. Science. 1995;268:415. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 32.Diener TO, editor. The Viroids. Plenum Press; 1987. [Google Scholar]
- 33.Hanold D, Randles JW. Plant Dis. 1991;75:330. doi: 10.1094/PDIS.2003.87.7.875B. [DOI] [PubMed] [Google Scholar]
- 34.Southern EM, et al. Nucleic Acids Res. 1994;22:1368. doi: 10.1093/nar/22.8.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kool ET. In: Delivery Systems for Antisense Oligonucleotide Therapeutics. Akhtar S, editor. CRC Press; 1995. pp. 123–149. [Google Scholar]
- 36.Rowley PT, Thomas MA, Kosciolek BA, Kool ET. J. Cell. Biochem. 1994;18a:235. [Google Scholar]
- 37.Chaudhuri NC, Kool ET. J. Chem. Soc. 117 (in press) [Google Scholar]
- 38.Chen J, Seeman NC. Nature. 1991;350:631. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Seeman NC. J. Am. Chem. Soc. 1994;116:1661. [Google Scholar]