Abstract
Highly oxygenated freshwater habitats in the High Paraná River (Argentina–Paraguay) were home to highly endemic snails of the genus Aylacostoma, which face extinction owing to the impoundment of the Yacyretá Reservoir in the 1990s. Two species, A. chloroticum and A. brunneum, are currently included in an ongoing ex situ conservation programme, whereas A. guaraniticum and A. stigmaticum are presumed extinct. Consequently, the validity and affinities of the latter two have remained enigmatic. Here, we provide the first molecular data on the extinct A. stigmaticum by means of historical DNA analysis. We describe patterns of molecular evolution based on partial sequences of the mitochondrial 12S ribosomal RNA gene from the extinct species and from those being bred within the ex situ programme. We further use this gene to derive a secondary structure model, to examine the specific status of A. stigmaticum and to explore the evolutionary history of these snails. The secondary structure model based on A. stigmaticum revealed that most polymorphic sites are located in unpaired regions. Our results support the view that the mitochondrial 12S region is an efficient marker for the discrimination of species, and the extinct A. stigmaticum is recognized here as a distinct evolutionary genetic species. Molecular phylogenetic analyses revealed a sister group relationship between A. chloroticum and A. brunneum, and estimated divergence times suggest that diversification of Aylacostoma in the High Paraná River might have started in the late Miocene via intra-basin speciation due to a past marine transgression. Finally, our findings reveal that DNA may be obtained from dried specimens at least 80 years after their collection, and confirms the feasibility of extracting historical DNA from museum collections for elucidating evolutionary patterns and processes in gastropods.
Introduction
Freshwater gastropods account for a disproportionate number of documented molluscan extinctions, which are the highest recorded for any major taxonomic group [1–3]. Freshwater molluscs have experienced severe declines as a consequence of a variety of human-mediated impacts, with many species imperilled or facing extinction [2–8]. Known examples include rivers with species-rich assemblages of molluscs such as the Pleuroceridae from the Mobile River basin in the southeastern United States and the Rissooidea from the Mekong River in Southeast Asia, where extensive impoundment of mainstem rivers has resulted in the loss of many species [1–3]. An emblematic example of human-mediated extinction within the Mollusca can be found among South American freshwater snails of the genus Aylacostoma Spix, 1827 (Caenogastropoda: Thiaridae) [9,10]. The members of this endemic genus have been known since the 19th century, but have received little attention since the original descriptions [10–13]. Nonetheless, some Aylacostoma species from the High Paraná River between Argentina and Paraguay have attracted the interest of researchers and conservation biologists owing to an impending extinction in response to human-mediated modifications to their habitat [9,10].
Historically, the highly oxygenated fresh waters near the Yacyretá–Apipé rapids in the High Paraná River were home to five Aylacostoma species: Aylacostoma guaraniticum (Hylton Scott, 1953), Aylacostoma chloroticum Hylton Scott, 1954, Aylacostoma stigmaticum Hylton Scott, 1954, and two others that have never been formally described [9,14–16]. The decline of this mollusc fauna began in the early 1990s in response to the impoundment of the Yacyretá Reservoir for a binational hydroelectric power plant between Argentina and Paraguay [10,13,16]. With the progressive filling of the reservoir, most Aylacostoma populations experienced drastic range reductions, with all species except A. chloroticum going extinct in the wild [13,16,17]. Conservation strategies were adopted during the initial filling stages in the 1990s through an ongoing ex situ conservation programme, known as the “Aylacostoma Project” hosted by the Universidad Nacional de Misiones (UNaM; Posadas, Argentina), in conjunction with the Museo Argentino de Ciencias Naturales (MACN; Buenos Aires, Argentina), and supported by the Entidad Binacional Yacyretá [18]. At present, only A. chloroticum and the recently described Aylacostoma brunneum Vogler & Peso, 2014 [13] are being maintained in captivity. The former species is restricted to only one known wild population in a small and fragile habitat under anthropogenic threat due to the construction of an artificial beach for human recreation. Wild populations of A. brunneum are presumed to have gone extinct after their habitats were flooded in 2011 during the final filling stage of the reservoir [13,16]. Aylacostoma guaraniticum and A. stigmaticum are not represented by any captive population and are presumed extinct [19,20].
Recent morphological and genetic studies investigating the taxonomic status and phylogeography of captive Aylacostoma populations from the High Paraná River, provided critical historical information for maintaining the integrity of evolutionarily lineages in the ex situ breeding program, as well as for future reintroductions and translocations [13,16]. However, owing to extinction, the validity and affinities of A. guaraniticum and A. stigmaticum have remained enigmatic, as the only material available for study in natural history collections dates back to the early 1930s, and is represented only by dried shells of the type series. Moreover, soft-part anatomy is poorly documented for A. guaraniticum [21], and is not available for A. stigmaticum whose radula was characterized for the first time from a dry syntype by Vogler [18].
In the present study, we provide the first molecular data for the extinct freshwater snail A. stigmaticum obtained from a dry syntype. Using information from the third domain of the mitochondrial 12S rRNA gene and a secondary structure model of this region, we assessed the evolutionary relationships among the extinct species and those successfully being bred in captivity. We further examined the validity of A. stigmaticum using two methods implemented for single locus data, and estimated divergence times in order to establish a timeframe for diversification. The goal of this study is to provide insights into the evolutionary history of Aylacostoma snails, to shed light on the possible scenarios involved in the diversification and evolution of this endemic genus in the High Paraná River.
Material and Methods
Samples and DNA extraction
Total genomic DNA of A. stigmaticum was obtained from a dry syntype housed in the malacological collection of the Museo de La Plata (MLP 10965; La Plata, Argentina) from which the radula was recovered by Vogler [18] following a modification of the non-destructive method described by Holznagel [22]. The shell was rinsed with deionized water, placed in a 10 ml tube, with 1.5 ml of NET buffer (10 mM Tris, 10 mM EDTA, 2% SDS) and 10 μl of proteinase K (20 mg/ml), and incubated at 37°C for seven days; an identical volume of proteinase K was added on the third day. After incubation, a gelatinous mass containing the radula was recovered from inside the shell. This mass was further incubated at 56°C with 500 μl of NET buffer and 10 μl of proteinase K for 24 hrs, eventually producing the clean radula. DNA was purified from the remaining solution by a threefold extraction, one with phenol and two with chloroform-isoamyl alcohol (24:1) followed by precipitation with cold isopropanol. DNA was then resuspended in 50 μl of 10 mM Tris/HCl. In addition, a genomic DNA collection of Aylacostoma specimens from the ongoing ex situ conservation programme at UNaM collected in the High Paraná River between 1994 and 2011 was made available at MLP by Vogler et al. [13]. We selected 13 samples from that collection belonging to A. chloroticum, A. brunneum and the outgroups Doryssa sp., Pachychilus nigratus (Poey, 1858), and Pachychilus laevissimus (Sowerby, 1825). Samples of A. chloroticum included representatives of both haplotype clades [16]. Collection information and GenBank accession numbers for the samples analysed is presented in Table 1.
Table 1. Collection information and GenBank accession numbers for the samples analysed herein.
| Species | Locality | Coordinates | Year | N | GenBank accession nos. |
|---|---|---|---|---|---|
| Aylacostoma stigmaticum | Isla Ibicuy, PY* | 27°17′56.76′′S; 56°3′28.44′′W** | 1934 | 1 | KU168372 |
| Aylacostoma brunneum | Ita Cuá, PY | 27°24′42.13′′S; 55°48′45.69′′W | 2007 | 1 | KU168374 |
| Río Beach, PY* | 27°24′29.83′′S; 55°49′32.94′′W | 2007 | 2 | KU168373, KU168375 | |
| Aylacostoma chloroticum | Candelaria, AR | 27°26′50.96′′S; 55°45′0.84′′W | 2008 | 4 | KU168376 –KU168379 |
| 2011 | 2 | KU168380, KU168381 | |||
| Río Beach, PY | 27°24′29.83′′S; 55°49′32.94′′W | 2007 | 1 | KU168382 | |
| Doryssa sp.*** | Venezuela | – | 2010 | 1 | KU168383 |
| Pachychilus laevissimus*** | Venezuela | – | 2010 | 1 | KU168384 |
| Pachychilus nigratus*** | Cuba | – | 1997 | 1 | KU168385 |
AR, Argentina; PY, Paraguay; N, number of specimens analysed.
*Type locality.
**The coordinates are approximate; only the locality name was reported in the original description and the precise location is uncertain.
***Outgroup species.
PCR amplification and sequencing
Repeated attempts to amplify partial fragments of the mitochondrial cytochrome c oxidase subunit I (COI) and cytochrome b (cyt b) genes of A. stigmaticum following the methodology of Vogler et al. [13] were unsuccessful. Consequently, the mitochondrial 12S rRNA gene (hereafter 12S) was selected for further characterization based on previous research showing its potential utility in other gastropods (e.g. [23–25]). Partial sequences of the 12S gene were amplified with the 12SF (5′-AAC TCA AAG GAC TTG GCG GTG C-3′) and 12SR (5′-GTT TTT TTA CTT TCA AGT CCT CC-3′) primers [24]. PCR was performed in a total volume of 50 μl containing 30–50 ng of template DNA, 0.5 μM of each primer, 1X PCR buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, and 1.5 U Platinum Taq polymerase (Invitrogen, Brazil). The thermocycling profile was 1 min at 94°C followed by 40 cycles of 30 s at 94°C, 30 s at 55°C, 1 min at 72°C. Amplifications were run on a T18 thermocycler (Ivema Desarrollos). Success of PCR reactions was verified by agarose gel electrophoresis. Owing to the co-amplification of nonspecific fragments, PCR products were purified from 1.5% (w/v) agarose gel through the use of a Zymoclean Gel DNA Recovery Kit (Zymo Research, Orange, California). After purification, both DNA strands were then directly cycle sequenced (Macrogen Inc, Seoul, Korea). The resulting sequences were trimmed to remove the primers, and the consensus sequences compared with reference sequences in GenBank through the use of the BLASTN algorithm [26] to rule out potential contamination. Multiple alignment was performed with LocARNA through the Freiburg RNA Tools webserver (http://rna.informatik.uni-freiburg.de) [27,28] with manual corrections as necessary.
Sequence data and phylogenetic analyses
Several strategies were used to quantify and understand the distribution of polymorphisms in Aylacostoma. The number and nucleotide composition of haplotypes in Aylacostoma species were analysed in BioEdit 7.0.9 [29]. Nucleotide substitutions were examined in relation to conserved sequence motifs, alignment and secondary structure of the third domain of the 12S gene predicted for molluscs and other invertebrates. A secondary structure model was derived following the template proposed by Hickson et al. [23] and contrasted by comparison with structural diagrams for other molluscs (e.g. [30]). Genetic distances were estimated in MEGA 6.06 [31] using the number of differences (p) and the Kimura´s two-parameter (K2P) substitution model. Phylogenetic analyses were performed using neighbour-joining (NJ), maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI). The NJ analysis was conducted with MEGA 6.06 with the K2P model. The MP analysis was carried out using PAUP*4.0b10 [32] via the branch and bound algorithm (for smaller datasets) with gaps treated as missing and character states treated as unordered and equally weighted. For the ML analysis, the optimal model of nucleotide substitution (TrN+G) was selected with the likelihood-ratio test by means of the corrected Akaike Information Criterion (AICc) as implemented in jModelTest 2.1.7 [33]. The ML analysis was conducted in PhyML [34] via the Phylemon2 webserver (http://phylemon.bioinfo.cipf.es) [35]. Nodal support values were assessed by bootstrapping with 1000 (NJ, MP) and 100 (ML) replicates [36]. The BI was performed with MrBayes 3.2.6 [37] with the parameters from the best model (HKY+G) as identified by jModelTest under the Bayesian information criterion. Two runs were performed simultaneously with four Markov chains for 1000000 generations, sampling every 100 generations, with a burn-in of 10%.
Species delimitation
Two approaches were used to explore species boundaries in the Aylacostoma dataset: the Automatic Barcode Gap Discovery (ABGD) method [38], and the K/θ method [39,40]. The ABGD is a non-tree-based distance method that has been used to define primary species hypotheses [41–44]. The aligned dataset (excluding outgroups) was analysed via the ABGD web-server (http://wwwabi.snv.jussieu.fr/public/abgd/), with default parameters using the K2P model. The K/θ method was conducted to assess the status of Aylacostoma species under the Evolutionary Genetic Species Concept (EGSC) [39,40]. This method, based on population genetic theory, requires a gene tree to identify putative sister clades and distance matrices to estimate genetic variation within and between clades [42]. It has recently been used to describe A. brunneum based on partial sequences of the COI gene, and was suggested by Vogler et al. [13] as a useful framework for comprehensively reviewing the status of the species in this genus. The K/θ method was performed following the procedures described in Schön et al. [45] and Birky [40]. In order to fulfil the K/θ criteria, sister clades must have K/θ ≥ 4 to be considered different species with probability ≥ 0.95 ([40] and references therein). Mean pairwise differences between clades were estimated in MEGA 6.06.
Divergence time estimates
In order to estimate relative divergence times, the molecular clock hypothesis was tested in MEGA 6.06 using Tajimas´s non-parametric relative rate test [46], and by means of a likelihood-ratio test on ML trees with and without enforcing a molecular clock. Divergence time (T) was estimated following Vogler et al. [16] as T = Da/2μ, where Da is the net nucleotide divergence [47] and 2μ indicates the divergence rate (hereafter net divergence approach). Net divergence and standard errors (SE) were estimated in MEGA 6.06 under the K2P model with 1000 bootstrap replicates. 95% confidence intervals (CI) were calculated as ±1.96 SE of the net distances [48]. Divergence times were also estimated using a Bayesian approach implemented in BEAST 1.8.3 [49] under the HKY+G model (as identified by jModelTest under the Bayesian information criterion), assuming a strict clock and the Yule process to model speciation. The posterior distribution of divergence times was obtained by a MCMC sampling run of 100000000 generations, sampling every 1000 generations [50]. Convergence was confirmed using Tracer 1.6.0 [51,52]. Estimation of the time to the most recent common ancestors (TMRCAs), and associated 95% high posterior density (HPD) intervals were obtained after removing a burn-in of 10%. For both time estimation approaches, we used a divergence rate of 0.6% per million year based on Rumbak et al. [53], which has been widely used in phylogenetic and phylogeographic studies of gastropods (e.g. [54,55]).
Results
Sequence data and phylogenetic analyses
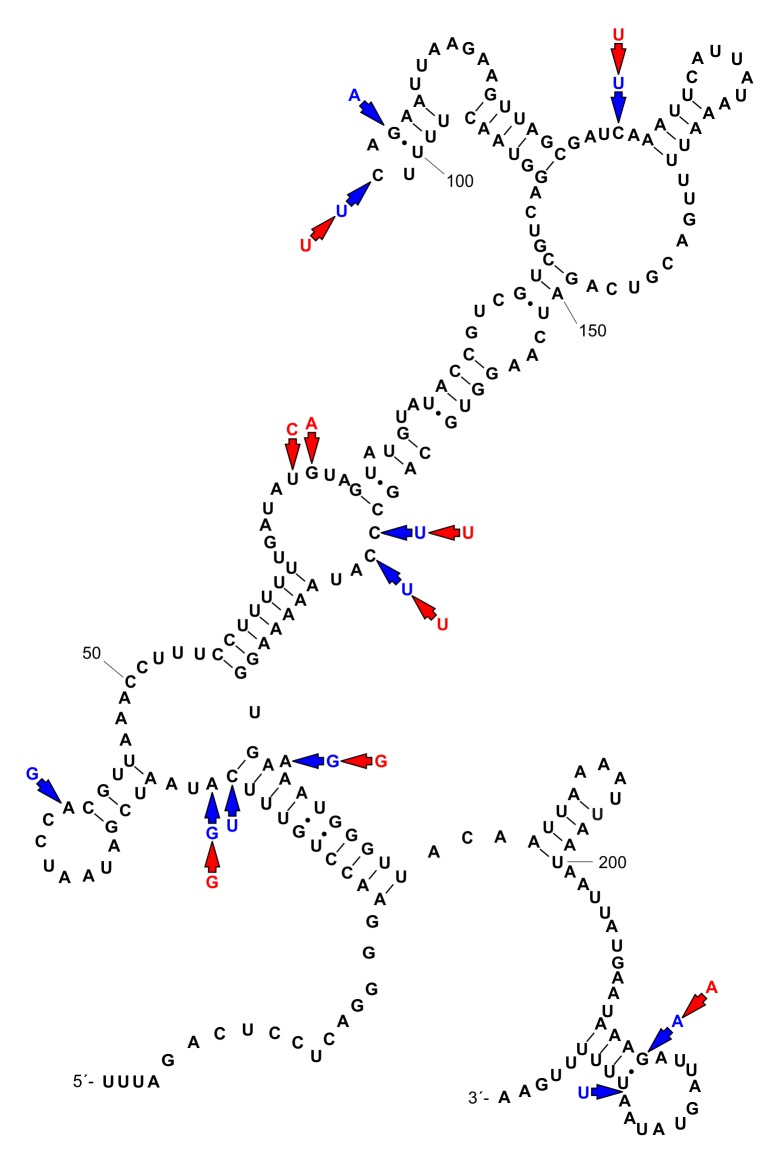
Partial 12S sequences of Aylacostoma individuals from the High Paraná River consisted of 233 base pairs (bp) for A. stigmaticum and A. brunneum, and 234 bp for A. chloroticum. There were 13 variable positions in the sequence alignment (234 bp). Each species was characterized by a single haplotype. Aylacostoma stigmaticum differed from A. brunneum by nine nucleotides, and from A. chloroticum by ten nucleotides and one indel. Aylacostoma brunneum and A. chloroticum differed in five nucleotides and one indel (Table 2). Base frequencies, as well as AT and GC content are shown in Table 3. The secondary structure was conserved amongst the three species with ten polymorphic sites occurring in unpaired regions; the three remaining variable positions involved stems and represented alternative basepairing not affecting the secondary structure (Fig 1). Sequence divergences are presented in Table 4.
Table 2. Polymorphic positions based on a 234 bp fragment of the 12S mt rRNA gene for Aylacostoma species.
| 27 | 28 | 42 | 67 | 68 | 102 | 104 | 123 | 163 | 164 | 177 | 214 | 225 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aylacostoma stigmaticum | C | A | A | T | G | C | G | C | C | C | A | G | – |
| Aylacostoma brunneum | ∙ | G | ∙ | C | A | T | ∙ | T | T | T | G | A | – |
| Aylacostoma chloroticum | T | G | G | ∙ | ∙ | T | A | T | T | T | G | A | T |
Numbers indicate the position of variable sites. Aylacostoma stigmaticum is shown as reference sequence; dot indicates identity with the reference sequence, dash represent a gap.
Table 3. Nucleotide composition of 12S mt rRNA sequences for Aylacostoma species.
| A | C | G | T | AT content | GC content | |
|---|---|---|---|---|---|---|
| Aylacostoma stigmaticum | 81 (34.76%) | 33 (14.17%) | 39 (16.74%) | 80 (34.33%) | 69.10% | 30.90% |
| Aylacostoma brunneum | 81 (37.76%) | 30 (12.88%) | 39 (16.74%) | 83 (35.62%) | 70.39% | 29.61% |
| Aylacostoma chloroticum | 80 (34.19%) | 28 (11.97%) | 40 (17.09%) | 86 (36.75%) | 70.94% | 29.06% |
Fig 1. Secondary structure model of the third domain of the 12S mt rRNA gene for Aylacostoma stigmaticum.
Mutational changes for A. brunneum and A. chloroticum are indicated by red and blue arrows, respectively (see Table 2).
Table 4. Genetic distances between Aylacostoma species.
Uncorrected (below the diagonal) and corrected (K2P; above the diagonal) distances are shown.
| Aylacostoma stigmaticum | Aylacostoma brunneum | Aylacostoma chloroticum | |
|---|---|---|---|
| Aylacostoma stigmaticum | – | 0.040200 | 0.044873 |
| Aylacostoma brunneum | 0.038627 | – | 0.021933 |
| Aylacostoma chloroticum | 0.042918 | 0.021459 | – |
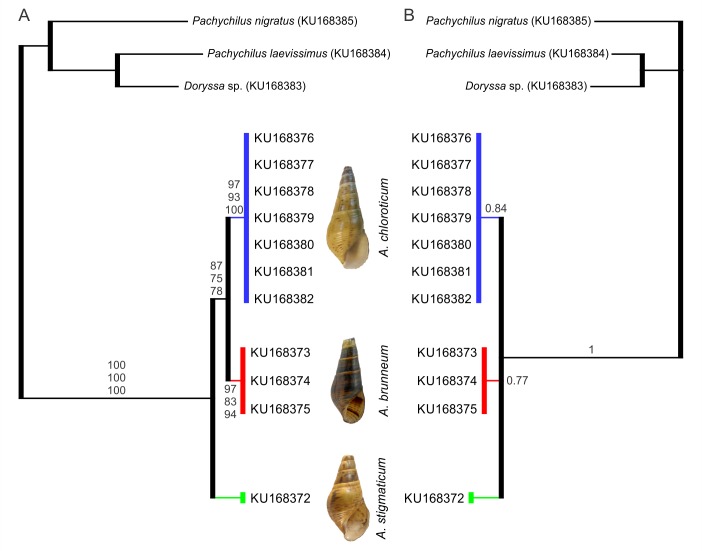
Results of the NJ, MP, ML and BI analyses all supported Aylacostoma as monophyletic with high bootstrap and posterior probability support (Fig 2). This group was further separated into three phylogenetic lineages, which represented A. chloroticum, A. brunneum and A. stigmaticum. In the results of the NJ, MP, and ML analyses, A. chloroticum and A. brunneum were supported as sister taxa, while in the results of the BI analysis, the relationships among species were unresolved.
Fig 2. Phylogenetic trees of Aylacostoma species from the High Paraná River based on partial sequences of the of 12S mt rRNA gene.
A, neighbour-joining tree. Bootstrap values for NJ, MP and ML trees, respectively, are shown above and below the branches. B, Bayesian consensus tree with posterior probabilities. Numbers within groups are GenBank accession numbers.
Species delimitation
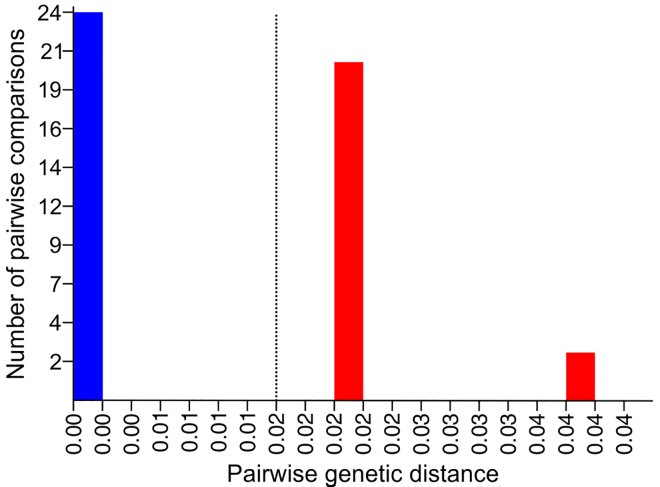
The ABGD approach revealed a trimodal pairwise genetic distance (K2P) distribution (Fig 3) and resulted in seven sequence partitions that were stable across three groups (A. chloroticum, A. brunneum, and A. stigmaticum) at prior maximum intraspecific divergence values of 0.0215 and below (Table 5). The results of the K/θ method are shown in Table 6. All of the K/θ ratios were higher than 4 and the EGSC criteria were clearly fulfilled. Thus, A. chloroticum, A. brunneum, and A. stigmaticum were recognized as distinct evolutionary genetic species.
Fig 3. Frequency distribution of pairwise genetic distances for the 12S mt rRNA gene in the ABGD analysis.
Pairwise distances were calculated using the K2P model. Dashed line corresponds to maximum value of intraspecific genetic divergences that resulted in stable candidate species.
Table 5. Results of ABGD analysis for the 12S mt rRNA dataset using the K2P model to calculate pairwise distances.
| Partition no. | Groups | P-value |
|---|---|---|
| 1 | 3 | 0.001000 |
| 2 | 3 | 0.001668 |
| 3 | 3 | 0.002783 |
| 4 | 3 | 0.004642 |
| 5 | 3 | 0.007743 |
| 6 | 3 | 0.012915 |
| 7 | 3 | 0.021544 |
P, prior maximum intraspecific divergence.
Table 6. K/θ ratio calculations for Aylacostoma 12S mt rRNA sequences.
| θ | K | K/θ ratio | n1, n2 | |
|---|---|---|---|---|
| A. chloroticum–A. brunneum | 0.0043165 | 0.0219333 | 5.08 | 7, 3 |
| A. brunneum–A. stigmaticum | 0.0043165 | 0.0402002 | 9.31 | 3, 1 |
| A. chloroticum–A. stigmaticum | 0.0014333 | 0.0448731 | 31.31 | 7, 1 |
θ, mean pairwise sequence difference within a clade; K, mean pairwise sequence difference between clades; n1, n2, number of sequences within each of the clades compared.
Divergence time estimates
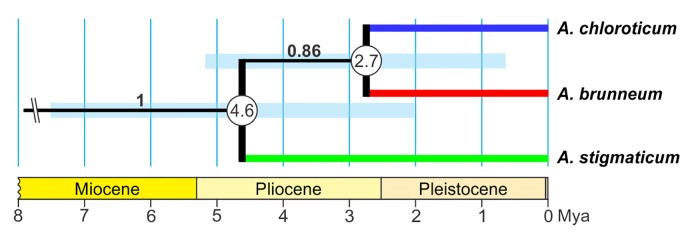
Tajima’s relative rate test and likelihood-ratio test indicated a clock like behaviour for the 12S dataset. Divergence between A. stigmaticum and the common ancestor of A. chloroticum and A. brunneum is inferred to have occurred roughly 6.57 (net divergence)– 4.64 (Bayesian) million years ago (mya), between the late Miocene and mid-Pliocene (Fig 4, Table 7). The maximum time interval for this divergence is estimated to be 10.63–2.04 mya (Table 7 and S1 Table). Subsequently, A. brunneum and A. chloroticum are estimated to have split between 3.75 (net divergence)– 2.71 (Bayesian) mya, between the mid-Pliocene and early Pleistocene (Fig 4, Table 7). The maximum time interval inferred for this split is estimated to be 0.54–6.97 mya (Table 7 and S1 Table).
Fig 4. Divergence times for Aylacostoma species obtained from BEAST analysis of the 12S mt rRNA dataset.
Numbers within circles indicate divergence times in millions of years. Bars at each node represent 95% confidence intervals. Posterior probabilities are shown on branches. Time axis (in millions of years) adapted from the International Chronostratigraphic Chart [56].
Table 7. Divergence times among Aylacostoma species based on net divergence and Bayesian approaches.
| Net divergence approach | Bayesian approach | |||
|---|---|---|---|---|
| Node | T = Da/2μ | 95% CI | Mean | 95% HPD |
| A. stigmaticum / A. chloroticum–A. brunneum | 6.57 | 2.50–10.63 | 4.64 | 2.04–7.56 |
| A. chloroticum / A. brunneum | 3.75 | 0.54–6.97 | 2.71 | 0.67–5.21 |
T, divergence time; Da, net nucleotide divergence; 2μ, divergence rate; CI, confidence interval; HPD, highest posterior density interval.
Discussion
Material deposited in natural history collections represent a valuable, but limited, resource for providing critical anatomical, morphological, ecological and genetic information on extinct species [57,58]. Historical DNA has enabled ecological, evolutionary, taxonomic and conservation studies, including for example, the identification of “extinct species” in captivity misassigned to extant taxa, refinement of evolutionary relationships, as well as the identification of cryptic diversity [59–61].
As for most invertebrates, genomic quality tissues are not usually available in museum collections, as most specimens were not preserved for this purpose [58,59]. Acquiring genetic information from shells is possible but has remained challenging as protocols for extracting DNA from empty shells have met with limited success, and in some cases requires complete dissolution of the shells [58,62]. In the present study, dissolution of shells of the extinct A. stigmaticum was not possible, since the only material available for study was a name-bearing type. Consequently, DNA was obtained here in a similar way as described by Caldeira et al. [63] by means of an extensive incubation period, after which the shell was recovered and returned to the MLP collection. From such DNA, it was possible to amplify and generate partial sequences of the 12S gene, although we failed to amplify the COI and cyt b regions. These results are in agreement with expectations for DNA obtained from historical material which is expected to be highly degraded, and PCR amplification generally restricted to short amplicons (<200 bp) [58,59]. Amplification of the standard COI barcoding region in molluscs has proven to be particularly problematic or impossible with historical material using standard primers [64].
Although failure to amplify the COI and cyt b genes for A. stigmaticum prevent us from making comparisons with available sequence data for Aylacostoma, comparisons of 12S sequences yielded results almost identical to those found for COI and cyt b by Vogler et al. [13] in terms of molecular identification while showing lower interspecies variability (ca. 2.1% for 12S vs 4.41% for COI and 6.65% for cyt b) reflecting its slower substitution rate [65,66]. Not unexpectedly, then, 12S did not capture the haplotype clade diversity in A. chloroticum previously characterized with the COI gene [13,16]; consequently, this marker appears unsuitable for exploring intraspecific variation in these species.
Consistent with the findings of Lydeard et al. [67,68] and Ramírez & Ramírez [69] for the mitochondrial LSU rRNA gene (16S) of molluscs, the secondary structure model derived here demonstrated that most of the sequence variation among the Aylacostoma species was allocated in loops. As this is the first such structural model for Aylacostoma, comparisons with other congeners are presently not possible, although our results suggest that the 12S region and the secondary structure model based on the extinct A. stigmaticum have considerable potential for further comparative analyses of structural changes within the genus and other molluscan taxa.
Despite the apparent lack of utility of the 12S gene to assess intraspecific variability in Aylacostoma, our findings support this region as an efficient marker for the discrimination of species. This was demonstrated herein by means of two species delimitation approaches, where three genetic lineages were identified by the ABGD method, all of which fulfilled the criteria of the Evolutionary Genetic Species Concept. Consequently, this confirms A. stigmaticum as a distinct evolutionary genetic species, and the previous treatment of A. chloroticum and A. brunneum as evolutionary genetic species based on COI data [13]. Interestingly, the validity of the extinct A. stigmaticum was questioned by de Castellanos [21] on the basis of shared features of the shell with the other species inhibiting the High Paraná region. The original description of A. stigmaticum was based solely on shell features: a conical to ovate shell, yellow horn colour, low spire, last whorl somewhat convex; surface almost smooth, last whorl sculptured by low spiral cords, a spiral band of reddish spots, and the presence of irregular black spots on the surface of all specimens, with the latter being the most conspicuous feature [15,18]. Until now, apart from the fewer number of whorls, its relatively low spire and the inflation of the body whorl, the distinctive coloration pattern represents the most notable distinguishing feature of A. stigmaticum, whereas A. chloroticum is characterized by greenish-yellow to greenish-brown shells, with some specimens bearing minute, dark reddish brown, spiral spots, while A. brunneum has dark brown shells with alternating lighter bands, delicately decorated with minute, dark reddish brown, regularly spaced spiral spots [13]. Moreover, the present results complement those of Vogler et al. [70] who conducted a geometric morphometric analysis of available museum material for A. guaraniticum, A. chloroticum and A. stigmaticum documenting significant differences in shell shape, in which A. stigmaticum was found to possess a comparatively more globose shell and more ovate aperture. However, in that study affinities could not be assessed based only on overall similarity of the shell, as convergence in shell shape and ecophenotypic variation could not be rejected as confounding factors. Here, with the exception of the results of the BI analysis which were unresolved, phylogenetic analyses consistently supported a sister group relationship between A. chloroticum and A. brunneum, with the extinct A. stigmaticum grouping outside. It is important to note that the sequences of A. chloroticum presented an indel, treated as “missing” in the phylogenetic analyses, which generated a somewhat overestimated similarity among the species. DNA sequences for the extinct A. guaraniticum are still unavailable and further research is required to understand the evolutionary framework shaping diversification of Aylacostoma in the High Paraná River. However, by placing the present results into the context of previous research, the extinct A. stigmaticum provides interesting new insights into several aspects of the evolution of the High Paraná River species.
The Paraná River, the second largest of South American rivers, becomes the High Paraná River from the former Guairá Falls along the Brazil–Paraguay border, now flooded by the Itaipú Reservoir, which comprised a series of imposing falls with walls 100 m high [71]. Downstream, the river runs along the Argentina–Paraguay border along a fault line across a broad basaltic plateau known as Alto Paraná Encajonado [72], and continues without major changes until near 56°W, where it turns westward. From this point and before the impoundment of the Yacyretá Reservoir, the course historically formed a large and complex system of extensive anastomosing meanders and more than 300 islands, including many deeper passages and rapids in a floodplain 25–30 km wide extending to the Yacyretá–Apipé rapids, now the site of the Yacyretá dam [9,71,73,74]. This unique stretch of river has been considered as a biogeographic crossroads by Arzamendia & Giraudo [75] and is currently recognized as a transition zone between the Atlantic Forest and Chaco biogeographical provinces. Although the origins of its endemic molluscan fauna have been little investigated, bearing in mind that all Aylacostoma species from Argentina–Paraguay have been described from such a particular stretch of the river within a narrow distribution range, the perception of A. stigmaticum as a distinct, endemic species, further highlights that the now flooded convergence zone of the High Paraná River and the Alto Paraná Encajonado was a micro-hotspot supporting a highly endemic molluscan fauna [76,77].
Geomorphological history has been implicated in playing a major role in the evolution of Aylacostoma species in the High Paraná River [9,10,16]. In this context, divergence times obtained with 12S for the split between A. chloroticum and A. brunneum yielded an equivalent time estimate to that reported by Vogler et al. [16] using the COI gene, and suggest a divergence between the mid-Pliocene and early Pleistocene. The split between A. stigmaticum and the common ancestor of A. chloroticum and A. brunneum likely occurred roughly 6.57–4.64 mya between the late Miocene and mid-Pliocene. These divergence estimates are consistent with known episodes in the geomorphological history of the High Paraná River, which may have driven the intra-basin diversification of Aylacostoma. During the middle and late Miocene, the “Paranense Sea” is recognized as one of the most widespread Atlantic marine transgressions that covered parts of Argentina, Uruguay, and southern portions of Brazil, Bolivia and Paraguay [78–80]. Miocene marine transgressions are considered to have played a significant role in species origin, diversification and distribution patterns of South America diversity, particularly among its freshwater fauna [81–84]. Geological evidence supports two cycles of marine transgression into the Paraná basin in the late Miocene, the first between 15–13 mya and the second between 10–5 mya [78]. The latter coincides with our estimates for the split between A. stigmaticum and the common ancestor of A. chloroticum and A. brunneum. Consequently, as for the fish of the Paraná River [84], it can be hypothesized that marine incursion into the Paraná basin fragmented the range of a continuously distributed ancestor promoting speciation. On the other hand, as has been suggested by Vogler et al. [16], the more recent divergence between A. chloroticum and A. brunneum may have occurred in a large and heterogeneous alluvial fan complex of the Paleoparaná [85] which formed after the withdrawal of the “Paranense Sea”, through the colonization of transitional environments, in which geographic fragmentation and ecological specialization promoted differentiation. Thus, Aylacostoma snails from the High Paraná River may represent a species flock as the result of a geographically localized adaptive radiation during the late Miocene and Pliocene. However, as stated by Vogler et al. [16] further paleontological studies are required to understand the likely complex interplay between spatial, biological and ecological factors driving speciation of Aylacostoma in this region [86].
Finally, the data presented herein establish an evolutionary framework for future comparative analyses of Aylacostoma and other South American thiarids, in which most of the species are known primarily from shells in natural history collections. Our results reveal that DNA can be obtained from dried specimens of freshwater species at least 80 years after their collection, similar to previous findings in land snails. As demonstrated here historical DNA can provide invaluable information for elucidating evolutionary patterns and processes, and we suggest this resource be more frequently incorporated into the malacological toolbox for further studies aimed at reconstructing the tempo and mode of evolution in gastropods.
Supporting Information
See text for details.
(DOCX)
Acknowledgments
We thank Dr. Cristina Damborenea and Dr. Gustavo Darrigran for authorizing us to test DNA extraction from the type series of A. stigmaticum housed in Museo de La Plata. We thank Dr. Geerat Vermeij and Dr. Frank Wesselingh as reviewers for providing constructive suggestions to improve the manuscript.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1. Lydeard C, Cowie RH, Ponder WF, Bogan AE, Bouchet P, Clark SA, et al. The global decline of nonmarine mollusks. Bioscience. 2004; 54:321–330. [Google Scholar]
- 2.Strong EE, Gargominy O, Ponder WF, Bouchet P. Global diversity of gastropods (Gastropoda; Mollusca) in freshwater. Hydrobiologia. 2008; 595:149–166. [Google Scholar]
- 3.Johnson PD, Bogan AE, Brown KM, Burkhead NM, Cordeiro JR, Garner JT, et al. Conservation status of freshwater gastropods of Canada and the United States. Fisheries. 2013; 38:247–282. [Google Scholar]
- 4.Strayer DL. Challenges for freshwater invertebrate conservation. J North Am Benthol Soc. 2006; 25:271–287. [Google Scholar]
- 5.Régnier C, Fontaine B, Bouchet P. Not knowing, not recording, not listing: numerous unnoticed mollusk extinctions. Conserv Biol. 2009; 23:1214–1221. 10.1111/j.1523-1739.2009.01245.x [DOI] [PubMed] [Google Scholar]
- 6.Geist J. Strategies for the conservation of endangered freshwater pearl mussels (Margaritifera margaritifera L.): a synthesis of conservation genetics and ecology. Hydrobiologia. 2010; 644:69–88. [Google Scholar]
- 7.Whelan NV, Johnson PD, Harris PM. Rediscovery of Leptoxis compacta (Anthony, 1854) (Gastropoda: Cerithioidea: Pleuroceridae). PLoS ONE. 2012; 7:e42499 10.1371/journal.pone.0042499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Régnier C, Bouchet P, Hayes KA, Yeung NW, Christensen CC, Chung DJD, et al. Extinction in a hyperdiverse endemic Hawaiian land snail family and implications for the underestimation of invertebrate extinction. Conserv Biol. 2015; 29:1715–1723. 10.1111/cobi.12565 [DOI] [PubMed] [Google Scholar]
- 9.Quintana MG. Aylacostoma in Yacyretá, South America. Tentacle. 1997; 7:17–20. [Google Scholar]
- 10.Quintana MG, Mercado Laczkó AC. Caracoles de los rápidos en Yacyretá. Ciencia Hoy. 1997; 7:22–31. [Google Scholar]
- 11.Simone LRL. Phylogenetic analysis of Cerithioidea (Mollusca: Caenogastropoda) based on comparative morphology. Arq Zool. 2001; 36:147–263. [Google Scholar]
- 12.Simone LRL. Land and freshwater molluscs of Brazil. São Paulo: EGB, Fapesp; 2006. [Google Scholar]
- 13.Vogler RE, Beltramino AA, Peso JG, Rumi A. Threatened gastropods under the evolutionary genetic species concept: redescription and new species of the genus Aylacostoma (Gastropoda: Thiaridae) from High Paraná River (Argentina-Paraguay). Zool J Linn Soc. 2014; 172:501–520. [Google Scholar]
- 14.Hylton Scott MI. El género Hemisinus (Melaniidae) en la costa fluvial argentina (Mol. Prosobr.). Physis. 1953; 20:438–443. [Google Scholar]
- 15.Hylton Scott MI. Dos nuevos melánidos del Alto Paraná (Mol. Prosobr.). Neotropica. 1954; 1:45–48. [Google Scholar]
- 16.Vogler RE, Beltramino AA, Strong EE, Peso JG, Rumi A. A phylogeographical perspective on the ex situ conservation of Aylacostoma (Thiaridae, Gastropoda) from the High Paraná River (Argentina–Paraguay). Zool J Linn Soc. 2015; 174:487–499. [Google Scholar]
- 17.Vogler RE. Aylacostoma chloroticum Hylton Scott, 1954: antecedentes de la especie. Amici Molluscarum. 2012; 20:43–46. [Google Scholar]
- 18.Vogler RE. The radula of the extinct freshwater snail Aylacostoma stigmaticum (Caenogastropoda: Thiaridae) from Argentina and Paraguay. Malacologia. 2013; 56:329–332. [Google Scholar]
- 19.Peso JG, Costigliolo Rojas C, Molina MJ. Aylacostoma stigmaticum Hylton Scott, 1954: antecedentes de la especie. Amici Molluscarum. 2013; 21:43–46. [Google Scholar]
- 20.Peso JG, Molina MJ, Costigliolo Rojas C. Aylacostoma guaraniticum (Hylton Scott, 1953): antecedentes de la especie. Amici Molluscarum. 2013; 21:39–42. [Google Scholar]
- 21.de Castellanos ZJA. La familia Thiaridae Morrison 1952 en la Argentina In: Ringuelet RA, editor. Fauna de agua dulce de la República Argentina. Buenos Aires: Fundación para la Educación, la Ciencia y la Cultura; 1981. pp. 7–18. [Google Scholar]
- 22.Holznagel WE. A nondestructive method for cleaning gastropod radulae from frozen, alcohol fixed, or dried material. Am Malacol Bull. 1998; 14:181–183. [Google Scholar]
- 23.Hickson RE, Simon C, Cooper A, Spicer GS, Sullivan J, Penny D. Conserved sequence motifs, alignment, and secondary structure for the third domain of animal 12s rRNA. Mol Biol Evol. 1996; 13:150–169. [DOI] [PubMed] [Google Scholar]
- 24.Facon B, Pointier JP, Glaubrecht M, Poux C, Jarne P, David P. A molecular phylogeography approach to biological invasions of the New World by parthenogenetic Thiarid snails. Mol Ecol. 2003; 12:3027–3039. [DOI] [PubMed] [Google Scholar]
- 25.Collado GA, Valladares MA, Méndez MA. Hidden diversity in spring snails from the Andean Altiplano, the second highest plateau on Earth, and the Atacama Desert, the driest place in the world. Zool Stud. 2013; 52:50. [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990; 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 27.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010; 38:W373–W377. 10.1093/nar/gkq316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Will S, Joshi T, Hofacker IL, Stadler PF, Backofen R. LocARNA-P: Accurate boundary prediction and improved detection of structural RNAs. RNA. 2012; 18:900–914. 10.1261/rna.029041.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999; 41:95–98. [Google Scholar]
- 30.Medina M, Walsh PJ. Molecular systematics of the order Anaspidea based on mitochondrial DNA sequence (12S, 16S, and COI). Mol Phylogenet Evol. 2000; 15:41–58. 10.1006/mpev.1999.0736 [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland: Sinauer Associates; 2002. [Google Scholar]
- 33.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012; 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003; 52:696–704. [DOI] [PubMed] [Google Scholar]
- 35.Sánchez R, Serra F, Tárraga J, Medina I, Carbonell J, Pulido L, et al. Phylemon 2.0: a suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 2011; 39:W470–W474. 10.1093/nar/gkr408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985; 39:783–791. [DOI] [PubMed] [Google Scholar]
- 37.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012; 61:539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puillandre N, Lambert A, Brouillet S, Achaz G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol. 2012; 21:1864–1877. 10.1111/j.1365-294X.2011.05239.x [DOI] [PubMed] [Google Scholar]
- 39.Birky CW Jr, Adams J, Gemmel M, Perry J. Using population genetic theory and DNA sequences for species detection and identification in asexual organisms. PLoS ONE. 2010; 5:e10609 10.1371/journal.pone.0010609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Birky CW Jr. Species detection and identification in sexual organisms using population genetic theory and DNA sequences. PLoS ONE. 2013; 8:e52544 10.1371/journal.pone.0052544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jörger KM, Norenburg JL, Wilson NG, Schrödl M. Barcoding against a paradox? Combined molecular species delineations reveal multiple cryptic lineages in elusive meiofaunal sea slugs. BMC Evol Biol. 2012; 12:245 10.1186/1471-2148-12-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fontaneto D, Flot JF, Tang CQ. Guidelines for DNA taxonomy, with a focus on the meiofauna. Mar Biodivers. 2015; 45:433–451. [Google Scholar]
- 43.Shrestha Y, Wirshing HH, Harasewych MG. The genus Cerion (Gastropoda: Cerionidae) in the Florida Keys. PLoS ONE. 2015; 10:e0137325 10.1371/journal.pone.0137325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evangelisti F, Bonfitto A, Morassi M, Sabelli B. How many native Cerithium species in the Mediterranean Sea? An integrative taxonomic approach. J Molluscan Stud. 2016; 82:292–304. [Google Scholar]
- 45.Schön I, Pinto RL, Halse S, Smith AJ, Martens K, Birky CW Jr. Cryptic species in putative ancient asexual darwinulids (Crustacea, Ostracoda). PLoS ONE. 2012; 7:e39844 10.1371/journal.pone.0039844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993; 135:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 48.Brunhoff C, Galbreath KE, Fedorov VB, Cook JA, Jaarola M. Holarctic phylogeography of the root vole (Microtus oeconomus): implications for late Quaternary biogeography of high latitudes. Mol Ecol. 2003; 12:957–968. [DOI] [PubMed] [Google Scholar]
- 49.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012; 29:1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collado GA, Valladares MA, Méndez MA. Unravelling cryptic species of freshwater snails (Caenogastropoda, Truncatelloidea) in the Loa River basin, Atacama Desert. Syst Biodivers. 2016; 14:417–429. [Google Scholar]
- 51.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer version 1.6. 2014; Available: http://beast.bio.ed.ac.uk/Tracer. Accessed 01 August 2016
- 52.Greve C, Hutterer R, Groh K, Haase M, Misof B. Evolutionary diversification of the genus Theba (Gastropoda: Helicidae) in space and time: a land snail conquering islands and continents. Mol Phylogenet Evol. 2010; 57:572–584. 10.1016/j.ympev.2010.08.021 [DOI] [PubMed] [Google Scholar]
- 53.Rumbak E, Reid DG, Thomas RH. Reconstruction of phylogeny of 11 species of Littorina (Gastropoda, Littorinidae) using mitochondrial DNA sequence data. Nautilus. 1994; 108 (Supplement 2):91–97. [Google Scholar]
- 54.Ketmaier V, Giusti F, Caccone A. Molecular phylogeny and historical biogeography of the land snail genus Solatopupa (Pulmonata) in the peri-Tyrrhenian area. Mol Phylogenet Evol. 2006; 39:439–451. 10.1016/j.ympev.2005.12.008 [DOI] [PubMed] [Google Scholar]
- 55.Ketmaier V, Manganelli G, Tiedemann R, Giusti F. Peri-Tyrrhenian phylogeography in the land snail Solatopupa guidoni (Pulmonata). Malacologia. 2010; 52:81−96. [Google Scholar]
- 56.International Commission on Stratigraphy. International chronostratigraphic chart v 2016/04. 2016; Available: http://www.stratigraphy.org/index.php/ics-chart-timescale. Accessed 01 August 2016
- 57.Whelan NV. Radular morphology of extinct pleurocerids (Gastropoda: Cerithioidea: Pleuroceridae). Am Malacol Bull. 2015; 33:1–6. [Google Scholar]
- 58.Villanea FA, Parent CE, Kemp BM. Reviving Galápagos snails: ancient DNA extraction and amplification from shells of probably extinct Galápagos endemic land snails. J Molluscan Stud. 2016; 82:449–456. [Google Scholar]
- 59.Wandeler P, Hoeck PEA, Keller LF. Back to the future: museum specimens in population genetics. Trends Ecol Evol. 2007; 22:634–642. 10.1016/j.tree.2007.08.017 [DOI] [PubMed] [Google Scholar]
- 60.Gubili C, Robinson CEC, Cliff G, Wintner SP, de Sabata E, De Innocentiis S, et al. DNA from historical and trophy samples provides insights into white shark population origins and genetic diversity. Endanger Species Res. 2015; 27:233–241. [Google Scholar]
- 61.Russello MA, Poulakakis N, Gibbs JP, Tapia W, Benavides E, Powell JR, et al. DNA from the past informs ex situ conservation for the future: an “extinct” species of Galápagos tortoise identified in captivity. PLoS ONE. 2010; 5:e8683 10.1371/journal.pone.0008683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andree KB, López MA. Species identification from archived snail shells via genetic analysis: a method for DNA extraction from empty shells. Molluscan Res. 2013; 33:1–5. [Google Scholar]
- 63.Caldeira RL, Jannotti-Passos LK, Lira PM, Carvalho OS. Diagnostic of Biomphalaria snails and Schistosoma mansoni: DNA obtained from traces of shell organic materials. Mem Inst Oswaldo Cruz. 2004; 99:499–502. [DOI] [PubMed] [Google Scholar]
- 64.Jaksch K, Eschner A, Rintelen TV, Haring E. DNA analysis of molluscs from a museum wet collection: a comparison of different extraction methods. BMC Res Notes. 2016; 9:348 10.1186/s13104-016-2147-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueller RL. Evolutionary rates, divergence dates, and the performance of mitochondrial genes in Bayesian phylogenetic analysis. Syst Biol. 2006; 55:289–300. 10.1080/10635150500541672 [DOI] [PubMed] [Google Scholar]
- 66.Machida RJ, Kweskin M, Knowlton N. PCR Primers for Metazoan mitochondrial 12S ribosomal DNA sequences. PLoS ONE. 2012; 7:e35887 10.1371/journal.pone.0035887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lydeard C, Holznagel WE, Schnare MN, Gutell RR. Phylogenetic analysis of molluscan mitochondrial LSU rDNA sequences and secondary structures. Mol Phylogenet Evol. 2000; 15:83–102. 10.1006/mpev.1999.0719 [DOI] [PubMed] [Google Scholar]
- 68.Lydeard C, Holznagel WE, Ueshima R, Kurabayashi A. Systematic implications of extreme loss reduction of mitochondrial LSU rRNA helical-loop structures in gastropods. Malacologia. 2002; 44:349–352. [Google Scholar]
- 69.Ramírez J, Ramírez R. Analysis of the secondary structure of mitochondrial LSU rRNA of Peruvian land snails (Orthalicidae: Gastropoda). Rev Peru Biol. 2010; 17:53–57. [Google Scholar]
- 70.Vogler RE, Beltramino AA, Gutiérrez-Gregoric DE, Peso JG, Griffin M, Rumi A. Threatened Neotropical mollusks: analysis of shape differences in three endemic snails from High Paraná River by geometric morphometrics. Rev Mex Biodivers. 2012; 83:1045–1052. [Google Scholar]
- 71.Bonetto AA. The Paraná River system In: Davies B, Walker KF, editors. The ecology of river systems. Dordrecht: Dr. W. Junk Publishers; 1986. pp. 541–555. [Google Scholar]
- 72.Matteucci SD, Morello J, Rodríguez AF, Mendoza NE. El Alto Paraná Encajonado argentino-paraguayo: mosaicos de paisaje y conservación regional. Buenos Aires: Ediciones FADU-UNESCO; 2004. [Google Scholar]
- 73.Davidson H. Informe de una espedicion al Alto Paraná para estudiar las mejoras necesarias en el “Salto Grande de Apipé”. Buenos Aires: Establecimiento tipográfico de La Pampa; 1882. [Google Scholar]
- 74.Neiff JJ. Las grandes unidades de vegetación y ambiente insular del río Paraná en el tramo Candelaria-Itá Ibaté. Rev Asoc Cienc Nat Litoral. 1986; 17:7–30. [Google Scholar]
- 75.Arzamendia V, Giraudo AR. A panbiogeographical model to prioritize areas for conservation along large rivers. Divers Distrib. 2012; 18:168–179. [Google Scholar]
- 76.Rumi A, Gutiérrez Gregoric DE, Núñez V, César II, Roche MA, Tassara MP, et al. Freshwater Gastropoda from Argentina: species richness, distribution patterns, and an evaluation of endangered species. Malacologia. 2006; 49:189–208. [Google Scholar]
- 77.Gutiérrez Gregoric DE, de Lucía M. Freshwater gastropods diversity hotspots: three new species from the Uruguay River (South America). PeerJ. 2016; 4:e2138 10.7717/peerj.2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hernández RM, Jordan TE, Dalenz Farjat A, Echavarría L, Idlemand BD, Reynolds JH. Age, distribution, tectonics, and eustatic controls of the Paranense and Caribbean marine transgressions in southern Bolivia and Argentina. J South Am Earth Sci. 2005; 19:495–512. [Google Scholar]
- 79.Brea M, Zucol AF. The Paraná-Paraguay Basin: geology and paleoenvironments In: Albert JS, Reis RE, editors. Historical biogeography of Neotropical freshwater fishes. Berkeley: University of California Press; 2011. pp. 69–87. [Google Scholar]
- 80.Candela AM, Bonini RA, Noriega JI. First continental vertebrates from the marine Paraná Formation (Late Miocene, Mesopotamia, Argentina): Chronology, biogeography, and paleoenvironments. Geobios. 2012; 45:515–526. [Google Scholar]
- 81.Hubert N, Renno JF. Historical biogeography of South American freshwater fishes. J Biogeogr. 2006; 33:1414–1436. [Google Scholar]
- 82.Hubert N, Duponchelle F, Nuñez J, Garcia-Davila C, Paugy D, Renno JF. Phylogeography of the piranha Serrasalmus and Pygocentrus: implications for the diversifications of the Neotropical ichthyofauna. Mol Ecol. 2007; 16:2115–2136. 10.1111/j.1365-294X.2007.03267.x [DOI] [PubMed] [Google Scholar]
- 83.Fuchs J, Chen S, Johnson JA, Mindell DP. Pliocene diversification within the South American Forest falcons (Falconidae: Micrastur). Mol Phylogenet Evol. 2011; 60: 398–407. 10.1016/j.ympev.2011.05.008 [DOI] [PubMed] [Google Scholar]
- 84.Cardoso YP, Almirón A, Casciotta J, Aichino D, Lizarralde MS, Montoya-Burgos JI. Origin of species diversity in the catfish genus Hypostomus (Siluriformes: Loricariidae) inhabiting the Paraná river basin, with the description of a new species. Zootaxa. 2012; 3453:69–83. [Google Scholar]
- 85.Herbst R. La Formación Ituzaingó (Plioceno). Estratigrafía y distribución. Ser Correl Geol. 2000; 14:181–190. [Google Scholar]
- 86.Glaubrecht M. Towards solving Darwin’s “mystery”: speciation and radiation in lacustrine and riverine freshwater gastropods. Am Malacol Bull. 2011; 29:187–216. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See text for details.
(DOCX)
Data Availability Statement
All relevant data are within the paper.