Abstract
Background
Metabolomics represents a promising approach for discovering novel targets and biomarkers in head and neck squamous cell carcinoma (HNSCC). Here we used metabolomics to identify oral metabolites associated with HNSCC.
Methods
Tumor and adjacent normal tissue from surgical resections and presurgical oral washes as well as oral washes were collected from healthy participants. Metabolites extractions of these samples were analyzed by liquid chromatography-mass spectroscopy (LC/MS), LC/MS/MS and gas chromatography-MS (GC/MS).
Results
Among 28 samples obtained from 7 HNSCC cases and 7 controls, 422 metabolites were detected (269 identified and 153 unidentified). Oral washes contained 12 and 23 metabolites in healthy controls and HNSCC patients, respectively, with phosphate and lactate being the most abundant. Small molecules related to energy metabolism were significantly elevated in HNSCC patients compared to controls. Levels of beta-alanine, alpha-hydroxyisovalerate, tryptophan, and hexanoylcarnitine were elevated in HNSCC oral washes compared to healthy controls (range 7.8-12.2-fold). Resection tissues contained 22 metabolites, of which eight were overproduced in tumor by 1.9- to 12-fold compared to controls. TCA cycle analogs 2-hydroxyglutarate (2-HG) and 3-GMP were detected exclusively in tumor tissues. Targeted quantification of 2-HG in a representative HNSCC patient showed increase in tumor tissue (14.7 μg/mL), but undetectable in normal tissue. Moreover, high levels of 2-HG were detected in HNSCC cell lines but not in healthy primary oral keratinocyte cultures.
Conclusions
Oral metabolites related to energy metabolism were elevated in HNSCC, and acylcarnitine and 2HG may have potential as non-invasive biomarkers. Further validation in clinical studies is warranted.
Keywords: HNSCC, Metabolomics, 2-Hydroxyglutarate, Biomarker, Hexanoylcarnitine
Highlights
-
•
Metabolomics identified HNSCC-associated oral metabolites in tissues and oral wash.
-
•
Oral metabolites related to energy metabolism were elevated in HNSCC.
-
•
2-hydroxyglutarate was detected in HNSCC tumor but not in adjacent normal tissues.
-
•
2-hydroxyglutarate and acylcarnitine may have potential as non-invasive biomarkers.
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide, accounting for > 600.000 new cases/year [1]. HNSCC affects squamous cells lining the oral cavity, nasopharynx, pharynx, and larynx. Despite the existence of curative treatment modalities for HNSCC, clinical outcome remains poor, with < 50% surviving 5 years. Affected individuals usually present at advanced stages with multiple sites of nodal metastatic spread. High mortality is due to high rates of loco-regional recurrence (25%) and developing a second primary malignancy (25%). Moreover, distant metastatic disease is uniformly fatal.
Despite major achievements in HNSCC clinical management and research over the last 30 years, overall outcome does not reflect these achievements for the majority of patients with advanced disease. One reason for this lack of progress is availability of diagnostic tools. In both primary and recurrence diagnosis, early detection leads to decreased mortality and morbidity, since treatment of advanced stage HNSCC can lead to loss of swallowing and speech, as well as significant disfigurement. Direct laryngoscopy and biopsy, the current gold standard of HNSCC diagnosis is time-consuming and requires general anesthesia. Moreover, primary sites, especially in the tonsil and base of tongue, are often not found due to the constraints of anatomy and limitations of gross visual inspection. Therefore, identifying feasible candidate approaches to the screening for early detection of new or recurrent HNSCC would be clinically advantageous and transformative to surgical and oncologic practice.
Another obstacle to achieving progress in HNSCC treatment is the lack of correlation between identified molecular targets and the efficacy of targeted treatment. For example, epidermal growth factor receptor (EGFR) overexpression has been identified in 10-30% of HNSCC tumors [2]; however, multiple clinical trials testing treatment regimens containing EGFR inhibitors have not shown correlation with biomarkers including EGFR overexpression [3], [4], [5], [6]. The implication of these data is that key links in the pathogenesis and progression of HNSCC remain to be elucidated.
Metabolomics (simultaneous global analysis of small molecule metabolites and metabolic patterns) is emerging as a useful tool to identify diagnostic biomarkers and drug targets. Metabolic profiles of biofluids can be altered by a variety of physiological/pathological processes, and therefore global changes in such profiles may signal the presence of a particular disease state [7], [8], [9]. Oral or serum levels of metabolites in HNSCC cells have previously been profiled and compared with healthy volunteers [10], [11], [12], [13]. In the current study, we obtained sets of matched “Trios” of samples (resection tissue of tumor and adjacent normal tissue, and oral wash) from HNSCC patients to gain further insight into their metabolomic profiles. We conducted the current exploratory study using metabolomics to identify metabolites present at differential levels in oral wash of patients with HNSCC compared to matched non-diseased participants as well as in tumor tissue compared to matched normal oral tissue.
2. Methods
2.1. Study participants
Table 1.
Demographic summary of enrolled patients with HNSCC.
| ID | Age | Sex | Ethnicity | Tumor location | TNM stage | Prior chemo/radiotherapy | Tobacco exposure | Alcohol exposure |
|---|---|---|---|---|---|---|---|---|
| 192 | 73 | M | WHITE | Retromolar trigone | T2 N0 M0 | No/yes | Never | No |
| 198 | 72 | F | White | Tongue | T3 N0 M0 | No/no | Never | No |
| 205 | 63 | M | White | Supraglottis | T3 N2c M1 | No/no | 1 packs/d, for 40 yrs.-quit | No |
| 206 | 50 | F | White | Supraglottis | T2 N2c M0 | Yes/yes | 4-5 cigarettes/d | 5-15 cans of beer/week |
| 207 | 51 | M | White | Lip | T4 N0 M0 | No/no | 1 packs/d, for 30 yrs | 12 pack/wk |
| 210 | 66 | M | White | Larynx | T3 N3 M0 | No/no | 2 packs/d, for 50 yrs | No |
| 212 | 50 | F | White | Floor of mouth | T4 N0 M0 | No/yes | 0.5 packs/d, for 27 yrs | 2 beers/d |
2.2. Sample collection
Two types of samples were collected: tissue from surgical resections and oral washes. For each HNSCC patient, tissue samples of approximately 50 mg each were sterilely obtained from both tumor and adjacent normal tissue approximately 2 cm away from the tumor. Tissue was flash frozen and stored at -80C until extraction. Thawed tissues were homogenized with stainless steel beads in a TissueLyserII (Qiagen, Valencia, CA) and sonicated with a Sonic Dismembrator Model 100 (Fisher Scientific, Waltham, MA), centrifuged at 12,000 rpm for 10 min, with the resulting supernatants used for metabolomic analyses. Oral washes were obtained from HNSCC patients and corresponding matched healthy controls. Oral wash supernatants were extracted with acetonitrile, and then analyzed for the presence of metabolites. Metabolite profiles from both tissues and oral washes were performed in collaboration with Metabolon (Durham, NC).
2.3. Metabolomics analyses
Levels of metabolites were determined using a MS/MS, GC/MS platform as described previously by our group [14].
2.4. Sample processing
Samples were subjected to a series of organic and aqueous extractions to remove the protein fraction while allowing maximum recovery of small molecules, and processed using the automated MicroLab STAR® system (Hamilton Company, Reno, NV). Recovery standards were added prior to the extraction process for quality control (QC) purposes. The resulting extract was divided into two fractions: one for analysis by LC and the second for analysis by GC. Residual organic solvent was removed under vacuum, each sample was then frozen and dried under vacuum, and prepared for LC/MS or GC/MS. A small aliquot of each experimental sample for a specific matrix was obtained and pooled together as a “Client matrix” (CMTRX). Aliquots of these CMTRX samples were injected throughout the platform day run and served as technical replicates. Such analysis allows monitoring of variability in the quantitation of the detected metabolites in the samples. With this monitoring, a metric for overall process variability can be assigned for the platform's performance based on the quantitation of metabolites in the actual experimental samples.
2.5. LC/MS, LC/MS2
The LC/MS portion of the platform is based on a Waters Acquity UPLC and a Thermo-Finnigan LTQ mass spectrometer, which consists of an electrospray ionization (ESI) source and linear ion-trap (LIT) mass analyzer. The sample extract was split into two aliquots, dried, then reconstituted in acidic or basic LC-compatible solvents, each of which contain 11 or more injection standards at fixed concentrations. One aliquot was analyzed using acidic positive ion optimized conditions and the other using basic negative ion optimized conditions in two independent injections using separate dedicated columns. Extracts reconstituted in acidic conditions were gradient eluted using water and methanol both containing 0.1% formic acid, while the basic extracts, which also used water/methanol, contained 6.5 mM ammonium bicarbonate. The MS analysis alternates between MS and data-dependent MS2 scans using dynamic exclusion.
2.6. Mass determination and MS/MS fragmentation (LC/MS), (LC/MS/MS), GC/MS
The LC/MS accurate mass portion of the platform was based on a Waters Acquity UPLC and a Thermo-Finnigan LTQ-FT mass spectrometer, which has a LIT front end and a Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer backend. For ions with counts > 2 million, an accurate mass measurement can be performed. Accurate mass measurements can be made on the parent ion as well as fragments. The typical mass error is < 5 ppm. Ions with < 2 million counts require a greater amount of effort to characterize. Fragmentation spectra (MS/MS) are typically generated in data-dependent manner, but if necessary, targeted MS/MS can be employed, such as in the case of lower level signals. The samples destined for GC/MS analysis were redried under vacuum desiccation for a minimum of 24 h prior to being derivatized under dried nitrogen using bistrimethyl-silyl-triflouroacetamide (BSTFA). The GC column used was 5% phenyl and the temperature ramp was from 40° to 300 °C in a 16-min period. Samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization. The instrument was tuned and calibrated for mass resolution and mass accuracy daily. The information output from the raw data files was automatically extracted as discussed below.
2.7. Bioinformatics
The informatics system consisted of four major components, the Laboratory Information Management System (LIMS), the data extraction and peak-identification software, data processing tools for QC and compound identification, and a collection of information interpretation and visualization tools. The hardware and software foundations for these informatics components were the LAN backbone, and a database server running Oracle 10.2.0.1 Enterprise Edition.
2.8. Compound identification
Biochemicals were identified by comparison to library entries of purified standards or recurrent unknown entities. Approximately 1500 commercially available purified standard biochemicals have been acquired and registered into LIMS for distribution to both the LC and GC platforms, for determination of their analytical characteristics. The combination of chromatographic properties and mass spectra give an indication of a match to the specific compound or an isobaric entity. Additional chemical entities can be identified by virtue of their recurrent nature (both chromatographic and mass spectral). These biochemicals have the potential to be identified by future acquisition of a matching purified standard or by classical structural analysis.
2.9. Curation and normalization
A variety of curation procedures were carried out to ensure that a high-quality data set was made available for statistical analysis and data interpretation. The QC and curation processes were designed to ensure accurate and consistent identification of true chemical entities, and to remove those representing system artifacts, mis-assignments, and background noise. Library matches for each compound were checked for each sample and corrected if necessary. For studies spanning multiple days, a data normalization step was performed to correct variation resulting from instrument inter-day tuning differences. Essentially, each compound was corrected in run-day blocks by registering the medians to equal 1.00 and normalizing each data point proportionately (termed the “block correction”). For studies that do not require > 1 day of analysis, no normalization was necessary, other than for purposes of data visualization.
2.10. Sequencing analysis of IDH (isocitrate dehydrogenase) genes
Genomic DNA was extracted from normal and tumor tissue of HNSCC patients using a previously described protocol [15]. Mutation screening utilized PCR amplification of genomic DNA followed by direct sequencing of IDH1 and IDH2 in both forward and reverse directions using the ABI3730x1 sequencer (Applied Biosystems, Foster City, CA) with previously published primers [16]. Variant classification was performed using MutationTaster2 [17].
2.11. Quantitation of 2-HG in HNSCC tissue samples and cell lines
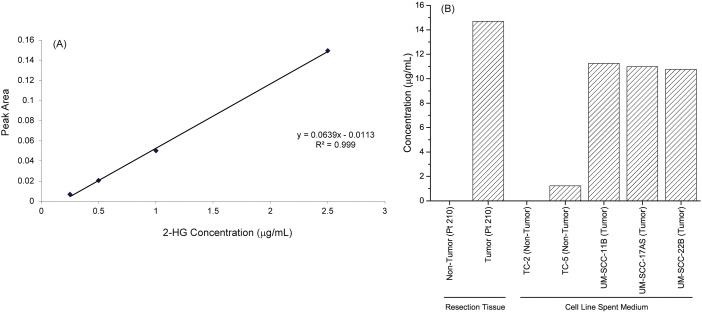
Since 2-HG was found to be unique to tumor tissue from HNSCC patients on global metabolic profiling, we independently confirmed its levels in tissue samples obtained from a representative HNSCC patient (#210). Additionally we assayed 2HG levels in spent media of three HNSCC cell lines, UM-SCC-11B, UM-SCC-17as, and UM-SCC-22B, and two primary epithelial cell cultures obtained from healthy individuals. UM-SCC cell lines were a gift from the Thomas Carey lab, University of Michigan. Cell lines were grown in DMEM with FBS and penicillin/streptomycin and collected at 80-95% confluency following trypsinization. The resulting cell suspensions were centrifuged, and the supernatants collected for metabolite assays. Sample preparation was carried out as described earlier [18]. Briefly, 500 μL of extracted supernatant was added to 50% acetonitrile (v/v) to precipitate the hydroxyl acid fraction. The precipitate was resuspended in 75% isopropanol (v/v), and samples were dried for GC/MS with negative electrospray ionization (GCTOF). A four-point calibration curve was generated using pure 2-HG (Sigma Aldrich, St. Louis) and used to determine the concentration of 2-HG in test samples.
2.12. Statistical analysis
Metabolomic data were analyzed using the R statistical computing language [19], and packages “lattice” [20] and “ggplot2” [21]. SPSS (ver. 22) was used to perform non-parametric tests (Kruskal-Wallis, Wilcoxon). On initial analysis, citrate was found to be the most abundant metabolite in samples obtained from HNSCC and non-diseased samples (≥ 98%) and its levels did not vary significantly between the two groups (P > 0.05). This finding served as an internal control confirming quality of metabolite extraction and analytic pipeline given that our tissue samples were extracted with citrate buffer. Final analyses of the metabolite dataset were completed after removal of citrate levels. A P-value of < 0.05 was considered statistically significant for all comparisons, and was adjusted for multiple testing.
3. Results
3.1. Clustering of metabolites in oral wash and resection tissues
Global metabolomic profiles were determined and compared across disease status (cancer vs. non-diseased) for each sample type (oral wash and tissue samples). Among the 28 samples obtained from the enrolled 7 HNSCC cases and 7 controls, 422 total metabolites were detected, comprising 269 identified metabolites and 153 unidentified molecules (Supplementary Table 1). Comparison of oral wash and tissue extract samples by hierarchical clustering showed that they grouped separately, thus these tissue types were considered separately in subsequent analyses.
3.2. Metabolomic profiling of oral wash samples
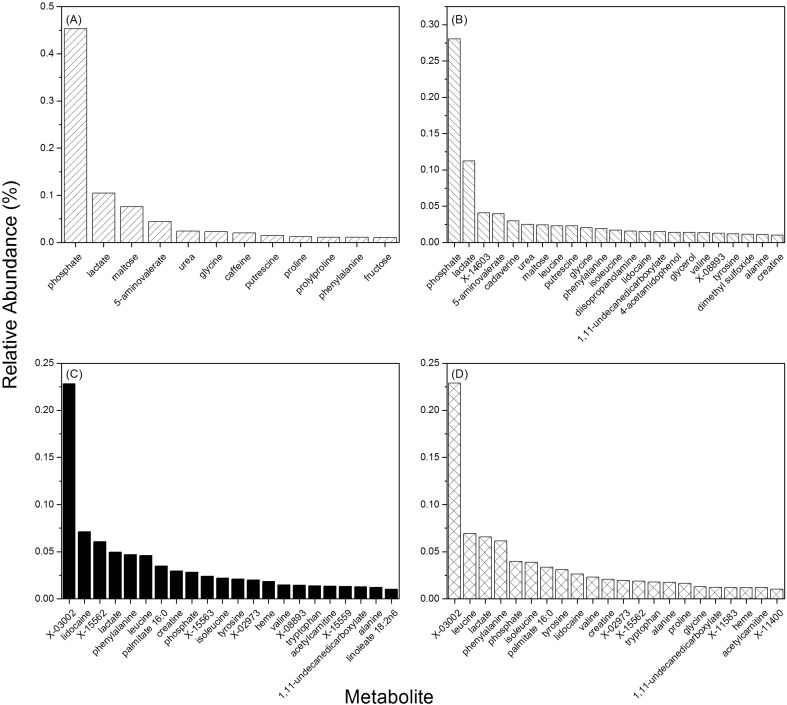
Fig. 1.
Metabolites with abundance ≥ 1% in oral wash and resection tissue. (A) Oral wash from non-diseased or (B) HNSCC patients; (C) Adjacent normal tissue or (D) tumor tissues for surgical resection of HNSCC patients.
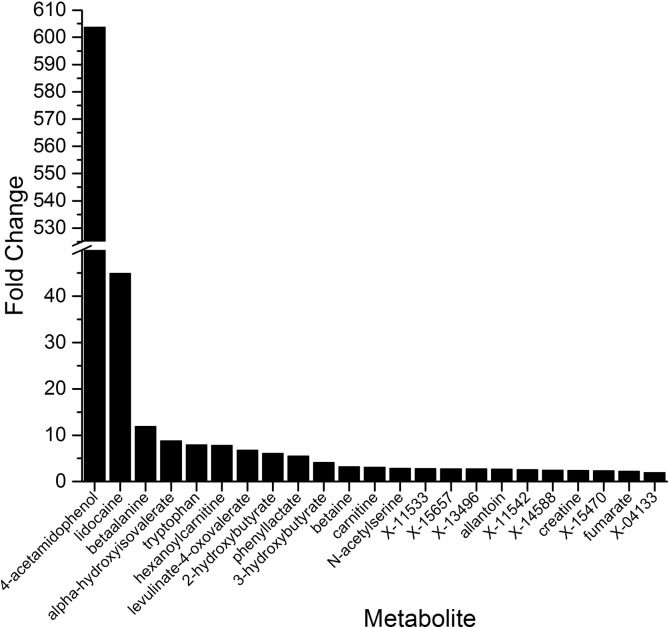
Fig. 2.
Fold-change in levels of metabolites detected in oral wash of HNSCC patients compared to oral wash of non-diseased individuals. Bars represent HNSCC:normal ratio of metabolite levels (P ≤ 0.038 for all comparisons; P-value range = 0.038-5.83E-41).
3.3. Metabolomic profiling of HNSCC tissue
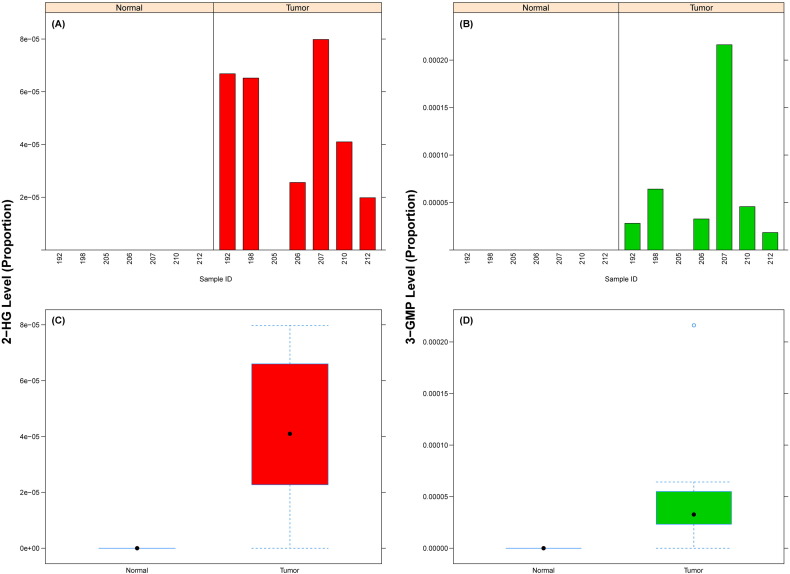
Fig. 3.
Relative levels of (A,C) 2-hydroxyglutarate (2-HG) and (B,D) glycerol-3-monophopsphate (3-GMP) in tissue samples. (A,B) levels across all samples, and (C,D) box-plots showing summary distribution for each group (Normal or Tumor).
Fig. 4.
Directed measurement of 2-hydroxyglutarate (2-HG). (A) Standard curve using pure 2-HG, (B) 2-HG levels in tissue samples from a representative patient (Pt. 210), and in spent media of non-tumor and tumor cell lines.
Table 2.
Significantly different metabolites between HNSCC tumor and normal adjacent tissue.
| Metabolite | Normal tissue |
Tumor tissue |
P-value | Fold change† | ||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| Guanosine3monophosphate 3GMP⁎ | 0.000% | 0.000% | 0.006% | 0.007% | 0.0041 | – |
| 2-Hydroxyglutarate⁎ | 0.000% | 0.000% | 0.004% | 0.003% | 0.0041 | – |
| N-Acetylputrescine | 0.004% | 0.007% | 0.047% | 0.066% | 0.0261 | 12.7 |
| Adenosine 3-monophosphate | 0.010% | 0.009% | 0.090% | 0.072% | 0.0041 | 9.1 |
| X-15564 | 0.009% | 0.014% | 0.083% | 0.109% | 0.0111 | 9.1 |
| X-15500 | 0.003% | 0.003% | 0.012% | 0.011% | 0.0381 | 4.0 |
| X-11400 | 0.290% | 0.187% | 1.029% | 0.484% | 0.0111 | 3.5 |
| Uracil | 0.030% | 0.041% | 0.077% | 0.062% | 0.0381 | 2.6 |
| Spermidine | 0.292% | 0.097% | 0.665% | 0.419% | 0.0381 | 2.3 |
| C-Glycosyl tryptophan | 0.035% | 0.009% | 0.066% | 0.021% | 0.0071 | 1.9 |
| X-16200 | 0.179% | 0.103% | 0.056% | 0.092% | 0.0171 | 0.3 |
| X-15559 | 1.331% | 0.829% | 0.396% | 0.810% | 0.0381 | 0.3 |
| X-08893 | 1.438% | 0.655% | 0.401% | 0.771% | 0.0111 | 0.3 |
Unique to tumor tissue.
Fold change - HNSCC:Normal.
3.4. Presence of 2-hydroxyglutarate in cancer samples is not related to known mutations in IDH genes
Since production of 2-hydroxyglutarate has been linked to specific somatic mutations in the IDH1 (R132) and IDH2 (R172 and R140) genes [24], [26], we sequenced exon 2 of IDH1 and exon 4 of IDH2 in tissue samples collected from HNSCC patients to determine whether production of 2-HG was associated with mutation in the IDH genes. No somatic mutations were present in these two exons. In order to rule out novel mutations in these genes, we additionally sequenced the remaining exons of both genes and found no somatic mutations. Moreover, sequencing of 14 additional HNSCC patient samples revealed a single somatic variant in either IDH1 or IDH2 in only 3 patients. Two of these somatic variants are predicted to be disease-causing (IDH1, c.494 T > G, p.V165G; IDH2, c.1166A > C, p.Q389P), while the third variant is predicted to be benign (IDH1, c.18 T > G, p.S6R). None of these mutations are reported in the somatic COSMIC mutation database or found in the germline dbSNP variant database.
4. Discussion
The metabolomic profile of HNSCC is an untapped biologic resource for a cancer not only with recently increasing incidence but also that would highly benefit from advances in risk assessment and early detection. In this pilot study, we found levels of several known metabolites that were elevated in both oral wash and tissue samples from HNSCC patients compared to non-diseased controls. Additionally, we found that levels of acylcarnitines were elevated in oral washes of HNSCC patients. Lastly, this is the first report to our knowledge that 2-HG, a known oncometabolite reported in leukemias and gliomas, is uniquely present in tumor tissues of HNSCC and undetectable in normal adjacent tissue.
Among HNSCC-associated elevated metabolites, we found beta-alanine, betaine, hexanoylcarnitine, allantoin, phenyllactate in oral washes, and spermidine and C-glycosyl tryptophan (C-glyTrp) in tissues. Normally associated with disorders in energy metabolism [27], oxidative status [28], and aging [29], more recently these metabolites have been associated with breast (beta-alanine) and liver cancers [30], [31]. Beta-alanine is a component of naturally occurring peptides (carnosine, anserine) and pantothenic acid (Vitamin B-5), and displays a 2.4-fold change between ER- and ER + breast cancer [30]. Furthermore, metabolomic profiling in breast cancer identified ER- disease as a preferential target for novel glutaminase inhibitors, which have proved to have antiprofilerative activity in triple-negative breast cancer cell lines [32]. Hexanoylcarnitine is a medium-chain acylcarnitine, and elevation of this metabolite has been associated with pathologic disturbances in energy production and in intermediary metabolism, as is seen in the congenital condition known as medium-chain acyl-CoA dehydrogenase deficiency [27]. Incidentally, we found elevated levels of acetaminophen and lidocaine in oral wash of HNSCC patients but not controls, which is not surprising since these are commonly used analgesic and anesthetic drugs and would be common exposures in a population undergoing evaluation for head and neck cancer.
The finding of the oncometabolite 2-HG as uniquely expressed in HNSCC tissue but not adjacent normal tissue is novel for this cancer, and opens a new avenue for exploration of oncogenic pathways for therapeutic exploitation in HNSCC. Even more interesting is that IDH mutations do not appear to be responsible for the production of 2-HG in HNSCC tumor, and thus we conclude that tumor-specific production of 2-HG must have another etiology in HNSCC. None of the previously reported dominant negative mutations in IDH1/2 were found in our HNSCC tumors. While we found a few putative somatic variants in other regions of IDH1 and IDH2, not only were these variants rare, they appear to be deleterious mutations which would abrogate the function of IDH protein completely rather than cause the aberrant function which produces 2-HG. These rare mutations do not explain the consistent 2-HG expression in all of our tumor samples. Intriguingly, 2-HG is a known bacterial product of 2-hydroxyglutarate synthase [33]. Further investigation is needed to identify the source of 2-HG which is exclusive to HNSCC tumor tissue in our study. Given the lack of explanative somatic mutations in IDH1 and IDH2 as well as microbiome shifts reported in the HNSCC microbiome between tumor and normal tissue [34], bacteria are a prime suspect for the differential production of 2-HG in HNSCC. Another potential source of 2-HG may be from upregulated conversion of α-ketoglutarate.
Previous studies support using metabolomics as an initial step to identify promising cancer-associated metabolites. Identified metabolites from this type of study serve two purposes: first, as candidates for screening and diagnosis, and second, to illuminate novel oncogenic pathways as therapeutic targets. For example Yan et al. [35] used metabolomics to report that 14 salivary metabolites are associated with oral squamous cell carcinoma, 13 with lichen planus, and 11 with leukoplakia. However, these investigators did not report the identities of these metabolites. Separate studies have also suggested the association of acylcarnitines with bladder cancer [36], [37], as well as with esophageal squamous cell carcinoma [38]. Since our results also reveal that acylcarnitines are significantly elevated in HNSCC, this class of metabolites may have potential as a biomarker for epithelial-derived cancers.
Given that a biomarker that predicts disease is usually not a single molecule, but rather a panel of several molecules [39], a combination of identified metabolites may be more robust for earlier HNSCC diagnosis, particularly applicable to high risk populations. Such a panel could also be candidates for a screening study involving precancerous tissue. Of our identified candidates, hexanoylcarnitine and 2-HG hold the most promise. These two metabolites demonstrated levels that were either significantly higher (hexanoylcarnitine) or uniquely present (2-HG) in HNSCC samples. Moreover, each metabolite has significance in different sample types – hexanoylcarnitine in oral wash and 2-HG in tumor tissue. These findings open an attractive possibility whereby hexanoylcarnitine can be used as a noninvasive screening test for HNSCC. While histopathology is clearly the gold standard of diagnosis once tissue is obtained, the use of 2-HG as a diagnostic or pre-diagnostic, could be imagined in biopsies involving minute fragments of tissue, as metabolomic assays can be done with smaller tissue volumes and do not require a biopsy to be structurally intact.
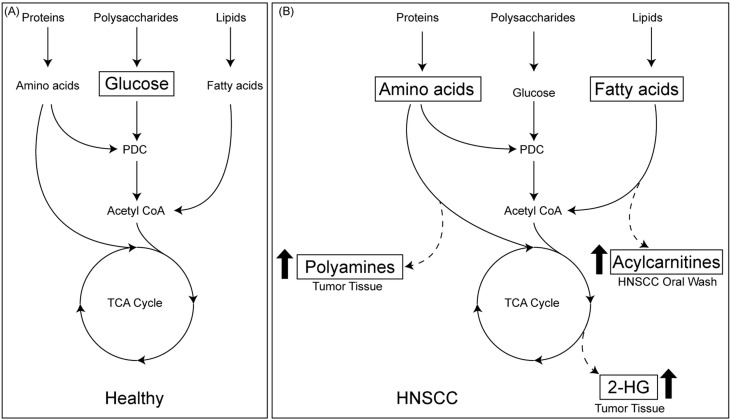
Fig. 5.
Schema showing HNSCC-associated changes in metabolic flux. (A) Metabolic processes in healthy, non-cancer cells, with energy flow driven primarily by glucose metabolism (glycolysis, TCA cycle). In cancer cells (B), levels of acyl carnitines and polyamines are elevated, suggesting amino acid and fatty acid metabolism to be major sources of energy production. Cancer cells also produce 2HG, indicating specific variations in the TCA cycle.
In this exploratory study, we identified multiple known metabolites that are differentially expressed between normal and tumor tissue, as well as between oral washes from HNSCC patients and controls. Importantly, we report for the first time in HNSCC the exclusively somatic expression of the oncometabolite 2-HG. Moreover, as opposed to all the previously reported cancers that express 2-HG, we found that IDH mutations are not responsible for 2-HG production in this cancer. Additionally, we detected several unidentified metabolites, which may have potential as biomarkers. The role of these molecules in cancer and their association with disease remains to be investigated. Confirmation of these HNSCC-specific metabolites in an independent large series would provide not only effective targets for early detection, but also may yield molecular insights into preventative or therapeutic targeting, particularly in high risk patients.
The following are the supplementary data related to this article.
Raw data and testing platform.
Relative abundance of metabolites in tested samples.
Significantly different metabolites in oral wash samples.
Transparency document
Transparency document.
Acknoweldgements
This study was funded, in part, by grants from the NIH [R01DE21544 to CE; R01DE024228 to MAG and PKM; RO1DE17846 and the Oral HIV AIDS Research Alliance (OHARA, BRS-ACURE-S-11-000049-110229) to MAG, and R21EY021303 and R21AI074077 to PKM]. We also acknowledge support from the NIH-funded Skin Diseases Research Center at Case (NIAMS P30 AR039750) and Dr. Thomas McCormick for providing the epithelial cells from healthy volunteers (obtained after informed consent). PF is an Ambrose Monell Foundation Cancer Genomic Medicine Clinical Fellow. CE is the Sondra J. and Stephen R. Hardis Endowed Chair in Cancer Genomic Medicine at the Cleveland Clinic, and an ACS Clinical Research Professor.
Footnotes
The Transparency document associated with this article can be found, in online version.
Contributor Information
Charis Eng, Email: engc@ccf.org.
Mahmoud A Ghannoum, Email: Mahmoud.Ghannoum@Case.edu.
References
- 1.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Leemans C.R., Braakhuis B.J., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 3.Le Tourneau C., Siu L.L. Molecular-targeted therapies in the treatment of squamous cell carcinomas of the head and neck. Curr. Opin. Oncol. 2008;20:256–263. doi: 10.1097/CCO.0b013e3282f9b575. [DOI] [PubMed] [Google Scholar]
- 4.Le Tourneau C., Vidal L., Siu L.L. Progress and challenges in the identification of biomarkers for EGFR and VEGFR targeting anticancer agents. Drug Resist. Updat. 2008;11:99–109. doi: 10.1016/j.drup.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Le Tourneau C., Chen E.X. Molecularly targeted agents in the treatment of recurrent or metastatic squamous cell carcinomas of the head and neck. Hematol. Oncol. Clin. North Am. 2008;22:1209–1220. doi: 10.1016/j.hoc.2008.08.002. (ix) [DOI] [PubMed] [Google Scholar]
- 6.Bonner J.A., Harari P.M., Giralt J., Cohen R.B., Jones C.U., Sur R.K., Raben D., Baselga J., Spencer S.A., Zhu J., Youssoufian H., Rowinsky E.K., Ang K.K. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11:21–28. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- 7.Halama A. Metabolomics in cell culture–a strategy to study crucial metabolic pathways in cancer development and the response to treatment. Arch. Biochem. Biophys. 2014;564:100–109. doi: 10.1016/j.abb.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Green D.R., Galluzzi L., Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345:1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang J., Liu L., Wang W., Xu H., Wu C., Xu J., Liu C., Long J., Ni Q., Yu X. Metabolic tumor burden: a new promising way to reach precise personalized therapy in PDAC. Cancer Lett. 2015;359:165–168. doi: 10.1016/j.canlet.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Gupta A., Gupta S., Mahdi A.A. (1)H NMR-derived serum metabolomics of leukoplakia and squamous cell carcinoma. Clin. Chim. Acta. 2015;441:47–55. doi: 10.1016/j.cca.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Wang J., Christison T.T., Misuno K., Lopez L., Huhmer A.F., Huang Y., Hu S. Metabolomic profiling of anionic metabolites in head and neck cancer cells by capillary ion chromatography with Orbitrap mass spectrometry. Anal. Chem. 2014;86:5116–5124. doi: 10.1021/ac500951v. [DOI] [PubMed] [Google Scholar]
- 12.Tripathi P., Kamarajan P., Somashekar B.S., MacKinnon N., Chinnaiyan A.M., Kapila Y.L., Rajendiran T.M., Ramamoorthy A. Delineating metabolic signatures of head and neck squamous cell carcinoma: phospholipase A2, a potential therapeutic target. Int. J. Biochem. Cell Biol. 2012;44:1852–1861. doi: 10.1016/j.biocel.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei J., Xie G., Zhou Z., Shi P., Qiu Y., Zheng X., Chen T., Su M., Zhao A., Jia W. Salivary metabolite signatures of oral cancer and leukoplakia. Int. J. Cancer. 2011;129:2207–2217. doi: 10.1002/ijc.25881. [DOI] [PubMed] [Google Scholar]
- 14.Ghannoum M.A., Mukherjee P.K., Jurevic R.J., Retuerto M., Brown R.E., Sikaroodi M., Webster-Cyriaque J., Gillevet P.M. Metabolomics Reveals Differential Levels of Oral Metabolites in HIV-Infected Patients: Toward Novel Diagnostic Targets. OMICS. 2011;15 doi: 10.1089/omi.2011.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bebek G., Koyuturk M., Price N.D., Chance M.R. Network biology methods integrating biological data for translational science. Brief. Bioinform. 2012;13:446–459. doi: 10.1093/bib/bbr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons D.W., Jones S., Zhang X., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I.M., Gallia G.L., Olivi A., McLendon R., Rasheed B.A., Keir S., Nikolskaya T., Nikolsky Y., Busam D.A., Tekleab H., Diaz L.A., Jr., Hartigan J., Smith D.R., Strausberg R.L., Marie S.K., Shinjo S.M., Yan H., Riggins G.J., Bigner D.D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V.E., Kinzler K.W. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 18.Fiehn O., Kind T. Metabolite profiling in blood plasma. In: Weckwerth W., editor. Metabolomics. Humana Press; 2007. pp. 3–17. [DOI] [PubMed] [Google Scholar]
- 19.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2016. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 20.Sarkar D. Springer; New York: 2008. Lattice: Multivariate Data Visualization with R. [Google Scholar]
- 21.Wickham H. Springer; New York: 2009. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 22.Ward P.S., Patel J., Wise D.R., Abdel-Wahab O., Bennett B.D., Coller H.A., Cross J.R., Fantin V.R., Hedvat C.V., Perl A.E., Rabinowitz J.D., Carroll M., Su S.M., Sharp K.A., Levine R.L., Thompson C.B. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losman J.A., Kaelin W.G., Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cairns R.A., Iqbal J., Lemonnier F., Kucuk C., de Leval L., Jais J.-P., Parrens M., Martin A., Xerri L., Brousset P., Chan L.C., Chan W.-C., Gaulard P., Mak T.W. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012 doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kloosterhof N.K., Bralten L.B.C., Dubbink H.J., French P.J., van den Bent M.J. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? Lancet Oncol. 2011;12:83–91. doi: 10.1016/S1470-2045(10)70053-X. [DOI] [PubMed] [Google Scholar]
- 26.Guo C., Pirozzi C.J., Lopez G.Y., Yan H. Isocitrate dehydrogenase mutations in gliomas: mechanisms, biomarkers and therapeutic target. Curr. Opin. Neurol. 2011;24:648–652. doi: 10.1097/WCO.0b013e32834cd415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt-Sommerfeld E., Penn D., Rinaldo P., Kossak D., Li B.U., Huang Z.H., Gage D.A. Urinary medium-chain acylcarnitines in medium-chain acyl-CoA dehydrogenase deficiency, medium-chain triglyceride feeding and valproic acid therapy: sensitivity and specificity of the radioisotopic exchange/high performance liquid chromatography method. Pediatr. Res. 1992;31:545–551. doi: 10.1203/00006450-199206000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Il'yasova D., Scarbrough P., Spasojevic I. Urinary biomarkers of oxidative status. Clin. Chim. Acta. 2012;413:1446–1453. doi: 10.1016/j.cca.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menni C., Kastenmuller G., Petersen A.K., Bell J.T., Psatha M., Tsai P.C., Gieger C., Schulz H., Erte I., John S., Brosnan M.J., Wilson S.G., Tsaprouni L., Lim E.M., Stuckey B., Deloukas P., Mohney R., Suhre K., Spector T.D., Valdes A.M. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 2013;42:1111–1119. doi: 10.1093/ije/dyt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budczies J., Brockmoller S.F., Muller B.M., Barupal D.K., Richter-Ehrenstein C., Kleine-Tebbe A., Griffin J.L., Oresic M., Dietel M., Denkert C., Fiehn O. Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: alterations in glutamine and beta-alanine metabolism. J. Proteome. 2013;94:279–288. doi: 10.1016/j.jprot.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Pellanda H. Betaine homocysteine methyltransferase (BHMT)-dependent remethylation pathway in human healthy and tumoral liver. Clin. Chem. Lab. Med. 2013;51:617–621. doi: 10.1515/cclm-2012-0689. [DOI] [PubMed] [Google Scholar]
- 32.Gross M.I., Demo S.D., Dennison J.B., Chen L., Chernov-Rogan T., Goyal B., Janes J.R., Laidig G.J., Lewis E.R., Li J., Mackinnon A.L., Parlati F., Rodriguez M.L., Shwonek P.J., Sjogren E.B., Stanton T.F., Wang T., Yang J., Zhao F., Bennett M.K. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 33.Reeves H.C., Ajl S.J. Alpha-hydroxyglutaric acid synthetase. J. Bacteriol. 1962;84:186–187. doi: 10.1128/jb.84.1.186-187.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bebek G., Bennett K.L., Funchain P., Campbell R., Seth R., Scharpf J., Burkey B., Eng C. Microbiomic subprofiles and MDR1 promoter methylation in head and neck squamous cell carcinoma. Hum. Mol. Genet. 2012;21:1557–1565. doi: 10.1093/hmg/ddr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan S.K., Wei B.J., Lin Z.Y., Yang Y., Zhou Z.T., Zhang W.D. A metabonomic approach to the diagnosis of oral squamous cell carcinoma, oral lichen planus and oral leukoplakia. Oral Oncol. 2008;44:477–483. doi: 10.1016/j.oraloncology.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Jin X., Yun S.J., Jeong P., Kim I.Y., Kim W.J., Park S. Diagnosis of bladder cancer and prediction of survival by urinary metabolomics. Oncotarget. 2014;5:1635–1645. doi: 10.18632/oncotarget.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Z., Lin L., Gao Y., Chen Y., Yan X., Xing J., Hang W. Bladder cancer determination via two urinary metabolites: a biomarker pattern approach. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.007922. M111.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J., Chen Y., Zhang R., Song Y., Cao J., Bi N., Wang J., He J., Bai J., Dong L., Wang L., Zhan Q., Abliz Z. Global and targeted metabolomics of esophageal squamous cell carcinoma discovers potential diagnostic and therapeutic biomarkers. Mol. Cell. Proteomics. 2013;12:1306–1318. doi: 10.1074/mcp.M112.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaddurah-Daouk R., Kristal B.S., Weinshilboum R.M. Metabolomics: a global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data and testing platform.
Relative abundance of metabolites in tested samples.
Significantly different metabolites in oral wash samples.
Transparency document.